Abstract
PIDD (p53-induced protein with a death domain [DD]), together with the bipartite adapter protein RAIDD (receptor-interacting protein-associated ICH-1/CED-3 homologous protein with a DD), is implicated in the activation of pro–caspase-2 in a high molecular weight complex called the PIDDosome during apoptosis induction after DNA damage. To investigate the role of PIDD in cell death initiation, we generated PIDD-deficient mice. Processing of caspase-2 is readily detected in the absence of PIDDosome formation in primary lymphocytes. Although caspase-2 processing is delayed in simian virus 40–immortalized pidd−/− mouse embryonic fibroblasts, it still depends on loss of mitochondrial integrity and effector caspase activation. Consistently, apoptosis occurs normally in all cell types analyzed, suggesting alternative biological roles for caspase-2 after DNA damage. Because loss of either PIDD or its adapter molecule RAIDD did not affect subcellular localization, nuclear translocation, or caspase-2 activation in high molecular weight complexes, we suggest that at least one alternative PIDDosome-independent mechanism of caspase-2 activation exists in mammals in response to DNA damage.
Introduction
PIDD (p53-induced protein with a death domain [DD]) was identified as one of many transcriptional targets that may mediate apoptosis induction by the tumor suppressor p53 (Lin et al., 2000). PIDD is widely expressed in various organs and cell types and is characterized by the presence of certain structural motives, including Leu-rich repeats, ZU5 domains, and a C-terminal DD (Tinel and Tschopp, 2004). DD-containing proteins play important roles in the formation of multimeric signaling complexes that regulate diverse cellular responses, including cytokine secretion, nuclear factor κB (NF-κB) activation, cell survival, and apoptosis (Reed et al., 2004). RAIDD (receptor-interacting protein-associated ICH-1/CED-3 homologous protein with a DD), a bipartite adapter molecule, was identified as a possible interaction partner of PIDD (Tinel and Tschopp, 2004). RAIDD contains a caspase recruitment domain as well as a C-terminal DD (Duan and Dixit, 1997). The caspase recruitment domain of RAIDD was previously described to interact with pro–caspase-2 (Duan and Dixit, 1997) and has been implicated in the activation of this ill-defined initiator caspase thought to be required for DNA damage–induced apoptosis in certain tumor cells and the metabolic death of oocytes (Degterev and Yuan, 2008). A trimolecular complex containing PIDD, RAIDD, and caspase-2 was subsequently identified as the long-sought-after activation platform of caspase-2 and dubbed the PIDDosome (Tinel and Tschopp, 2004). This complex appears to form spontaneously in cell extracts from different cell lines upon temperature shift in vitro, but formation in extracts from cells undergoing apoptosis after DNA damage has not been detected so far (Read et al., 2002). A second, PIDD-containing multimeric protein complex was described shortly thereafter that contains the Ser-Thr kinase RIP-1 (receptor-interacting protein 1) and IKK-γ/NF-κB essential modulator, the regulatory subunit of the IκB kinase complex, which were both implicated in the activation of NF-κB signaling after genotoxic stress (Janssens et al., 2005). Subsequent analysis revealed that PIDD possesses autoproteolytic activity, facilitating the generation of two different active protein fragments, termed PIDD-C and PIDD-CC. Although PIDD-CC was proposed to be required for activation of caspase-2 and cell death, PIDD-C was suggested to activate NF-κB, DNA repair, and survival after low grade DNA damage (Tinel et al., 2007).
Consistent with a proapoptotic role for PIDD in p53-induced cell death, its overexpression facilitated caspase-2 activation and cell death induction in response to DNA damage in HeLa cells (Tinel and Tschopp, 2004), and RNA interference or antisense oligonucleotides targeting PIDD mRNA delayed cell death induced by overexpression of p53 in H1299 colon cancer (Baptiste-Okoh et al., 2008) or K562 myelogenous leukemia cells, respectively (Lin et al., 2000). Interestingly, a correlation between apoptotic index and PIDD expression was recently reported in oral squamous cell carcinoma patient samples, whereas no correlation was found regarding the p53 mutation status in these tumors (Bradley et al., 2007). The latter finding is consistent with the observation that the basal expression of PIDD does not depend on the presence of functional p53, suggesting multiple modes of regulation of PIDD protein expression (Cuenin et al., 2008). Although all of these findings support a critical role for PIDD in cell death and caspase-2 activation, targeting RAIDD or PIDD expression by siRNA failed to interfere with caspase-2 processing in response to 5-fluoruracil treatment in HCT-116 colon carcinoma cells (Vakifahmetoglu et al., 2006). In addition, thymocytes and fibroblasts from mice lacking RAIDD were reported to respond normally to cell death induction by DNA-damaging agents or TNF (Berube et al., 2005). However, cell death induced by overexpression of PIDD in fibroblasts strictly depended on the presence of RAIDD but not caspase-2, suggesting that PIDD can also engage cell death in a manner independent of this initiator caspase (Berube et al., 2005). To investigate the relevance of PIDD for caspase-2 activation and cell death under physiological conditions, we have generated a PIDD-deficient mouse model by homologous recombination in embryonic stem cells.
Results
Generation of PIDD-deficient mice
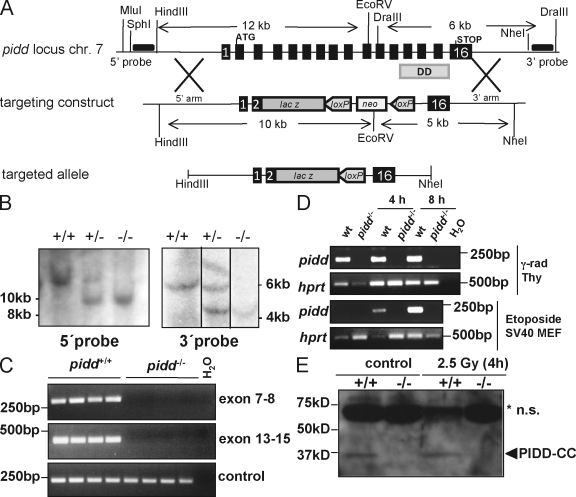
Our targeting strategy aimed to replace exons 3–15 of the PIDD locus, replacing all relevant protein domains with the open reading frame of β-galactosidase followed by a loxP element-flanked neomycin resistance cassette (Fig. 1 A). Correct targeting of the pidd locus was confirmed by Southern blot analysis and exon-specific PCR on genomic DNA derived from embryonic stem cells and, subsequently, tail tissue, mouse embryonic fibroblasts (MEFs), and liver derived from wild-type, pidd+/−, and pidd−/− mice (Fig. 1, B and C; and not depicted). RT-PCR analysis was performed to confirm the absence of pidd mRNA in thymocytes and SV40-immortalized MEFs (Fig. 1 D). Finally, to ascertain lack of protein, a polyclonal antiserum was generated using the DD of PIDD (aa 770–910) as immunogen. Reactivity of the affinity-purified IgG fraction was confirmed in MEFs that overexpress a Flag-tagged version of mouse PIDD (mPIDD). Three protein fragments corresponding to the full-length protein, processed PIDD-C, and PIDD-CC were detected by the anti-Flag antibody and the purified anti-PIDD IgG fraction (Fig. S1 and not depicted). Transient cotransfection of 293T cells with an expression construct encoding mPIDD together with two different lentiviral vectors encoding alternative short hairpin RNAs targeting mPIDD confirmed specificity of the purified antiserum (Fig. S1 and not depicted). Western blot analysis performed on thymocyte extracts derived from wild-type and pidd−/− mice revealed that PIDD-CC appears to be the only detectable form (Fig. 1 E). Efforts to identify endogenous full-length protein or PIDD-C by immunoblotting or immunoprecipitation were repeatedly unsuccessful, suggesting that PIDD may be rapidly autoprocessed in thymocytes (unpublished data).
Figure 1.
Targeting the pidd locus in mice. (A) Schematic representation of the pidd/lrdd gene locus on mouse chromosome 7 (chr 7). Exons 3–15 of the pidd gene were replaced by the open reading frame of β-galactosidase. The loxP element-flanked neomycin selection marker cassette was removed in vivo by cre-mediated deletion. The locations of the 5′ and 3′ probes used for Southern blot analysis are indicated as horizontal black bars. (B) Southern blot analysis on genomic DNA derived from wild-type, pidd+/−, and pidd−/− mice. (C) Exon-specific PCR using three different primer pairs spanning exons 7–8 or 13–15 confirmed correct targeting of the pidd gene. (D) RT-PCR was performed on cDNA generated from total RNA derived from thymocytes (top two panels) that were left untreated or exposed to 2.5 Gy of γ irradiation or SV40-immortalized wild-type (wt) and PIDD-deficient MEFs (bottom two panels) cultured in the absence or presence of etoposide to confirm absence of pidd mRNA. (E) Western blot analysis on thymocyte extracts derived from wild-type and pidd−/− mice before and after exposure to γ irradiation in vitro confirmed absence of PIDD at the protein level. The asterisk indicates nonspecific (n.s.) bands.
Mice lacking both alleles of PIDD were born in normal Mendelian frequency and did not show fertility problems, gender bias, or any other overt phenotype. Furthermore, histological assessment failed to reveal any gross abnormalities, and pidd−/− mice show normal lymphocyte development and subset distribution in the relevant primary and secondary lymphatic organs (Fig. S2 A and not depicted). Collectively this demonstrates that PIDD is dispensable for embryonic development and postnatal life in unstressed animals.
Loss of PIDD fails to protect lymphocytes from apoptosis induction
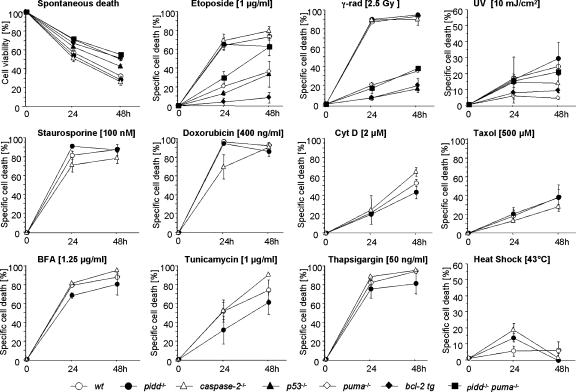
To investigate the role of PIDD in cell death induction, we isolated primary thymocytes as well as mature T and B cells from spleen and granulocytes from bone marrow by FACS sorting from age-matched wild-type, pidd−/−, and caspase-2−/− mice. Lymphocytes overexpressing antiapoptotic Bcl-2 in all hematopoietic cells and cells lacking p53 or its proapoptotic target gene puma−/−, a BH3-only Bcl-2 family member (Villunger et al., 2003), were included in some of our assays as internal controls. Thymocytes were put in culture without further treatment (spontaneous death) or exposed to DNA damage triggered by topoisomerase II inhibitor etoposide, the DNA-intercalating drug doxorubicin, or γ or UV irradiation. In parallel, cells were also exposed to heat stress or were treated with the broad-spectrum kinase inhibitor staurosporine, the ER stressors tunicamycin, brefeldin A, or thapsigargin as well as cytochalasin D or taxol, both of which disrupted cytoskeletal structures, or to death receptor agonists Fas ligand or TNF-α (Table I). Cell viability was monitored over time using Annexin V/propidium iodide (PI) staining and flow cytometric analysis. Loss of PIDD failed to protect thymocytes (Fig. 2), mature T and B cells, or granulocytes (not depicted) from any of the tested cell death stimuli. In contrast, overexpression of Bcl-2 or loss of p53 or puma each efficiently protected thymocytes and mature T and B cells from DNA damage–induced apoptosis (Fig. 2 and not depicted), which is consistent with previous results (Villunger et al., 2003; Erlacher et al., 2006). To test whether the proposed PIDD–RAIDD–caspase-2 apoptosis pathway could act in a redundant manner with the Bcl-2–regulated one, we also assessed survival in thymocytes lacking PIDD and p53 up-regulated modulator of apoptosis (Puma) simultaneously. However, the degree of protection from apoptosis induced upon DNA damage was comparable between puma−/− and pidd−/−puma−/− double-deficient thymocytes (Fig. 2), arguing against such redundancy. To exclude the possibility that loss of PIDD may have an impact on the survival of lymphocytes in vivo, masked by the lack of coregulatory factors in our in vitro analysis, we also exposed wild-type and pidd−/− mice to graded doses of whole body irradiation (2.5 and 5.0 Gy) and evaluated lymphocyte cellularity as well as subset distribution of immature and mature T and B cells in thymus, bone marrow, and spleen 20 h after irradiation. Consistent with our findings in vitro, loss of PIDD did not enhance the survival of lymphocytes after irradiation in vivo (Fig. S2 A and not depicted).
Table I.
Summary of stimuli and final concentrations used to induce cell death
| Stimulus | Primary thymocytes | Splenic B cells | Splenic T cells | Bone marrow granulocytes | SV40-immortalized MEFs | Primary MEFs |
| Etoposide | 1 µg/ml | 1 µg/ml | 1 µg/ml | 1–10 µg/ml | 10 – 50 µg/ml | 10–50 µg/ml |
| Doxorubicin | 400 ng/ml | 400 ng/ml | 400 ng/ml | 400 ng/ml | ND | ND |
| Cisplatin | 25–100 µM | 50 µM | 50 µM | 50 µM | 20 µM | ND |
| Staurosporine | 100 and 500 nM | 100 nM | 100 nM | 100 and 500 nM | 50 nM | ND |
| Ionomycin | 1 µg/ml | ND | ND | ND | ND | ND |
| PMA | 10 ng/ml | ND | ND | ND | ND | ND |
| Velcade | 50 ng/ml | 50 ng/ml | 50 ng/ml | ND | ND | ND |
| Trichostatin A | 50 nM | 50 nM | 50 nM | ND | ND | ND |
| Dexamethasone | 10−7 M | 10−7 M | 10−7 M | ND | ND | ND |
| TNF-α | 10 ng/ml | 10 ng/ml | 10 ng/ml | 10 ng/ml | 30 ng/ml | ND |
| Cycloheximide | 5 µg/ml | 5 µg/ml | 5 µg/ml | 5 µg/ml | 5 µg/ml | ND |
| Super Fas ligand | 10 ng/ml | 10 ng/ml | 10 ng/ml | 10 ng/ml | ND | ND |
| Cytochalasin D | 2 µM | ND | ND | ND | 20 µM | 20 µM |
| Taxol | 500 µM | ND | ND | ND | 100 µM | 100 µM |
| Vincristine | ND | ND | ND | ND | ND | 50 µg/ml |
| Tunicamycin | 0.1–5 µg/ml | 1 µg/ml | 1 µg/ml | ND | 1.2 µg/ml | ND |
| Brefeldin A | 1.25 and 2.5 µg/ml | ND | ND | ND | 1 µg/ml | ND |
| Thapsigargin | 0.05–0.1 µg/ml | ND | ND | ND | 50 µg/ml | 50 µg/ml |
Figure 2.
PIDD is dispensable for cell death induction in lymphocytes. Thymocytes derived from mice of the indicated genotypes were put in culture without further treatment (spontaneous cell death) or exposed to apoptosis inducers as indicated over time. Cell death was monitored by Annexin V–FITC/PI staining and flow cytometric analysis at the indicated time points. Data are means ± SD from four to eight independent experiments per genotype, with the exception of pidd−/−puma−/− (n = 2). The extent of apoptosis induced specifically by different stimuli was calculated by the equation (induced apoptosis − spontaneous cell death)/(100 − spontaneous cell death). No significant differences in cell death induction were observed between wild-type and PIDD- and caspase-2–deficient cells treated with any of the indicated stimuli by analysis of variance.
Because PIDD has been implicated in caspase-2–mediated apoptosis as well as NF-κB activation, enabling DNA repair (Janssens et al., 2005), we investigated the impact of loss of PIDD on clonal survival after DNA damage in primary as well as SV40-immortalized MEFs transiently exposed to heat shock, etoposide, UV radiation (UVR) or γ irradiation. Loss of PIDD or caspase-2 did not influence residual survival of MEFs observed upon low dose of UVR or transient exposure to 43°C. Higher doses of UVR, etoposide treatment, or transient exposure to 45°C did not allow clonal survival of primary (Fig. S3) or immortalized cells (Fig. S4) independent of their genotypes.
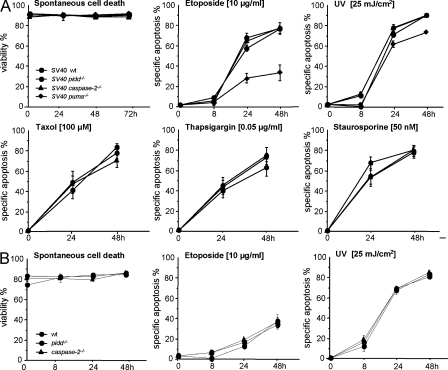
To address the possible relevance of PIDD for transient, short-term survival, primary as well as SV40-immortalized MEFs were analyzed for their initial cell death responses. MEFs from wild-type, pidd−/−, caspase-2−/−, or puma−/− mice (Fig. 3 A) as well as primary low passage wild-type and pidd−/− or caspase-2−/− MEFs (Fig. 3 B) were exposed to DNA damage induced by etoposide, UV irradiation, serum deprivation, heat stress, ER stress, cytochalasin D, taxol, or staurosporine (Table I), but, again, neither loss of PIDD nor of caspase-2 protected these cells significantly from any of these apoptotic stimuli. In contrast, loss of puma transiently protected MEFs from the effects of DNA damage (Fig. 3 A and not depicted).
Figure 3.
Normal cell death responses in primary and SV40-immortalized MEFs. (A and B) SV40-immortalized MEFs (A) and primary MEFs (B) from mice of the indicated genotypes were put in culture without further treatment (spontaneous cell death) or exposed as indicated over time. Cell death was monitored by Annexin V–FITC/PI staining and flow cytometric analysis at the indicated time points. Data are means ± SD from three independent experiments performed in duplicate using SV40-immortalized MEFs and two to three independent experiments performed in duplicate using primary MEFs. The extent of apoptosis induced specifically by different stimuli was calculated by the equation (induced apoptosis − spontaneous cell death)/(100 − spontaneous cell death). wt, wild type.
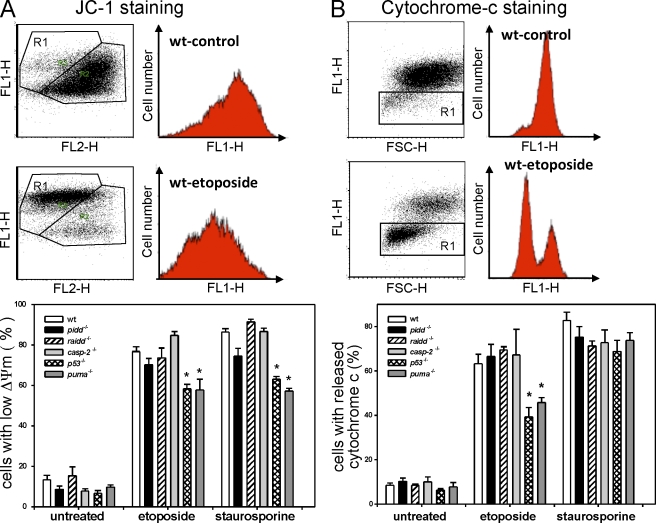
To gain insight into possible minor mechanistic differences in apoptosis induction too subtle to be captured by Annexin V/PI staining, we also investigated the impact of PIDD deficiency on mitochondrial membrane depolarization using JC-1 staining (Fig. 4 A) and by monitoring cytochrome c release (Fig. 4 B). In SV40-immortalized fibroblasts (Fig. 4) or thymocytes (not depicted) exposed to DNA damage, none of these parameters was significantly altered as a result of the absence of PIDD or caspase-2, but, again, loss of p53 or Puma partially blocked these events upon DNA damage. To backup our observations, excluding a role for the PIDDosome in cell death induction, we also included raidd−/− MEFs in our analysis. Consistent with our results using PIDD-deficient MEFs, raidd−/− MEFs also responded like wild-type cells to cell death induction by etoposide or staurosporine treatment (Fig. 4, A and B).
Figure 4.
Impact of PIDDosome loss on mitochondrial function and cytochrome c release. (A) Mitochondrial membrane potential was determined by JC-1 staining, and representative FACS blots of control and etoposide-treated SV40-immortalized MEFs are depicted. The bar graph shows quantification of FACS analysis using JC-1 staining of SV40-immortalized MEFs of the indicated genotypes 24 h after treatment. (B) Cytochrome c release was monitored in untreated or etoposide- or staurosporine-treated SV40-immortalized MEFs after 24 h by flow cytometric analysis. Representative FACS blots of control and etoposide-treated SV40-immortalized MEFs are depicted. The bars represent the percentage of cells that released cytochrome c. (A and B) The bar graphs represent the means ± SEM of n ≥ 3 independent experiments per genotype and treatment. *, P < 0.05 compared with wild type (wt).
Caspase-2 processing is normal in primary lymphocytes but delayed in immortalized MEFs lacking PIDD
To investigate the potential role of PIDD and RAIDD in caspase-2 processing, which can occur independent of dimerization-induced activation, we first analyzed caspase-2 cleavage (processing) in protein lysates extracted from primary thymocytes and SV40-immortalized MEFs derived from wild-type, PIDD-deficient, and RAIDD-deficient mice upon DNA damage. Again, cells overexpressing Bcl-2 and deficient for p53 or puma were included in our analysis as controls. By using a monoclonal antibody (clone 11B4) that recognizes the large subunit of caspase-2, we were able to detect its pro-form (50 kD) as well as an ∼32-kD fragment that arises from intrasubunit cleavage between the p20 and the p10 subunit during autoprocessing, after prior removal of the caspase recruitment domain, or processing and the large subunit only (p20), which is generated by subsequent autoproteolysis or proteolysis of the pro-domain (O'Reilly et al., 2002). Because the generation of these fragments can also be caused by caspase-3–mediated cleavage of caspase-2, it has to be pointed out in this study that the appearance of processed caspase-2 fragments in Western blotting analysis does not necessarily reflect its true dimerization-induced activation.
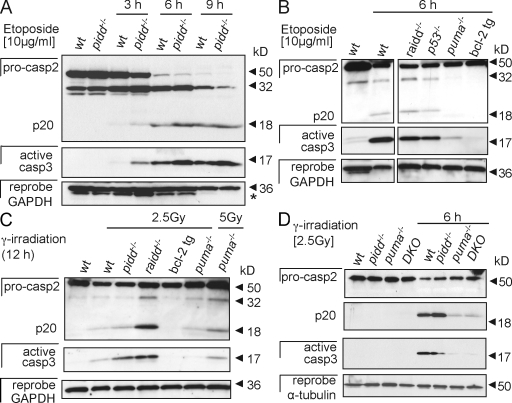
In primary thymocytes (Fig. 5) and splenocytes (not depicted), cleaved caspase-2 was readily detectable after exposure of cells to etoposide treatment or γ irradiation. However, processing occurred with similar efficiency in wild-type and PIDD-deficient cells and coincided with the appearance of the active form of caspase-3, indicating that the classical mitochondrial outer membrane permeabilization (MOMP)–apoptosome-mediated apoptosis pathway was active either preceding caspase-2 cleavage or caspase-2 may have been activated in a dimerization-dependent manner in a parallel pathway (Fig. 5 A). However, because processing of both caspases was efficiently blocked in thymocytes overexpressing Bcl-2 (Fig. 5, B and C), we conclude that caspase-2 processing requires MOMP and cytochrome c release, at least under the conditions tested in this study. Thymocytes lacking Puma showed a significant delay but, in contrast to Bcl-2–overexpressing cells, not a complete block in caspase-2 processing (Fig. 5, B and C), indicating that additional effector proteins such as Noxa or Bim contribute to DNA damage–induced thymocyte apoptosis, as shown previously (Erlacher et al., 2006; Michalak et al., 2008). Importantly, caspase-2 processing occurred with similar decreased efficiency in thymocytes lacking Puma or PIDD and Puma simultaneously, demonstrating that caspase-2 cleavage during DNA damage–induced thymocyte apoptosis triggered by γ irradiation (Fig. 5 D) or etoposide treatment (not depicted) requires MOMP and that PIDD does not contribute to caspase-2 processing under these conditions, not even in a redundant manner with the Bcl-2–regulated activation pathway.
Figure 5.
Caspase-2 processing occurs in thymocytes lacking the PIDDosome. (A) Thymocytes isolated from wild-type (wt) and pidd−/− mice were cultured without further treatment or stimulated with etoposide. Caspase-2 processing and cleavage of effector caspase-3 was monitored over time by Western blot analysis. The asterisk indicates a residual caspase-2 signal remaining upon reprobing with anti-GAPDH antibody. (B and C) Caspase-2 and -3 processing in etoposide-treated (B) or γ-irradiated thymocytes (C) of the indicated genotypes. tg, transgenic. (D) Thymocytes isolated from wild-type, pidd−/−, puma−/−, and pidd−/−puma−/− mice were cultured without further treatment or after exposure to γ irradiation for 6 h. Caspase-2 processing and cleavage of effector caspase-3 were monitored by Western blot analysis. After detection of caspase-2, membranes were first stripped and reprobed with anti–active caspase-3 antibody and finally probed without stripping with an antibody recognizing GAPDH or α-tubulin to compare protein loading. DKO, double knockout.
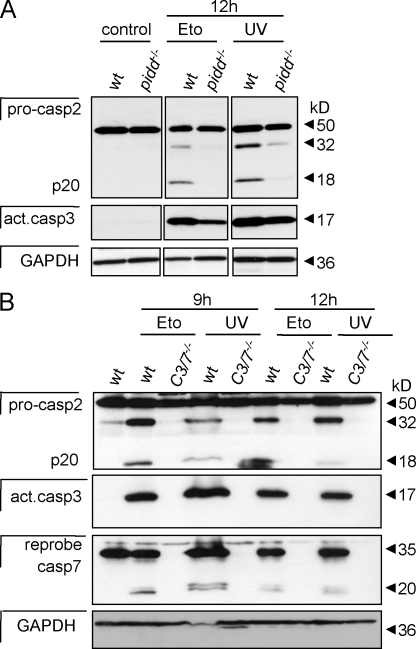
Somewhat surprisingly, in contrast to our observations in primary lymphocytes, caspase-2 processing was delayed in SV40-immortalized MEFs lacking PIDD. This coincided with slightly reduced levels of activated caspase-3 (Fig. 6 A) and could be interpreted in a manner that caspase-2 may be able to act upstream of MOMP or in a parallel PIDD-dependent pathway in immortalized MEFs, as previously reported for certain tumor cell lines by others (Robertson et al., 2002, 2004). To investigate whether caspase-2 processing can occur independent of effector caspase activation in SV40-immortalized MEFs and, thus, MOMP, we investigated processing of caspase-2 in response to DNA damage in MEFs lacking both caspase-3 and -7. However, in these cells, caspase-2 cleavage did not occur in response to etoposide treatment or UV irradiation (Fig. 6 B). This demonstrates that processing of caspase-2, usually observed during DNA damage–induced apoptosis, depends on the presence of caspase-3 and/or -7 and is a secondary event downstream of effector caspase activation.
Figure 6.
Caspase-2 processing is delayed in PIDD-deficient MEFs but still requires effector caspase activation. (A) Immortalized wild-type (wt) and pidd−/− SV40-MEFs were exposed to 50 µg/ml etoposide (Eto) or 5 mJ/cm2 UVR, and lysates were immunoblotted for caspase-2 processing, stripped, and reprobed with anti–active caspase-3 antibody followed by a reprobing with anti-GAPDH. (B) Wild-type and caspase-3/7 double-deficient SV40-MEFs were exposed to DNA damage as in A, and lysates were immunoblotted for caspase-2 processing. After detection of caspase-2, membranes were first stripped and reprobed with anti–active caspase-3 antibody followed by an antibody recognizing GAPDH to compare protein loading. Finally, membranes were stripped before detection of caspase-7.
As previously pointed out by others, fluorescence-based caspase-substrate cleavage assays cannot be used to quantify caspase-2 activity (McStay et al., 2008), and, in fact, we observed that the supposedly caspase-2–specific substrate Ac-VDVAD-AMC was equally well processed in caspase-2–deficient and wild-type MEFs or thymocytes (unpublished data). Therefore, we limited our efforts to investigate the proteolytic activity of caspase-3/7 during cell death induction in extracts derived from wild-type, pidd−/−, caspase-2−/−, or p53−/− SV40-immortalized MEFs after treatment with etoposide, serum deprivation, or exposure to UV irradiation. Caspase-3/7 activity (Ac-DEVD-AMC cleavage) was comparable in wild-type and caspase-2– and PIDD-deficient cell lysates, whereas lysates from p53-deficient cells showed reduced activity in response to etoposide treatment at the time of analysis (Fig. S5 A). A time course comparing caspase-3/7–like activities in etoposide-treated thymocytes expressing or lacking PIDD also failed to reveal significant differences between genotypes (Fig. S5 B).
Collectively, our findings suggest that PIDD is not required to trigger caspase-2 processing in response to DNA damage in primary lymphocytes and that caspase-2 cleavage is effectively prevented by the maintenance of mitochondrial integrity by Bcl-2 family proteins. However, in SV40-immortalized MEFs lacking PIDD, caspase-2 processing is less effective, perhaps as the result of impaired p53 function, albeit still depends on effector caspase activation upon DNA damage. Regardless, reduced caspase-2 activation in the absence of PIDD does not translate into increased short-term (Figs. 2 and 3) or long-term clonal survival in response to the stimuli tested in this study (Figs. S3 and S4).
Caspase-2 multimerization and activation can occur in the absence of the PIDDosome
As mentioned in the previous section, monitoring cleavage of initiator caspases is not indicative for their activation in response to external signals because this can also be mediated by activated effector caspases (Riedl and Salvesen, 2007). Dimerization and subsequent autoprocessing in high molecular weight complexes such as the Apaf-1 containing apoptosome, the death-inducing signaling complex, or the PIDDosome are considered the initial activating events for pro–caspase-9 or -8 or pro–caspase-2 (Riedl and Salvesen, 2007).
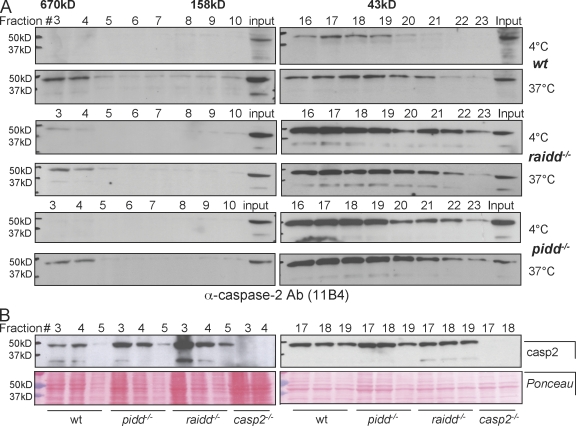
Recruitment of pro–caspase-2 into a high molecular weight complex of ≥670 kD, together with the components of the PIDDosome, RAIDD and PIDD, has been reported to occur spontaneously upon cell rupture followed by a temperature shift in HeLa or 293T cell extracts (Tinel and Tschopp, 2004). To investigate whether caspase-2 can still be recruited into this high molecular weight fraction in the absence of components of the PIDDosome, we performed gel filtration analysis on extracts derived from wild-type, pidd−/−, or raidd−/− SV40-immortalized MEFs before and after temperature shift. Immunoblotting of eluted fractions after size-exclusion chromatography revealed that a high molecular weight complex of ≥670 kD containing caspase-2 is formed in all genotypes analyzed upon incubation of cell extracts at 37°C (Fig. 7 A). To monitor for quantitative differences in recruitment, we also directly compared caspase-2 levels in the high molecular weight fractions of all genotypes investigated, but comparable amounts were detected in wild-type, RAIDD- and PIDD-deficient extracts (Fig. 7 B). Together, this suggests that the formation of this complex containing multimerized caspase-2 can occur independently of PIDD or RAIDD.
Figure 7.
The PIDDosome is not required for the formation of high molecular weight complexes containing caspase-2. (A) Lysates from wild-type (wt), raidd−/−, and pidd−/− SV40-immortalized MEFs were incubated at 4°C or 37°C for 1 h and subjected to size-exclusion chromatography, and fractions were precipitated, separated by SDS-PAGE, and immunoblotted with an anti–caspase-2 antibody (Ab). (B) Fractions 3–5, containing complexes ≥600 kD, and fractions 17–19, containing proteins of a molecular weight range of ∼75–40 kD, of all indicated genotypes were analyzed for the relative caspase-2 expression level. Ponceau staining of the membranes was used to compare protein loading (bottom).
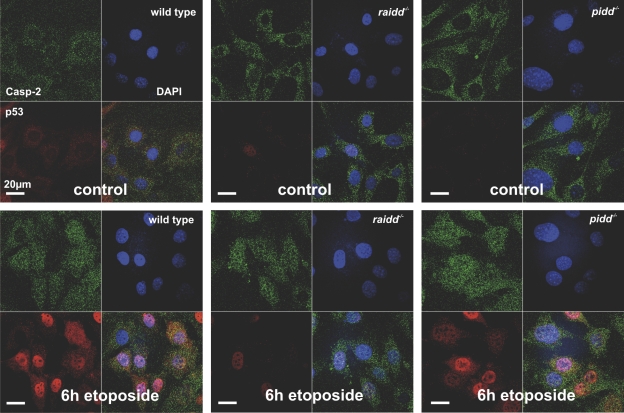
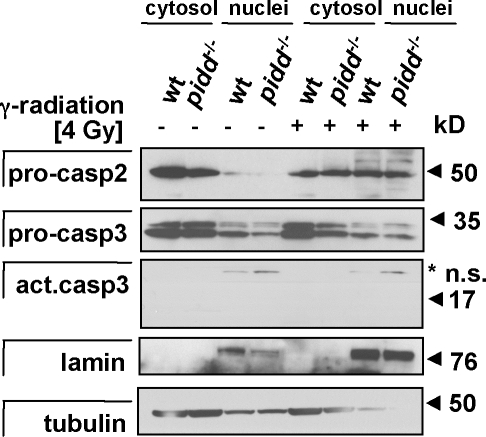
Finally, we also investigated subcellular localization of caspase-2 in MEFs lacking PIDD or RAIDD before and after DNA damage by immunostaining and laser-scanning microscopy. Endogenous caspase-2 was found mainly in the cytoplasm in untreated MEFs, but a significant portion appeared to translocate into the nucleus after etoposide treatment, which is consistent with previous results (Colussi et al., 1998). However, loss of raidd or pidd did not influence the subcellular distribution or translocation of caspase-2 in response to DNA damage, as assessed by immunofluorescence analysis (Fig. 8). In addition, we monitored cytoplasmic to nuclear translocation of caspase-2 in wild-type and PIDD-deficient MEFs after exposure to γ irradiation by Western blot analysis but again failed to observe significant differences between genotypes regarding caspase-2 nuclear translocation (Fig. 9). In summary, these results indicate that an additional platform for caspase-2 activation exists in mammalian cells or that caspase-2 can multimerize and autoprocess in the absence of such a platform.
Figure 8.
Subcellular localization of caspase-2 in the absence of PIDD or RAIDD. Localization of endogenous caspase-2 (green) was detected in wild-type SV40-immortalized MEFs and MEFs lacking pidd or raidd before (control) or after drug treatment (50 µg/ml etoposide). Cells were costained for p53 (red) and the nucleus detecting fluorochrome DAPI (blue). Overlays of all staining are depicted (merge).
Figure 9.
Nuclear translocation of caspase-2 upon DNA damage. Cytosolic and nuclear extracts from wild-type (wt) and PIDD-deficient MEFs were generated from untreated cultures or 4 h after exposure to 4 Gy of γ irradiation. Extracts were separated by SDS-PAGE, and caspase-2 was detected by immunoblotting. Membranes were reprobed with antilamin- and antitubulin-specific antibodies to assess cross-contamination of fractions as well as anti–active caspase-3 to monitor for effector caspase activation. The asterisk indicates nonspecific (n.s.) bands.
Discussion
We have generated mice that lack the p53-induced protein PIDD and found that its absence does not interfere with normal mouse development, caspase-2 activation, or cell death induction by several different apoptotic stimuli. Formation of the PIDDosome has previously been suggested to be rate limiting for activation of caspase-2 and p53-induced cell death in several transformed human cancer cell lines (Tinel and Tschopp, 2004; Baptiste-Okoh et al., 2008). Our study clearly indicates that PIDD and caspase-2 are dispensable for apoptosis induction in response to DNA damage, growth factor deprivation, ER stress, disruption of cytoskeletal architecture, and pan-kinase inhibition as well as Fas ligand– or TNF-α–induced cell death in lymphocytes and SV40-transformed MEFs (Figs. 2 and 3). Loss of mitochondrial membrane potential or release of cytochrome c was also not delayed in cells lacking caspase-2, RAIDD, or PIDD (Fig. 4). Collectively, this demonstrates that the PIDDosome is not required for efficient apoptosis induction in response to a wide range of stimuli, including DNA damage (Table I). It is unclear at present whether apoptosis induced by overexpression of p53 is impaired in PIDD-deficient cells, as suggested by knockdown experiments in H1299 carcinoma cells (Baptiste-Okoh et al., 2008), but stabilization of p53 by adding the Mdm2 inhibitor Nutlin-3a killed PIDD-deficient MEFs as effectively as wild-type cells while sparing p53−/− MEFs, arguing against a prominent role for PIDD in p53-induced apoptosis (unpublished data).
Caspase-2 processing occurred with comparable kinetics in wild-type, raidd−/−, and pidd−/− thymocytes after DNA damage and was always observed concomitantly with the appearance of the activated form of caspase-3 (Fig. 5). Similar observations have been made in HCT116 cells treated with PIDD- or RAIDD-specific siRNA, but the cleavage of caspase-2 observed in response to 5-fluoruracil treatment in these cells was attributed to incomplete knockdown of the PIDDosome components (Vakifahmetoglu et al., 2006). However, our observations reported in this study suggest that caspase-2 cleavage occurs secondary to MOMP and activation of the classical apoptosome during apoptosis induced by DNA damage. Consistently, all of these events were blocked by preserving mitochondrial integrity, either by overexpression of Bcl-2 or loss of Puma (Fig. 5). Importantly, PIDD also does not act in a redundant manner with the Bcl-2–regulated pathway of caspase-2 processing, as its cleavage, as well as cell death, is comparable in puma−/− and pidd−/−puma−/− cells (Figs. 2 and 5). Our data on caspase-2 processing are in line with the observation that its cleavage depends on the presence of Apaf-1 and caspase-9 in primary lymphocytes (O'Reilly et al., 2002; Marsden et al., 2004) and cannot be used as a readout for its activation during apoptosis induction and contradict reports where activation of caspase-2 was positioned upstream of MOMP and cytochrome c release in response to DNA damage in tumor cell lines such as Jurkat or HeLa cells or when recombinant caspase-2 was added directly to isolated mitochondria (Guo et al., 2002; Robertson et al., 2002, 2004). Primary MEFs lacking caspase-2 were also reported to resist cell death induced by disruption of the cytoskeleton (Ho et al., 2008), which we failed to observe in SV40-immortalized MEFs (Fig. 3 A). All of these differences may be inherent to the cell types analyzed and/or the mode of immortalization/transformation, which can affect the apoptotic response of cells in a highly diverse manner. In line with the latter, we also observed that caspase-2 processing was delayed in pidd−/− SV40-immortalized MEFs (Fig. 6 A). This phenomenon was observed in independent clones derived from individual mouse embryos (unpublished data). Nevertheless, MEFs of both genotypes were equally sensitive to apoptosis in response to all stimuli tested (Fig. 3, A and B). Similar observations have been previously reported using MEFs lacking RAIDD (Berube et al., 2005). Nevertheless, cleavage of caspase-2 upon DNA damage strictly depended on the presence of activated effector caspase-3 and/or -7 (Fig. 6 B). All together, this suggests that the PIDDosome may contribute to caspase-2 processing after DNA damage in an effector caspase-dependent manner. Although counterintuitive at first sight, effector caspases appear to possess the potential to influence events considered to occur upstream of mitochondria such as the activation and translocation of Bax (Lakhani et al., 2006). Therefore, we propose that modulation of caspase-2 activity via the PIDDosome does not contribute to apoptosis but may rather influence other events triggered by DNA damage such as NF-κB activation, cell cycle arrest, and/or DNA repair. Consistently, a role for PIDD and caspase-2 in G2/M checkpoint control and nonhomologous end joining of double-strand breaks was reported recently (Shi et al., 2009). However, our experiments investigating the role of PIDD in NF-κB activation upon DNA damage are currently too preliminary to draw firm conclusions.
Finally, we also excluded that multimerization-induced activation of caspase-2 in high molecular weight complexes depends on the presence of the PIDDosome because gel filtration analysis revealed that such caspase-2–containing complexes can also form in the absence of the PIDDosome upon temperature shift (Fig. 7, A and B) but contain the active form of the protease (Read et al., 2002). It will be important to demonstrate that such a high molecular weight complex containing caspase-2 actually forms in whole cells in vivo upon apoptosis induction or may only be triggered in vitro upon temperature shift. Finally, subcellular localization of caspase-2 before and after exposure of cells to DNA damage was not affected by loss of PIDD or RAIDD, but, interestingly, caspase-2 accumulated in the nucleus of SV40-MEFs upon etoposide treatment before detectable signs of apoptosis and the activation of effector caspases (Figs. 8 and 9).
Collectively, our findings suggest that at least one alternative PIDDosome-independent mechanism of caspase-2 activation exists in mammals. Furthermore, caspase-2 dimerization and activation may even occur in the absence of any activating platform, as observed upon overexpression in bacteria. A local and transient increase in protein concentration either alone or in combination with one of the many described posttranslational modifications (Krumschnabel et al., 2009) may suffice to promote its full activation. Finally, we postulate that caspase-2 activation by PIDD does not contribute to apoptosis induction but regulates events triggered in response to DNA damage such as cell cycle arrest or repair.
Materials and methods
Generation of pidd−/− mice and other mouse strains used
All animal experiments were performed in accordance with the Austrian Tierversuchsgesetz (in accordance with Austrian legislation BGB1 Nr. 501/1988 i.d.F. 162/2005) and have been granted by the Bundesministerium für Bildung, Wissenschaft und Kultur. To generate a pidd-targeting construct, C57BL/6 genomic DNA was isolated and subcloned from the Bac clone RPCI-23:141F19 (Roswell Park Cancer Institute), harboring the entire pidd locus (Fig. 1). Exons 3–15 in pidd were replaced with the open reading frame encoding β-galactosidase followed by a loxP element-flanked neomycin cassette. Two correctly targeted embryonic stem cell clones were identified by Southern blot analysis and subsequently injected into C57BL/6 blastocysts and delivered by CD1 surrogate mothers to generate chimeric mice. Chimaeras derived from both independent embryonic stem cell clones were mated with wild-type C57BL/6 mice. Germline transmission was only obtained from one of these clones, and heterozygous offspring were used for the generation of pidd−/− mice. The neomycin resistance marker was subsequently removed by crossing to female C57BL/6 cre deleter mice (Schwenk et al., 1995).
With the exception of the in vivo irradiation experiments conducted with mice on mixed genetic background (B6/129SV; 1:1), all experiments shown were performed on pidd−/− cells from mice that were backcrossed onto C57BL/6 genetic background for eight generations. The generation of caspase-2−/− (provided by D.L. Vaux, LaTrobe University, Melbourne, Australia; O'Reilly et al., 2002), puma−/− (provided by A. Strasser, Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia; Villunger et al., 2003), and vav-bcl-2 transgenic mice (provided by J. Adams, Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia; Ogilvy et al., 1999), all maintained on C57BL/6 genetic background, has been previously described (Villunger et al., 2003). p53-deficient mice on C57BL/6 background were provided by M. Serrano (Centro Nacional de Investigaciones Oncológicas, Madrid, Spain). Mice deficient for raidd (Berube et al., 2005) were provided by T.W. Mak (University of Toronto, Toronto, Canada) and obtained on a mixed genetic background (∼72% C57BL/6 background). Caspase-3/7 double-deficient MEFs were provided by R. Flavell (Yale School of Medicine, New Haven, CT).
Southern blotting and PCR analysis
To confirm correct targeting of embryonic stem cells and deletion of the pidd gene in tissues, 20 µg total genomic DNA was digested with the appropriate restriction enzymes (DraIII [New England Biolabs, Inc.] for the 3′ probe and MluI and EcoRV [Promega] for the 5′ probe) overnight. Samples of DNA were size fractioned in 0.7% agarose gels in Tris-acetate-EDTA buffer, depurinated in 0.25 M HCl, denatured in 0.4 M NaOH, and transferred in the same buffer onto Hybond N+ nylon membranes. Filters were probed with 5′ and 3′ external probes or a neomycin cassette–specific probe in Church buffer at 65°C overnight. Membranes were washed in 40 mM Na-phosphate buffer containing 1% SDS at 65°C and exposed in a phosphoimager (Typhoon; GE Healthcare) for up to 2 d.
Littermates from heterozygous (pidd+/−) intercrosses were genotyped by PCR using primer pairs specific for the wild-type or the targeted pidd allele and the following cycle conditions: 4 min at 94°C; 40 s at 94°C, 30 s at 55°C, and 60 s at 72°C for 30 cycles; and 5 min at 72°C. Wild-type allele primers used were forward, 5′-ACTTCTCCTGGTACTGGCTCTG-3′; and reverse, 5′-AAGGCTGCAAAGAACTTCTCAC-3′. Primers used for the targeted allele (LacZ) were forward, 5′-TGCCACTCGCTTTAATGATG-3′; and reverse, 5′-CTCCACAGTTTCGGGTTTTC-3′. The wild-type and targeted pidd alleles were amplified in separate reactions yielding PCR products of 259 bp and 420 bp in size, respectively.
Primers for RT-PCR were designed exon specific for exon 7–8 with forward, 5′-TCGCTGTCGTGAGGTAGTTG-3′, and reverse, 5′-GAGAAGTGCTCCCTCTGGTG-3′; and for exon 13–15 with forward, 5′-GGAAGCTCAGGATGTTCG-3′, and reverse 5′-GGTTCAGAGGCATCAAGGAG-3′. RT-PCR was performed using the following cycle conditions: 5 min at 94°C; 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C for 30 cycles; and 7 min at 72°C.
Cell culture and reagents
MEFs were isolated from embryonic day 13.5 embryos after trypsin digestion and cultured in DME containing 250 µM L-Gln (Invitrogen), penicillin/streptomycin (Sigma-Aldrich), and 10% FCS. All MEFs used were derived from mouse mutants maintained on C57BL/6 genetic background with the exception of raidd−/− MEFs, which were derived from mice maintained on mixed genetic background (∼72% B6). Primary MEFs used for experiments were always of low passage (less than five). Alternatively, MEFs were immortalized by standard procedures using retroviruses encoding the SV40 large T antigen.
Primary hemopoietic cells were cultured in RPMI-1640 medium (PAA), 250 µM L-Gln, 50 µM 2-mercaptoethanol, nonessential amino acids (Invitrogen), penicillin/streptomycin, and 10% FCS (PAA). For the induction of cell death, the following reagents were used: Super Fas ligand (Enzo Biochem, Inc.) at 10 ng/ml, staurosporine (Sigma-Aldrich) at 100 and 500 nM, ionomycin (Sigma-Aldrich) at 1 µg/ml, PMA (Sigma-Aldrich) at 10 ng/ml, VP16 (Sigma-Aldrich) at 1, 10, or 50 µg/ml, doxorubicin (Sigma-Aldrich) at 400 ng/ml, cisplatin (Sigma-Aldrich) at 25, 50, and 100 µM, the ER stressors tunicamycin (Sigma-Aldrich) at 0.1–5 µg/ml, brefeldin A at 1.25 and 2.5 µg/ml, and thapsigargin (Sigma-Aldrich) at 0.05–50 µg/ml, taxol (Sigma-Aldrich) at 100–500 µM, cytochalasin D (Sigma-Aldrich) at 2 µM, vincristine (EMD) at 50 µg/ml, glucocorticoid dexamethasone (Sigma-Aldrich) at 10−7 M, trichostatin A (Sigma-Aldrich) at 50 nM, bortezomib (Velcade; Millennium Pharmaceuticals, Inc.) at 50 ng/ml, and TNF (PeproTech) at 10 ng/ml in the absence and presence of cycloheximide (Sigma-Aldrich) at 5 µg/ml. γ irradiation was performed at 1.25, 2.5, and 5 Gy, and UV-C radiation was performed at 5, 10, and 25 mJ/cm2.
Generation of an mPIDD-reactive antiserum
A 6× His-tagged murine PIDD-DD fragment (aa 770–910) was produced in Escherichia coli (strain BL21) from an IPTG-inducible pET21 expression vector and purified by column chromatography using Ni–nitrilotriacetic acid agarose beads. New Zealand white rabbits were immunized three times with 0.5 mg recombinant protein in Freund's adjuvants. After confirming serum reactivity with overexpressed mPIDD, the serum was affinity purified using a Sepharose column (GE Healthcare) coupled with PIDD-DD. Sera were mixed with PBS in a ratio of 10:1 and loaded onto the column, washed with 5 bed volumes of buffer A (20 mM Tris, 5 mM EDTA, 500 mM NaCl, and 0.1% NP-40, pH 8.0) followed by 5 bed volumes of buffer B (20 mM Tris, 5 mM EDTA, and 150 mM NaCl, pH 8.0). Next, 0.1 M Gly, pH 2.8, was added to the column to elute the IgG fraction, which was collected and neutralized by adding 0.1 vol of 1 M Tris buffer, pH 8.8.
Apoptosis assay
Primary thymocytes were cultured at a density of 7.5 × 105/ml in 96-well plates and treated with a broad range of apoptotic stimuli. After treatment, cells were costained with FITC-conjugated Annexin V and 5 µg/ml PI and viability was assessed by flow cytometric analysis. SV40-immortalized MEFs were cultured at a density of 40,000 cells in 12-well plates. To determine cellular viability, cells were stained with FITC-conjugated Annexin V and 5 µg/ml PI and analyzed by flow cytometry.
Quantification of mitochondrial membrane potential
SV40-immortalized MEFs were seeded into 6-well plates at ∼120,000 cells per well and grown overnight before being stimulated with relevant cell death inducers for up to 24 h. Cells were harvested by trypsinization, washed once with excess PBS, resuspended in DME medium containing 4 µM JC-1 (5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine iodide; Sigma-Aldrich), and incubated for 20 min at 37°C in the dark. Cells were washed once again with PBS and resuspended in 200 µl PBS. Alterations in mitochondrial membrane potential were then estimated from changes in cell populations with high red and low green fluorescence (high mitochondrial potential) to cells with low red and high green fluorescence (low mitochondrial potential) as determined by FACS analysis.
Quantification of cytochrome c release
SV40-immortalized MEFs were grown to subconfluence on 6-cm dishes, stimulated as indicated, trypsinized, washed in PBS, and permeabilized by the addition of digitonin in 50 µg/ml PBS for 5 min at 4°C. After removing digitonin by centrifugation, cells were fixed with 4% PFA for 20 min at RT. Thereafter, cells were washed three times with PBS, and nonspecific binding was blocked using PBS containing 3% BSA and 0.05% saponin. Subsequently, cells were incubated with anti–cytochrome c antibody (diluted to 1:200 in blocking buffer; clone 7H8.2C12; BD) at 4°C overnight. Cells were washed three times in PBS, and Alexa Fluor 488–labeled anti–mouse (Invitrogen) was used as secondary antibody at a dilution of 1:100 in blocking solution and incubated for 1 h at RT. After three further washing steps, cells were resuspended in PBS, and cytochrome c release was determined by flow cytometric analysis.
Biochemical fractionation and immunoblot analysis
Cells were harvested and lysed for 1 h on ice in lysis buffer (1% CHAPS, 20 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 137 mM KCl, 1 mM EDTA, 1 mM EGTA, 0.5 mM PMSF, 5 mM NaF, and 2 mM Na3VO4; Roche), and the protein concentration was determined using Bradford reagent (Bio-Rad Laboratories). Protein was separated by SDS-PAGE, transferred to a nitrocellulose membrane, and incubated with the relevant antibodies (caspase-2 [11B4; Enzo Biochem, Inc.]; cleaved caspase-3 [Asp175; 5A1], caspase-3 [8G10], and caspase-7 [cs-9492; all from Cell Signaling Technology]; glyceraldehyde-3-phosphate dehydrogenase [GAPDH; 71.1; Sigma-Aldrich]; and α-tubulin [DM1A; Santa Cruz Biotechnology, Inc.]). Blots were visualized by enhanced chemiluminescence (GE Healthcare).
Size chromatography and gel filtration
SV40-immortalized MEFs were washed in PBS and resuspended in a hypotonic buffer containing 20 mM Hepes-KOH, 10 mM KCl, 1 mM MgCl2, 1 mM EDTA, 1 mM EGTA, pH 7.5, and Complete Protease Inhibitor Cocktail (Roche). Resuspended cells were subjected to three rounds of freeze thawing using liquid nitrogen. Cellular debris was removed by centrifugation at 10,000 g for 20 min at 4°C followed by filtration through a 0.5-µm mesh. Cell extracts were incubated at 37°C or 4°C for 60 min and then subjected to size exclusion chromatography. Treated lysates were fractionated using a fast protein liquid chromatography protein purification system on a sorbent (Ultrogel AcA 34; Pall Life Sciences) in a column (Tricorn10/600; GE Healthcare) at 4°C. The column was equilibrated with hypotonic buffer, 1.5–2 mg protein lysates was applied and eluted from the column with the same buffer at a constant flow rate of 0.5 ml/min, and 1-ml fractions were collected. The column was calibrated with GE Healthcare gel filtration standards containing bovine thyroglobulin (670 kD), aldolase (158 kD), conalbumin (75 kD), and ovalbumin (43 kD). Each fraction from the gel filtration was precipitated with TCA at a final concentration of 13% (vol/vol), and samples were incubated for 5 min at −20°C followed by 15 min at 4°C. After centrifugation at 10,000 g for 20 min at 4°C, pellets were dissolved in 50 µl of 2× Laemmli buffer and 10 µl of 1 M Tris-HCl, pH 8.8. 20-µl samples were resolved on a 4–20% SDS–polyacrylamide gel and transferred to a nitrocellulose membrane. Western blot analysis was conducted according to a standard protocol using the aforementioned caspase-2 antibody provided by D.C. Huang and L. O'Reilly (Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia).
Localization of endogenous caspase-2
SV40-immortalized MEFs were plated onto glass cover slides overnight, exposed to the desired stimulus, washed once with PBS, and then fixed with 2% PFA at RT for 20 min. To permeabilize the cells, PFA was replaced with 70% prechilled ethanol for 5 min at RT. After three washes with PBS, unspecific binding sites were blocked by incubation with 8% BSA (in PBS) for 1 h at RT. Incubation with primary antibodies (1:200 anti–caspase-2 [clone 11B4] and 1:100 anti-p53 [Pab246; Santa Cruz Biotechnology, Inc.], both in blocking solution) was performed at 4°C overnight followed by three washings with PBS for 10 min each. After incubation with secondary antibodies (1:100 in blocking solution; Alexa Fluor 488–labeled anti–rat and Alexa Fluor 546–labeled anti–mouse; Invitrogen) for 1 h at RT, cells were again washed with PBS, counterstained with 2 µg/ml DAPI, and washed and fixed with Vectashield antifade mounting medium (Vector Laboratories). Images were taken at RT with a confocal laser-scanning microscope (Axiovert 100M; Carl Zeiss, Inc.) equipped with LSM 510 acquisition software version 2.8 (Carl Zeiss, Inc.) and a 63× NA 1.4 oil immersion lens at a resolution of 1024 × 1024 pixels, with pinholes set to acquire images <1 µm thick. The adjustment of brightness and contrast to enhance visibility of details was performed using the LSM Image Browser software version 4.2 (Carl Zeiss, Inc.), and the assembly of images, labeling, and export to tiff format were performed in Draw X4 (Corel).
Nuclear extracts
To separate cytoplasmatic and nuclear fractions, SV40-MEFs were harvested after 4 Gy of γ radiation at the indicated time points. Cells were lysed in a cytosolic lysis buffer containing 10 mM Hepes, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT, and 0.5 mM PMSF. Nuclei were pelleted by centrifugation and were lysed in a nuclear buffer containing 20 mM Hepes, pH 7.9, 0.4 M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, and 1 mM PMSF during a 30-min incubation at 4°C. Nuclear extracts were centrifuged to obtain the solubilized nuclear fraction.
Statistical analysis
Statistical analysis was performed using the unpaired Student's t test or analysis of variance, where indicated, and applying the Stat-view 4.1 software program (Abacus Concepts). P-values of <0.05 were considered to indicate statistically significant differences.
Transient transfection of 293T cells
Specificity of the affinity-purified antiserum was confirmed on extracts from 293T cells overexpressing mPIDD alone or together with an mPIDD-specific short hairpin RNA construct (Tinel and Tschopp, 2004) using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instruction. Lysates were generated 24 h after transfection (see Biochemical fractionation and immunoblot analysis) and subjected to SDS-PAGE separation and immunoblotting. Lysates from MEFs stably transfected with mPIDD served as a positive control.
Whole body irradiation experiments
PIDD-deficient mice and littermate controls, both on a mixed genetic background (B6/SV129; 1:1), were exposed to graded doses of γ irradiation (2.5–5 Gy) at the age of 8–10 wk. Tissues were collected 20 h after irradiation to prepare single-cell suspensions. Cells were counted in a chamber hemocytometer (Neubauer), and aliquots were stained using fluorochrome-conjugated cell surface marker–specific antibodies followed by flow cytometric analysis (see next section).
Immunofluorescence staining, flow cytometric analysis, and cell sorting
Single-cell suspensions from peripheral blood, bone marrow, lymph nodes, spleen, and thymus were surface stained with monoclonal antibodies conjugated with FITC, R-phycoerythrin (PE), allophycocyanin, or biotin (Invitrogen). The monoclonal antibodies used and their specificities are RA3-6B2, anti-B220; GK1.5, anti-CD4; YTS169, anti-CD8; RB6-8C5, anti–Gr-1; R2/60, anti-CD43; 5.1, anti-IgM; 11/26C, anti-IgD; MI/70, anti–Mac-1; Ter119, anti-erythroid cell surface marker; T24.31.2, anti–Thy-1; IM7, anti-CD44; H57-59, anti–TCR-β (all from eBioscience); 7G6, anti-CD21; and B3B4, anti-CD23 (both from BD). Biotinylated antibodies were detected using streptavidin-R-PE (Dako) or streptavidin-vPE-Cy7 (BD). Flow cytometric analysis was performed using a cell analyzer (FACScalibur; BD). The sorting of cells was performed using a cell sorter (FACSvantage; BD).
Clonal survival of MEFs
Colony formation of primary or SV40-immortalized MEFs was assessed by seeding an increasing numbers of cells per well (6,000, 12,000, and 18,000 cells) 24 h before DNA damage or exposure to heat shock. To induce heat shock, cells were incubated for 60 min in a tissue culture CO2 incubator (CB150; Binder) set to 43°C or 45°C and then grown under standard conditions. Etoposide was removed after 24 h of incubation. Surviving cells were stained 2 wk later using crystal violet.
Caspase activity assay
Cells were lysed in extraction buffer (10 mM Hepes, 1.5 mM MgCl2, 300 mM sucrose, 10 mM KCl, 10 mM DTT, 0.5% NP-40, and Complete Protease Inhibitor Cocktail, pH adjusted to 7.0) for 30 min on ice and spun down for 10 min at 10.000 g, and the supernatant was stored at −20°C until use. 30 µl of the supernatant (containing ∼15 µg protein) was transferred into the well of a 96-well plate, and 100 µl of capsase-3 assay buffer (50 mM Hepes-KOH, pH 7.4, 2 mM EDTA, 10% sucrose (wt/vol), 0.1% CHAPS (wt/vol), pH adjusted to 7.4, and 5 mM DTT added freshly from a 1-M stock), including 100 µM freshly added caspase-3 substrate (Ac-DEVD-AMC; Enzo Biochem, Inc.), was added. Substrate cleavage activity was then measured in a fluorescence microplate reader (Ascent Fluoroscan; Titertek) by following fluorescence changes over time with excitation and emission wavelengths set at 355 nm and 460 nm, respectively. Enzyme activities were normalized to sample protein contents determined by a Bradford assay.
Online supplemental material
Fig. S1 shows the specific reactivity of the anti-PIDD polyclonal antiserum. Fig. S2 shows that loss of PIDD does not impair lymphocyte development or survival in vivo. Fig. S3 shows the colony formation of primary MEFs in the absence of PIDD. Fig. S4 shows the colony formation of SV40-MEFs in the absence of PIDD. Fig. S5 shows the quantification of caspase activity in the absence of PIDD. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200811105/DC1.
Acknowledgments
We thank K. Rossi and M. Saurwein for animal husbandry as well as C. Soratroi and I. Gaggl for excellent technical assistance. We also thank R. Flavell, T.W. Mak, M. Serrano, A. Strasser, D.C. Huang, J. Adams, D.L. Vaux, and L. O'Reilly for providing mice, MEFs, or reagents and P. Lukas for enabling irradiation experiments as well as R. Fässler for enabling blastocyst injections.
This work was supported by grants and fellowships from the Austrian Science Fund (FWF; START Y212-B13 and SFB021 to A. Villunger) and the Tiroler Wissenschaftsfond (to C. Manzl). F. Bock is funded by the Marie Curie Research Training Network ApopTrain.
Footnotes
Abbreviations used in this paper: DD, death domain; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MEF, mouse embryonic fibroblast; MOMP, mitochondrial outer membrane permeabilization; mPIDD, mouse PIDD; NF-κB, nuclear factor κB; PE, phycoerythrin; PI, propidium iodide; Puma, p53 up-regulated modulator of apoptosis; UVR, UV radiation.
References
- Baptiste-Okoh N., Barsotti A.M., Prives C. 2008. A role for caspase 2 and PIDD in the process of p53-mediated apoptosis.Proc. Natl. Acad. Sci. USA. 105:1937–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube C., Boucher L.M., Ma W., Wakeham A., Salmena L., Hakem R., Yeh W.C., Mak T.W., Benchimol S. 2005. Apoptosis caused by p53-induced protein with death domain (PIDD) depends on the death adapter protein RAIDD.Proc. Natl. Acad. Sci. USA. 102:14314–14320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley G., Tremblay S., Irish J., MacMillan C., Baker G., Gullane P., Benchimol S. 2007. The expression of p53-induced protein with death domain (Pidd) and apoptosis in oral squamous cell carcinoma.Br. J. Cancer. 96:1425–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colussi P.A., Harvey N.L., Kumar S. 1998. Prodomain-dependent nuclear localization of the caspase-2 (Nedd2) precursor. A novel function for a caspase prodomain.J. Biol. Chem. 273:24535–24542 [DOI] [PubMed] [Google Scholar]
- Cuenin S., Tinel A., Janssens S., Tschopp J. 2008. p53-induced protein with a death domain (PIDD) isoforms differentially activate nuclear factor-kappaB and caspase-2 in response to genotoxic stress.Oncogene. 27:387–396 [DOI] [PubMed] [Google Scholar]
- Degterev A., Yuan J. 2008. Expansion and evolution of cell death programmes.Nat. Rev. Mol. Cell Biol. 9:378–390 [DOI] [PubMed] [Google Scholar]
- Duan H., Dixit V.M. 1997. RAIDD is a new ‘death’ adaptor molecule.Nature. 385:86–89 [DOI] [PubMed] [Google Scholar]
- Erlacher M., Labi V., Manzl C., Bock G., Tzankov A., Hacker G., Michalak E., Strasser A., Villunger A. 2006. Puma cooperates with Bim, the rate-limiting BH3-only protein in cell death during lymphocyte development, in apoptosis induction.J. Exp. Med. 203:2939–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Srinivasula S.M., Druilhe A., Fernandes-Alnemri T., Alnemri E.S. 2002. Caspase-2 induces apoptosis by releasing proapoptotic proteins from mitochondria.J. Biol. Chem. 277:13430–13437 [DOI] [PubMed] [Google Scholar]
- Ho L.H., Read S.H., Dorstyn L., Lambrusco L., Kumar S. 2008. Caspase-2 is required for cell death induced by cytoskeletal disruption.Oncogene. 27:3393–3404 [DOI] [PubMed] [Google Scholar]
- Janssens S., Tinel A., Lippens S., Tschopp J. 2005. PIDD mediates NF-kappaB activation in response to DNA damage.Cell. 123:1079–1092 [DOI] [PubMed] [Google Scholar]
- Krumschnabel G., Sohm B., Bock F., Manzl C., Villunger A. 2009. The enigma of caspase-2: the laymen's view.Cell Death Differ. 16:195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhani S.A., Masud A., Kuida K., Porter G.A., Jr., Booth C.J., Mehal W.Z., Inayat I., Flavell R.A. 2006. Caspases 3 and 7: key mediators of mitochondrial events of apoptosis.Science. 311:847–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Ma W., Benchimol S. 2000. Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis.Nat. Genet. 26:122–127 [DOI] [PubMed] [Google Scholar]
- Marsden V.S., Ekert P.G., Van Delft M., Vaux D.L., Adams J.M., Strasser A. 2004. Bcl-2-regulated apoptosis and cytochrome c release can occur independently of both caspase-2 and caspase-9.J. Cell Biol. 165:775–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay G.P., Salvesen G.S., Green D.R. 2008. Overlapping cleavage motif selectivity of caspases: implications for analysis of apoptotic pathways.Cell Death Differ. 15:322–331 [DOI] [PubMed] [Google Scholar]
- Michalak E.M., Villunger A., Adams J.M., Strasser A. 2008. In several cell types tumour suppressor p53 induces apoptosis largely via Puma but Noxa can contribute.Cell Death Differ. 15:1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly L.A., Ekert P., Harvey N., Marsden V., Cullen L., Vaux D.L., Hacker G., Magnusson C., Pakusch M., Cecconi F., et al. 2002. Caspase-2 is not required for thymocyte or neuronal apoptosis even though cleavage of caspase-2 is dependent on both Apaf-1 and caspase-9.Cell Death Differ. 9:832–841 [DOI] [PubMed] [Google Scholar]
- Ogilvy S., Metcalf D., Print C.G., Bath M.L., Harris A.W., Adams J.M. 1999. Constitutive bcl-2 expression throughout the hematopoietic compartment affects multiple lineages and enhances progenitor cell survival.Proc. Natl. Acad. Sci. USA. 96:14943–14948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read S.H., Baliga B.C., Ekert P.G., Vaux D.L., Kumar S. 2002. A novel Apaf-1-independent putative caspase-2 activation complex.J. Cell Biol. 159:739–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.C., Doctor K.S., Godzik A. 2004. The domains of apoptosis: a genomics perspective.Sci. STKE. doi:10.1126/stke.2392004re9 [DOI] [PubMed] [Google Scholar]
- Riedl S.J., Salvesen G.S. 2007. The apoptosome: signalling platform of cell death.Nat. Rev. Mol. Cell Biol. 8:405–413 [DOI] [PubMed] [Google Scholar]
- Robertson J.D., Enoksson M., Suomela M., Zhivotovsky B., Orrenius S. 2002. Caspase-2 acts upstream of mitochondria to promote cytochrome c release during etoposide-induced apoptosis.J. Biol. Chem. 277:29803–29809 [DOI] [PubMed] [Google Scholar]
- Robertson J.D., Gogvadze V., Kropotov A., Vakifahmetoglu H., Zhivotovsky B., Orrenius S. 2004. Processed caspase-2 can induce mitochondria-mediated apoptosis independently of its enzymatic activity.EMBO Rep. 5:643–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F., Baron U., Rajewsky K. 1995. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells.Nucleic Acids Res. 23:5080–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Vivian C.J., Lee K.J., Ge C., Morotomi-Yano K., Manzl C., Bock F., Sato S., Tomomori-Sato C., Zhu R., et al. 2009. DNA-PKcs-PIDDosome: a nuclear caspase-2-activating complex with role in G2/M checkpoint maintenance.Cell. 136:508–520 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tinel A., Tschopp J. 2004. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress.Science. 304:843–846 [DOI] [PubMed] [Google Scholar]
- Tinel A., Janssens S., Lippens S., Cuenin S., Logette E., Jaccard B., Quadroni M., Tschopp J. 2007. Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-kappaB pathway.EMBO J. 26:197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakifahmetoglu H., Olsson M., Orrenius S., Zhivotovsky B. 2006. Functional connection between p53 and caspase-2 is essential for apoptosis induced by DNA damage.Oncogene. 25:5683–5692 [DOI] [PubMed] [Google Scholar]
- Villunger A., Michalak E.M., Coultas L., Mullauer F., Bock G., Ausserlechner M.J., Adams J.M., Strasser A. 2003. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa.Science. 302:1036–1038 [DOI] [PubMed] [Google Scholar]