Abstract
The complete RNA genome sequence of avian paramyxovirus (APMV) serotype 2, strain Yucaipa isolated from chicken has been determined. With genome size of 14,904 nucleotides (nt), strain Yucaipa is consistent with the “rule of six” and is the smallest virus reported to date among the members of subfamily Paramyxovirinae. The genome contains six non-overlapping genes in the order 3′-N-P/V-M-F-HN-L-5′. The genes are flanked on either side by highly-conserved transcription start and stop signals and have intergenic sequences varying in length from 3 to 23 nt. The genome contains a 55 nt leader sequence at 3′ end and a 154 nt trailer sequence at 5′ end. Alignment and phylogenetic analysis of the predicted amino acid sequences of strain Yucaipa proteins with the cognate proteins of viruses of all of the five genera of family Paramyxoviridae showed that APMV-2 strain Yucaipa is more closely related to APMV-6 than APMV-1.
Keywords: Avian Paramyxoviruses, APMV-2 strain Yucaipa, Complete genome sequence, Phylogenetic analysis
1. Introduction
Paramyxoviridae is a large and diverse family whose members have been isolated from many species of avian, terrestrial, and aquatic animals around the world (Lamb and Parks, 2007; Wang and Eaton, 2001). Paramyxoviruses are pleomorphic, enveloped, cytoplasmic viruses that have a non-segmented negative-strand RNA genome. Paramyxoviruses are divided into two subfamilies, Paramyxovirinae and Pneumovirinae, based on structure, genome organization, and sequence relatedness (Lamb et al., 2005). Subfamily Paramyxovirinae comprises five genera; Respirovirus (including Sendai virus [SeV] and human parainfluenza virus types 1 and 3 [HPIV-1 and -3]), Rubulavirus (including simian virus type 5 [SV5], mumps virus [MuV], and human parainfluenza virus types 2 and 4 [HPIV-2 and -4]), Morbillivirus (including measles [MeV] and canine distemper [CDV] viruses), Henipavirus (including Hendra [HeV] and Nipah [NiV] viruses), and Avulavirus (comprising the nine serotypes of avian paramyxoviruses [APMV-1 to -9]). Subfamily Pneumovirinae contains two genera, Pneumovirus (comprising human respiratory syncytial virus [HRSV] and its animal counterparts) and Metapneumovirus (comprising human metapneumovirus [HMPV] and its avian counterpart [AMPV].
All paramyxoviruses examined to date encode a major nucleocapsid protein (N) that binds the entire length of the genomic and antigenomic RNAs, a nucleocapsid phosphoprotein (P) that is a polymerase co-factor, a large protein (L) that is the major polymerase subunit and bears catalytic domains, a matrix protein (M) that lines the inner surface of the envelope, a fusion glycoprotein (F) that is a surface antigen that mediates viral penetration and syncytium formation and a major glycoprotein (G) or hemagglutinin-neuraminidase (HN) glycoprotein that is a second surface antigen and mediates attachment. Most members of subfamily Paramyxovirinae engage in RNA editing, whereby a specific motif in the P gene directs non-templated addition of one or more nucleotides to a proportion of P transcripts. This produces mRNA subpopulations containing frameshifts whereby the reading register is shifted to access one or more alternate internal ORFs to produce chimeric proteins in which the upstream end is encoded by the P ORF and the downstream end is encoded by the alternative ORF. The most notable of these proteins is one called V, in which the downstream domain contains a highly conserved cysteine motif.
Genus Avulavirus contains all of the paramyxoviruses that have been isolated from avian species except for avian metapneumovirus. The disease potential of many avulaviruses is not known. These viruses have been classified into nine serotypes based on hemagglutination inhibition (HI) and neuraminidase inhibition (NI) tests (Alexander, 2003). The cross-HI and -NI tests also indicated that APMV isolates could be organized into two broad subgroups; the first subgroup consisting of APMV-2 and -6 and the second subgroup consisting of APMV-1, -3, -4, -7, -8 and -9 (Lipkind and Shihmanter, 1986). Not much is known about APMV-5. The many strains of Newcastle disease virus (NDV) comprise APMV-1. Since NDV is an important cause of disease in chickens, APMV-1 is the most extensively characterized serotype of the APMVs. Very little is known about the molecular biology and pathogenesis of serotypes 2-9.
The first isolate of APMV-2 was obtained in 1956 in Yucaipa, California from a diseased chicken that was also infected with infectious laryngotracheitis virus (Bankowski et al., 1960). Since then many APMV-2 strains have been isolated from chickens, turkeys and feral birds across the globe (Alexander et al., 1982; Asahara et al., 1973; Collings et al., 1975; Fleury et al., 1979; Goodman et al., 1988; Lang et al., 1975; Lipkind et al., 1979; Lipkind et al., 1982; Mbugua et al., 1985; Nymadawa et al., 1977; Shihmanter et al., 1997; Weisman et al., 1984; Zhang et al., 2006; Zhang et al., 2007). Serological survey of chickens and turkeys has shown a wide prevalence of APMV-2 in the United States. The infection is more prevalent in turkeys than chickens (Bankowski et al., 1968). APMV-2 viruses also have been frequently isolated from passerine and psittacine birds. Surveillance of wild birds has indicated that APMV-2 viruses are more frequent in passerines (Alexander, 1986; Senne et al., 1983). APMV-2 infection was shown to affect hatchability and poult yield in turkeys (Bankowski et al., 1981). Experimental infection with this virus caused mild disease in chickens (Alexander, 1980). To date, the complete genome sequences of a number of strains of APMV-1 (Krishnamurthy and Samal, 1998; de Leeuw and Peeters, 1999) and of a single strain of APMV-6 (Chang et al., 2001) have been published. As a first step towards characterizing the molecular genetics and pathogenesis of APMV-2, we have sequenced the complete genome of APMV-2, strain Yucaipa. The sequence information of strain Yucaipa was compared with that of the other APMV serotypes and other paramyxoviruses in order to describe its genomic structure and phylogenetic relationship.
2. Methods
2.1. Virus and cells
APMV-2 strain Yucaipa was obtained from the National Veterinary Services Laboratory, Ames, Iowa. The virus was grown in nine-day-old embryonated, specific pathogen free (SPF) chicken eggs. Hemagglutination (HA) titers were determined using 0.5 % chicken RBC at room temperature. Replication of the virus was evaluated in nine different cell lines: BHK21, Baby Hamster Kidney; BT, Bovine Turbinate; DF1, chicken embryo fibroblast; HEp2, human cervical carcinoma; MDCK, Madin Darby Canine Kidney; PK15, Pig Kidney; QT35, Quail fibrosarcoma; RK13, Rabbit Kidney cells; and Vero, African green monkey kidney. The DF1 and QT35 cells were grown in Dulbecco's minimum essential medium containing 10% fetal calf serum (FCS), while the other cells were grown in Eagle's minimum essential medium (MEM) containing 10% FCS, all in a 37°C incubator with 5% CO2. Each cell type was grown as a monolayer and infected with a 10-3 dilution of 210 HA units of egg-grown APMV-2 strain Yucaipa with and without 10% allantoic fluid supplementation in the medium, which provides a source of protease for cleavage of the F protein. The cells were observed daily for cytopathic effects (CPE) and HA titers were recorded every 24 h until the fifth day. A total of three passages of virus were made in each cell type. The ability of the virus to produce plaques was tested in the different cell lines under various conditions, including 1% methylcellulose, 1% low melting agar, and 0.8% noble agar with or without magnesium sulphate (25mM) and 1% diethylaminoethyl dextran (30μg/ml). Plaques were visualized by staining with either crystal violet or neutral red.
2.2. Viral RNA isolation and sequence analysis
APMV-2 strain Yucaipa RNA was isolated from the allantoic fluid of virus-infected eggs using RNeasy kit (QIAGEN, USA) according to the manufacturer's instructions. The complete genome sequence exclusive of the termini was determined using a combination of three different strategies. First, the nucleotide sequences of the N genes of all the available rubulaviruses and avulaviruses were aligned to identify a consensus sequence that was used to design the forward primer N-451 (5′-GAAGATGATGCACCAGAAGA, numbered according to the consensus sequence). A reverse primer was designed from the APMV-2 F gene sequence that was available in GenBank (Accession no. AF422844) (F-127r, 5′-ACTGCGATGGTCCCTGTGAG, numbered according to the Yucaipa strain F gene sequence). This yielded a part of sequence upstream of F gene. Similarly, L genes of rubulaviruses and avulaviruses were aligned to design two reverse degenerate primers in the conserved regions of L gene; L-5544r, 5′-NGGNCCRAARTGNCKYTGNGGNGGRTT (N=A/C/G/T, R=A/G, K=G/T, Y=C/T) and L-6960r, 5′-NSWRTARTANCCYTTNGCNGCRTTNCCDATNGT (N=A/C/G/T, S=G/C, W=A/T, R=A/G, Y=C/T, D=G/A/T). APMV-2 L gene-specific forward primers were designed from the partial sequence that was available in GenBank (Accession no. AF515835). The PCR using these primers resulted in multiple bands which upon cloning and sequencing yielded different regions of L gene. Second, we designed a gene-start forward primer (5′GGAAAACTTGGGGGCGACA) containing the presumptively-conserved gene-start sequence at its 3′ end (underlined) and a reverse primer (5′TTTTTTCTTAAACCAGGCTTC) with the presumptively-conserved gene-end sequence at its 5′ end (underlined). Reverse transcription (RT) with the gene-start forward primer and PCR with the same forward primer and the gene-end reverse primer resulted in multiple fragments which upon sequencing yielded different regions of all the viral genes. Finally, as the third strategy, most of the L gene was sequenced by primer walking originating in the partial sequence of the upstream end of L available in GenBank. Briefly, cDNA synthesized from an RT reaction with an L gene-specific forward primer was purified by ethanol precipitation and a 3′ poly C tail was added with terminal deoxynucleotidyl transferase (Invitrogen). The dC-tailed cDNA was amplified by PCR using an L gene-specific forward primer and a poly dG-containing reverse primer. The PCR-amplified products were cloned and sequenced. The sequence from one round of cloning was used to design the forward primer for the next round of RT-PCR. All the primers were purchased from Integrated DNA technologies, USA. The RT reactions were performed with the SuperScript II RT kit (Invitrogen, USA) and PCR was performed in 50 μl reactions using Takara LA taq (Takara Bio, USA), both according to the manufacturer's instructions. The general conditions for PCR were 95°C for 5 min, 25 cycles of 95°C for 1 min (denaturation), 56°C for 1 min (annealing) and 72°C for 1 min (extension), followed by 72°C for 10 min (final extension). The PCR fragments were cloned in TOPO TA cloning kit (Invitrogen, USA). In addition, selected PCR products were purified by agarose gel electrophoresis and sequenced directly. DNA sequencing was carried out using BigDye® Terminator v3.1 cycle sequencing kit (Applied Biosystems, USA) and an ABI PRISM 3100 Avant Genetic Analyser (Applied Biosystems). Every nucleotide in the genome was sequenced at least three times and once directly from RT-PCR product without cloning, thus ensuring a consensus sequence.
2.3. Determination of the sequences of genome termini
The sequences of the 3′ and 5′ ends of the virus genome were obtained by ligation of a RNA oligonucleotide to viral RNA and cDNA, respectively, as described (Troutt et al., 1992). To determine the 3′ end of viral RNA, a 5′-phosphorylated and 3′ blocked RNA oligonucleotide (5′phos-CCAAAACGCCAUUUCCACCUUCUCUUC 3′-blocked), was ligated to viral RNA. Briefly, 8 μl of viral RNA (1-5μg) and 1 μl of RNA oligonucleotide (50 pmol) were denatured at 65°C for five min and snap frozen on dry ice. The ligation reaction was carried out overnight at 16°C with 4 μl of 10× T4 RNA ligase buffer, 10 units of T4 RNA ligase (Promega, USA), 1 mM hexamine cobalt chloride, 10 μg/ml BSA, 25% (w/v) of PEG 8000 and RNase-free water to make a 40 μl reaction mixture. Ligation was terminated by heating to 65°C for 20 min. The ligated RNA was precipitated following the protocol described in the GeneRacer kit (Invitrogen, USA), RT was carried out with an adaptor primer (5′ GAAGAGAAGGTGGAAATGGCGTTTTGG) complimentary to the RNA oligonucleotide, as described (Li et al., 2005). PCR was performed with the same primer together with a reverse primer within N gene, N287 (5′ GGATCGCCCCTTGTCTCAT). To determine the 5′ end, viral RNA was reverse transcribed using an L gene-specific primer L-5.7 (5′-AAGAGTTTGACAGGGGGATGC). The cDNA was ligated to the RNA oligonucleotide following the same procedure as described above. The ligated cDNA was amplified by PCR using an L gene-specific forward primer, L-5.9 (5′-GGCTTGATATACACCGGAACTCGT, which anneals to sequence downstream of L 5.7), together with the adaptor primer. The PCR products were cloned into the TOPO TA cloning vector and sequenced, and also were directly sequenced.
2.4. Sequence and phylogenetic tree analysis
Sequence compilation and prediction of ORFs were carried out using the SEQMAN and EDITSEQ programs in the LASERGENE (DNASTAR) software package (www.dnastar.com). The search for homologous protein sequences in GenBank was done using the BLAST program in the same package. Phylogenetic analysis was carried out using T-Coffee, (Tree-based consistency objective function for alignment evaluation), a multiple sequence alignment program. The phylogenetic trees were drawn using the same program and applying the “average distance using percentage identity” method (Notredam, 2000).
2.5. Database accession numbers
The complete genome sequence of APMV-2 strain Yucaipa has been deposited in GenBank under accession number EU338414. Accession numbers for other sequences used in this study are given below. For some viruses, individual gene sequences were used since full-length genome sequences were unavailable. They are indicated by the abbreviated gene letter in parentheses following the GenBank accession number. Virus sequences used were as follows: AMPV, NC_007652; APMV-1 (NDV) strain Beaudette C (for the 3′ leader sequence, see reference Krishnamurthy and Samal, 1998), AF064091 (N), X60599 (P/V), X04687 (M), X04719 (F), X04355 (HN), and X05399 (L); APMV-3, AY137207; APMV-4, D14031 (HN gene); APMV-6, AY029299; CDV, AF014953; HeV, AF017149; HMPV, NC_004148; HPIV-2, X57559; HPIV-3, AB012132; HRSV, AF013254; MeV, AB016162; MuV, AB040874; MrV, D13990 (F and HN genes); NiV, AF212302; SeV, AB005795.
3. Results
3.1. Growth characteristics of APMV-2 strain Yucaipa
APMV-2 strain Yucaipa yielded a titer of 210-212 HA units in nine-day-old embryonated SPF chicken eggs four days post inoculation. Nine different cell culture systems each representing a different species of origin were evaluated to determine the cell type(s) that can support the growth of APMV-2 to high titers as well as whether or not added protease is required for replication. Each of the nine cell types supported the growth of APMV-2, as determined by the observable CPE and HA activity of the infected cell culture supernatant. The HA titers were the same with and without 10% allantoic fluid supplementation of the media, and varied from 23 to 29 HA units among the cell types. The peak HA titers of the different cell lines tested in HA units were BHK21- 29, BT – 24, DF1-28, Hep2-26, MDCK – 24, PK15 – 25, QT35-27, RK13 – 24, Vero - 26. The virus grew most efficiently in BHK21, QT35 and DF1 cell lines, representing hamsters, quail, and chicken, respectively. In general, there was not a strong host range restriction in vitro, and each of the cell lines was able to execute efficient cleavage of the F protein without added protease, even at low moi (10-6 dilution of 210 HA units of the virus). Virus replication in all cell types was detected even at passage 1, but the CPE was more pronounced in subsequent passages. The general CPE observed in all the cell types involved rounding of cells and detachment of dead cells. Interestingly, syncytia formation, which is the hallmark of many paramyxoviruses, was absent. The virus failed to produce plaques in any cell line despite the use of different overlay media and plaque assay conditions. Examination of infected cell culture supernatant by electron microscopy confirmed the presence of virus particles whose morphology was characteristic of family Paramyxoviridae. The viruses observed by negative staining were pleomorphic, enveloped and 150-200 nm in size.
3.2. Determination of complete genome sequence of AMPV-2 strain Yucaipa
The genome of APMV-2 strain Yucaipa consists of 14,904 nt (GenBank accession number EU338414) and thus is the smallest among the members of subfamily Paramyxovirinae reported to date. The genome length is a multiple of six, conforming to the “rule of six” common to members of subfamily Paramyxovirinae (Calain and Roux, 1993; Samal and Collins, 1996). The genome organization of Yucaipa virus is 3′N-P/V-M-F-HN-L-5′, resembling that of NDV. The percentage of the genome that encodes protein is 92.37%, the same as the average coding percentage (92%) of other members of subfamily Paramyxovirinae (Miller et al., 2003). The length, position, and characteristics of the six genes and their intergenic sequences (IGS) are summarized in Table 1a and described in detail below.
Table 1.
Table 1a. Molecular features of genes and their proteins products of APMV-2 strain Yucaipa and subunit hexamer phasing position at gene start.
Table 1b. Gene start, Gene end and intergenic sequences of APMV-2 strain Yucaipa
| mRNA features (nt) | Deduced protein | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genes | Hexamer Phasing position at gene start | Total Length | 5′ UTR | ORF | 3′ UTR | Intergenic sequence (nt) | Size (aa) | MW (Da) | pI |
| N | 2 | 1547 | 85 | 1374 | 88 | 7 | 457 | 50481.19 | 5.321 |
| P/V (P) | 2 | 1379 | 71 | 1200 | 108 | 7 | 399 | 42280.21 | 5.557 |
| P/V (V) | 2 | 1380 | - | - | - | - | 232 | 25134.59 | 5.508 |
| P/V (W) | 2 | 1381 | - | - | - | - | 207 | 22168.30 | 6.456 |
| M | 2 | 1280 | 42 | 1110 | 128 | 23 | 369 | 40417.07 | 9.254 |
| F | 3 | 1707 | 54 | 1611 | 42 | 9 | 536 | 57692.77 | 5.099 |
| HN | 3 | 1899 | 76 | 1743 | 80 | 3 | 580 | 63869.87 | 7.667 |
| L | 3 | 6834 | 21 | 6729 | 84 | – | 2242 | 252621.16 | 7.421 |
| Genes | Gene End | IGS | Gene Start | ||||||
| / N | C5GCUGUAG | ||||||||
| N / P | AAAUUCU6 | CCUGGGA | C5GCUUCAA | ||||||
| P / M | AAAUUGU6 | CUUCAAA | C5GCUUCAG | ||||||
| M / F | UAAUUCU6 | GAAUUGAUGUAUUGAAGUUGUAA | C5GCUGUCG | ||||||
| F / HN | AAAUUCU6 | AACCUUCCA | C5GCUGUCG | ||||||
| HN / L | AAAUUCU6 | GAA | C5GCUUACG | ||||||
| L / | UAAUUCU6 | ||||||||
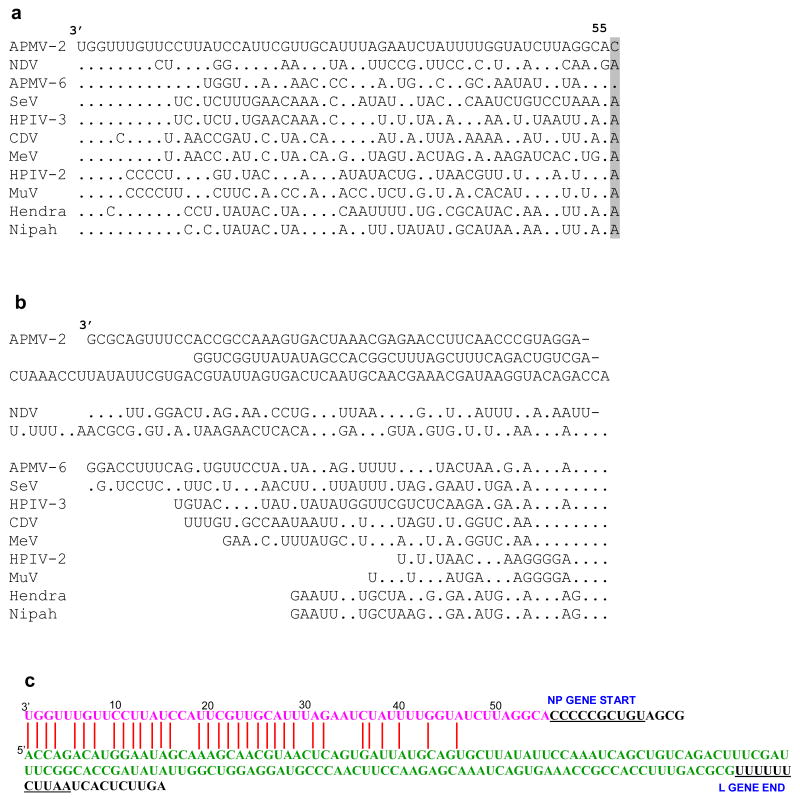
The 3′ leader sequence of APMV-2 strain Yucaipa is 55 nt, a length that generally is conserved among almost all of the members of subfamily Paramyxovirinae (Fig. 1a). The length of the 5′ trailer sequence is 154 nt, a property that is variable among the members of Paramyxovirinae (Fig. 1b). The first four nucleotides at the 3′ end of the leader (3′-UGGU) and the 5′ end of the trailer (5′-ACCA) sequences are identical in all Paramyxovirinae members (Fig. 1). The first 8 nucleotides of the leader (3′-UGGUUUGU) are conserved exactly in the avulaviruses (APMV-1 and APMV-6) and respiroviruses (BPIV-3, HPIV-3 and SeV) (Fig. 1), but are less well conserved compared with the other genera. The comparable sequences at the 5′ end of the genome were somewhat less conserved but showed a similar pattern among the different genera. The sequences of the 34 nucleotides at the 3′ leader and 5′ trailer termini are complementary, suggestive of conserved elements in the 3′ promoter regions of the genome and antigenome (Fig. 1c). We also identified a 3 time repeated motif (73UUCGGC78, 79UAGAGC84, 85UCUAGC90) in the N gene and another 3 time repeated motif (14832CUUUCG14827, 14826AUUUCG14821, 14820GCACCG14815) in the 5′ end of the genome. Thus, as seen in some paramyxoviruses, strain Yucaipa also has a bipartite promoter.
Fig. 1.
Alignment of the (a) 3′-leader and N gene start sequence (b) 5′-trailer regions of APMV-2 strain Yucaipa with other paramyxoviruses of subfamily Paramyxovirinae. Dots indicate identity with the assignment in APMV-2 (top line). The gene-start signal of the N gene of APMV-2 and APMV-6 are unique starting with C residue and they are shaded. (c) Complementary nucleotides between the 3′-leader and 5′-trailer regions of APMV-2 strain Yucaipa. Abbreviations: NDV, Newcastle disease virus (AMPV-1); SeV, Sendai virus; HPIV-3, human parainfluenza virus types 3; CDV, canine distemper virus; MeV, measles virus; MuV, mumps virus; HeV, Hendra virus; NiV, Nipah virus.
3.3. Sequences of transcription gene-start, gene-end, and intergenic sequences
The conserved gene-start sequence of APMV-2 strain Yucaipa is 3′-C5GCUG(U)U(C/A) while the conserved gene-end sequence is 3′-A(U)AAUUC(G)U6 (Table 1b). Thus, the first nucleotide of the mRNAs of APMV-2 strain Yucaipa (the gene start signal) is 5′-G (mRNA-sense) compared to A, for most of the other members of Paramyxovirinae mRNA have an A residue (Fig 1a), which also is the nucleotide assignment at the 5′ end of the genome and antigenome. Three other viruses in Paramyxovirinae that have G residue at the 5′ end of their mRNAs are Menangle, Tioman and APMV-6 (Wang et al., 2003). The APMV-2 strain Yucaipa IGS vary in length between 3 and 23 nt (Table 1b), whereas the IGS of other members of Paramyxovirinae are up to 45 nt in length (Wang et al., 2003), and they all end with an A residue (Table 1b), as observed in many paramyxoviruses (Collins et al., 1986; Crowley et al., 1988; Kawano et al., 1991; Chang et al., 2001), but otherwise there were no evident conserved IGS sequence motifs. The hexamer phasing positions of the gene start sequences of APMV-2 strain Yucaipa are 2, 2, 2, 3, 3 and 3 (Table 1), which are different from those of the viruses within the genus Avulavirus namely, APMV-1 (2, 4, 3, 3, 2 and 5) and APMV-6 (2, 2, 2, 2, 2, 4 and 4) (Kolakofsky et al., 1998), while all the members of a particular genus within the family share the same pattern of hexamer phasing positions (Wang et al., 2003).
3.4. The nucleoprotein (N) gene
The N gene is 1547 nt long with a major ORF of 1374 nt. The encoded protein is 457 amino acids (aa) long, with a predicted molecular weight (Mr) of 50,481 kDa. The N protein of strain Yucaipa has 55.8% and 41.3% amino acid sequence identity, respectively, with that of APMV-6 and APMV-1 of genus Avulavirus. The extent of amino acid sequence identity with members of the other genera of subfamily Paramyxovirinae decreased in the order: rubulaviruses (36.5 - 41.4 %), henipaviruses (28.9-29.4 %), morbilliviruses (23.7-24.2 %), and respiroviruses (17.3-19.7 %). An amino acid sequence motif that is highly conserved in the N proteins of members of Paramyxovirinae and is thought to be involved in N-N self assembly, F-X4-Y-X3-Φ-S-Φ-A-M-G, where X represents any amino acid residue and Φ represents an aromatic amino acid residue (Morgan, 1991), is seen within the central domain of the strain Yucaipa N protein (324FAPANFSTLYSYAMG338). In SeV and in other paramyxoviruses, residue Y260, needed for N protein binding with RNA, and residue F324, needed for correct self-assembly (Myers et al., 1997), are also conserved at the same amino acid sequence number in strain Yucaipa.
3.5. The phosphoprotein (P) gene and P/V/W editing
The P gene is 1379 nt long with a major ORF of 1200 nt. The P protein is encoded by the unedited mRNA; the addition of a single G residue to the editing site would yield a predicted V protein; and the addition of 2 G residues would yield a predicted W protein, as is the case with NDV (Steward et al., 1993). The putative editing site of the strain Yucaipa P gene is 5′-AAAAAGGGG (mRNA sense) at nt position 2090-2102 in the viral RNA genome, while the P gene editing sequence of NDV and other paramyxoviruses with similar coding strategy is AAAAA(A)GGG (Fig. 2). The P protein is 399 amino aa long, with a predicted Mr of 42.28 KDa; the V protein is 232 aa long with a Mr of 25.13 KDa; the predicted W protein would be 207 aa length with a Mr of 22.16 KDa, The amino acid sequence of strain Yucaipa P protein has 28.1% and 27.5% identity, respectively, with that of APMV-1 and APMV-6 of genus Avulavirus. The extent of amino acid sequence identity with the P proteins of members of the other genera of Paramyxovirinae decreased in the order: rubulaviruses (23-28%), morbilliviruses (15.8-16%), henipaviruses (14.9-15.4 %), and respiroviruses (8.6-9.3%). The V protein has 34.2% and 34.4% amino acid sequence identity with that of APMV-1 and Porcine Rubulavirus (PoRV), respectively, and has 17 conserved residues with other paramyxoviruses including 7 cysteine residues in the C terminal portion that resemble the zinc-finger like motif found in other paramyxoviruses (Fig 3).
Fig. 2.
RNA editing sites in the P genes of selected members of the five genera of subfamily Paramyxoviridae, shown as positive (mRNA) sense sequence. The number of inserted G nucleotides necessary to yield V (or, in the case of Rubulavirus, P) is indicated.
Fig. 3.
Sequence alignment of the C-terminal end of the V proteins of representative members of subfamily Paramyxovirinae. Dots indicate identity with the assignment in AMPV-2 (top line), and the seven characteristic cysteine residues are shaded grey. Note that at two positions, the assignments W and D that are conserved in the other viruses are not conserved in APMV-2 strain Yucaipa (bold and underlined S and P assignments in the AMPV-2 sequence).
3.6. The matrix protein (M) gene
The M gene is 1280 nt long with a major ORF of 1110 nt. The encoded protein is 369 aa long, with a predicted Mr of 40.41 KDa. The matrix protein showed 42.5% and 31.2% amino acid sequence identity with those of APMV-6 and APMV-1, respectively. The extent of amino acid sequence identity with members of the other genera of Paramyxovirinae decreased in the order: rubulaviruses (30%), morbilliviruses (18.5 - 21.8 %), henipaviruses (18.2 %). and respiroviruses (16.4 -18.1 %).
3.7. The fusion protein (F) gene
The F gene is 1707 nt with a major ORF of 1611 nt. The F protein is 536 aa long with a predicted Mr of 57.69 KDa. Among the available paramxyxovirus sequences, the F protein of Yucaipa was most closely related (80.6%) to that of Murayama virus (MrV), a monkey paramyxovirus that is antigenically related to Yucaipa virus and for which the F and HN sequences are available (Nishikawa et al., 1977, GenBank accession no. D13990). In contrast, the Yucaipa virus F protein was less closely related to APMV-1 and APMV-6 (42.2% and 49.8 % identity, respectively). The extent of amino acid sequence identity with members of the other genera of Paramyxovirinae decreased in the order: rubulaviruses (29.4-31.2%), henipaviruses (27.5-27.7 %), morbilliviruses (21.9-26.4%), and respiroviruses (22.5-22.8%). The first 18 aa of the Yucaipa virus F protein are highly hydrophobic and are predicted to contain the signal sequence.
Compared to the Yucaipa virus F gene sequence available in GenBank (Accession no. AF422844), the present consensus sequence has nucleotide differences in F gene at positions 4309, 4310, 4786, 5099 and 5489 (assignments of G, C, C, C and T in our sequence compared to C, G, A, T and C in the previous sequence). This resulted in one amino acid difference at position 140, which is alanine in the present sequence and glutamic acid in the previous sequence. The alignment of the F protein cleavage sites of APMV-1, -2, -6 and MrV are shown in Fig. 4. The putative F protein cleavage site contained a monobasic residue but a phenylalanine at the beginning of the F1 subunit.
Fig. 4.
Comparison of the F protein cleavage site (left of arrow) and resulting N-terminus of the F1 subunit (right of arrow) of the sequences available for members of genus Avulavirus. Basic amino acids are underlined, and residues conserved with that of virulent NDV are shaded. Both APMV-2 strain Yucaipa and Murayama virus (an APMV-2-like virus)have two basic amino acids in the cleavage site like avirulent NDV but have the phenylalanine residue at the N terminus of F1 like that of virulent NDV.
3.8. The hemagglutination-neuraminidase protein (HN) gene
The HN gene is 1899 nt long with a single ORF of 1743 nt. The encoded protein is 580 aa long with predicted Mr of 63.86 KDa. The strain Yucaipa HN protein has 75% amino acid sequence identity with that of MrV and 43.6% and 36% identity with that of APMV-6 and APMV-1. There was a lower level of sequence identity with the HN proteins of rubulaviruses (29%), respiroviruses (21-23.7%), morbilliviruses (10.4-11.6%) and with the attachment protein of Henipaviruses (17.6-18.2%). By aligning the HN sequence of APMV-1 and APMV-2 strain Yucaipa, the 6 conserved neuraminidase active sites were identified as R175, E400, R415, R505, Y533, E554 equivalent to R174, E401, R416, R498, Y526, E547 of NDV (Langedijk et al., 1997). The hexapeptide NRKSCS, thought to form part of the sialic acid binding site is present at aa positions 235 to 240 (Mirza et al., 1994). Five potential N-linked glycosylation sites are found, at N 120, N 279, N 346, N 391, N 488, compared to six potential sites in NDV Beaudette C strain at N119, N341, N433, N481, N500 and N538. The HN protein of strain Yucaipa has all the 11 conserved cysteine residues in the region corresponding to the globular head, as also found in NDV. The sequence that we determined for the HN gene has two differences compared to the Yucaipa virus HN gene sequence available in GenBank (Accession no. AF422844). One involves position 6774 (residue T compared to residue G in the previous sequence) resulting in a histidine in our sequence compared to glutamine in the previous sequence. The second difference was found at position 7768 (T to G), which did not result in an amino acid coding change.
3.9. The large polymerase protein (L) gene
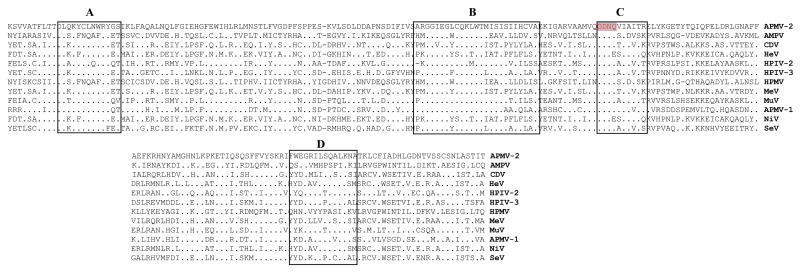
The L gene is 6834 nt long, with a 6729 nt long ORF. The L protein is 2242 aa long with predicted Mr of 252.62 KDa. The Yucaipa L protein has 44.1% and 39.6% amino acid sequence identity with that of APMV-6 and APMV-1. The extent of amino acid sequence identity with the L proteins of members of the other general of Paramyxovirinae decreased in the order: rubulaviruses (37.5%), respiroviruses (31.2%), morbilliviruses (30.3 %), and henipaviruses (25.2-25.7 %). The six stronglyconserved linear domains of L proteins of nonsegmented negative-strand RNA viruses (Poch et al., 1990) are also identified within the L protein of strain Yucaipa (Fig. 5). The conserved GDNQ sequence motif within domain III concerned with L protein transcription activity (Schnell and Conzelmann, 1995) was found in the L protein of strain Yucaipa at aa positions 774 to 777.
Fig.5.
Amino acid alignment of domain III of the L proteins of selected members of family Paramyxoviridae. The putative polymerase core motifs (A, B, C and D) are boxed within domain III (Poch et al., 1990). Amino acid identity with APMV-2 (top line) is indicated by dots, while gaps introduced to maximize amino acid identity are indicated by dashes. A conserved GDNQ motif implicated in L protein transcription activity (Schnell and Conzelmann, 1995) is shaded.
3.10. Phylogenetic analysis
Phylogenetic trees were generated from amino acid sequence alignments of the N, P, M, F, HN and L proteins of strain Yucaipa with the cognate proteins of prototype viruses of all the five genera of family Paramyxoviridae. The phylogenetic trees clearly indicate the close genetic relationship between APMV-2 strain Yucaipa and APMV-6, and strongly supporting the classification of APMV-1, APMV-2, APMV-3, APMV-4 and APMV-6 under the genus Avulavirus. The phylogenetic trees for the proteins F and HN of members of family Paramyxoviridae are shown in the fig 6.
Fig. 6. Phylogenetic analysis of F and HN proteins of members of family Paramyxoviridae.
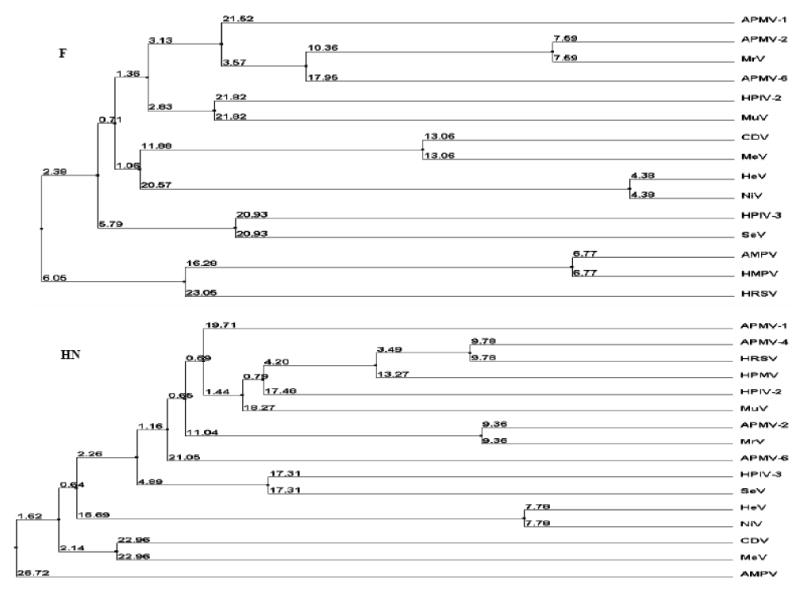
The phylogenetic trees were constructed by average distance using percentage identity in the T-COFFEE program. The numbers represent the amino acid sequence distance between the viruses.
4. Discussion
Nine serological types of avian paramyxoviruses have been isolated around the world. The disease potential and molecular features of these viruses are mostly unknown apart from APMV-1. It is important to characterize these common viruses. Here, we present the complete genome sequence of APMV-2 strain Yucaipa. APMV-2 strain Yucaipa has the shortest genome in subfamily Paramyxovirinae (14,904 nt) described to date, being 276 nt shorter than the next smallest genome, that of PoRV. The pattern of sequence relatedness clearly places APMV-2 in genus Avulavirus, consistent with the International Committee on Taxonomy of Viruses statement that the amino acid sequence relationships are the main criteria for grouping viruses into genera within the family Paramyxoviridae (Lamb et al., 2000). This is offered with the caveat that most of the serotypes of Avulavirus remain to be sequenced, and so the extent of diversity within the genus is unknown. Sequence identity between Yucaipa virus and members of the other genera of subfamily Paramyxovirinae was greatest with rubulaviruses and usually (except for the L protein) was least with the respiroviruses, and was intermediate with the morbilliviruses and henipaviruses.
The classification of Yucaipa virus in Avulavirus also is supported by (i) the absence of a C protein, which is present in respiroviruses, morbilliviruses, and henipaviruses and is encoded by an alternative ORF in the P gene, (ii) the presence of intergenic regions of nonconserved length and sequence, as are found in avulaviruses and rubulaviruses but not in respiroviruses, morbilliviruses, or henipaviruses, and (iii) the pattern of P/V RNA editing in which the nonedited mRNA encodes P and an edited version encodes V, which distinguishes avulaviruses and the other members of subfamily Pneumovirinae from rubulaviruses. Whereas most members of subfamily Paramyxovirinae initiate their mRNAs with an A residue, Yucaipa virus is predicted to use G, a feature that is shared with APMV-6 (but APMV-1), the rubulaviruses, Tioman and Menangle viruses, and HRSV and HMPV of subfamily Pneumovirinae.
The paramyxovirus F protein is synthesized as an inactive precursor (F0) and is cleaved to two biologically-active disulfide-bonded F1–F2 subunits by host protease (Lamb and Parks, 2007). The F protein cleavage site is a well-characterized determinant of NDV pathogenicity in chickens. Virulent NDV strains typically contain a polybasic cleavage site that contains the preferred recognition site for furin (R-X-K/R-R↓), which is an intracellular protease that is present in most cells. This provides for efficient cleavage in a wide range of tissues, making it possible for virulent strains to spread systemically. In contrast, avirulent NDV strains typically have basic residues at the −1 and −4 positions relative to the cleavage site and depend on secretory protease (or, in cell culture, added trypsin) for cleavage. This limits the replication of avirulent strains to the respiratory and enteric tracts where the secretory protease is found. The putative cleavage site of strain Yucaipa F protein (DKPASR↓F) has basic residues (underlined), which is similar but not identical to the pattern of avirulent NDV strains. Conversely, the F1 subunit of Yucaipa virus begins with a phenylalanine residue, as is characteristic of virulent NDV strains, rather than a leucine reside, as seen in most avirulent NDV strains (Collins et al., 1993). We found that the Yucaipa virus replicated in a wide range of cells without the addition of exogenous protease, and the inclusion of protease did not improve the efficiency of replication. This is incongruent with the observation that the F protein cleavage site is not polybasic and does not conform to the preferred furin motif. Thus, the Yucaipa virus is an example of paramyxovirus in which efficient intracellular cleavage occurs in the absence of an apparent furin motif. As another example, whereas wild type SeV contains a single basic residue at the cleavage site (GAPQSR↓) and is strictly dependent on added trypsin for infectivity in vitro, a number of experimentally-derived mutants of SeV have been described that are trypsin-independent and yet have not acquired a furin site (Okada et al., 1998). Sequence analysis identified a number of mutations occurring upstream of the cleavage site, one being a S-to-P substitution at position –2 relative to the cleavage site and another involving the loss of an upstream glycosylation site. It may be that these mutations altered the local protein structure to make the cleavage site more accessible and thus more readily cleaved. Certain naturally occurring isolates of HPIV-3 contain glutamate at the –2 position (DPRTKR↓), thus lacking a furin site, whereas other isolates possess a furin site. The two types of isolates appeared to replicate with equal efficiency in vitro and in the respiratory tract of rhesus monkeys (Coelingh and Winter, 1990). Yet another example involves NiV and HeV, in which intracellular cleavage does not depend on a furin site, and indeed does not even require a basic residue in the –1 position (Moll et al., 2004). This suggests that some paramyxovirus F proteins can be cleaved by proteases in addition to the furin- or trypsin-related ones. However, the study with the SeV mutants indicated above showed that, while a number of mutants lacking the preferred furin cleavage site were competent for efficient multi-cycle replication in vitro without added protease, some were restricted in their ability to form plaques and spread systemically. Similarly, the lack of a furin site might explain the inability of Yucaipa virus to form plaques and might also correlate with reduced virulence in birds.
The F and HN proteins of strain Yucaipa were more closely related to those of MrV (80.6% and 75% amino acid sequence identity (respectively) than those of any other paramyxovirus, including NDV. MrV is a paramyxovirus that was isolated in 1973 from cynomolgous monkeys experiencing mild respiratory tract disease (Nishikawa et al., 1977). MrV exhibited no serological relationship with mammalian paramyxoviruses, but crossreacted with APMV-2 strain Yucaipa. The high level of sequence relatedness between the F and HN gene and proteins of Yucaipa virus and MrV (F and HN are the only sequences available for MrV) provide strong support for the interpretation that these viruses indeed are members of the same serotype, APMV-2. Avulavirus appears to contain at least one virus that can infect and cause disease in a non-avian host. Yucaipa virus also was shown to infect and cause disease in a non-avian host, namely guinea pigs. These observations support the previous suggestion that MrV might have evolved by adapting in monkeys after infection with APMV-2 strain Yucaipa or a similar avian AMPV-2 strain (Kusagawa et al., 1993). It will be interesting to sequence the complete genome of MrV and other APMV-2 strains (Ozdemir et al, 1990) and compare it with that of strain Yucaipa, as well as to compare the relative permissiveness of avian and mammalian hosts for these viruses.
Acknowledgments
We thank Daniel Rockemann and all our laboratory members for their excellent technical assistance and help. We also thank Hamp Edwards for performing the electron micrograph and Ireen Dryburgh-Barry for proof reading of this manuscript. “This research was supported by NIAID contract no. N01A060009 (85% support) and NIAID, NIH Intramural Research Program (15% support). The views expressed herein do not necessarily reflect the official policies of the Department of Health and Human Services; nor does mention the trade names, commercial practices or organizations imply endorsement by the U.S. Government.”
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander DJ. Avian Paramyxoviruses. The Vet Bull. 1980;50:737–751. [Google Scholar]
- Alexander DJ, Allan WH, Parsons G, Collins MS. Identification of paramyxoviruses isolated from birds dying in quarantine in Great Britain. Vet Rec. 1982;111:571–574. [PubMed] [Google Scholar]
- Alexander DJ, Pattisson M, Macpherson Avian paramyxoviruses of PMV-3 serotype in British turkeys. Avian Pathol. 1983;12:469–482. doi: 10.1080/03079458308436192. [DOI] [PubMed] [Google Scholar]
- Alexander DJ. The classification, host range and distribution of avian paramyxoviruses. In: McFerran JB, McNulty MS, editors. Acute viral infections of Poultry. 1986. pp. 52–66. [Google Scholar]
- Alexander DJ. Newcastle disease and other avian Paramyxoviridae infections. In: Calnek BW, editor. Diseases of Poultry. 10th. Iowa State University Press; Ames: 1997. pp. 541–569. [Google Scholar]
- Alexander DJ. Avian Paramyxoviruses 2-9. In: Saif YM, editor. Diseases of poultry. 11th. Iowa State University Press; Ames: 2003. pp. 88–92. [Google Scholar]
- Asahara T, Yoshimura M, Tusubaki S, Yamagamt T, Aoi T, Ide S, Masu S. Isolation in Japan of a virus similar to myxovirus Yucaipa (MVY) Bull Azabu Vet Coll. 1973;26:67–81. [Google Scholar]
- Bankowski RA, Corstert RE, Clark CT. Isolation of an unidentified agent from the respiratory tract of chickens. Science. 1960;132:292–293. doi: 10.1126/science.132.3422.292. [DOI] [PubMed] [Google Scholar]
- Bankowski RA, Conrad RD, Reynolds B. Avian Influenza A and paramyxoviruses complicating respiratory diagnosis in poultry. Avian Dis. 1968;12:259–278. [PubMed] [Google Scholar]
- Bankowski RA, Almquist J, Dombrucki J. Effect of paramyxovirus Yucaipa on fertility, hatchability, and poult yield of turkeys. Avian Dis. 1981;25:517–520. [PubMed] [Google Scholar]
- Calain P, Roux L. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol. 1993;67:4822–4830. doi: 10.1128/jvi.67.8.4822-4830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PC, Hsieh ML, Shien JH, Graham DA, Lee MS, Shieh HK. Complete nucleotide sequence of avian paramyxovirus type 6 isolated from ducks. J Gen Virol. 2001;82:2157–2168. doi: 10.1099/0022-1317-82-9-2157. [DOI] [PubMed] [Google Scholar]
- Coelingh KVW, Winter CC. Naturally occurring Human Parainfluenza Type 3 viruses exhibit divergence in amino acid sequence of their fusion protein neutralization epitopes and cleavage sites. J Virol. 1990;64:1329–1334. doi: 10.1128/jvi.64.3.1329-1334.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collings DF, Fitton J, Alexander DJ, Harkness JW, Pattison M. Preliminary characterization of a paramyxovirus isolated from a parrot. Res Vet Sci. 1975;19:219–221. [PubMed] [Google Scholar]
- Collins MS, Bashiruddin JB, Alexander DJ. Deduced amino acid sequences at the fusion protein cleavage site of Newcastle disease viruses showing variation in antigenicity and pathogenicity. Arch Virol. 1993;128:363–370. doi: 10.1007/BF01309446. [DOI] [PubMed] [Google Scholar]
- Collins PL, Dickens LE, Buckler-White A, Olmsted RA, Spriggs MK, Camargo E, Coelingh KV. Nucleotide sequences for the gene junctions of human respiratory syncytial virus reveal distinctive features of intergenic structure and gene order. Proc Natl Acad Sci USA. 1986;83:4594–4598. doi: 10.1073/pnas.83.13.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley JC, Dowling PC, Menonna J, Silverman JI, Schuback D, Cook SD, Blumberg BM. Sequence variability and function of measles virus 3′ and 5′ ends and intercistronic regions. Virology. 1988;164:498–506. doi: 10.1016/0042-6822(88)90564-8. [DOI] [PubMed] [Google Scholar]
- de Leeuw O, Peeters BJ. Complete nucleotide sequence of Newcastle disease virus: evidence for the existence of a new genus within the subfamily Paramyxovirinae. J Gen Virol. 1999;80:131–136. doi: 10.1099/0022-1317-80-1-131. [DOI] [PubMed] [Google Scholar]
- Fleury HJA, Alexander DJ. Isolation of twenty-three Yucaipa-like viruses from 616 wild birds in Senegal, West Africa. Avian Dis. 1979;23:742–744. [PubMed] [Google Scholar]
- Goodman BB, Hanson RP. Isolation of avian paramyxovirus-2 from domestic and wild birds in Costa Rica. Avian Dis. 1988;32:713–717. [PubMed] [Google Scholar]
- Kawano M, Okamoto K, Bando H, Kondo K, Tsurudome M, Komada H, Nishio M, Ito Y. Characterization of the human parainfluenza type 2 virus gene encoding the L protein and the intergenic sequences. Nucleic Acids Res. 1991;19:2739–2746. doi: 10.1093/nar/19.10.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolakofsky D, Pelet T, Garcin D, Hausmann S, Curran J, Roux L. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol. 1998;72:891–899. doi: 10.1128/jvi.72.2.891-899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy S, Samal SK. Nucleotide sequences of the trailer, nucleocapsid protein gene and intergenic regions of Newcastle disease virus strain Beaudette C and completion of the entire genome sequence. J Gen Virol. 1998;79:2419–2424. doi: 10.1099/0022-1317-79-10-2419. [DOI] [PubMed] [Google Scholar]
- Kusagawa S, Komada H, Mao X, Kawano M, Nishikawa F, Tsurudome M, Matsumura H, Ohta H, Yuasa T, Nishio M, Ito Y. Antigenic and molecular properties of Murayama virus isolated from cynomolgus monkeys: the virus is closely related to avian paramyxovirus type 2. Virology. 1993;194:828–32. doi: 10.1006/viro.1993.1325. [DOI] [PubMed] [Google Scholar]
- Lamb RA, Collins PL, Kolakofsky D, Melero JA, Nagai Y, Oldstone MBA, Pringle CR, Rima BK. Family Paramyxoviridae. In: van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, Mc-Geoch DJ, Pringle CR, Wickner RB, editors. Virus Taxonomy: Classification and Nomenclature of Viruses. The Seventh report of the International Committee for Taxonomy of Viruses. San Diego: Academic Press; 2000. pp. 549–56l. [Google Scholar]
- Lamb RA, Parks GD. Paramyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th. Lippincott Williams and Wilkins; Philadelphia: 2007. pp. 1449–1496. [Google Scholar]
- Lamb RA, Collins PL, Kolakofsky D, Melero JA, Nagai Y, Oldstone MBA, Pringle CR, Rima BK. Family Paramyxoviridae. In: Fauquet CM, editor. Virus Taxonomy: The Classification and Nomenclature of Viruses. The Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier Acedemic Press; 2005. pp. 655–668.pp. 655–668. [Google Scholar]
- Lang G, Gagnon A, Howell J. Occurrence of paramyxovirus Yucaipa in Canadian poultry. Can Vet J. 1975;16:233–237. [PMC free article] [PubMed] [Google Scholar]
- Langedijk JP, Daus FJ, van Oirschot JT. Sequence and structure alignment of Paramyxoviridae attachment proteins and discovery of enzymatic activity for a morbillivirus hemagglutinin. J Virol. 1997;71:6155–6167. doi: 10.1128/jvi.71.8.6155-6167.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Yu M, Zhang H, Wang HY, Wang LF. Improved rapid amplification of cDNA ends (RACE) for mapping both the 5′ and 3′ terminal sequences of paramyxovirus genomes. J Virol Methods. 2005;130:154–156. doi: 10.1016/j.jviromet.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Lipkind M, Weisman Y, Shihmanter E, Shoham D, Aronovici A. The isolation of Yucaipa-like paramyxoviruses from epizootics of a respiratory disease in turkey poultry farms in Israel. Vet Rec. 1979;105:577–578. [PubMed] [Google Scholar]
- Lipkind M, Weisman Y, Shihmanter E, Shoham D, Aronovici A. Isolation of Yucaipa-like avian paramyxovirus from a wild mallard duck (Anas platyrhinchos) wintering in Israel. Vet Rec. 1982;110:15–16. doi: 10.1136/vr.110.1.15. [DOI] [PubMed] [Google Scholar]
- Lipkind M, Shihmanter E. Antigenic relationships between avian paramyxoviruses. I. Quantitative characteristics based on hemagglutination and neuraminidase inhibition tests. Arch Virol. 1986;89:89–111. doi: 10.1007/BF01309882. [DOI] [PubMed] [Google Scholar]
- Mbugua HCW, Karstad L. Isolation of avian paramyxoviruses (Yucaipa-like) from wild birds in Kenya, 1980–1982. J Wildl Dis. 1985;21:52–54. doi: 10.7589/0090-3558-21.1.52. [DOI] [PubMed] [Google Scholar]
- Miller PJ, Boyle DB, Eaton BT, Wang LF. Full-length genome sequence of Mossman virus, a novel paramyxovirus isolated from rodents in Australia. Virology. 2003;317:330–344. doi: 10.1016/j.virol.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Mirza AM, Deng R, Iorio RM. Site-directed mutagenesis of a conserved hexapeptide in the paramyxovirus hemagglutininneuraminidase glycoprotein: Effects on antigenic structure and function. J Virol. 1994;68:5093–5099. doi: 10.1128/jvi.68.8.5093-5099.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll M, Diederich S, Klenk HD, Czub M, Maisner A. Ubiquitous activation of the Nipah virus fusion protein does not require a basic amino acid at the cleavage site. Journal of Virology. 2004;78:9705–9712. doi: 10.1128/JVI.78.18.9705-9712.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan EM. Evolutionary relationships of paramyxovirus nucleocapsid-associated proteins. In: Kingsbury DW, editor. The Paramyxoviruses. Plenum Press; New York: 1991. pp. 163–179. [Google Scholar]
- Myers TM, Pieters A, Moyer SA. A highly conserved region of the Sendai virus nucleocapsid protein contributes to the NP-NP binding domain. Virology. 1997;229:322–335. doi: 10.1006/viro.1996.8429. [DOI] [PubMed] [Google Scholar]
- Nishikawa F, Sugiyama T, Suzuki K. A new Paramyxovirus isolated from cynomolgus monkeys. Jpn J Med Sci Biol. 1977;30:191–204. doi: 10.7883/yoken1952.30.191. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins D, Heringa J. T-Coffee: A novel method for multiple sequence alignments. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Nymadawa P, Konstantinow-siebelist I, Schulze P, Starke G. Isolation of paramyxoviruses from free-flying birds of the order Passeriformes in German Democratic Republic. Acta Virol. 1977;56:345–351. [PubMed] [Google Scholar]
- Ozdemir I, Russell PH, Collier J, Alexander DJ, Manvell RJ. Monoclonal antibodies to avian paramyxovirus type 2. Avian Pathol. 1990;19:395–400. doi: 10.1080/03079459008418689. [DOI] [PubMed] [Google Scholar]
- Okada H, Seto JT, McQueen NL, Klenk HD, Rott R, Tashiro M. Determinants of pantropism of the F1-R mutant of Sendai virus: specific mutations involved are in the F and M genes. Arch Virol. 1998;143:2343–2352. doi: 10.1007/s007050050465. [DOI] [PubMed] [Google Scholar]
- Poch O, Blumberg BM, Bougueleret L, Tordo N. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J Gen Virol. 1990;71:1153–1162. doi: 10.1099/0022-1317-71-5-1153. [DOI] [PubMed] [Google Scholar]
- Samal SK, Collins PL. RNA replication by a respiratory syncytial virus RNA analog does not obey the rule of six and retains a nonviral trinucleotide extension at the leader end. J Virol. 1996;70:5075–5082. doi: 10.1128/jvi.70.8.5075-5082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell M, Conzelmann KK. Polymerase activity of in vitro mutated rabies L protein. Virology. 1995;214:522–530. doi: 10.1006/viro.1995.0063. [DOI] [PubMed] [Google Scholar]
- Senne DA, Pearson JE, Miller LD, Gustafson GA. Virus isolations from pet birds submitted for importation into the United States. Avian Dis. 1983;27:731–744. [PubMed] [Google Scholar]
- Shihmanter E, Weisman Y, Manwell R, Alexander DJ, Lipkind M. Mixed paramyxovirus infection of wild and domestic birds in Israel. Vet Microbiol. 1997;58:73–78. doi: 10.1016/s0378-1135(97)00147-8. [DOI] [PubMed] [Google Scholar]
- Steward M, Vipond IB, Millar NS, Emmerson PT. RNA editing in Newcastle disease virus. J Gen Virol. 1993;74:2539–2547. doi: 10.1099/0022-1317-74-12-2539. [DOI] [PubMed] [Google Scholar]
- Troutt AB, Mcheyzer-Williams MG, Pulendran B, Nossal GJ. Ligation-anchored PCR: A simple amplification technique with single-sided specificity. Proc Natl Acad Sci USA. 1992;89:9823–9825. doi: 10.1073/pnas.89.20.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LF, Eaton BT. Emerging paramyxoviruses. Infect Dis Rev. 2001;3:52–69. [Google Scholar]
- Wang LF, Chua KB, Yu M, Eaton BT. Genome Diversity of Emerging Paramyxoviruses. Curr Genomics. 2003;4:263–273. [Google Scholar]
- Wang LF, Hansson E, Yu M, Chua KB, Mathe N, Crameri G, Rima BK, Moreno-Lopez J, Eaton BT. Full-length genome sequence and genetic relationship of two paramyxoviruses isolated from bat and pigs in the Americas. Arch Virol. 2007;152:1259–1271. doi: 10.1007/s00705-007-0959-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman Y, Aronovici A, Malkinson M, Shihmanter E, Lipkind M. Isolation of paramyxoviruses from pigeons in Israel. Vet Rec. 1984;115:605. doi: 10.1136/vr.115.23.605-a. [DOI] [PubMed] [Google Scholar]
- Zhang GZ, Zhao JX, Wang HW, Yang AM, Bu CY, Wang M. Isolation, identification and comparison of four isolates of avian paramyxovirus serotype 2 in China. Avian Dis. 2006;50:386–390. doi: 10.1637/7502-010906R1.1. [DOI] [PubMed] [Google Scholar]
- Zhang GZ, Zhao JX, Wang M. Serological Survey on Prevalence of Antibodies to Avian Paramyxovirus Serotype 2 in China. Avian Dis. 2007;51:137–139. doi: 10.1637/0005-2086(2007)051[0137:SSOPOA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]