Abstract
The protein deacetylase SIRT1 has been implicated in a variety of cellular functions, including development, cellular stress responses, and metabolism. Increasing evidence suggests that similar to its counterpart, Sir2, in yeast, Caenorhabditis elegans, and Drosophila melanogaster, SIRT1 may function to regulate life span in mammals. However, SIRT1's role in cancer is unclear. During our investigation of SIRT1, we found that c-Myc binds to the SIRT1 promoter and induces SIRT1 expression. However, SIRT1 interacts with and deacetylates c-Myc, resulting in decreased c-Myc stability. As a consequence, c-Myc's transformational capability is compromised in the presence of SIRT1. Overall, our experiments identify a c-Myc–SIRT1 feedback loop in the regulation of c-Myc activity and cellular transformation, supporting/suggesting a role of SIRT1 in tumor suppression.
Introduction
The Sir2 protein deacetylase has emerged as an important regulator of aging in yeast, Caenorhabditis elegans, and Drosophila melanogaster (Guarente and Picard, 2005; Longo and Kennedy, 2006). The conserved role of Sir2 in extending life span in lower organisms raises the possibility that the mammalian orthologue of Sir2 may also play a similar role. In mammals, there are seven homologues of Sir2, SIRT1–7, of which SIRT1 is most similar to the yeast Sir2 and is considered to be the orthologue of yeast Sir2. SIRT1 has been shown to regulate multiple cellular functions, including cellular stress response, cell differentiation, development, and metabolism (Michan and Sinclair, 2007). Activators of SIRT1 improve cellular mitochondria function and protect mice from metabolic diseases (Baur et al., 2006; Lagouge et al., 2006; Milne et al., 2007). Furthermore, SIRT1 transgenic mice also display beneficial phenotypes similar to mice on a calorie-restricted diet (leaner and more metabolically active; Bordone et al., 2007), supporting an antiaging role of SIRT1.
Recent studies also revealed a paradoxical role of SIRT1 in tumorigenesis, possibly as a result of SIRT1's inhibitory effect toward tumor suppressors such as p53. Inhibition of SIRT1 by small molecule inhibitors or down-regulation of SIRT1 by siRNA results in arrested cell growth and apoptosis in several tumor cell lines, including breast, lung, and colon cancer lines, suggesting a role for SIRT1 in tumor cell growth (Ford et al., 2005; Heltweg et al., 2006; Ota et al., 2006; Lain et al., 2008). In addition, inhibition of SIRT1 results in reactivation of tumor suppressor gene transcription in human breast and colon cancer lines (Pruitt et al., 2006), suggesting a role for SIRT1 in silencing tumor suppressor genes. Together, these studies suggest a possible role of SIRT1 in promoting tumorigenesis. However, recent work with SIRT1 transgenic and knockout mice also suggests a role for SIRT1 in tumor suppression (Firestein et al., 2008; Wang et al., 2008). Therefore, the biological effects of SIRT1 are complex. In this study, we show that c-Myc and SIRT1 form a negative feedback loop that inhibits c-Myc–induced cellular transformation. These results support a tumor suppression function of SIRT1.
Results and discussion
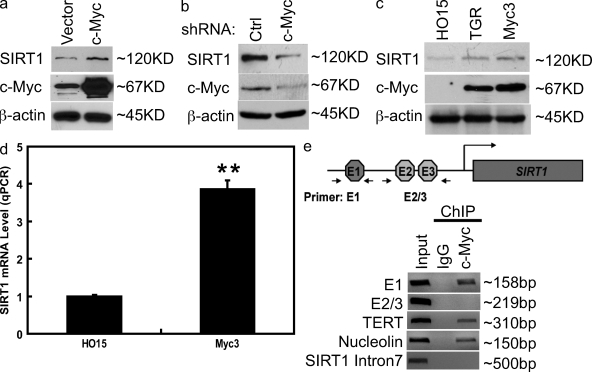
During our investigation of SIRT1 regulation, we found that there are three potential c-Myc–binding sites (E-box) localized at the SIRT1 promoter. This finding led us to hypothesize that c-Myc may regulate SIRT1 expression. Indeed, overexpression of c-Myc increased SIRT1 expression (Fig. 1 a). However, down-regulation of c-Myc decreased SIRT1 expression (Fig. 1 b). Furthermore, SIRT1 expression is higher in c-Myc–proficient fibroblasts (TGR and Myc3) than that of Myc-deficient cells (HO15; Fig. 1 c). These results suggest that c-Myc may act to enhance SIRT1 expression. To investigate whether c-Myc regulates SIRT1 expression at the transcriptional level, we performed quantitative RT-PCR (QRT-PCR) of SIRT1 transcripts. As shown in Fig. 1 d, there were more SIRT1 transcripts in c-Myc–proficient cells than those of c-Myc–deficient cells, suggesting that c-Myc induces SIRT1 transcription. Finally, to directly demonstrate that c-Myc binds at the SIRT1 promoter, we performed chromatin immunoprecipitation (ChIP) assays using anti–c-Myc antibodies. As shown in Fig. 1 e, Myc binds one E-box (E1) but not other E-boxes (E2 and E3) of the SIRT1 promoter. Overall, our results suggest that c-Myc promotes SIRT1 expression by elevating SIRT1 transcription.
Figure 1.
c-Myc induces SIRT1 expression. (a) Control vector or constructs encoding Flag–c-Myc were transfected into 293T cells. 48 h later, cell lysates were examined by Western blotting. (b) HeLa cells were transfected with control (ctrl) shRNA or c-Myc shRNA. 72 h later, proteins were examined by Western blotting. (c) HO15 (c-Myc−/−), TGR (c-Myc+/+), and Myc3 (HO15 cells reconstituted with c-Myc) were lysed, and the cell lysates were subjected to Western blotting. (d) mRNA from HO15 and Myc3 were subjected to QRT-PCR. Values represent the relative induction of SIRT1 mRNA normalized to GAPDH. Data represent the mean of three determinations ± SEM. **, P < 0.01 by two-tailed Student's t test. (e) ChIP assays were performed using anti–c-Myc antibody. The TERT and nucleolin promoter primers were used as a positive control, and SIRT1 intron 7 primers were used as a negative control.
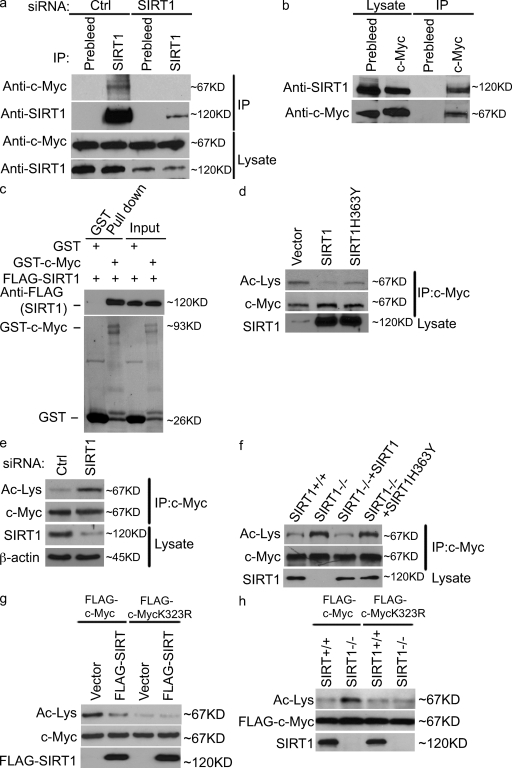
Interestingly, we also found that SIRT1 coimmunoprecipitated with c-Myc in vivo (Fig. 2 a). To confirm the specificity of this interaction, we used SIRT1 siRNA to down-regulate SIRT1. c-Myc failed to coimmunoprecipitate with SIRT1 in cells transfected with SIRT1 siRNA (Fig. 2 a), confirming the specificity of the c-Myc–SIRT1 interaction. Furthermore, immunoprecipitation of c-Myc also pulled down SIRT1 (Fig. 2 b). To examine whether the SIRT1–c-Myc interaction is direct, we incubated purified c-Myc and SIRT1 under cell-free conditions and found that SIRT1 interacts with c-Myc in vitro, suggesting that SIRT1 directly interacts with c-Myc (Fig. 2 c).
Figure 2.
SIRT1 interacts with c-Myc and deacetylates c-Myc at K323. (a and b). HeLa cells were transfected with control (ctrl) or SIRT1 siRNA (a) or left untransfected (b). Cell lysates were subjected to immunoprecipitation (IP) and immunoblotting with the indicated antibodies. (c) Purified Flag-SIRT1 was incubated with recombinant GST or GST–c-Myc coupled to glutathione-Sepharose. Proteins retained on the beads were blotted with the indicated antibodies. (d) The indicated constructs were transfected into 293T cells. 48 h later, cell lysates were subjected to immunoprecipitation and immunoblotting with the indicated antibodies. (e) Cells were transfected with control or SIRT1 siRNA. 72 h later, the acetylation (Ac) level of c-Myc was detected as in d. (f) SIRT1+/+, SIRT1−/−, and SIRT1−/− reconstituted with SIRT1 and SIRT1-H363Y were lysed, and the acetylation level of c-Myc were detected as in d. (g) The indicated constructs were transfected into cells. 48 h later, cell lysates were subjected to immunoprecipitation and immunoblotting with the indicated antibodies. (h) SIRT1+/+ and SIRT1−/− cells were transfected with the indicated expressing constructs. 48 h later, cell lysates were subjected to immunoprecipitation immunoblotting with the indicated antibodies. To compare the acetylation of c-Myc, the levels of c-Myc were equalized in d–h.
We next investigated the functional significance of the c-Myc–SIRT1 interaction. Because SIRT1 is a protein deacetylase, it is possible that SIRT1 could regulate c-Myc activity by deacetylating c-Myc. As shown in Fig. 2 d, overexpression of wild-type (WT) SIRT1, but not catalytically inactive SIRT1-H363Y, resulted in hypoacetylation of c-Myc. Conversely, down-regulation of SIRT1 using SIRT1 siRNA resulted in c-Myc hyperacetylation (Fig. 2 e). Finally, SIRT1−/− cells showed that c-Myc hyperacetylation compared with SIRT1+/+ cells; reconstitution of SIRT1−/− cells with WT SIRT1, but not inactive SIRT1-H363Y, decreased c-Myc acetylation (Fig. 2 f). These results suggest that SIRT1 deacetylates c-Myc in cells.
We further explored potential SIRT1 deacetylation sites of c-Myc. Previous studies have identified several potential acetylation sites of c-Myc (Patel et al., 2004; Faiola et al., 2005). We found that Lys323 (K323) is a major SIRT1 deacetylation site because overexpression of SIRT1 was unable to reduce c-MycK323R acetylation as it was with the WT c-Myc. Thus, although the c-MycK232R mutant is not able to be acetylated, SIRT1 overexpression was not able to reduce the acetylation status of the mutant below that of basal levels (Fig. 2 g). Confirming this result, the loss of SIRT1 did not increase acetylation levels of the K323R mutant (Fig. 2 h). These results suggest that K323 of c-Myc is the major SIRT1 deacetylation site.
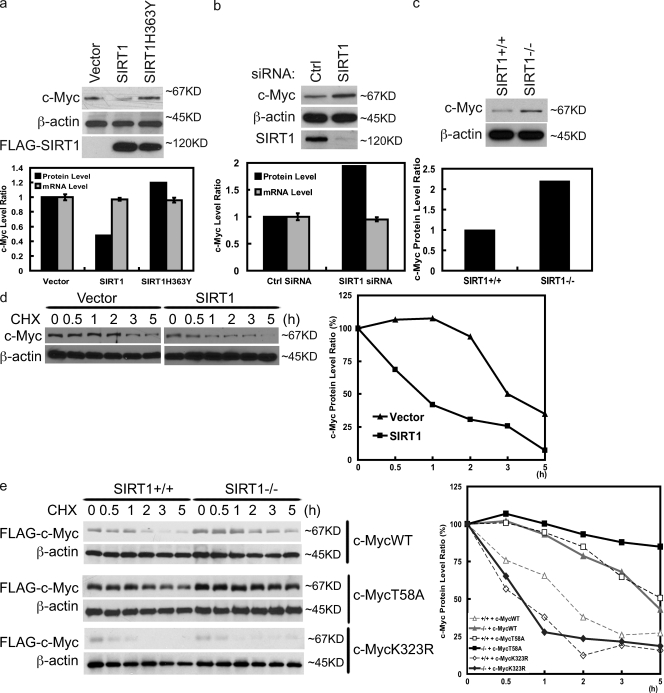
c-Myc has previously been shown to be acetylated, which enhances c-Myc stability (Vervoorts et al., 2003; Patel et al., 2004; Faiola et al., 2005). Because SIRT1 deacetylates c-Myc, we hypothesized that SIRT1 affects c-Myc stability. In support of our hypothesis, overexpression of SIRT1 decreased the c-Myc protein level without affecting the c-Myc mRNA level (Fig. 3 a). Conversely, down-regulation of SIRT1 increased c-Myc expression with no effect on c-Myc transcripts (Fig. 3 b). To confirm these findings, we used SIRT1+/+ and SIRT1−/− mouse embryonic fibroblasts (MEFs). As shown in Fig. 3 c, there was increased expression of c-Myc in SIRT1−/− cells. Reconstitution of WT SIRT1, but not catalytically inactive SIRT1, decreased c-Myc expression (Fig. S1 a). These results suggest that SIRT1 regulates c-Myc expression at the posttranscriptional level. We next examined whether SIRT1 regulates c-Myc stability in the presence of cycloheximide, which blocks protein translation. As shown in Fig. 3 d, overexpression of SIRT1 decreased c-Myc stability. Conversely, loss of SIRT1 expression enhanced c-Myc stability (Fig. S1 b). Reconstitution of SIRT1−/− cells with WT SIRT1, but not catalytically inactive SIRT1, restored the c-Myc half-life to the level of WT MEFs (Fig. S1 b). These results suggest that SIRT1 negatively regulates c-Myc stability.
Figure 3.
SIRT1 decreases c-Myc stability. (a) The indicated constructs were transfected into 293T cells. 48 h later, proteins and mRNA were extracted and subjected to Western blotting or QRT-PCR. (b) HeLa cells were transfected with control (ctrl) or SIRT1 siRNA. 72 h later, proteins and mRNA were extracted and subjected to Western blotting or QRT-PCR. (a and b) Quantification of c-Myc protein and transcript levels is shown in the bottom panels. (c) Cell lysates from SIRT1+/+ or SIRT1−/− were blotted with the indicated antibodies. (bottom) Quantification of c-Myc protein levels is shown. (d) The indicated constructs were transfected into 293T cells. 20 h later, cells were treated with 0.1 mg/ml cycloheximide (CHX) and harvested at the indicated time. Immunoblots of c-Myc and β-actin protein at the indicated times are shown. (right) Quantification of the c-Myc levels is shown. (e) SIRT1+/+ and SIRT1−/− were transfected with the indicated constructs. c-Myc stability was then examined as in d. (right) Quantification of the Flag–c-Myc levels is shown. Error bars represent SEM.
Previous studies have established that the F-box protein FBW7 binds to phospho-Thr58 of c-Myc and targets it to the SKP1–Cul1–F-box ubiquitin ligase complex for degradation (Welcker et al., 2004; Yada et al., 2004). It is possible that SIRT1 regulates c-Myc stability through FBW7. We used the MycT58A mutant to investigate this possibility. As shown in Fig. 3 e, MycT58A is more stable than WT c-Myc, which is consistent with previous findings (Welcker et al., 2004; Yada et al., 2004). However, SIRT1 still decreased MycT58A stability, suggesting that SIRT1 and FBW7 regulate c-Myc stability through distinct pathways.
Because we have shown that K323 is a major SIRT1 deacetylation site of c-Myc (Fig. 2, g and h), we further examined whether SIRT1 regulates c-Myc stability through K323. As shown in Fig. 3 e, MycK323 was less stable than WT c-Myc. Although SIRT1 decreased the stability of WT c-Myc, it had no effect toward MycK323R. These results suggest that SIRT1 regulates c-Myc stability by deacetylating K323.
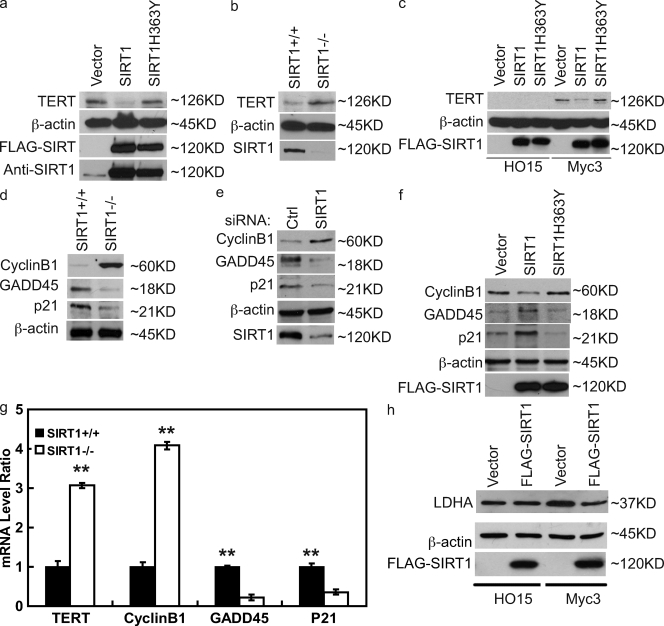
We next investigated how SIRT1 affects c-Myc function by examining the expression of c-Myc target genes. We first examined the expression of telomerase reverse transcriptase gene (TERT), which is a direct target gene of c-Myc (Greenberg et al., 1999; Wu et al., 1999). Consistent with its negative regulation of c-Myc, overexpression of SIRT1 decreased TERT expression (Fig. 4 a). However, overexpression of the catalytically inactive SIRT1 mutant SIRT1-H363Y did not affect TERT expression. Furthermore, TERT expression is clearly higher in SIRT1−/− cells than that of SIRT1+/+ cells (Fig. 4 b), confirming that SIRT1 negatively regulates TERT expression.
Figure 4.
SIRT1 inhibits Myc target gene expression through c-Myc. (a) 293T cells were transfected with the indicated constructs. 48 h later, cells were lysed, and cell lysates were blotted with the indicated antibodies. (b) Cell lysates from SIRT1+/+ or SIRT1−/− MEFs were blotted with the indicated antibodies. (c) HO15 and Myc3 cells were infected with retrovirus control or retrovirus encoding SIRT1 and SIRT1-H363Y. 48 h later, cells were lysed, and cell lysates were blotted with the indicated antibodies. (d) Cell lysates from SIRT1+/+ and SIRT1−/− were blotted with the indicated antibodies. (e) HeLa cells were transfected with control (ctrl) or SIRT1 siRNA. 72 h later, cells were lysed, and cell lysates were blotted with the indicated antibodies. (f) 293T cells were transfected with the indicated constructs. 48 h later, cell lysates were blotted with the indicated antibodies. (g) SIRT1+/+ and SIRT1−/− were lysed, and mRNA were extracted and subjected to QRT-PCR. Values represent the relative induction normalized to GAPDH. Data represent the mean of three determinations ± SEM. **, P < 0.01 by two-tailed Student's t test. (h) HO15 and Myc3 cells were infected with retrovirus control or retrovirus encoding SIRT1. 48 h later, cells were lysed, and cell lysates were blotted with the indicated antibodies.
Because SIRT1 could regulate multiple transcriptional factors, we decided to confirm that SIRT1 regulates TERT expression through c-Myc. We found little expression of TERT in Myc−/− HO15 cells. However, HO15 cells stably reconstituted with c-Myc (Myc3) showed enhanced TERT expression (Fig. 4 c). Importantly, overexpression of WT SIRT1, but not catalytically inactive SIRT1, suppressed TERT expression, suggesting that SIRT1 negatively regulates TERT expression through c-Myc.
To further confirm the inhibitory effects of SIRT1 toward c-Myc, we also examined the expression of other c-Myc target genes. c-Myc has been shown to activate cyclin B1 expression and suppress Gadd45 and p21 expression (Adhikary and Eilers, 2005). We found increased expression of cyclin B1 and decreased expression of Gadd45 and p21 in SIRT1−/− cells and cells transfected with SIRT1 siRNA (Fig. 4, d and e). However, overexpression of WT SIRT1, but not catalytically inactive SIRT1, results in decreased cyclin B1 expression and increased Gadd45 and p21 expression (Fig. 4 f). Furthermore, QRT-PCR results showed increased transcripts of cyclin B1 and decreased transcripts of Gadd45 and p21 in SIRT1−/− cells (Fig. 4 g). In addition, Myc has also been shown to regulate cancer cell energy metabolism by inducing genes such as lactate dehydrogenase A (LDHA) and mitochondrial serine hydroxymethyltransferase (mSHMT) expression (Shim et al., 1997, 1998; Nikiforov et al., 2002). We also found that SIRT1 decreased Myc-induced LDHA expression by Western analysis and both LDHA and mSHMT expression by QRT-PCR (Fig. 4 h; and Fig. S2, a and b, respectively). Overall, these results strongly suggest that SIRT1 negatively regulates c-Myc activity.
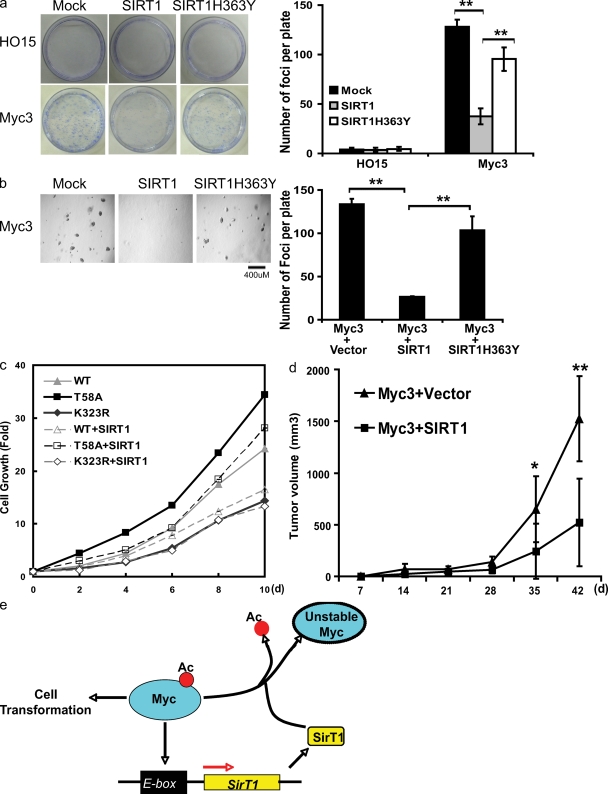
The inhibitory effect of SIRT1 on c-Myc raised the possibility that SIRT1 could negatively regulate cell growth and transformation. To specifically examine how SIRT1 affects c-Myc–induced cell growth, we used Myc−/− HO15 cells, and HO15 rat fibroblast cells stably reconstituted with c-Myc (Myc3). As shown in Fig. 5 a, expression of c-Myc greatly increased colony formation. However, coexpression of WT SIRT1, but not catalytically inactive SIRT1, with c-Myc significantly decreased c-Myc–induced colony formation. Furthermore, we performed soft agar assays to examine how SIRT1 affects anchorage-independent cell growth. As shown in Fig. 5 b, Myc3 cells displayed anchorage-independent cell growth in soft agar. However, expression of SIRT1 significantly decreased the colony formation in soft agar, whereas catalytically inactive SIRT1 only slightly affected colony formation. These results suggest that SIRT1 inhibits c-Myc–dependent cellular transformation. To confirm that SIRT1 suppresses cell growth by deacetylating c-Myc, we used the MycK323R mutant to perform cell growth experiments. As shown in Fig. 5 c, although SIRT1 could suppress WT c-Myc and MycT58A-induced cell growth, it had no effect on MycK323R-induced cell growth. These results suggest that SIRT1 exerts its antitransformation effects by deacetylating K323 of Myc. SIRT1 also decreased MycT58A-induced cell growth and colony formation of FBW7+/+ and FBW7−/− cells (Fig. S3 a), suggesting that SIRT1 regulates c-Myc activity independently of the FBW7 pathway.
Figure 5.
SIRT1 inhibits c-Myc–induced cellular transformation. (a) Colony formation assays were performed using HO15 and Myc3 Rat1 cells infected with retrovirus encoding SIRT1 and SIRT1-H363Y. Colony formation was examined 2 wk later. The results shown are mean ± SEM and are representative of three independent assays. (b) Soft agar colony formation assay was performed using Myc3 Rat1 cells infected with retrovirus encoding SIRT1 and SIRT1-H363Y. The figure is representative of three independent assays. (c) HO15 cells were stably transfected with the indicated vectors. Cell growth was quantified at the indicated time. (d) Myc3 cells were transfected with Ras-encoding constructs together with vector or SIRT1-encoding constructs. Transfected cells were implanted into nude mice, and tumor formation was monitored. *, P < 0.05; **, P < 0.01 by two-tailed Student's t test. (e) Model of how c-Myc–SIRT1 loop regulates cell growth. Ac, acetylation.
Finally, to confirm the role of SIRT1 in suppressing cellular transformation in vivo, we stably expressed Ras and SIRT1 in Myc3 cells. Cells were then implanted into athymic nude mice. As shown in Fig. 5 d and Fig. S3 b, overexpression of SIRT1 significantly inhibited tumorigenesis in nude mice. Collectively, these results demonstrate a tumor suppression role of SIRT1 in vitro and in vivo.
In this study, we present evidence that c-Myc induces SIRT1 expression, and SIRT1 in turn deacetylates and down-regulates c-Myc, resulting in decreased c-Myc target gene (i.e., TERT) expression and cellular transformation (Fig. 5 e). Consequently, this Myc-SIRT1 negative feedback loop can restrain Myc's transformational activities. Although it has been established that SIRT1 has several important roles in metabolism and cellular stress response, its role in tumorigenesis remains controversial because both pro- and antitumorigenesis effects of SIRT1 have been suggested. It is possible that SIRT1's role in tumorigenesis is context dependent. For example, senescence is important for suppression of tumor initiation; however, it is thought to drive tumor growth by secreting inflammatory factors and tissue-disrupting enzymes (Campisi, 2005). It is possible that SIRT1 inhibits the initiation of tumor in premalignant cells by inhibiting c-Myc. However, cancer cells could become addicted to SIRT1 as a result of SIRT1's antiapoptosic functions. In support of this notion, inhibition of SIRT1 induces cell cycle arrest and death only in cancer cells but not noncancerous cells (Ford et al., 2005). Therefore, SIRT1 might have both tumor suppression and promotion roles depending on the stage of cell transformation.
Feedback mechanisms have previously been demonstrated for the regulation of Myc activity. For example, Myc induced expression of ribosomal protein L11, which in turn binds Myc and blocks Myc transcriptional activity (Dai et al., 2007), forming a negative feedback loop. Myc also induces L23 expression, which blocks Miz1 function and p21 and p16 transcription, thereby acting as a positive feedback loop to further potentiate Myc's ability to promote cell growth (Wanzel et al., 2008). We showed that through a negative feedback loop, SIRT1 could decrease the expression of c-Myc target genes (e.g., TERT) and increase expression of tumor suppressors (e.g., p21 and Gadd45). Our results are consistent with previous findings that SIRT1 can limit replicative life span (Chua et al., 2005) and TERT expression (Narala et al., 2008).
In addition, through this Myc-SIRT1 feedback loop, SIRT1 could also modulate cells' energy metabolism. It has been established that in addition to regulating cell cycle progression and apoptosis, c-Myc also regulates energy metabolism through the induction of LDHA and mSHMT (Shim et al., 1997; Nikiforov et al., 2002), which are important for cellular transformation. We showed that SIRT1 also blocks Myc-mediated expression of LDHA and mSHMT, providing another route by which SIRT1 regulates glucose metabolism in addition to its known role in regulating glucose metabolism. One can also postulate that Myc-induced redox metabolism could change the NAD/NADH ratio, which in turn affects SIRT1 activity. This is another possible feedback mechanism that requires further investigation.
In summary, our experiments suggest that a c-Myc–SIRT1 negative feedback loop suppresses cell growth and transformation. These experiments also provide a rationale using SIRT1 activators as cancer prevention agents in the future.
Materials and methods
Cell culture
HEK293T and HeLa cells were cultured in RPMI with 10% FBS. HO15, TGR, Myc3, and Rat1 cells were provided by J. Sedivy (Brown University, Providence, RI). SIRT1 MEFs were provided by C. Deng (National Institutes of Health, Bethesda, MD). Rat1 and MEF cells were cultured in DME with 15% FBS.
Plasmids
S/Flag/streptavidin-binding peptide (SBP)–tagged SIRT1 and c-Myc were cloned into pIRES2-EGFP (Clontech Laboratories, Inc.). S/Flag/SBP-SIRT1 was cloned into pDEST8 for expression of GST fusion protein in insect cells (sf9 cells). c-Myc was cloned into pGEX4T-1 for GST–c-Myc production. pcDNA3.1/HisMyc-SIRT1 and SIRT1-H363Y were provided by W. Gu (Columbia University, New York, NY). SIRT1 retroviruses were constructed using the Gateway System (Invitrogen). Deletion mutants were generated by site-directed mutagenesis (Agilent Technologies).
RNAi
SIRT1 siRNA was purchased from Thermo Fisher Scientific. Transfection was performed twice 24 h apart with 200 nM siRNA using Oligofectamine (Invitrogen). Myc short-hairpin RNA (shRNA) was described previously (Popov et al., 2007) and obtained from Addgene, Inc.
Preparation and infection of retrovirus
Packaging cell line BOSC23 was transfected with the retrovirus pBabe–S/Flag/SBP-SIRT1. Media were changed 24 h later and collected 48 or 72 h after transfection. The media were filter sterilized (0.45-µm filter) and used to infect SIRT−/− MEF, HO15, or Myc3 Rat1 cells. Infected cells were selected with 2 µg/ml puromycin (Sigma-Aldrich). Resistant clones were picked and expanded for further analysis.
ChIP assay
ChIP assay was performed by cross-linking ∼2 × 107 cells in 1% formaldehyde for 5 min and sonicating until the bulk of DNA was 300–600 bp in size. Chromatin was immunoprecipitated with anti–c-Myc antibody, washed, and the DNA–protein cross-links were reversed. The recovered DNA was amplified by 30 cycles of PCR with the following primers: E1, 5′-AGGCCAAGTCATTTCCTTCC-3′ and 5′-ACCTTTGACGTGGAGGTTTG-3′; E2/3, 5′-GGAGCGGTAGACGCAACA-3′ and 5′-CTTCCAACTGCCTCTCTGG-3′; TERT promoter, 5′-GGCCGGGCTCCCAGTGGATTC-3′ and 5′-CAGCGGGGAGCGCGCGGCATCG-3′; nucleolin promoter, 5′-TTGCGACGCGTACGAGCTGG-3′ and 5′-ACTCCGACTAGGGCCGATAC-3′; and SIRT1 intron 7, 5′-TTCCTCCTCTGCCTCTCAAA-3′ and 5′-CTTGGGAGGAGAACTGCTTG-3′.
Western blots and antibodies
Cells lysis, immunoprecipitation, and immunoblotting were performed following standard procedures. SIRT1 antibodies were made as previously described (Kim et al., 2008). c-Myc antibodies were raised by immunizing rabbits with full-length GST–c-Myc. Antisera were affinity purified with AminoLink Plus Immobilization and Purification kit (Thermo Fisher Scientific). Anti-p21 antibody was provided by J. van Deursen (Mayo Clinic, Rochester, MN). Human TERT (H-231), Gadd45α (H-165), cyclin B1, and c-Myc were purchased from Santa Cruz Biotechnology, Inc. Anti–acetylated lysine antibody (#06-933) was purchased from Millipore, LDHA antibody was purchased from Cell Signaling Technology, and anti-Flag (M2) antibody was purchased from Sigma-Aldrich.
QRT-PCR
mRNA was isolated with PARIS kit (Applied Biosystems). QRT-PCR was performed using Brilliant II SYBR Green QRT-PCR Master Mix kit (Agilent Technologies). Primers (200 nM; QIAGEN) used include rat SIRT1, human c-Myc, mouse TERT, mouse cyclin B1, mouse p21, mouse Gadd45, rat LDHA, rat mSHMT, human glyceraldehyde 3-phosphate dehydrogenase (GAPDH), mouse GAPDH, and rat GAPDH. Changes in gene expression were quantified based on the 2ΔCt value normalized to GAPDH.
Colony and soft agar colony formation assays
Cells were plated in 60- or 35-mm dishes. After 2 wk, cells were stained with 2% methylene blue, and colonies were counted.
The soft agar colony formation assay was performed as described previously (Shim et al., 1997). In brief, Myc3 Rat1 cells were infected with either control, SIRT1, or SIRT1-H363Y retroviruses. Cells were plated in 0.3% top agarose in 35-mm dishes and cultured for 2 wk. Colonies were counted at room temperature under a light microscope (ECLIPSE 80i; Nikon) using a 4× NA 0.10 objective lens (Nikon). Images were captured with a camera (SPOT 2 Megasample; Diagnostic Instruments, Inc.) and processed using SPOT software (version 4.6; Diagnostic Instruments, Inc.). Photoshop and Illustrator (Adobe) were used to generate figures.
Athymic nude mice tumor formation assay
Myc3 cells stably expressing H-Rasv12 together with vector or SIRT1 were injected subcutaneously and bilaterally into the dorsal left and right scapular areas of 5-wk-old male athymic recessive nude/nude mice (obtained from the National Cancer Institute) using 19-gauge needles. Each mouse received two injections of a 20-µl mixture of 2 ×106 cells in 100 µl 1× PBS and 100 µl growth factor–reduced Matrigel (BD). Tumor growth was monitored for 6 wk, and tumor volume was calculated as 0.5 × length × height × width. The tumors were surgically removed, weighed, and processed.
Online supplemental material
Fig. S1 shows that reconstitution of WT SIRT1, but not catalytically inactive SIRT1 in SIRT1−/− cells, decreased c-Myc stability. Fig. S2 shows that SIRT1 decreased Myc-induced LDHA and mSHMT expression. Fig. S3 shows that SIRT1 decreased colony formation of FBW7+/+ and FBW7−/− cells and tumor formation. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200809167/DC1.
Acknowledgments
We thank Drs. Chuxia Deng, Stephen Hann, Wei Gu, John Sedivy, Zhiguo Zhang, Martin Eilers, and Jan van Deursen for providing reagents used in this study.
This work was supported in part by grants from the Richard Schultz Foundation, the Fraternal Order of Eagles Cancer Research Fund, the Susan G. Komen Breast Cancer Foundation Research Grant, and the National Institutes of Health (grant to Z. Lou).
Footnotes
Abbreviations used in this paper: ChIP, chromatin immunoprecipitation; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; LDHA, lactate dehydrogenase A; MEF, mouse embryonic fibroblast; mSHMT, mitochondrial serine hydroxymethyltransferase; QRT-PCR, quantitative RT-PCR; SBP, streptavidin-binding peptide; shRNA, short-hairpin RNA; TERT, telomerase reverse transcriptase gene; WT, wild type.
References
- Adhikary S., Eilers M. 2005. Transcriptional regulation and transformation by Myc proteins.Nat. Rev. Mol. Cell Biol. 6:635–645 [DOI] [PubMed] [Google Scholar]
- Baur J.A., Pearson K.J., Price N.L., Jamieson H.A., Lerin C., Kalra A., Prabhu V.V., Allard J.S., Lopez-Lluch G., Lewis K., et al. 2006. Resveratrol improves health and survival of mice on a high-calorie diet.Nature. 444:337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordone L., Cohen D., Robinson A., Motta M.C., van Veen E., Czopik A., Steele A.D., Crowe H., Marmor S., Luo J., et al. 2007. SIRT1 transgenic mice show phenotypes resembling calorie restriction.Aging Cell. 6:759–767 [DOI] [PubMed] [Google Scholar]
- Campisi J. 2005. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors.Cell. 120:513–522 [DOI] [PubMed] [Google Scholar]
- Chua K.F., Mostoslavsky R., Lombard D.B., Pang W.W., Saito S., Franco S., Kaushal D., Cheng H.L., Fischer M.R., Stokes N., et al. 2005. Mammalian SIRT1 limits replicative life span in response to chronic genotoxic stress.Cell Metab. 2:67–76 [DOI] [PubMed] [Google Scholar]
- Dai M.S., Arnold H., Sun X.X., Sears R., Lu H. 2007. Inhibition of c-Myc activity by ribosomal protein L11.EMBO J. 26:3332–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faiola F., Liu X., Lo S., Pan S., Zhang K., Lymar E., Farina A., Martinez E. 2005. Dual regulation of c-Myc by p300 via acetylation-dependent control of Myc protein turnover and coactivation of Myc-induced transcription.Mol. Cell. Biol. 25:10220–10234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein R., Blander G., Michan S., Oberdoerffer P., Ogino S., Campbell J., Bhimavarapu A., Luikenhuis S., de Cabo R., Fuchs C., et al. 2008. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth.PLoS ONE. 3:e2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford J., Jiang M., Milner J. 2005. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival.Cancer Res. 65:10457–10463 [DOI] [PubMed] [Google Scholar]
- Greenberg R.A., O'Hagan R.C., Deng H., Xiao Q., Hann S.R., Adams R.R., Lichtsteiner S., Chin L., Morin G.B., DePinho R.A. 1999. Telomerase reverse transcriptase gene is a direct target of c-Myc but is not functionally equivalent in cellular transformation.Oncogene. 18:1219–1226 [DOI] [PubMed] [Google Scholar]
- Guarente L., Picard F. 2005. Calorie restriction–the SIR2 connection.Cell. 120:473–482 [DOI] [PubMed] [Google Scholar]
- Heltweg B., Gatbonton T., Schuler A.D., Posakony J., Li H., Goehle S., Kollipara R., Depinho R.A., Gu Y., Simon J.A., Bedalov A. 2006. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes.Cancer Res. 66:4368–4377 [DOI] [PubMed] [Google Scholar]
- Kim J.E., Chen J., Lou Z. 2008. DBC1 is a negative regulator of SIRT1.Nature. 451:583–586 [DOI] [PubMed] [Google Scholar]
- Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., et al. 2006. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha.Cell. 127:1109–1122 [DOI] [PubMed] [Google Scholar]
- Lain S., Hollick J.J., Campbell J., Staples O.D., Higgins M., Aoubala M., McCarthy A., Appleyard V., Murray K.E., Baker L., et al. 2008. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator.Cancer Cell. 13:454–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo V.D., Kennedy B.K. 2006. Sirtuins in aging and age-related disease.Cell. 126:257–268 [DOI] [PubMed] [Google Scholar]
- Michan S., Sinclair D. 2007. Sirtuins in mammals: insights into their biological function.Biochem. J. 404:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne J.C., Lambert P.D., Schenk S., Carney D.P., Smith J.J., Gagne D.J., Jin L., Boss O., Perni R.B., Vu C.B., et al. 2007. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes.Nature. 450:712–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narala S.R., Allsopp R.C., Wells T.B., Zhang G., Prasad P., Coussens M.J., Rossi D.J., Weissman I.L., Vaziri H. 2008. SIRT1 acts as a nutrient-sensitive growth suppressor and its loss is associated with increased AMPK and telomerase activity.Mol. Biol. Cell. 19:1210–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov M.A., Chandriani S., O'Connell B., Petrenko O., Kotenko I., Beavis A., Sedivy J.M., Cole M.D. 2002. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism.Mol. Cell. Biol. 22:5793–5800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota H., Tokunaga E., Chang K., Hikasa M., Iijima K., Eto M., Kozaki K., Akishita M., Ouchi Y., Kaneki M. 2006. Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MAPK signaling in human cancer cells.Oncogene. 25:176–185 [DOI] [PubMed] [Google Scholar]
- Patel J.H., Du Y., Ard P.G., Phillips C., Carella B., Chen C.J., Rakowski C., Chatterjee C., Lieberman P.M., Lane W.S., et al. 2004. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60.Mol. Cell. Biol. 24:10826–10834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov N., Wanzel M., Madiredjo M., Zhang D., Beijersbergen R., Bernards R., Moll R., Elledge S.J., Eilers M. 2007. The ubiquitin-specific protease USP28 is required for MYC stability.Nat. Cell Biol. 9:765–774 [DOI] [PubMed] [Google Scholar]
- Pruitt K., Zinn R.L., Ohm J.E., McGarvey K.M., Kang S.H., Watkins D.N., Herman J.G., Baylin S.B. 2006. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation.PLoS Genet. 2:e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H., Dolde C., Lewis B.C., Wu C.S., Dang G., Jungmann R.A., Dalla-Favera R., Dang C.V. 1997. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth.Proc. Natl. Acad. Sci. USA. 94:6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H., Chun Y.S., Lewis B.C., Dang C.V. 1998. A unique glucose-dependent apoptotic pathway induced by c-Myc.Proc. Natl. Acad. Sci. USA. 95:1511–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vervoorts J., Luscher-Firzlaff J.M., Rottmann S., Lilischkis R., Walsemann G., Dohmann K., Austen M., Luscher B. 2003. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP.EMBO Rep. 4:484–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.H., Sengupta K., Li C., Kim H.S., Cao L., Xiao C., Kim S., Xu X., Zheng Y., Chilton B., et al. 2008. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice.Cancer Cell. 14:312–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanzel M., Russ A.C., Kleine-Kohlbrecher D., Colombo E., Pelicci P.G., Eilers M. 2008. A ribosomal protein L23-nucleophosmin circuit coordinates Miz1 function with cell growth.Nat. Cell Biol. 10:1051–1061 [DOI] [PubMed] [Google Scholar]
- Welcker M., Orian A., Jin J., Grim J.E., Harper J.W., Eisenman R.N., Clurman B.E. 2004. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation.Proc. Natl. Acad. Sci. USA. 101:9085–9090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K.J., Grandori C., Amacker M., Simon-Vermot N., Polack A., Lingner J., Dalla-Favera R. 1999. Direct activation of TERT transcription by c-MYC.Nat. Genet. 21:220–224 [DOI] [PubMed] [Google Scholar]
- Yada M., Hatakeyama S., Kamura T., Nishiyama M., Tsunematsu R., Imaki H., Ishida N., Okumura F., Nakayama K., Nakayama K.I. 2004. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7.EMBO J. 23:2116–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]