Abstract
The 2 micron plasmid of Saccharomyces cerevisiae uses the Kip1 motor, but not the functionally redundant Cin8 motor, for its precise nuclear localization and equal segregation. The timing and lifetime of Kip1p association with the plasmid partitioning locus STB are consistent with Kip1p being an authentic component of the plasmid partitioning complex. Kip1–STB association is not blocked by disassembling the mitotic spindle. Lack of Kip1p disrupts recruitment of the cohesin complex at STB and cohesion of replicated plasmid molecules. Colocalization of a 2 micron reporter plasmid with Kip1p in close proximity to the spindle pole body is reminiscent of that of a CEN reporter plasmid. Absence of Kip1p displaces the plasmid from this nuclear address, where it has the potential to tether to a chromosome or poach chromosome segregation factors. Exploiting Kip1p, which is subsidiary to Cin8p for chromosome segregation, to direct itself to a “partitioning center” represents yet another facet of the benign parasitism of the yeast plasmid.
Introduction
The yeast 2 micron plasmid is a circular multicopy selfish DNA element in the nucleus that propagates itself without conferring any apparent advantage to its host (Velmurugan et al., 2003; Jayaram et al., 2004; Ghosh et al., 2006). At its steady-state copy number of ∼40–60 per cell, the plasmid poses no significant impediment to the host's fitness. The genetic organization of the plasmid is an efficient evolutionary design for stable persistence at a high, but regulated, copy number.
The 2 micron circle genome can be divided into a partitioning system and a copy number control system. The copy number control system is called into play only to counter rare missegregation events. The resulting drop in plasmid molecules in one of the two daughter cells is corrected by DNA amplification mediated by the plasmid coded Flp site-specific recombinase (Futcher, 1986; Volkert and Broach, 1986). Tight negative regulation of amplification safeguards against runaway increase in copy number.
The partitioning system consists of plasmid coded proteins Rep1p and Rep2p and a cis-acting locus STB located near the replication origin. Despite its multicopy status, the 2 micron plasmid is partitioned as one clustered entity consisting of ∼3–5 closely knit plasmid foci. This reduction in copy number causes stable plasmid propagation to be absolutely dependent on the Rep–STB system. The nearly chromosome-like stability of the plasmid results from the ability of the partitioning system to couple plasmid segregation to chromosome segregation (Velmurugan et al., 2000; Mehta et al., 2002). A critical feature of this coupling is the Rep1p- and Rep2p-mediated recruitment of the yeast cohesin complex to STB, followed by one-to-one segregation of duplicated plasmid clusters to daughter cells (Mehta et al., 2002). Single-copy plasmid derivatives reveal that cohesin recruitment results in sister plasmid cohesion in metaphase cells (Ghosh et al., 2007). Hitherto known features of 2 micron circle segregation are accommodated by a chromosome tethering model in which sister plasmid clusters hitchhike to opposite cell poles on sister chromatids (Mehta et al., 2002). However, chromosome-independent, though cohesin-mediated, plasmid segregation cannot be ruled out entirely.
The integrity of the mitotic spindle is a critical requirement for plasmid segregation. The key spindle-dependent steps are: tight clustering of plasmid foci, precise nuclear positioning of the cluster in proximity to the spindle pole body, localization of the plasmid in chromosome spreads, association of the histone H3 variant Cse4p with STB, and assembly of the cohesin complex at STB (Velmurugan et al., 2000; Mehta et al., 2002; Hajra et al., 2006; Ghosh et al., 2007). Every known step of the plasmid partitioning pathway, except for the association of the Rep proteins and components of the RSC2 chromatin remodeling complex with STB, is blocked by disrupting the spindle (Mehta et al., 2005; unpublished data). Apparently, spindle-mediated organization and localization of the plasmid cluster are early events in assembling the partitioning complex. However, the molecular mechanisms underlying this spatial control have remained unclear.
The mitotic spindle is a dynamic molecular machine that, in conjunction with associated motor and nonmotor proteins, orchestrates faithful chromosome segregation (Manning and Compton, 2008). Five of the six kinesin-related motor proteins of Saccharomyces cerevisiae have demonstrated roles in spindle function (Hildebrandt and Hoyt, 2000). Of these, Cin8p and Kip1p (kinesin-5 family) are entirely nuclear, Kar3p (kinesin-14 family) and Kip3p (kinesin-8 or -13 family) are nuclear as well as cytoplasmic, and Kip2p (kinesin-7 family) is exclusively cytoplasmic. S. cerevisiae also harbors a single cytoplasmic dynein. These spindle-associated motors generate pulling and pushing forces that control spindle length as well as nuclear migration, cross-link and bundle microtubules, and regulate microtubule dynamics to promote synchronized poleward movement of sister chromatids.
All four nuclear motors of S. cerevisiae perform distinct functions related to kinetochore organization and dynamics (Tytell and Sorger, 2006). Cin8p and Kip1p are responsible for the typical bi-lobed metaphase configuration of kinetochores. Kip3p promotes depolymerization of kinetochore microtubules during anaphase and translocation of sister kinetochores in opposite directions. Kar3p, which localizes primarily to the spindle pole, also localizes preferentially to kinetochores detached from the spindle. Kar3p facilitates the lateral movement of kinetochores toward the spindle pole after their capture by microtubules (Tanaka et al., 2005). This function is important during early S phase, when centromeres become dissociated from the spindle during their replication (Kitamura et al., 2007).
The nuclear localization of the 2 micron plasmid is similar to that of centromeres. Furthermore, the 2 micron plasmid resembles CEN plasmids or chromosomes in the dynamics and kinetics of segregation (Velmurugan et al., 2000; Ghosh et al., 2007). The multiple functional contributions of motor proteins at kinetochores suggested to us that the role of the spindle in 2 micron circle partitioning may be mediated through one or more of these motors. A spindle-associated motor protein may deliver the plasmid cluster to a locale where it could tether to a chromosome and/or access host factors required for plasmid partitioning. In support of this hypothesis, we find that the Kip1p nuclear motor (1) interacts with the plasmid partitioning system, (2) associates with STB de novo during the G1–S window of the cell cycle, (3) promotes centromere-like location of the plasmid cluster in close proximity to the spindle pole body, (4) assists recruitment of Cse4p and the cohesin complex at STB, (5) fosters cohesion between replicated plasmid sisters, and (6) promotes, likely through steps 1–5, equal plasmid segregation.
Results
General experimental features
The 2 micron reporter plasmids contained the replication origin and the STB locus in their native context. In [cir+] strains, Rep1 and Rep2 proteins provided by the endogenous 2 micron circles supported their segregation. In [cir0] strains (lacking the Rep proteins), they were akin to ARS plasmids, containing a replication origin but lacking STB. In several experiments, the copy number of the STB-containing plasmids was reduced close to one by introducing a centromere (CEN3) into them (Ghosh et al., 2007). The CEN could be conditionally inactivated by driving transcription through it from the inducible GAL promoter. In [cir+] strains, in the presence of glucose or raffinose, the CEN and STB segregation mechanisms were active in the STB-CEN reporter plasmids. In galactose, segregation was strictly controlled by the Rep–STB system. In [cir0] host strains, the CEN pathway was active in the presence of glucose or raffinose; in galactose, neither the CEN nor the Rep–STB pathway was operational. None of the reporter plasmids have the requisite genomic organization for copy number amplification by Flp (Futcher, 1986; Volkert and Broach, 1986).
A 2 micron circle–derived high-copy reporter plasmid missegregates in the absence of Kip1p
As noted, the mitotic spindle has a strong influence on multiple aspects of 2 micron plasmid segregation (Velmurugan et al., 2000; Yang et al., 2004; Mehta et al., 2005; Hajra et al., 2006). Could a motor protein mediate capture of the 2 micron cluster by the spindle and/or its localization, facilitating its functional interaction with chromosomes or a specific sub-nuclear structure? We tested whether equal plasmid segregation is affected by the absence of individual nuclear motors: Cin8p, Kip1p, Kar3p, and Kip3p.
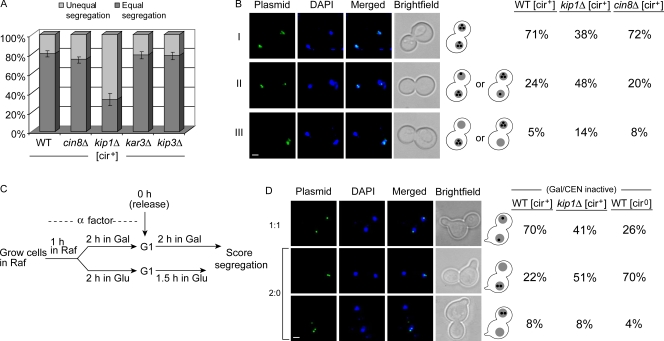
We initially followed the segregation of a multicopy reporter plasmid pSV1 harboring the 2 micron origin, STB, and a LacO256 array (Mehta et al., 2002). In a [cir+] host strain expressing GFP-LacI, pSV1 revealed itself as a cluster of ∼3–5 fluorescent foci in the nucleus. A preculture of this tester strain, grown overnight under selection for pSV1 and the GFP-LacI expression plasmid, was diluted into fresh medium and grown for two generations under conditions that induce GFP-LacI expression. Plasmid fluorescence was scored in the subset of anaphase or post-anaphase cells, namely, large-budded cells displaying two equal DAPI-staining zones separated into the mother and daughter compartments. Equal segregation of the plasmid, indicated by equal number of fluorescent foci in the two compartments (n:n), was not significantly different between the wild-type strain and its cin8Δ, kar3Δ, and kip3Δ derivatives (Fig. 1 A). However, the kip1Δ strain displayed a large increase in the frequency of unequal plasmid segregation (n:≠n). For a subset of the assays, we verified that results from counting foci and measuring fluorescence intensities were concordant (unpublished data). Earlier analysis showed that total fluorescence intensity, which reflects the number of foci, gives a reliable estimate of plasmid copy number (Velmurugan et al., 2000).
Figure 1.
Faithful segregation of STB reporter plasmids is dependent on the Kip1p motor protein. (A) Segregation data for a fluorescence-tagged multicopy reporter plasmid in [cir+] strains were averaged from three separate experiments (≥100 cells per experiment). Error bars represent standard deviations from the mean. (B) The types of segregations for the indicated strains, assayed separately from A, were: equal (I), unequal (II), and failed segregation (III). Within II and III, segregations favoring the mother or daughter were not binned into distinct groups. Bar, 2 µm. (C) The scheme for assaying the segregation of a single-copy reporter plasmid is illustrated. The host strain, arrested in G1 using α factor, was further conditioned in glucose or galactose in the presence of the pheromone before being released into the cell cycle in glucose (CEN active) or galactose (CEN inactive; STB active). (D) The data shown are for cells released into the cell cycle in galactose. Bar, 2 µm. Results for cells released into glucose are given in Fig. S2. Individual values in B and D were derived by scoring a minimum of 100 cells.
The data in Fig. 1 B subdivide segregation into three classes: equal (I), unequal (II), and failed segregation (III). This assay, done independently of those summarized in Fig. 1 A, compared only the kip1Δ and cin8Δ strains to wild type. Class III segregation (n:0) was rare in the kip1Δ [cir+] strain, the segregation defect being largely of the unequal type (class II) (Fig. 1 B, middle column). The distributions of the three classes were more or less the same for the wild-type and cin8Δ strains (Fig. 1 B, columns 1 and 3). Within classes II and III, there was a clear mother bias (Murray and Szostak, 1983) for plasmid retention in all strains. The Rep–STB system is believed to alleviate this bias.
A CEN reporter plasmid pSV2 (Velmurugan et al., 2000) did not show a higher incidence of missegregation when Kip1p function was ablated (Fig. S1 A). Chromosomes do not suffer a significantly higher loss rate in a kip1Δ strain, presumably because Cin8p can functionally substitute for Kip1p (Hoyt et al., 1992). The reported chromosome loss frequency of ∼1 × 10−4 (a loss rate of 5.3 × 10−6 per generation, assuming 15 generations of growth) would not have registered in our assay.
A cloned copy of KIP1, expressed in the kip1Δ strain, restored normal segregation of the STB reporter plasmid (Fig. S1 B). A mutant clone, with a nonfunctional Walker A motif (Walker et al., 1982), failed to do so. The mutant Kip1p caused a metaphase delay in cycling cells, perhaps due to a partially dominant “rigor” binding phenotype similar to that described for a P-loop mutant of Kar3p that shows enhanced microtubule association (Meluh and Rose, 1990). The plasmid partitioning function of Kip1p apparently requires its locomotive activity or at least its dynamic interaction with the spindle.
The above findings suggest that Kip1p promotes equal segregation of the 2 micron plasmid. Because counting of closely clustered foci can be somewhat subjective, the segregation assay was revised using a single-copy derivative of an STB reporter plasmid (see below).
A single-copy STB plasmid reporter shows a high loss rate when deprived of Kip1p
The potential impact of kip1Δ on native 2 micron circle stability is expected to be mollified by high plasmid copy number coupled with the action of the amplification system. At low copy number, and in the absence amplification, lack of Kip1p should manifest a strong effect on plasmid propagation. To test this notion, we followed the transmittance of a unit copy 2 micron reporter plasmid in the kip1Δ strain.
The assays were performed with the CEN-STB reporter pSG1, fluorescence tagged by LacO256–GFP–LacI interaction (Ghosh et al., 2007). As pointed out, plasmid segregation could be forced to become entirely Rep–STB dependent by galactose driven transcription through CEN. The experimental scheme for assaying plasmid segregation in cells released from G1 arrest is outlined in Fig. 1 C. Note that the reporter was present within the pool of endogenous 2 micron plasmids that supplied the Rep1p and Rep2p proteins. A missegregating single-copy reporter, and the associated fluorescence signal, would be confined entirely to one cell compartment.
Absence of Kip1p caused a marked increase in 2:0 segregation frequency when pSG1 partitioning in the [cir+] host was under Rep–STB control (Fig. 1 D, column 2). 2:0 segregation in galactose in a KIP1 [cir0] derivative of the host, lacking Rep proteins, provides a frame of reference (Fig. 1 D, column 3). Plasmid segregation was not affected by kip1Δ when pSG1 centromere was maintained functional in glucose (Fig. S2 A). The elevated 2:0 pSG1 segregation due to kip1Δ was also evident in a [cir0] host expressing Rep1p plus Rep2p in a galactose inducible fashion (Fig. S2 B, column 4). Thus, the single-copy reporter segregating on its own or as part of a plasmid cluster is subject to the Kip1p effect.
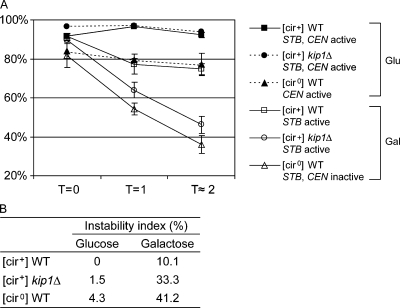
The microscopy assay was further corroborated by a plating-out assay (see Materials and methods). Aliquots from G1 cells at the start of the assay (T = 0) and after the first and second cell cycles (T = 1 and 2, respectively) were scored for plasmid-bearing and plasmid-free cells (Fig. 2 A). Unbudded cells accounted for 85–90% of the population at T = 1 and 2, and their haploid DNA content was verified by FACS analysis (unpublished data). In glucose-grown cells, with an active pSG1 centromere, the plasmid loss rates per generation were quite small without functional Kip1p or in the absence of the Rep proteins ([cir0] background) (Fig. 2 B). In galactose-grown cells, with the pSG1 centromere inactivated, the plasmid loss rate per generation became significantly large in the kip1Δ [cir+] strain. The loss rate was even higher in the [cir0] host strain expressing active Kip1p. In the wild-type [cir+] strain, pSG1 was less stable in galactose-grown cells than in glucose-grown cells. This result is consistent with previous observations that centromere based plasmid partitioning is more efficient than Rep–STB-based partitioning.
Figure 2.
Loss rates of a single-copy reporter plasmid in wild-type and kip1Δ strains. (A) Plasmid loss at the end of the first two generations after release of cells from G1 arrest (see Fig. 1 C) was estimated by scoring cells for the retention of the TRP1 marker harbored by the reporter plasmid. Results were obtained from three independent assays, and plotted with error bars to indicate standard deviations from the mean. (B) The rate of plasmid loss per cell division event (instability index) 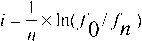 , where n = number of generations; f0 = fraction of plasmid bearing cells at time zero; fn = fraction of plasmid bearing cells after n generations (Murray and Cesareni, 1986).
, where n = number of generations; f0 = fraction of plasmid bearing cells at time zero; fn = fraction of plasmid bearing cells after n generations (Murray and Cesareni, 1986).
Segregation of the single-copy reporter plasmid confirms the inference from Fig. 1 (A and B) that the Kip1 motor plays a role in the equal segregation of the 2 micron plasmid.
Kip1p associates with STB in a Rep protein–dependent manner
A role for Kip1p in 2 micron circle segregation suggests possible interaction between Kip1p and the plasmid partitioning system. We tested, by yeast monohybrid and chromatin immunoprecipitation (ChIP) assays, whether Kip1p is associated with the STB locus. These assays, except for a small subset that will be pointed out below, were performed in [cir+] host strains. The native 2 micron circle served as the source of Rep1 and Rep2 proteins, and as the reporter for the STB locus in ChIP assays.
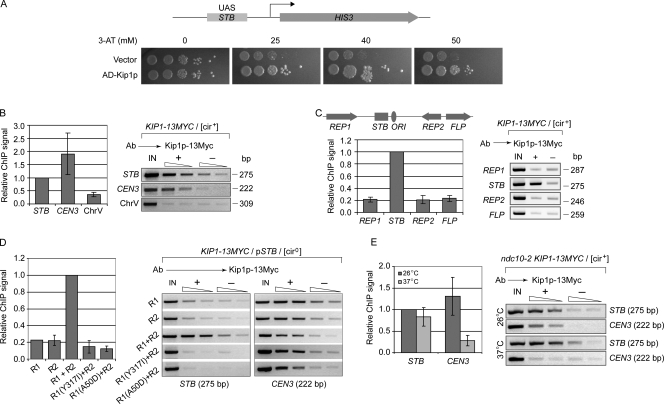
The chromosomally integrated HIS3 reporter for the monohybrid assay (Yang et al., 2004) harbored, upstream of its basal promoter, a 375-bp regulatory cassette spanning the STB locus (Velmurugan et al., 1998). Challenge with the His3p inhibitor 3-AT (3-aminotriazole) revealed a clear-cut growth advantage for the strain when a hybrid protein containing the Gal4p transcriptional activation domain fused to the N terminus of Kip1p was expressed in it (Fig. 3 A). The outcome is consistent with interaction between Kip1p and STB yielding enhanced transcriptional output from the reporter.
Figure 3.
The Kip1 motor interacts specifically with the STB locus. (A) In the monohybrid assay, the STB locus served as the upstream activator sequence (UAS) for a chromosomally integrated HIS3 reporter cassette. AD-Kip1p refers to the hybrid protein containing the Gal4p activation domain fused to Kip1p. Spots from left to right denote 10-fold serial dilutions of an initial inoculum of ∼5 × 104 cells in the leftmost spot. (B–E) ChIP assays were performed using antibodies directed against Myc-tagged Kip1p. The native 2 micron circle (B, C, and E) or a plasmid derived from it (pSTB; D) served as the reporter. In C, the relative locations of REP1, REP2, FLP, STB, and the replication origin (ORI) are schematically indicated on a linear representation of the 2 micron circle genome. The ChIP signals were normalized as follows: B and C, STB signal = 1; D, STB signal under Rep1p plus Rep2p supplementation = 1; E, STB signal from the 26°C samples = 1. The error bars represent standard deviations from the mean for three independent assays. IN, input template DNA controls; +, experimental samples; −, mock-immunoprecipitated (no antibody) negative controls.
ChIP assays directed at Kip1p, fused to the Myc epitope at the C terminus, revealed its association with STB and CEN3 sequences but not an arm sequence from chromosome V that binds the cohesin complex (Fig. 3 B). The association between Kip1p and STB was specific; no association was detected at REP1, REP2, and FLP regions of the 2 micron circle genome (Fig. 3 C).
To address the possible dependence of Kip1p–STB interaction on the Rep proteins, the ChIP assays were performed in a [cir0] strain harboring an STB reporter plasmid. The Rep proteins were expressed individually or in combination from chromosomally integrated cassettes by galactose induction. No association of Kip1p with STB was detected in the absence of the Rep proteins or in the presence of only one of the two; however, simultaneous presence of Rep1p and Rep2p restored Kip1p–STB association (Fig. 3 D).
Rep1p mutations have been described that selectively abrogate its interaction with STB (class I mutants) or with Rep2p (class II mutants) (Yang et al., 2004). Class I mutants retain interaction with Rep2p, class II mutants with STB. Neither a class I mutant (Rep1p (Y317I)) nor a class II mutant (Rep1p (A50D)) was able to sustain Kip1p–STB association when partnered with wild-type Rep2p (Fig. 3 D). As expected, the interaction between Kip1p and CEN3 was not affected by Rep1p mutations or by absence of the Rep proteins.
When kinetochore function was inactivated by the ndc10-2 mutation, association of Kip1p with CEN3 was blocked at the nonpermissive temperature (37°C) (Fig. 3 E); however, there was no effect on Kip1p association with STB.
The sum of the data from the monohybrid and ChIP analyses reveal specific association of Kip1p with STB in a Rep1p- and Rep2p-assisted manner. Interactions of Rep1p with both STB and Rep2p have to be preserved for this association, which is independent of the association between Kip1p and centromeres.
Cell cycle timing of Kip1p–STB association; kip1Δ ablates cohesin assembly at STB and leads to failure of plasmid cohesion
The de novo assembly of the partitioning complex at STB is triggered as cells exit G1 and enter S phase. The association of Rep proteins, Cse4p, and components of the RSC2 chromatin remodeling complex with STB is renewed during this window of the cell cycle (Huang et al., 2004; Yang et al., 2004; Hajra et al., 2006). The assembly of the cohesin complex at STB also occurs early in S phase (Mehta et al., 2002), and is critically dependent on functional Rep proteins, Cse4p, and the RSC2 complex. Is the association between Kip1p and STB cell cycle dependent? If so, how is the timing of this association related to that of the other components of the plasmid partitioning complex?
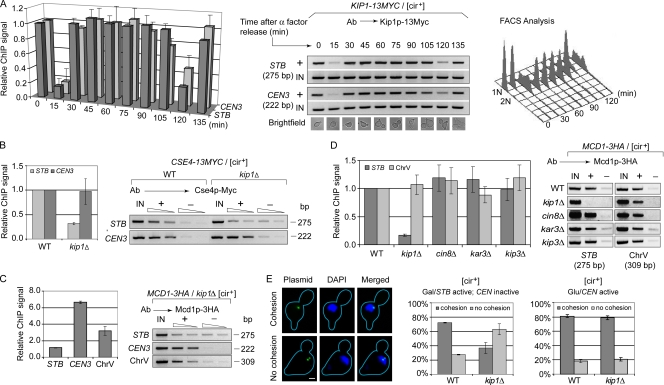
According to ChIP analysis, Kip1p was associated with STB in G1, but exited from STB briefly during transition to S (Fig. 4 A). The association was reestablished in early S, and persisted through the remainder of the cell cycle and into G1 of the next cell cycle. The transient disassociation and reassociation were repeated as cells proceeded once again into S phase. The cell cycle dependence of Kip1p–STB association was similar to that of Kip1p–CEN3 association.
Figure 4.
Kip1p–STB association occurs de novo during each cell cycle; lack of Kip1p abrogates Cse4p and cohesin association with STB and cohesion between duplicated plasmid copies. (A–D) ChIP assays were performed using antibodies directed against Myc-tagged Kip1p or HA-tagged Mcd1p, as indicated above the respective panels. In A, the cell cycle progression was followed by light microscopy of cell morphology as well as by FACS analysis. The relative ChIP signals were normalized against 1 for STB and CEN at time zero (A) or in the wild-type host strain (B and D). In C, the STB signal was set to 1. (E) Cohesion between sister plasmids, coalescence of two fluorescent foci into a single dot, was assayed in metaphase cells (Ghosh et al., 2007). Two separate dots indicated lack of cohesion. In each individual assay, > 100 metaphase cells were examined. The datasets in A–E were the averaged values from three separate experiments.
In a kip1Δ background, the recruitment of Cse4p and assembly of cohesin at STB, but not at CEN, were reduced to nearly background values (Fig. 4, B and C). Individual deletions of genes for the other nuclear motors did not block cohesin recruitment at STB (Fig. 4 D). Consistent with the failure of cohesin recruitment, cohesion between sister copies of a CEN-STB reporter plasmid was disrupted in metaphase kip1Δ cells when the plasmid was under Rep–STB control for partitioning (Fig. 4 E). Cohesion was normal when the plasmid borne CEN was functional. Lack of Kip1p function did not abrogate the association of Rep1 and Rep2 proteins with STB (Fig. S3).
The cell cycle timing of Kip1p association with STB and the lifetime of this association are consistent with Kip1p being required for equal segregation of the 2 micron plasmid. The pattern is quite similar to what has been previously observed for the association of Rep1 and Rep2 proteins with STB (Yang et al., 2004). The failure of cohesin assembly at STB and loss of cohesion between replicated plasmids in the absence of Kip1p account, at least in part, for the increased rates of unequal plasmid segregation in the kip1Δ strain.
Interaction of Kip1p with Rep1p is not sensitive to spindle depolymerization
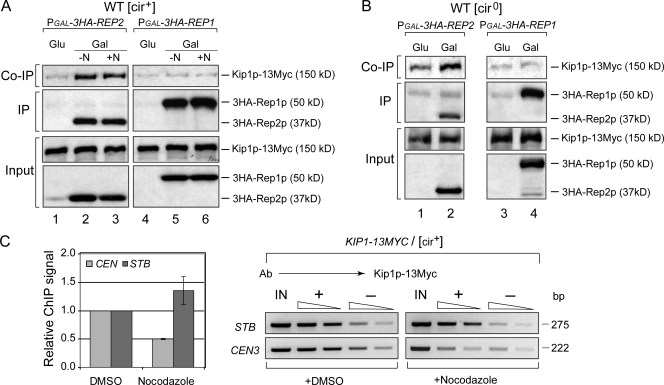
The Rep1 and Rep2 proteins interact with STB in the yeast nucleus independently of each other, and these interactions are resistant to spindle depolymerization (Velmurugan et al., 1998, 2000). However, integrity of the nuclear spindle is critical for the association of Cse4p and the cohesin complex with STB (Mehta et al., 2005; Hajra et al., 2006). Because the recruitment/establishment of Kip1p at STB requires both the Rep proteins, we wished to test whether Rep1p or Rep2p or both could interact with Kip1p, and what effect spindle integrity might have on such potential interaction(s).
Immunoprecipitation assays were performed in [cir+] or [cir0] strains engineered to express Kip1p–Myc from the native KIP1 promoter at the normal chromosomal locale. A Rep1p or a Rep2p derivative carrying tandem HA tags, three or six in Rep1p and three in Rep2p, at the N terminus was expressed inducibly from the GAL1 promoter harbored by a CEN expression vector. Immunoprecipitation of HA-Rep2 pulled down Kip1p in the extract from the [cir+] strain (Fig. 5 A; lane 2, top row). Their coimmunoprecipitation was insensitive to nocodazole treatment (Fig. 5 A; lane 3, top row) or absence of Rep1p ([cir0] cells) (Fig. 5 B; lane 2, top row). Immunoprecipitation of HA-Rep1p failed to bring down Kip1p in [cir+] as well as [cir0] extracts (Fig. 5 A; lanes 5 and 6, top row and Fig. 5 B; lane 4, top row). Rep2p–Kip1p interaction was verified by reverse coimmunoprecipitation using antibodies to Kip1p–Myc (Fig. S4). Consistent with Rep2p–Kip1p interaction being impervious to spindle disassembly, ChIP analysis revealed the association of Kip1p with STB in nocodazole treated cells (Fig. 5 C). By contrast, the association between Kip1p and CEN was greatly reduced by nocodazole.
Figure 5.
Kip1p interacts with Rep2p in a coimmunoprecipitation assay; Kip1p–Rep2p interaction and Kip1p–STB association are not disrupted by spindle disassembly. (A and B) Immunoprecipitations were performed in [cir+] or [cir0] cell extracts using antibodies to HA-tagged Rep1p or Rep2p expressed in a galactose-inducible manner. Extracts were prepared from cells in mid-log phase of growth (−N) or those treated with nocodazole (+N) and arrested in G2/M. Presence of Kip1p in the immunoprecipitates was probed by Western blotting using antibodies to Myc-tagged Kip1p. IP and Co-IP refer to immunoprecipitated and coimmunoprecipitated proteins, respectively. (C) ChIP signals were adjusted to a value of 1 for STB and CEN from the control samples (treated with DMSO, the solvent for nocodazole).
Interaction between Rep2p and Kip1p, independently of Rep1p, is compatible with the finding that the recruitment of Kip1p at STB is dependent on the Rep proteins. The inability of Rep1p to coimmunoprecipitate Kip1p suggests that the role of Rep1p in Kip1p–STB association may be indirect or that Kip1p interaction with Rep1p is less robust than that with Rep2p.
All nuclear motors can associate with STB; yet, only Kip1p association is insensitive to nocodazole
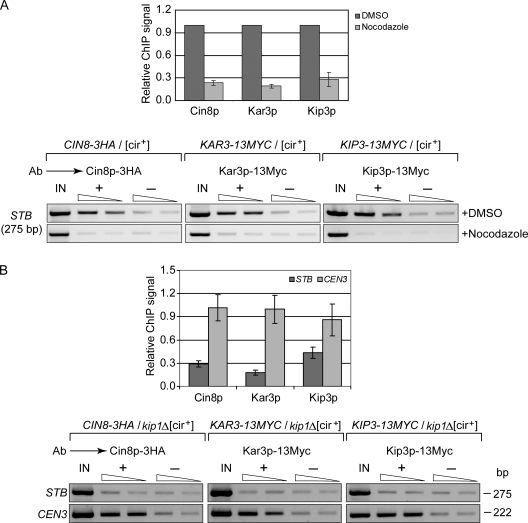
All four nuclear motor proteins in S. cerevisiae associate with centromeric DNA (He et al., 2000; Tanaka et al., 2005; Tytell and Sorger, 2006). Kip3p appears to be a core kinetochore component, its recruitment being independent of kinetochore–microtubule attachment (Tytell and Sorger, 2006). Association of Cin8p and Kip1p with kinetochores requires the Ndc80 complex, or kinetochore–microtubule attachment mediated through this complex. Kar3p localizes preferentially to kinetochores detached from the spindle. Based on functional similarities between CEN and STB plasmids in nuclear localization and segregation (Velmurugan et al., 2000; Mehta et al., 2002; Hajra et al., 2006; Ghosh et al., 2007), we wished to know whether the nuclear motors other than Kip1p can also be recruited to STB. In addition, we wished to test whether these potential associations are dependent on a functional mitotic spindle.
ChIP assays used a [cir+] host strain in which HA-tagged Cin8p and Myc-tagged Kar3p and Kip3p were expressed individually from native promoters and from normal chromosomal locales. All three proteins showed association with STB (Fig. 6 A). In nocodazole-treated cells (Fig. 6 A) or in the kip1Δ host strain, this association was no better than background (Fig. 6 B).
Figure 6.
Association of Cin8p, Kip3p, and Kar3p with STB is sensitive to spindle disassembly by nocodazole treatment or to kip1Δ. (A and B) ChIP assays were performed in [cir+] strains, wild type, or kip1Δ, using antibodies to the indicated Myc- or HA-tagged motor proteins. The epitope tags were positioned at the C terminus. The plotted STB signals were normalized by keeping those from DMSO treated (A) and wild-type (B) samples as 1.
The ChIP results suggest that association of Kip1p with STB, which does not require an intact spindle, precedes that of the other three motor proteins. If Kip1p guides the plasmid to the spindle, incidental association of the other motors with STB may follow. Kip1p is the only motor protein required for cohesin assembly at STB (Fig. 4 D) and stable plasmid propagation (Fig. 1); and Kip1p–STB association is insensitive to cin8Δ (unpublished data). Given their noninvolvement in plasmid segregation, Kip3p and Kar3p are also unlikely to influence Kip1p–STB association.
Localization of a 2 micron derived reporter plasmid with respect to Kip1p during the cell cycle and the effect of kip1Δ on the nuclear address of the plasmid
In metaphase cells, Kip1p associated with congressed centromeres shows a characteristic bi-lobed pattern along the short distance (1 to 1.2 µm) between duplicated spindle pole bodies (Tytell and Sorger, 2006). Inactivation of Ndc10p disrupts the bi-lobed organization of Kip1p without affecting its localization along spindle microtubules. As noted before, there is striking concordance between 2 micron reporter plasmids and a tagged chromosome or a centromere plasmid in the dynamics and timing of their segregation (Velmurugan et al., 2000; Ghosh et al., 2007). Because equal segregation of the 2 micron plasmid is dependent on Kip1p, we analyzed the localization of a single-copy reporter plasmid, conditioned to behave as a CEN, STB, or ARS plasmid, with respect to Kip1p during the cell cycle. We extended this analysis by estimating the potential displacement of the STB plasmid from its normal spindle pole proximal nuclear address (Velmurugan et al., 2000; Mehta et al., 2005) upon ablating Kip1p function.
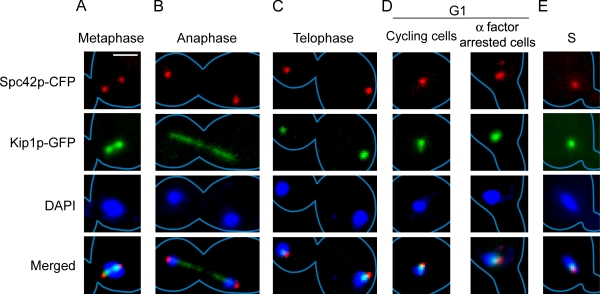
To set the frame of reference, we first mapped the relative locations of our primary landmarks, Kip1p and the spindle pole body, at different cell cycle stages. Kip1p was visualized by fusing GFP to its C terminus; the spindle pole body by tagging it with Spc42p harboring a C-terminal CFP fusion. Consistent with the findings of Tytell and Sorger (2006), metaphase cells were characterized by bi-lobed Kip1p, with the spindle pole bodies positioned at its outer edges (Fig. 7 A). In anaphase cells, Kip1p formed a linear array of puncta along the spindle from pole to pole (Fig. 7 B). In telophase cells, nearly all of the metaphase Kip1p puncta disappeared, except for two foci that remained associated with the spindle poles (Fig. 7 C). In G1 and early S phase cells, Kip1p formed a single focus that was coincident with or overlapped the unduplicated spindle pole body and the duplicated but bridged spindle pole bodies, respectively (Fig. 7, D and E).
Figure 7.
Localization of Kip1p with respect to spindle pole bodies at different cell cycle stages. Kip1p and Spc42p were visualized in cycling or α factor arrested yeast cells expressing these proteins as fusions to GFP and CFP, respectively. In the micrographs shown, cyan is pseudocolored in red. Cells displaying two clearly resolved spindle pole bodies in the mother cell compartment with a single unsegregated chromosome mass (DAPI) near the bud neck were deemed to be in metaphase (A). Large budded cells in which duplicated spindle pole bodies and chromosomes had segregated into opposite cell compartments were classified as anaphase cells if they contained a linear array of Kip1p puncta, signifying the disposition of the spindle (B). Those lacking such serial arrangement of puncta were classified as telophase cells (C). Unbudded cells (D) and cells with emerging buds (E) were categorized as G1 and S phase cells. For each cell cycle stage (A–E), at least 20 cells were examined, and the representative patterns are displayed here. Bar, 2 µm.
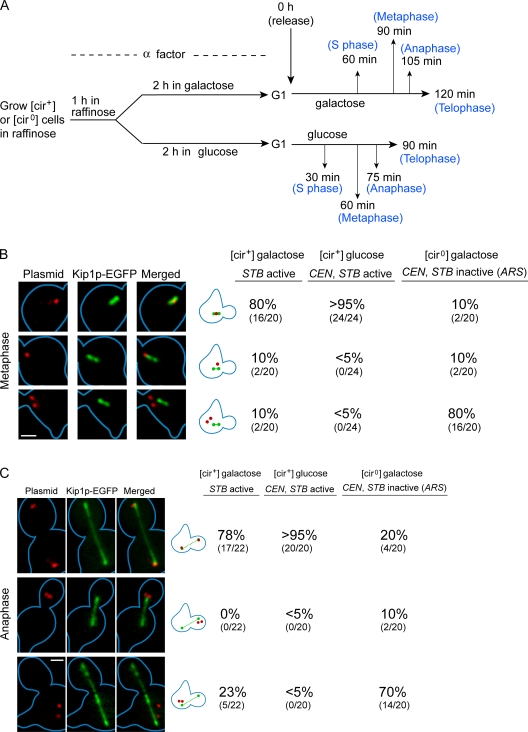
We then mapped the relative positions of the single-copy STB, CEN, and ARS reporter plasmids, fluorescence-tagged by [TetO]112–RFP–TetR interaction, within the context of the Kip1p reference frame. The data for metaphase and anaphase cells are presented in Fig. 8; those for cells at other stages are summarized in Fig. S5. Localization of a single plasmid dot (indicating cohesion of duplicated plasmid copies) at the center of the bi-lobed metaphase Kip1p pattern (Fig. 8 B, row 1) was observed in nearly all of the cells for the CEN plasmid, nearly 80% of the cells for the STB plasmid and only ∼10% of the cells for the ARS plasmid. In the predominant fraction of cells, the ARS plasmid was positioned away from Kip1p with the two-dot pattern (Fig. 8 B, row 3) in far excess of the one-dot pattern (Fig. 8 B, row 2). The preponderance of two dots is consistent with the lack of cohesion between sister copies of the ARS plasmid.
Figure 8.
Localization of a single-copy reporter plasmid with respect to Kip1p. (A) A single-copy reporter plasmid (tagged by red fluorescence) was subjected to conditions that selectively inactivate CEN or STB or both, as outlined schematically. After release into the cell cycle in glucose or galactose medium, cells were assayed by fluorescence microscopy at the indicated times. Kip1p was visualized by GFP tagging as described in Fig. 7. (B and C) Data from metaphase and anaphase cells are tabulated; those from the other cell cycle stages are given in the supplemental material (Fig. S5). Results when both CEN and STB were functional in the plasmid (middle column) or CEN alone was functional ([cir0]/glucose) were almost identical (not depicted). Bar, 2 µm.
The similarity between the CEN and STB reporter plasmids, and their contrast from the ARS reporter plasmid, in colocalization with Kip1p was also observed in cells at other stages of the cell cycle (Fig. 8 C and Fig. S5). In G1 and early S phase cells, the typical pattern for the CEN and STB reporters was a single plasmid dot coincident or coalesced with the Kip1 focus. The plasmid dot in the early S phase cells likely denotes primarily cohesed plasmid sisters along with perhaps a small fraction of the unreplicated plasmid. The ARS plasmid was seen as two distinct dots in roughly 60% of such cells. In anaphase cells (Fig. 8 C), the representative CEN and STB plasmid configuration comprised of two segregated plasmid dots, each positioned at either end of the linear punctate array of Kip1p foci. Telophase cells were characterized by CEN and STB plasmid dots coincident with or overlapping the two distal Kip1p foci that survived degradation or relocalization during anaphase. Anaphase and telophase cells also revealed a high rate of missegregation (2:0) for the ARS plasmid, with a clear segregation bias toward the mother cell.
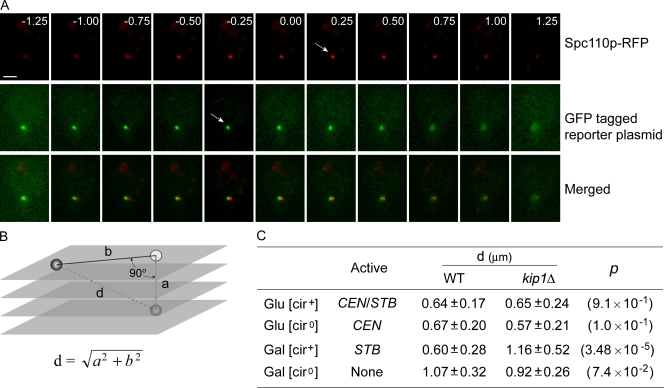
Finally, we examined the effect of kip1Δ on the location of the single-copy reporter plasmids, tagged in this instance by [LacO]256–LacI–GFP interaction, with respect to the spindle pole body in G1-arrested cells. The spindle pole body was visualized by expressing a hybrid version of Spc110p fused at its C terminus to RFP. The residence zones of the spindle pole body (red fluorescence) and plasmid (green fluorescence) were demarcated by Z-series optical sectioning of the yeast nucleus into consecutive 0.25-µm thick individual layers (Fig. 9 A). The vertical displacement “a” between the spindle pole body and the plasmid was estimated as the distance between the two positions that displayed the highest red and green fluorescence intensities, respectively. The horizontal displacement “b” between the spindle pole body and the plasmid was calculated by the MetaMorph software after the sections showing fluorescence signals were stacked, deconvoluted, and projected as 2D images (Fig. 9 B). The shortest plasmid to spindle pole distance was calculated as “d” = 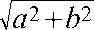 . For the STB plasmid, this distance was nearly doubled in the kip1Δ strain relative to that in the wild-type strain (Fig. 9 C). The lack of Kip1p function did not affect the “d” values for the CEN plasmid or the ARS plasmid or the plasmid in which both CEN and STB were active.
. For the STB plasmid, this distance was nearly doubled in the kip1Δ strain relative to that in the wild-type strain (Fig. 9 C). The lack of Kip1p function did not affect the “d” values for the CEN plasmid or the ARS plasmid or the plasmid in which both CEN and STB were active.
Figure 9.
Effect of kip1Δ on the distance between a single-copy reporter plasmid and the spindle pole body. The single-copy reporter plasmid was tagged by green fluorescence via [LacO]256–GFP–LacI interaction. The spindle pole body was tagged by cyan fluorescence (pseudocolored here in red) as described in Fig. 7. (A and B) The vertical displacement “a” between the plasmid and the spindle pole body was measured by Z-series sectioning of the nucleus. The data shown are for the wild-type strain. Their horizontal displacement “b” was derived by deconvolution and 2D projection. The distance “d” between the two was derived using “a” and “b”. (C) The values for “d” in the wild-type and kip1Δ strains were estimated under conditions that maintained either CEN or STB alone or both functional or both nonfunctional. Each “d” value was derived from at least 20 independent measurements. The values for the level of significance (p), calculated using the Student's t test, are given in parentheses. Bar, 2 µm.
An STB plasmid mimics a CEN plasmid in its nuclear positioning in the large majority of cells. This characteristic nuclear address of the STB plasmid is strongly dependent on Kip1p. Lack of Kip1p displaces the STB plasmid from its normal position of close proximity to the spindle pole body, and its nuclear location resembles that of an ARS plasmid.
Discussion
Co-opting by the 2 micron plasmid of Kip1p suggests a molecular explanation for prior observations that the integrity of the nuclear spindle is essential for several critical steps in the plasmid partitioning (Velmurugan et al., 2000; Mehta et al., 2005; Hajra et al., 2006; Ghosh et al., 2007). It is consistent with the tendency of the plasmid, in an mtw1 mutant strain, to cosegregate with the short spindle (and the attached subset of chromosomes) into the daughter cell (Mehta et al., 2005). A spindle-associated motor protein recruited to its partitioning locus may lead the plasmid to a nuclear locale where assembly of the partitioning complex and/or potential chromosome tethering can be accomplished.
The rarity of complete segregation failure (n:0) in a kip1Δ host relative to a [cir0] host (devoid of Rep proteins) is rather puzzling. Perhaps lack of Kip1p may so affect plasmid organization/localization as to mitigate constraints operating on one-to-one segregation of sister clusters. This partial alleviation of mother bias may skew the segregation defect toward n:≠n events and away from n:0 events.
Kip1p is an authentic component of the 2 micron plasmid partitioning system
The absolute dependence of Kip1p recruitment at STB on the Rep proteins and the cell cycle timing and duration of Kip1p–STB association are consistent with Kip1p being an authentic component of the 2 micron plasmid partitioning system. The transient dissociation-reassociation of Kip1p at STB during G1–S resembles the cell cycle dynamics of the Rep proteins at STB (Yang et al., 2004). Persistence of Kip1p–STB interaction for nearly the entire cell cycle is at odds with biochemical evidence for Kip1p degradation during metaphase to anaphase transition (Gordon and Roof, 2001). Our data argue that a small pool of Kip1p, bound to STB, CEN, and perhaps microtubules, escapes anaphase destruction. However, we cannot rule out that GFP or epitope tag present on Kip1p could have altered its stability.
Cin8p and, perhaps to a lesser extent, Kip1p, promote kinetochore clustering by cross-linking kinetochore microtubules (Tytell and Sorger, 2006). In principle, Kip1p tetramers may assist the Rep proteins in cross-linking (clustering) plasmid molecules via bridging interactions between their STB loci (Fig. 10 A). Association of Kip1p-bound plasmids with microtubules followed by their bundling can confer a second level of compaction on the plasmid cluster (Fig. 10, B and C). Spindle depolymerization, and loosening of plasmid adhesion, causes the cluster to occupy a wider residence zone (Velmurugan et al., 2000).
Figure 10.
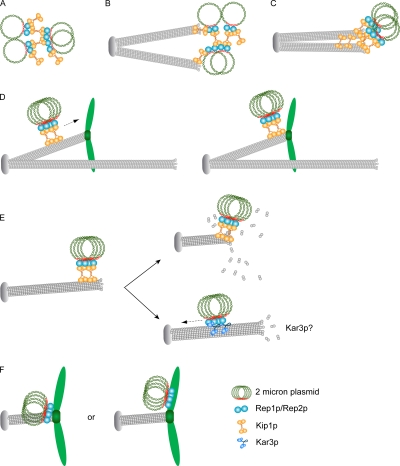
Possible roles for Kip1p in the 2 micron circle partitioning pathway. (A–C) organization, spindle association, and compaction of plasmid cluster. (D) Translocation toward the + end along a kinetochore microtubule. (E) Translocation toward the spindle pole by end-on movement or lateral movement after cargo transfer to Kar3p. (F) Localization of plasmid cluster in the vicinity of kinetochores or attachment of the cluster to a chromosome. See text for details.
Coimmunoprecipitation of Rep2p, but not Rep1p, with Kip1p is rather surprising. Note that Kip1p–STB association requires interactions of Rep1p with Rep2p and STB. Perhaps Rep1p–Kip1p interaction is transient or dynamic. Or, the plasmid partitioning complex may be assembled hierarchically from a set of sub-complexes. Interaction of a sub-complex containing Rep1p and Rep2p with STB may be a prerequisite for recruitment of a second sub-complex containing Rep2p and Kip1p, even though the two sub-complexes may not directly interact. For example, the former interaction could trigger remodeling of the STB chromatin to make it competent for the latter.
Kip1p-mediated plasmid capture by the spindle: transport of the 2 micron plasmid to its partitioning center?
We suggest the following plausible models for the action of Kip1p in 2 micron plasmid segregation (Fig. 10). Kip1p may promote capture of the plasmid cluster by the spindle and higher-order organization of the cluster (Fig. 10, A–C). Spindle–plasmid association could be augmented by interaction between independent pools of Rep1 and Rep2 proteins present on STB and on the spindle. The Rep proteins, localized primarily at or close to the spindle poles, form a decreasing concentration gradient toward the spindle midzone (Velmurugan et al., 2000). If the plasmid cluster contacts a kinetochore microtubule, the Kip1p motor can translocate it to the plus end where a chromosome is attached (Fig. 10 D). If contact is made with a nonkinetochore microtubule distal to the spindle pole, the cluster may be moved in the opposite direction by the minus end–directed Kar3p motor (Fig. 10 E, bottom right). Cargo transfer between motors is well documented in higher systems. Fusion between endosomes and influenza virus is preceded by intermittent movement of viral particles in the perinuclear region using microtubule-based plus and minus end–directed motilities (Lakadamyali et al., 2003). Recent evidence suggests that Kar3p associated with kinetochores transports chromosomes poleward laterally along microtubules (Tanaka et al., 2005, 2007; Kitamura et al., 2007). Alternatively, the Kip1p-associated plasmid cluster may approach the spindle pole by end-on movement through microtubule disassembly at the plus end (Fig. 10 E, top right). Regardless of the mechanism, localization in the immediate proximity of kinetochores would be conducive to either plasmid chromosome attachment (Fig. 10 F, right) or acquisition by the plasmid of chromosome segregation factors (Fig. 10 F, left).
Kinetochores transiently dissociate from the spindle during replication of centromere DNA, and reattach to the spindle within a short time (Kitamura et al., 2007). During recycling of the Rep1 and Rep2 proteins and of Kip1p at STB at the G1–S transition stage (Yang et al., 2004; Fig. 4 A), the plasmid cluster may briefly disengage from the spindle. Reattachment of the cluster to the spindle followed by its normal localization can be accomplished by mechanisms outlined in Fig. 10.
Similarities and contrasts of the partitioning mechanisms of the 2 micron plasmid, bacterial plasmids, and episomal viral genomes
The 2 micron plasmid and bacterial plasmid segregation systems display similarities in organization but not function. The type I and type II bacterial systems code for two partitioning (Par) proteins which interact with a plasmid “centromere”. One of the Par proteins is a DNA binding anchor with a Walker-box ATPase or an actin-like ATPase as its partner (Ebersbach and Gerdes, 2005). The type III system is based on an ancient tubulin homologue TubZ and perhaps a DNA binding partner protein (Larsen et al., 2007). Partitioning is accomplished through growing dynamic actin-like filaments, oscillations of Walker-ATPase filaments, or treadmilling of TubZ-GTPase filaments (Moller-Jensen et al., 2003; Ebersbach and Gerdes, 2005; Campbell and Mullins, 2007; Larsen et al., 2007).
Genomes of Epstein-Barr (EBV), Kaposi's sarcoma-associated herpes (KSHV) and bovine papilloma (BPV) viruses tether to host chromosomes for efficient partitioning (Botchan, 2004). Tethering is mediated by a virally encoded protein that interacts with the viral episome and with histones, host chromatin binding proteins, or a methyl-CpG binding protein (Shire et al., 1999; Wu et al., 2000; Kapoor et al., 2001; Krithivas et al., 2002; Viejo-Borbolla et al., 2005; Barbera et al., 2006; You et al., 2006). Human papilloma viruses (HPVs) use the viral E2 protein to associate with the spindle, suggesting a “viro-centromere” based segregation (Van Tine et al., 2004). Chromosome attachment through E2, in some cases specifically to pericentric regions, may constitute an alternative segregation mechanism (Oliveira et al., 2006). E2 localization adjacent to centromeres is reminiscent of the residence of the 2 micron plasmid and Rep proteins in proximity of spindle pole bodies.
Non-stochastic distribution of EBV into daughter cells is perhaps achieved through association of duplicated EBV genomes in pairs with sister chromatids (Kanda et al., 2007; Nanbo et al., 2007). This is analogous to cohesin mediated sister-to-sister segregation of the 2 micron plasmid (Ghosh et al., 2007). The cellular helicase ChlR1, which assists chromosomal loading of the papilloma E2 protein, is involved in sister chromatid cohesion (Parish et al., 2006). The S. cerevisiae homologue of ChlR1, Chl1p, also promotes cohesion, presumably through its interaction with replication factor C (RFC) and the cohesion establishment factor Ctf7p (Skibbens, 2004). There is no evidence for pairing of duplicated viral genomes by cohesin. However, once tethered to sister chromatids, they could potentially be paired by the cohesin bridge that holds sister chromatids together.
A mitotic kinesin-like protein MKlp2 appears to assist poleward translocation of papilloma viral genomes in association with microtubule plus ends (Yu et al., 2007). The potential functional similarity to the role of the mitotic spindle and the Kip1p motor in 2 micron plasmid segregation is self evident.
Despite individual mechanistic distinctions, segregation of bacterial plasmids, the yeast plasmid, and viral episomes exploits actin- or tubulin-based dynamic filaments either directly or through chromosome attachment.
Concluding remarks
Utilization of a nuclear motor for passage to a specific nuclear location reveals a novel aspect of the spatial regulation of 2 micron circle segregation. Although Cin8p and Kip1p serve redundant functions, Cin8p plays a more prominent role in chromosome segregation (Hoyt et al., 1992; Roof et al., 1992). Taking advantage of the lesser of the two motors is a shrewd strategy for a selfish DNA molecule whose propagation is inextricably linked to the host's fitness. Organizational and functional similarities and contrasts among partitioning systems of plasmids and viral episomes reveal direct or indirect reliance on a dynamic cytoskeletal structure. Whether these different systems arose as independent solutions dictated by specialized cellular contexts or share a common evolutionary descent followed by divergence under distinct host environments remains an open question.
Materials and methods
Strains and plasmids
Yeast strains and plasmids used in this study are listed in Table S1. The relevant genotypes of the strains and important attributes of the plasmids are briefly described and the appropriate references are given. Strains that contain endogenous 2 micron plasmids are designated as [cir+] and those that are cured of them are designated as [cir0].
Plasmid segregation assays
Multicopy reporter plasmid.
Yeast strains were grown overnight in selective media to maintain plasmids harbored by them. Cells were diluted into fresh medium to OD600 = ∼0.1 and allowed to go through two generations to an OD600 = ∼0.4. Fluorescence signals from the reporter plasmid was scored in anaphase/post-anaphase cells displaying two equally distributed DAPI staining zones in the mother and bud compartments. Procedures for fluorescence tagging of the reporter plasmid have been previously published (Velmurugan et al., 2000; Mehta et al., 2002).
Single-copy reporter plasmids.
The design of the CEN-STB unit copy reporter plasmids and the methodology for following their segregation have been described earlier (Ghosh et al., 2007). Cells harboring such a plasmid were first arrested in G1 using a factor. After their release from the pheromone to resume cell cycle in medium containing the appropriate carbon source, plasmid segregation was followed as described for the multicopy reporter plasmids.
Estimation of plasmid loss rates
The fractions of cells containing a reporter plasmid at the start of the experiment and after “n” generations of growth were estimated by plating them out on nonselective medium (∼200 per plate) and replicating grown colonies on to selective medium. The plasmid loss rate per generation was calculated according to Murray and Cesareni (1986).
Miscellaneous protocols
Chromatin immunoprecipitation (ChIP).
These analyses were performed as described previously (Hajra et al., 2006). The ndc10-1 strain grown to mid-log phase at 26°C was shifted to 37°C for 3 h for ChIP at the nonpermissive temperature. Serial dilutions of the template DNA were used in PCR reactions to obtain linear range of amplification. The relative ChIP signals were estimated by dividing the signal from a given sample by that from the corresponding mock immunoprecipitated negative control.
Standard immunoprecipitation.
These assays were performed according to West et al. (2007). Anti-HA and anti-Myc antibodies purchased from Covance were used for ChIP and immunoprecipitation after 1:100 dilution of the stock titer. For Western blotting, the antibody dilution was 1:1,000.
Fluorescence microscopy.
Observations were performed using an Olympus BX-60 microscope. Images were taken at room temperature at 100× (oil NA 1.30 objective) using a Photometrics Quantix camera from Roper Scientific, and then processed by MetaMorph (Universal Imaging Corporation) and PhotoShop CS (Adobe Systems, Inc.). Z-series sectioning of the yeast nucleus, deconvolution of the stacks, and their 2D projections were performed as detailed previously (Velmurugan et al., 2000; Mehta et al., 2005).
Other routine procedures
Treatment of cells with α factor for G1 arrest or with nocodazole for G2/M arrest, monohybrid assays, and FACS analysis were done according to published procedures (Velmurugan et al., 2000; Yang et al., 2004). Yeast and bacterial transformations, yeast genomic DNA and plasmid DNA preparation, curing of the 2 micron plasmid from [cir+] strains, culturing of yeast and bacteria and other general procedures were performed by following protocols listed on the Jayaram laboratory web page (http://www.sbs.utexas.edu/jayaram/jayaramlab_files/Protocols.htm).
Online supplemental material
Figure S1: segregation of a CEN plasmid is not significantly affected by kip1Δ; A P-loop mutant of Kip1p cannot support STB-based plasmid segregation. Figure S2: lack of KIP1 function affects segregation of a single-copy reporter plasmid when it is STB based and not when it is CEN based. Figure S3: association of Rep1p or Rep2p with STB, as assayed by chromatin immunoprecipitation (ChIP) is not affected by kip1Δ. Figure S4: immunoprecipitation of Kip1p brings down Rep2p with it. Figure S5: localization of a single-copy CEN-STB reporter plasmid with respect to Kip1p. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200810130/DC1.
Acknowledgments
We thank Peter Sorger (Harvard Medical School, Boston, MA) and Clarence Chan (University of Texas at Austin) for providing yeast strains and plasmids.
This work was supported by National Institutes of Health grant GM064363. Partial support came from the Robert F. Welch Foundation.
References
- Barbera A.J., Chodaparambil J.V., Kelley-Clarke B., Joukov V., Walter J.C., Luger K., Kaye K.M. 2006. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA.Science. 311:856–861 [DOI] [PubMed] [Google Scholar]
- Botchan M. 2004. Hitchhiking without covalent Integration.Cell. 117:280–281 [DOI] [PubMed] [Google Scholar]
- Campbell C.S., Mullins R.D. 2007. In vivo visualization of type II plasmid segregation: bacterial actin filaments pushing plasmids.J. Cell Biol. 179:1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G., Gerdes K. 2005. Plasmid segregation mechanisms.Annu. Rev. Genet. 39:453–479 [DOI] [PubMed] [Google Scholar]
- Futcher A.B. 1986. Copy number amplification of the 2 micron circle plasmid of Saccharomyces cerevisiae.J. Theor. Biol. 119:197–204 [DOI] [PubMed] [Google Scholar]
- Ghosh S.K., Hajra S., Paek A., Jayaram M. 2006. Mechanisms for chromosome and plasmid segregation.Annu. Rev. Biochem. 75:211–241 [DOI] [PubMed] [Google Scholar]
- Ghosh S.K., Hajra S., Jayaram M. 2007. Faithful segregation of the multicopy yeast plasmid through cohesin-mediated recognition of sisters.Proc. Natl. Acad. Sci. USA. 104:13034–13039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.M., Roof D.M. 2001. Degradation of the kinesin Kip1p at anaphase onset is mediated by the anaphase-promoting complex and Cdc20p.Proc. Natl. Acad. Sci. USA. 98:12515–12520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajra S., Ghosh S.K., Jayaram M. 2006. The centromere-specific histone variant Cse4p (CENP-A) is essential for functional chromatin architecture at the yeast 2-µm circle partitioning locus and promotes equal plasmid segregation.J. Cell Biol. 174:779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Asthana S., Sorger P.K. 2000. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast.Cell. 101:763–775 [DOI] [PubMed] [Google Scholar]
- Hildebrandt E.R., Hoyt M.A. 2000. Mitotic motors in Saccharomyces cerevisiae.Biochim. Biophys. Acta. 1496:99–116 [DOI] [PubMed] [Google Scholar]
- Hoyt M.A., He L., Loo K.K., Saunders W.S. 1992. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly.J. Cell Biol. 118:109–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Hsu J.-m., Laurent B.C. 2004. The RSC nucleosome-remodeling complex is required for cohesin's association with chromosome arms.Mol. Cell. 13:739–750 [DOI] [PubMed] [Google Scholar]
- Jayaram M., Yang X.M., Mehta S., Voziyanov Y., Velmurugan S. . The 2µm plasmid of Saccharomyces cerevisiae. 2004. Plasmid Biology. Funnell B.E., Phillips G.J., ASM Press, Washington, DC: 303–323 [Google Scholar]
- Kanda T., Kamiya M., Maruo S., Iwakiri D., Takada K. 2007. Symmetrical localization of extrachromosomally replicating viral genomes on sister chromatids.J. Cell Sci. 120:1529–1539 [DOI] [PubMed] [Google Scholar]
- Kapoor P., Shire K., Frappier L. 2001. Reconstitution of Epstein-Barr virus-based plasmid partitioning in budding yeast.EMBO J. 20:222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E., Tanaka K., Kitamura Y., Tanaka T.U. 2007. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae.Genes Dev. 21:3319–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krithivas A., Fujimuro M., Weidner M., Young D.B., Hayward S.D. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes.J. Virol. 76:11596–11604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakadamyali M., Rust M.J., Babcock H.P., Zhuang X. 2003. Visualizing infection of individual influenza viruses.Proc. Natl. Acad. Sci. USA. 100:9280–9285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen R.A., Cusumano C., Fujioka A., Lim-Fong G., Patterson P., Pogliano J. 2007. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis.Genes Dev. 21:1340–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A.L., Compton D.A. 2008. Structural and regulatory roles of non-motor spindle proteins.Curr. Opin. Cell Biol. 20:101–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S., Yang X.M., Chan C.S., Dobson M.J., Jayaram M., Velmurugan S. 2002. The 2 micron plasmid purloins the yeast cohesin complex: a mechanism for coupling plasmid partitioning and chromosome segregation? J. Cell Biol. 158:625–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S., Yang X.-M., Jayaram M., Velmurugan S. 2005. A novel role for the mitotic spindle during DNA segregation in yeast: promoting 2µm plasmid-cohesin association.Mol. Cell. Biol. 25:4283–4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P.B., Rose M.D. 1990. KAR3, a kinesin-related gene required for yeast nuclear fusion.Cell. 60:1029–1041 [DOI] [PubMed] [Google Scholar]
- Moller-Jensen J., Borch J., Dam M., Jensen R.B., Roepstorff P., Gerdes K. 2003. Bacterial mitosis: ParM of plasmid R1 moves plasmid DNA by an actin-like insertional polymerization mechanism.Mol. Cell. 12:1477–1487 [DOI] [PubMed] [Google Scholar]
- Murray A.W., Szostak J.W. 1983. Pedigree analysis of plasmid segregation in yeast.Cell. 34:961–970 [DOI] [PubMed] [Google Scholar]
- Murray J.A., Cesareni G. 1986. Functional analysis of the yeast plasmid partition locus STB.EMBO J. 5:3391–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbo A., Sugden A., Sugden B. 2007. The coupling of synthesis and partitioning of EBV's plasmid replicon is revealed in live cells.EMBO J. 26:4252–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira J.G., Colf L.A., McBride A.A. 2006. Variations in the association of papillomavirus E2 proteins with mitotic chromosomes.Proc. Natl. Acad. Sci. USA. 103:1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish J.L., Rosa J., Wang X., Lahti J.M., Doxsey S.J., Androphy E.J. 2006. The DNA helicase ChlR1 is required for sister chromatid cohesion in mammalian cells.J. Cell Sci. 119:4857–4865 [DOI] [PubMed] [Google Scholar]
- Roof D.M., Meluh P.B., Rose M.D. 1992. Kinesin-related proteins required for assembly of the mitotic spindle.J. Cell Biol. 118:95–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shire K., Ceccarelli D.F., Avolio-Hunter T.M., Frappier L. 1999. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance.J. Virol. 73:2587–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens R.V. 2004. Chl1p, a DNA helicase-like protein in budding yeast, functions in sister-chromatid cohesion.Genetics. 166:33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Mukae N., Dewar H., van Breugel M., James E.K., Prescott A.R., Antony C., Tanaka T.U. 2005. Molecular mechanisms of kinetochore capture by spindle microtubules.Nature. 434:987–994 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Kitamura E., Kitamura Y., Tanaka T.U. 2007. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles.J. Cell Biol. 178:269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tytell J.D., Sorger P.K. 2006. Analysis of kinesin motor function at budding yeast kinetochores.J. Cell Biol. 172:861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tine B.A., Dao L.D., Wu S.-Y., Sonbuchner T.M., Lin B.Y., Zou N., Chiang C.-M., Broker T.R., Chow L.T. 2004. Human papillomavirus (HPV) origin-binding protein associates with mitotic spindles to enable viral DNA partitioning.Proc. Natl. Acad. Sci. USA. 101:4030–4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velmurugan S., Ahn Y.-T., Yang X.-M., Wu X.-L., Jayaram M. 1998. The 2 micron plasmid stability system: analyses of the interactions among plasmid- and host-encoded components.Mol. Cell. Biol. 18:7466–7477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velmurugan S., Yang X.-M., Chan C.S.M., Dobson M., Jayaram M. 2000. Partitioning of the 2-µm circle plasmid of Saccharomyces cerevisiae: functional coordination with chromosome segregation and plasmid-encoded Rep protein distribution.J. Cell Biol. 149:553–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velmurugan S., Mehta S., Jayaram M. 2003. Selfishness in moderation: evolutionary success of the yeast plasmid.Curr. Top. Dev. Biol. 56:1–24 [DOI] [PubMed] [Google Scholar]
- Viejo-Borbolla A., Ottinger M., Bruning E., Burger A., Konig R., Kati E., Sheldon J.A., Schulz T.F. 2005. Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1.J. Virol. 79:13618–13629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkert F.C., Broach J.R. 1986. Site-specific recombination promotes plasmid amplification in yeast.Cell. 46:541–550 [DOI] [PubMed] [Google Scholar]
- Walker J.E., Saraste M., Runswick M.J., Gay N.J. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold.EMBO J. 1:945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West M., Hedges J.B., Lo K.Y., Johnson A.W. 2007. Novel interaction of the 60S ribosomal subunit export adapter Nmd3 at the nuclear pore complex.J. Biol. Chem. 282:14028–14037 [DOI] [PubMed] [Google Scholar]
- Wu H., Ceccarelli D.F., Frappier L. 2000. The DNA segregation mechanism of Epstein-Barr virus nuclear antigen 1.EMBO Rep. 1:140–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.-M., Mehta S., Uzri D., Jayaram M., Velmurugan S. 2004. Mutations in a partitioning protein and altered chromatin structure at the partitioning locus prevent cohesin recruitment by the Saccharomyces cerevisiae plasmid and cause plasmid missegregation.Mol. Cell. Biol. 24:5290–5303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J., Srinivasan V., Denis G.V., Harrington W.J., Jr., Ballestas M.E., Kaye K.M., Howley P.M. 2006. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes.J. Virol. 80:8909–8919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T., Peng Y.-C., Androphy E.J. 2007. Mitotic kinesin-like protein 2 binds and colocalizes with papillomavirus E2 during mitosis.J. Virol. 81:1736–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]