Abstract
We have developed a high-resolution fluorescence microscopy based on high-accuracy localization of photo-switchable fluorophores. In each imaging cycle, only a fraction of fluorophores were turned on, allowing their positions to be determined with nanometer accuracy. The fluorophore positions obtained from a series of imaging cycles were then used to reconstruct the overall image. An imaging resolution of 20 nm was demonstrated and this can in principle reach the molecular scale.
Fluorescence microscopy is widely used in molecular and cell biology for non-invasive, time-resolved imaging with high biochemical specificity. Despite these advantages, standard fluorescence microscopy is not useful for ultra-structural imaging due to a resolution limit set by the diffraction of light. Several approaches have been employed to break this diffraction limit, including near-field scanning optical microscopy (NSOM)1, multi-photon fluorescence2, stimulated emission depletion (STED)3, and saturated structured-illumination microscopy (SSIM)4. NSOM, STED and SSIM have all achieved lateral resolution of tens of nanometers, substantially lower than the wavelength of light, but each with its own limitations. NSOM is difficult to operate in a non-invasive mode and has a low imaging depth. Multi-photon microscopy, STED and SSIM are based on non-linear optical effects and typically require the use of high-intensity pulsed lasers, which can induce sample damage. Electron microscopy remains the method of choice for high resolution imaging of biological samples, and a continuing effort to develop existing4,5 and new techniques is needed to harness the benefits of fluorescence microscopy for ultra-resolution biological imaging.
Single-molecule detection offers new possibilities for obtaining sub-diffraction limit spatial resolution. The position of a single emitter can be determined to almost arbitrarily high accuracy if a sufficient number of photons are collected6-9. Based on this principle, fluorescence imaging with one nanometer accuracy (FIONA) has recently been demonstrated at the single-molecule level at room temperature9. Unfortunately, the localization accuracy of FIONA does not directly translate into imaging resolution since multiple emitters in close proximity will still be difficult to resolve. This issue has been addressed in part by taking advantage of photobleaching of single dyes or photoblinking of quantum dots to distinguish their individual signals10-13. These approaches were used to resolve 2 - 5 fluorophores within a diffraction limited spot, but extending these techniques to more than a few fluorophores remains a challenge.
In this work we report a novel high-resolution optical microscopy, stochastic optical reconstruction microscopy (STORM), in which a fluorescence image is constructed from high-accuracy localization of individual fluorescent molecules that are switched on and off using light of different colors. The STORM imaging process consists of a series of imaging cycles (Fig. 1a). In each cycle, only a fraction of the fluorophores in the field of view are switched on, such that each of the active fluorophores is optically resolvable from the rest, i.e. their images are not overlapping. This allows the position of these fluorophores to be determined with high accuracy. Repeating this process for multiple cycles, each causing a stochastically different subset of fluorophores to be turned on, enables the positions of many fluorophores to be determined and thus an overall image to be reconstructed. In the examples below we demonstrated an imaging resolution of approximately 20 nm, an improvement of 10 times over the resolution of conventional fluorescence microscopy, using a simple total-internal-reflection fluorescence microscope, low-power continuous-wave lasers, and a photoswitchable cyanine dye14,15.
Figure 1.
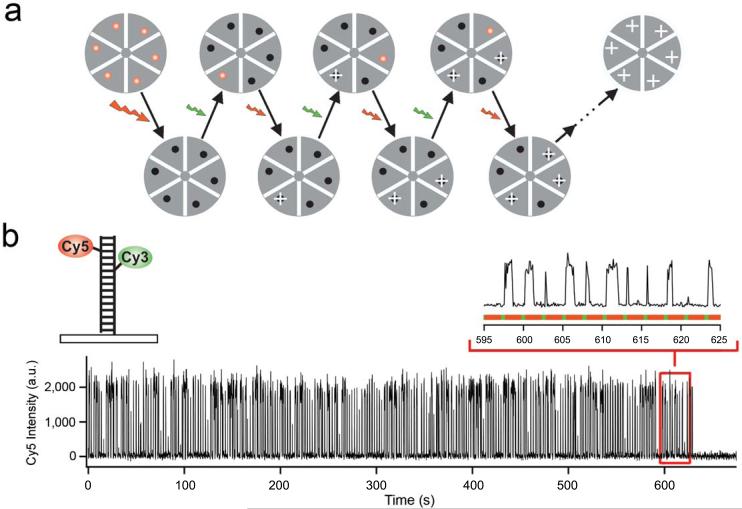
Stochastic optical reconstruction microscopy (STORM) with photo-switchable fluorophores. (a) The diagram shows a STORM imaging sequence using a hypothetical hexameric object labeled with red fluorophores that can be switched between a fluorescent and a dark state by a red and green laser, respectively. All fluorophores are first switched to the dark state by a strong red laser pulse. In each imaging cycle, a green laser pulse is used to switch on only a fraction of the fluorophores to give an optically resolvable set of active fluorophores. Next, under red illumination, these molecules emit fluorescence until they are switched off, allowing their positions (indicated by white crosses) to be determined with high accuracy. The overall image is then reconstructed from the fluorophore positions obtained from multiple imaging cycles. (b) A single Cy5 switch on DNA can be turned on and off for hundreds of cycles before being permanently photobleached. A red laser (633 nm, 30 W/cm2) is used to excite fluorescence (black line) from Cy5 and to switch Cy5 to the dark state. A green laser (532 nm, 1 W/cm2) is used to return Cy5 to the fluorescent state. The alternating red and green line indicates the laser excitation pattern. The recovery rate of Cy5 depends critically on the close proximity of Cy314.
We have recently reported that a cyanine dye, Cy5, can be switched between a fluorescent and a dark state in a controlled and reversible manner by light of different wavelengths14. Red laser light that produces fluorescent emission from Cy5 can also switch the dye to a stable dark state. Exposure to green laser light converts Cy5 back to the fluorescent state but the recovery rate depends critically on the close proximity of a secondary dye, Cy314. Hereafter, we refer to the Cy3-Cy5 dye pair simply as a switch. Under illumination conditions allowing single-molecule detection, we found that such a switch, when attached to nucleic acids or proteins, can be cycled on and off hundreds of times before permanent photobleaching occurs (Fig. 1b and Supplementary Fig. 1 online).
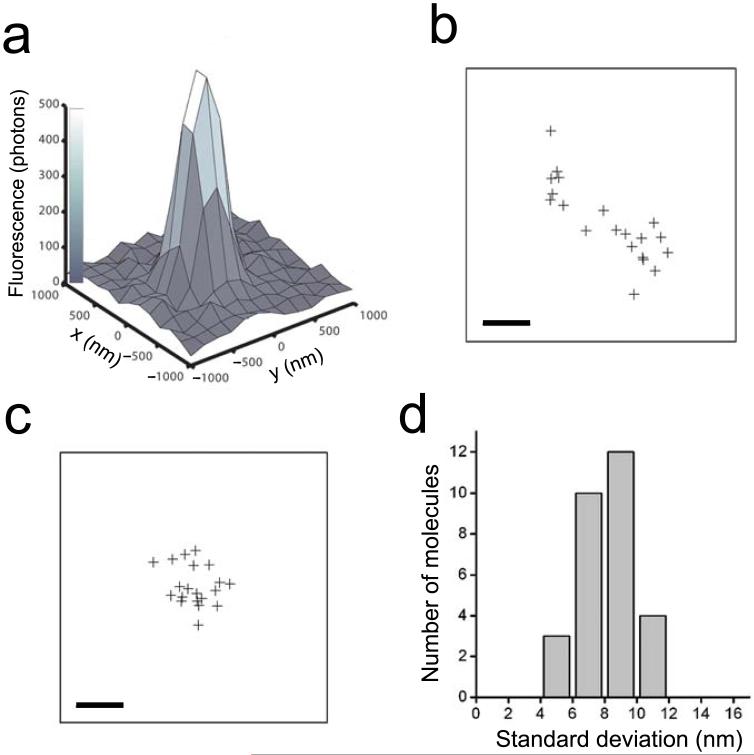
Here we demonstrate the concept of STORM using the cyanine switch, but in principle any suitably optically switched fluorophores could be used. The resolution of STORM is limited by the accuracy with which individual switches can be localized during a switching cycle. To determine the localization accuracy, we attached a switch to a short double-stranded DNA, which was surface-immobilized at low density so that single switches were clearly resolvable. These switches were periodically cycled on and off using green and red laser light, and the red laser also served to excite fluorescence from Cy5. No fluorescence from Cy3 was recorded in this process. The fluorescence image from a single switch gave a point-spread function shown in Fig. 2a. A Gaussian fit to this image was used to localize the position of the switch. The positions determined from multiple switching cycles showed a substantial spread (Fig. 2b), which was significantly reduced by correcting for sample drift over the course of the experiment (Fig. 2c, see Supplementary Methods online for details). The standard deviation of the drift-corrected positions obtained from 20 imaging cycles is on average 8 nm for individual switches (Fig. 2d). This value gives a measure of the uncertainty in the localization of a single switch per imaging cycle. Correspondingly, the experimentally measured spread of switch positions follows a Gaussian distribution with a FWHM of 18 nm, as expected from the 8 nm standard deviation (Supplementary Fig. 2 online). Therefore, under our current imaging conditions, two switches separated by 20 nm should be resolvable.
Figure 2.
The high localization accuracy of individual switches during each switching cycle defines the intrinsic resolution of STORM. (a) The point spread function (PSF) of the emission from a single switch on DNA during a single switching cycle. Fitting the PSF to a 2-dimensional Gaussian (not shown) gives the centroid position of the PSF. (b, c) The centroid positions of an individual switch determined in 20 successive imaging cycles before (b) and after (c) correction for sample drift. Scale bars: 20 nm. (d) A histogram of the standard deviation of centroid positions. The standard deviation is determined as (σx + σy)/2 for each switch using 20 imaging cycles, where σx and σy are the standard deviations of the centroid positions in the x and y dimensions. This histogram was constructed from 29 switches.
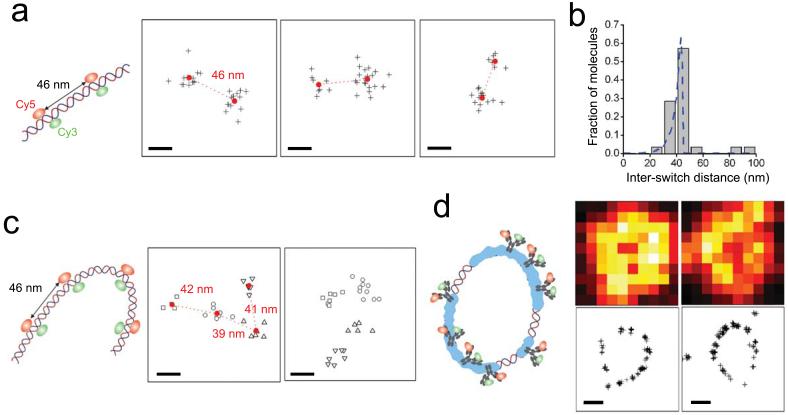
To demonstrate the capability of STORM to resolve fluorescent molecules in close proximity, we constructed standard samples of linear, double-stranded DNA labeled with multiple switches separated by a well-defined number of base pairs (see Supplementary Methods online). The DNA strands were also labeled with multiple biotins and attached to a high-density streptavidin layer (see Supplementary Methods online), increasing the likelihood of multiple attachments between the DNA and the surface so that the DNA is immobilized in the plane. We first examined DNA strands containing two switches separated by 135 base-pairs (Fig. 3a and Supplementary Fig. 3 online), corresponding to a length of 46 nm along the DNA contour. A STORM image obtained using the imaging procedure described above (see Supplementary Methods online for details) shows two clusters of measured switch positions, indicating that the two switches are well-resolved (Fig. 3a). The distance between the centers of the two clouds had a mean value of 41 nm (Figs. 3b), in quantitative agreement with the theoretical mean (40 nm) determined using the known contour and persistence lengths of the DNA sample (see Supplementary Methods online). We also imaged longer DNA samples labeled with four switches evenly separated by 46 nm along the contour. The STORM images indeed reveal four clusters of switch positions following a bent contour consistent with the persistence length of DNA and the engineered separation between the switches. These results indicate that STORM can image biological samples with sub-diffraction-limit resolution and that features separated by 40 nm are well within our resolving power.
Figure 3.
STORM can resolve structures with sub-diffraction-limit resolution. (a) STORM cleanly resolves two switches separated by a contour length of 46 nm on double-stranded DNA. The STORM images show two clearly-separated clusters of measured switch positions (crosses), each corresponding to a single switch. The center-of-mass position of each cluster is marked by a red dot. The inter-switch distances are 46 nm, 44 nm and 34 nm for these three examples. Scale bars: 20 nm. (b) Comparison between the inter-switch distances measured using STORM (grey column) and the predicted distance distribution considering the flexibility of DNA (dashed blue line). (c) STORM images of four switches attached to a double-stranded DNA, pair-wise separated by a contour length of 46 nm. The measured switch positions are clustered by an automated algorithm (see Supplementary Methods online) and different clusters are indicated by different symbols. Scale bars: 20 nm. (d) STORM images of RecA-coated circular plasmid DNA. The top panels show indirect immunofluorescence images with switch-labeled secondary antibody taken by a total internal reflection microscope. The bottom panels are the reconstructed STORM images of the same filaments. Scale bars: 300 nm.
A distinct advantage of STORM is its ability to localize a large number of switches within a diffraction-limited spot by cycling the switches on and off in a controlled manner, allowing this to be used as a general biological imaging technique. To demonstrate this capability, we prepared circular DNA plasmids coated with RecA protein and imaged them using indirect immunofluorescence with switch-labeled secondary antibody (Fig. 3d, see Supplementary Methods online). STORM images of the RecA filaments reveal their circular structure with greatly increased resolution when compared with wide-field images. Parts of the filament appear to be unlabeled or kinked, possibly corresponding to regions of the plasmid that remain uncoated by RecA.
In summary, we have demonstrated that STORM is capable of imaging biological structures with sub-diffraction limit resolution. The resolution of the technique is limited in principle only by the number of photons emitted per switch cycle, but not the wavelength of light. For the cyanine switch, we have detected approximately 3,000 photons per switching cycle, independent of red laser intensity (data not shown), predicting a theoretical localization accuracy of 4 nm8. The discrepancy between this theoretical prediction and our measured accuracy of 8 nm is most likely due to imperfect correction of stage drift and aberration due to focus drift in our measurements. This measured localization accuracy corresponds to an imaging resolution of approximately 20 nm. Indeed, fluorescent switches separated by ~ 40 nm have been clearly resolved (Fig. 3). The cyanine switches can be turned on and off reliably for hundreds of cycles before photobleaching, allowing STORM to be used for resolving structures with many fluorophores in a potentially time-resolved manner. We have shown that the circular structure of RecA filaments containing 10-20 switches can be resolved in a few minutes (See Supplementary Methods online). The imaging speed may be improved by increasing the switching rate through stronger excitation or fluorophores with faster switching kinetics. We expect that STORM will be a valuable tool for high-resolution fluorescence in situ hybridization (FISH) and immunofluorescence imaging. The STORM concept is also applicable to other photo-switchable fluorophores and fluorescent proteins, which will potentially allow high-resolution live-cell imaging with endogenous labels.
Supplementary Material
Acknowledgement
This work is supported by in part by the National Institute of Health, the Defense Advance Research Projects Agency and a Packard Science and Engineering Fellowship (to X.Z.). X.Z. is a Howard Hughes Medical Institute Investigator.
Footnotes
Competing interests statement The authors declare that they have no competing financial interests.
References
- 1.Pohl DW, Courjon D. Near field optics. Kluwer; Dordrecht: 1993. [Google Scholar]
- 2.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nat. Biotech. 2003;21:1368–1376. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 3.Hell SW. Toward fluorescence nanoscopy. Nat. Biotech. 2003;21:1347–1355. doi: 10.1038/nbt895. [DOI] [PubMed] [Google Scholar]
- 4.Gustafsson MGL. Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution. Proc. Natl. Acad. Sci., USA. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofmann M, Eggeling C, Jakobs S, Hell SW. Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins. Proc. Natl. Acad. Sci., USA. 2005;102:17565–17569. doi: 10.1073/pnas.0506010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gelles J, Schnapp BJ, Sheetz MP. Tracking kinesin-driven movements with nanometre-scale precision. Nature. 1988;331:450–453. doi: 10.1038/331450a0. [DOI] [PubMed] [Google Scholar]
- 7.van Oijen AM, Kohler J, Schmidt J, Muller M, Brakenhoff GJ. 3-Dimensional super-resolution by spectrally selective imaging. Chem. Phys. Lett. 1998;292:183–187. [Google Scholar]
- 8.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yildiz A, et al. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5 nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 10.Gordon MP, Ha T, Selvin PR. Single-molecule high-resolution imaging with photobleaching. Proc. Natl. Acad. Sci., USA. 2004;101:6462–6465. doi: 10.1073/pnas.0401638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu XH, Wu D, Mets L, Scherer NF. Nanometer-localized multiple single-molecule fluorescence microscopy. Proc. Natl. Acad. Sci., USA. 2004;101:11298–11303. doi: 10.1073/pnas.0402155101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lidke KA, Rieger B, Jovin TM, Heintzmann R. Super resolution by localization of quantum dots using blinking statistics. Optics Express. 2005;13:7052–7062. doi: 10.1364/opex.13.007052. [DOI] [PubMed] [Google Scholar]
- 13.Ram S, Ward ES, Ober RJ. Beyond Rayleigh's criterion: A resolution measure with application to single-molecule microscopy. Proc. Natl. Acad. Sci., USA. 2006;103:4457–4462. doi: 10.1073/pnas.0508047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bates M, Blosser TR, Zhuang X. Short-range spectroscopic ruler based on a single-molecule optical switch. Phys. Rev. Lett. 2005;94:108101. doi: 10.1103/PhysRevLett.94.108101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heilemann M, Margeat E, Kasper R, Sauer M, Tinnefeld P. Carbocyanine dyes as efficient reversible single-molecule optical switch. J. Am. Chem. Soc. 2005;127:3801–3806. doi: 10.1021/ja044686x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.