Abstract
We studied the function of the cyclin-dependent kinase Cdc28 (Cdk1) in the DNA damage response and maintenance of genome stability using Saccharomyces cerevisiae. Reduced Cdc28 activity sensitizes cells to chronic DNA damage, but Cdc28 is not required for cell viability upon acute exposure to DNA-damaging agents. Cdc28 is also not required for activation of the DNA damage and replication checkpoints. Chemical–genetic analysis reveals that CDC28 functions in an extensive network of pathways involved in maintenance of genome stability, including homologous recombination, sister chromatid cohesion, the spindle checkpoint, postreplication repair, and telomere maintenance. In addition, Cdc28 and Mre11 appear to cooperate to prevent mitotic catastrophe after DNA replication arrest. We show that reduced Cdc28 activity results in suppression of gross chromosomal rearrangements (GCRs), indicating that Cdc28 is required for formation or recovery of GCRs. Thus, we conclude that Cdc28 functions in a genetic network that supports cell viability during DNA damage while promoting the formation of GCRs.
Introduction
Maintaining a stable genome is critical for the development and well-being of all organisms. Failure to maintain genome stability is associated with a large variety of diseases, including not only sporadic and inherited cancers but also several neurological, neurodegenerative, neuromuscular, and aging disorders (Hanahan and Weinberg, 2000; Hasty et al., 2003; Pearson et al., 2005). Although genomic instability is well known to be associated with different diseases, our knowledge about the pathways that protect the organism against genomic instability remains incomplete. However, studies in the model organism Saccharomyces cerevisiae have begun to provide a comprehensive description of the pathways and mechanisms that prevent genome instability (for review see Kolodner et al., 2002). These include mechanisms that protect against reactive oxygen species (Huang and Kolodner, 2005), promote fidelity of DNA replication (Chen and Kolodner, 1999), function in the S-phase checkpoint response (Myung et al., 2001c; Myung and Kolodner, 2002), control telomere formation and maintenance (Pennaneach and Kolodner, 2004), and assemble newly replicated DNA into chromatin (Myung et al., 2003). In addition, the activity of Cdk1 is crucial to maintain a stable genome (Kitazono and Kron, 2002; Lengronne and Schwob, 2002; Tanaka and Diffley, 2002; Kitazono et al., 2003; Gibson et al., 2004).
Cdks govern cell cycle progression in eukaryotes. During each phase of the cell cycle, Cdks form a complex with specific cyclins that activate Cdks and help target them to their substrates (Bloom and Cross, 2007). A single Cdk, Cdc28, is sufficient for cell cycle progression in S. cerevisiae. Cdc28 controls a plethora of cell cycle–related processes, including specific transcriptional programs associated with each phase of the cell cycle, budding and cell morphogenesis, DNA replication, spindle pole body duplication, and mitotic spindle assembly (Kellogg, 2003; Bloom and Cross, 2007). Cdc28 is also involved in maintenance of telomeres, (Grandin and Charbonneau, 2003; Frank et al., 2006; Vodenicharov and Wellinger, 2006), contributing to telomere elongation by directly phosphorylating Cdc13 (Li et al., 2009).
Cdk activity is tightly regulated; several mechanisms, often referred to as checkpoints (Hartwell and Weinert, 1989), have evolved that target Cdk activity to control the cell cycle in response to environmental and endogenous stresses that might compromise cell viability. For example, cells arrest the cell cycle in response to DNA damage and replication stress (Paulovich et al., 1997; Santocanale and Diffley, 1998). The DNA damage and replication checkpoints are defined as the pathways that promote cell cycle delay or arrest in response to DNA damage or DNA replication stress (Hartwell and Weinert, 1989). In addition, the DNA damage response also involves processes such as recruitment of DNA repair factors (Lisby et al., 2004), stabilization of replication forks (Lopes et al., 2001), inhibition of late-firing origins of replication (Santocanale and Diffley, 1998), and cytoplasmic events, including cell morphogenesis (Enserink et al., 2006; Smolka et al., 2006) and nuclear positioning (Dotiwala et al., 2007). DNA replication stress and DNA damage induce activation of two phosphoinositide 3 kinase–related kinases, Tel1 and Mec1, which are similar to mammalian ataxia telangiectasia mutated (ATM) and ATM and Rad3 related (ATR). These function in the activation of downstream protein kinases, including Chk1 and Rad53 (S. cerevisiae Chk2). In Schizosaccharomyces pombe and higher eukaryotes, the DNA damage and DNA replication checkpoints inhibit Cdk activity to block cell cycle progression. In contrast, S. cerevisiae cells arrest with high Cdc28 activity upon genotoxic stress, and inhibition of Cdc28 activity is not essential for cell cycle arrest (Sorger and Murray, 1992). Instead, upon DNA damage or replication stress, S. cerevisiae cells directly target key processes involved in cell cycle progression, including inhibiting the firing of late replication origins and blocking mitosis by preventing precocious chromosome segregation through inhibition of Cin8 and Stu2 as well as by stabilizing Pds1 (Yamamoto et al., 1996a,b; Cohen-Fix and Koshland, 1997, 1999; Santocanale and Diffley, 1998; Sanchez et al., 1999; Krishnan et al., 2004).
The fact that S. cerevisiae cells arrest with high Cdc28 activity allows for a function of Cdc28 in the DNA damage response. Indeed, several studies found that Cdc28 has functions in the DNA damage checkpoint activation and response, which may involve direct phosphorylation of Rad9 and Srs2 (Li and Cai, 1997; Liberi et al., 2000; Ira et al., 2004; Barlow et al., 2008; Bonilla et al., 2008). Furthermore, Cdc28 is important for homologous recombination (HR) during mitosis as well as meiosis (Aylon et al., 2004; Ira et al., 2004; Henderson et al., 2006). DNA double-strand breaks (DSBs) can be repaired through HR or through nonhomologous end joining (NHEJ), and the choice of either of these pathways depends on the cell cycle: during G1 phase, haploid yeast cells repair DSBs through NHEJ because of the absence of a template for HR, whereas in S and G2/M phases, they preferentially make use of HR (Ira et al., 2004), using the sister chromatid as a template. Furthermore, although Cdc28 is active during the S and G2/M phases of the cell cycle, it is inactive during G1 phase because of low cyclin concentrations and a high abundance of the Cdk inhibitor (CKI) Sic1, and Cdc28 activity determines the mode of DSB repair because its activity is required for resection of the DSB (Ira et al., 2004), which is the first step in HR. The molecular target of Cdc28 in this process was recently identified as the nuclease Sae2, which is directly phosphorylated and activated by Cdc28 (Huertas et al., 2008). Efficient resection of a DSB may also involve additional factors such as the Mre11–Rad50–Xrs2 complex, the nucleases Dna2 and Exo1, and the helicase Sgs1 (Gravel et al., 2008; Mimitou and Symington, 2008; Zhu et al., 2008). The exposed single-stranded DNA (ssDNA) is subsequently bound by the replication protein A (RPA) complex, which is later replaced by Rad51. Rad52 then stimulates Rad51 to search for homologous sequences and is also involved in annealing the complementary ssDNA strands (Symington, 2002).
In this study, we analyzed the function of Cdc28 in the DNA damage response in more detail. We found that Cdc28 supports cell viability under conditions of chronic DNA damage, but it is not required for survival of acute genotoxic stress, and it does not appear to function as a direct regulator of the DNA damage and replication checkpoints. Furthermore, Cdc28 is part of an extensive genetic network of pathways involved in the maintenance of genome stability, and it cooperates with HR to prevent catastrophic mitotic progression after DNA replication arrest. Surprisingly, we found that Cdc28 activity is also required for formation of gross chromosomal rearrangements (GCRs). Therefore, Cdc28 maintains cell viability during DNA damage while contributing to the formation of genome rearrangements.
Results
Cdc28 promotes cell viability during chronic but not acute DNA damage and is not required for checkpoint activation
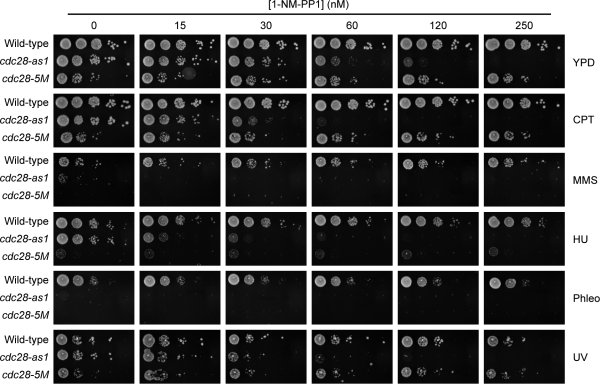
To test whether Cdc28 has a function in the response to DNA damage, we made use of two cdc28 mutants, cdc28-as1 (Bishop et al., 2000) and cdc28-5M (Li and Cai, 1997). The cdc28-as1 allele encodes a kinase with an enlarged ATP-binding pocket, allowing it to bind the nonhydrolyzable ATP analogue 1-NM-PP1, and treatment of cells with 1-NM-PP1 results in rapid and highly specific down-regulation of Cdc28 kinase activity (Bishop et al., 2000). However, it should be noted that cdc28-as1 is a hypomorphic allele because the kinase activity of this mutant is reduced by ∼20%, even in the absence of 1-NM-PP1 (Bishop et al., 2000). cdc28-5M encodes a temperature-sensitive form of Cdc28 with reduced kinase activity at the permissive temperature and further reduced kinase activity leading to lethality at elevated temperatures (Li and Cai, 1997). We spotted wild-type (WT) cells, cdc28-as1 mutants, and cdc28-5M mutants on yeast extract/peptone/dextrose (YPD) plates containing increasing but sublethal concentrations of 1-NM-PP1 in the absence or presence of various DNA-damaging agents. As expected, WT cells and cdc28-5M mutants were not affected by 1-NM-PP1, whereas the growth of cdc28-as1 mutants was slightly reduced at 60 and 120 nM 1-NM-PP1 (Fig. 1). However, these sublethal concentrations of 1-NM-PP1 greatly sensitized cdc28-as1 mutants to DNA-damaging agents like camptothecin (CPT; which inhibits DNA topoisomerase I), methylmethanesulfonate (MMS; an alkylating agent), hydroxyurea (HU; which depletes deoxynucleoside triphosphates, resulting in DNA replication arrest), and UV irradiation (Fig. 1 and Fig. S1 A). Furthermore, cdc28-as1 mutants were extremely sensitive to phleomycin (which induces free radical–mediated DNA damage, leading to single-strand breaks and DSBs [Sleigh, 1976]), even in the absence of 1-NM-PP1, which is likely caused by the fact that Cdc28-as1 has 20% lower kinase activity than WT Cdc28 (Bishop et al., 2000). cdc28-5M mutants were also very sensitive to MMS, HU, CPT, phleomycin, and UV irradiation (Fig. 1 and Fig. S1 A). These results show that Cdc28 activity is important for cell viability upon chronic exposure to various forms of DNA damage.
Figure 1.
Cdc28 functions in the DNA damage response. 10-fold dilutions of log-phase cultures were spotted on YPD supplemented with increasing concentrations of 1-NM-PP1 and fixed concentrations of 10 µg/ml CPT, 0.05% MMS, 100 mM HU, and 1 µg/ml phleomycin (Phleo) or irradiated with 100 J/m2 UV, as indicated.
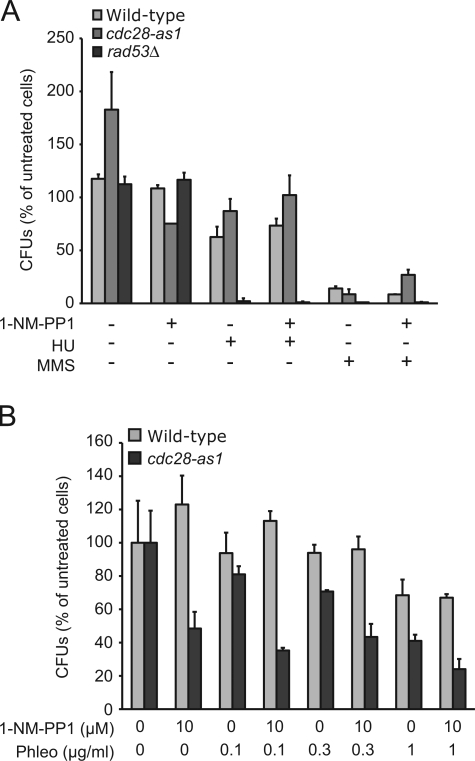
Unlike other eukaryotes, S. cerevisiae cells arrest with high levels of Cdc28 activity after treatment with DNA-damaging agents (Amon et al., 1992). Because Cdc28 activity is required for cell survival in the presence of different types of DNA-damaging agents, it is possible that Cdc28 functions as a checkpoint protein, something which has been suggested previously (Li and Cai, 1997; Ira et al., 2004; Barlow et al., 2008; Bonilla et al., 2008). Characteristically, checkpoint mutants do not recover from acute exposure to DNA-damaging agents as they fail to arrest the cell cycle and fail to stabilize replication forks (Lopes et al., 2001). To test whether Cdc28 has a checkpoint function, we first arrested WT cells and cdc28-as1 mutants in G1 phase with α factor and released them into YPD for 30 min to allow cells to go past Start (which is Cdc28 dependent) and enter S phase (which was confirmed by FACS analysis; unpublished data). We then added 1-NM-PP1 for 5 min before treating with either HU or MMS for 2 h. cdc28-as1 mutants were not more sensitive to killing by HU or MMS (Fig. 2 A) or phleomycin (Fig. 2 B) than WT cells. Similar results were obtained with cdc28-5M mutants (Fig. S1, B and C). In contrast, mutants lacking the checkpoint protein Rad53 did not survive acute treatment with these agents (Fig. 2 A).
Figure 2.
Cdc28 does not have a replication checkpoint function. (A) Cell survival after HU- or MMS-induced DNA damage does not depend on Cdc28 activity. WT cells or cdc28-as1 or rad53 mutants were treated with 10 µM 1-NM-PP1, 200 mM HU, or 0.05% MMS for 2 h, washed, and plated on YPD as described in Materials and methods. Cell survival was calculated as a percentage of untreated WT cells. (B) Cell survival after phleomycin (Phleo)-induced DNA damage does not depend on Cdc28 activity. WT cells or cdc28-as1 mutants were treated as in A with 1-NM-PP1 and increasing concentrations of phleomycin, and the percentage of cell survival was determined. (A and B) Error bars represent standard deviation. CFU, colony-forming unit.
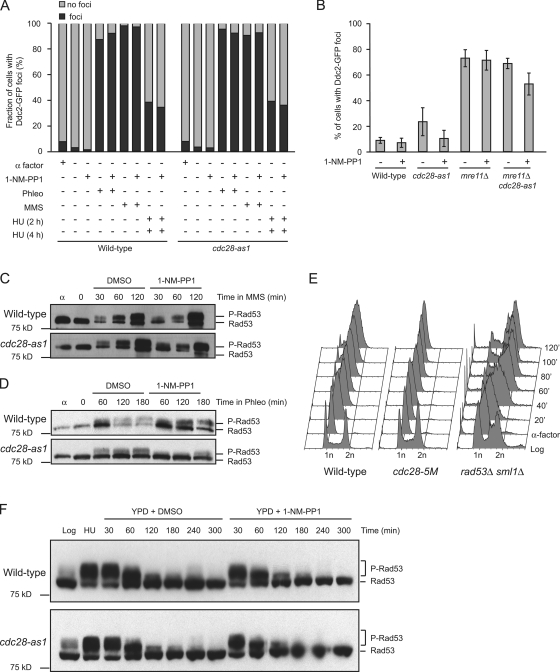
To more directly determine whether Cdc28 has an effect on checkpoint activation, we studied the formation of Ddc2 foci. Ddc2 functions in the initiation of DNA damage checkpoint activation by mediating the interaction between Mec1 and the proliferating cell nuclear antigen–like Rad17/Ddc1/Mec3 clamp (Majka et al., 2006). Recruitment of Ddc2 to sites of DNA damage can be visualized by fluorescence microscopy, and formation of Ddc2-GFP foci is used as a quantitative measure for checkpoint activation (Lisby et al., 2004). We arrested cells in G1 and released them into YPD for 30 min. We then added 1-NM-PP1 for 5 min to inactivate Cdc28-as1 before treating cells with phleomycin, HU, or MMS. As shown in Fig. 3 A, Cdc28 was not essential for the formation of Ddc2 foci, which is in accordance with previous findings (Barlow et al., 2008). However, although the frequency of Ddc2 focus formation was not affected by Cdc28 activity, we did notice that the intensity of Ddc2 foci was lower in 1-NM-PP1–treated cdc28-as1 mutants (unpublished data). We also studied the effect of Cdc28 on the formation of spontaneous rather than drug-induced Ddc2 foci in an mre11Δ mutant, which accumulates spontaneous DNA damage, resulting in high levels of genome rearrangements (Myung et al., 2001a). Although ∼10% of WT cells and cdc28-as1 mutants had at least one Ddc2 focus, nearly 80% of mre11Δ single mutants had Ddc2 foci (Fig. 3 B). mre11Δ cdc28-as1 double mutants were similar to mre11Δ single mutants, and inhibition of Cdc28-as1 activity by 1-NM-PP1 did not change the frequency of focus formation, although a reduction in the intensity of the foci was observed (unpublished data). We also monitored phosphorylation of Rad53 upon treatment with DNA-damaging agents in WT cells and in cdc28-as1 mutants. MMS-induced Rad53 activation was not inhibited by 1-NM-PP1 (Fig. 3 C), indicating that Cdc28 is not required for MMS-induced Rad53 activation, which is similar to the previously reported findings that 4-nitroquinoline 1-oxide–induced Rad53 activation is independent of Cdc28 (Ira et al., 2004) and that Cdc28 by itself is not essential for Rad53 activation (Barlow et al., 2008). Interestingly, we found that phleomycin-induced Rad53 phosphorylation was partially reduced in 1-NM-PP1–treated cdc28-as1 mutants, indicating that checkpoint activation by phleomycin-induced DNA damage may be partially dependent on Cdc28 (Fig. 3 D).
Figure 3.
Cdc28 has no major role in DNA damage–induced checkpoint activation. (A) DNA damage–induced formation of Ddc2 foci is independent of Cdc28. Cells were arrested with α factor and released into YPD or YPD containing 10 µM 1-NM-PP1 and the indicated DNA-damaging agents for 2 or 4 h, as indicated (see Materials and methods). Ddc2-GFP foci were visualized using fluorescence microscopy, and the percentage of cells with foci relative to the total cell population of that specific sample was calculated. Phleo, phleomycin. (B) Checkpoint activation upon endogenous DNA damage does not depend on Cdc28 activity. WT cells and cdc28-as1, mre11Δ, and cdc28-as1 mre11Δ mutants were arrested in α factor before being released into YPD supplemented with 10 µM 1-NM-PP1 for 2 h, and the percentage of cells with Ddc2-GFP foci were determined as described in A. Error bars represent standard deviation. (C) MMS-induced phosphorylation of Rad53 is independent of Cdc28. WT cells and cdc28-as1 mutants were arrested in α factor (α) and released into YPD for 30 min to allow cells to enter S phase. Then, either DMSO or 1-NM-PP1 was added to inactivate Cdc28-as1 (time point 0) followed by treatment with MMS for 30, 60, or 120 min. Cell lysates were analyzed by Western blotting using Flag antibodies to detect Rad53. (D) Phleomycin-induced phosphorylation of Rad53 is partially dependent on Cdc28. WT cells and cdc28-as1 mutants were arrested in α factor and released into YPD for 30 min to allow cells to enter S phase. Then, either DMSO or 1-NM-PP1 was added to inactivate Cdc28-as1 (time point 0) followed by treatment with phleomycin for 60, 120, or 180 min. Cell lysates were analyzed by Western blotting using Flag antibodies to detect Rad53. (E) Cdc28 is not required for MMS-induced S-phase arrest. WT cells and cdc28-5M and rad53Δ mutants were grown into log phase (log) or arrested in α factor before being released into YPD supplemented with 0.05% MMS. Samples were taken at the indicated time points and analyzed by FACS. (F) Checkpoint inactivation after replication arrest does not require Cdc28 activity. WT cells and cdc28-as1 mutants were arrested in HU for 3 h before HU was washed away, and cells were resuspended in YPD supplemented with either DMSO or 10 µM 1-NM-PP1. Samples were taken at the indicated time intervals, and Rad53 phosphorylation was analyzed as in C. P-Rad53, phospho-Rad53.
We also used FACS to determine whether Cdc28 is involved in the arrest of the cell cycle in response to MMS-induced DNA replication damage. We synchronized WT cells, rad53Δ mutants and cdc28-5M mutants in G1 and released them in YPD supplemented with 0.05% MMS. As shown in Fig. 3 E, WT cells arrested the cell cycle in the presence of MMS, whereas rad53Δ mutants, which are checkpoint defective, did not show slow progression through the cell cycle. cdc28-5M mutants, which are very sensitive to MMS even at the permissive temperature (Fig. 1), entered S phase somewhat later than WT cells after release from α factor but efficiently arrested the cell cycle, indicating that Cdc28 is not required for activation of the checkpoint.
Another reason why mutants with reduced Cdc28 activity might be sensitive to DNA damage is that they might not recover from checkpoint activation, which is similar to what has been described for mutants lacking the phosphatases Ptc2, Ptc3, and Pph3 that down-regulate the checkpoint by dephosphorylating Rad53 (Leroy et al., 2003; O'Neill et al., 2007). To test whether Cdc28 may have a function in recovery from DNA replication arrest, we arrested WT cells and cdc28-as1 mutants in S phase for 3 h with HU and released them into either YPD or YPD supplemented with 1-NM-PP1. As shown in Fig. 3 F, in WT cells, Rad53 was largely dephosphorylated 1 h after release from HU arrest and almost completely dephosphorylated 2 h after release, indicating down-regulation of the checkpoint. The degree of Rad53 dephosphorylation in cdc28-as1 mutants was identical to that of WT cells and was not impaired by 1-NM-PP1, indicating that Cdc28 is not required for turning off the checkpoint. In addition, FACS analysis showed that both WT cells and cdc28-as1 mutants had completed DNA replication 1 h after release from HU arrest, and this also was not affected by 1-NM-PP1 (unpublished data). Therefore, although Cdc28 is important for cell viability during chronic exposure to DNA damage, it is not important for cell survival after acute DNA damage, and, by itself, it does not appear to have a major role in either the activation or down-regulation of DNA damage or replication checkpoints.
Cdc28 functions with Mre11 in recovery from DNA replication arrest
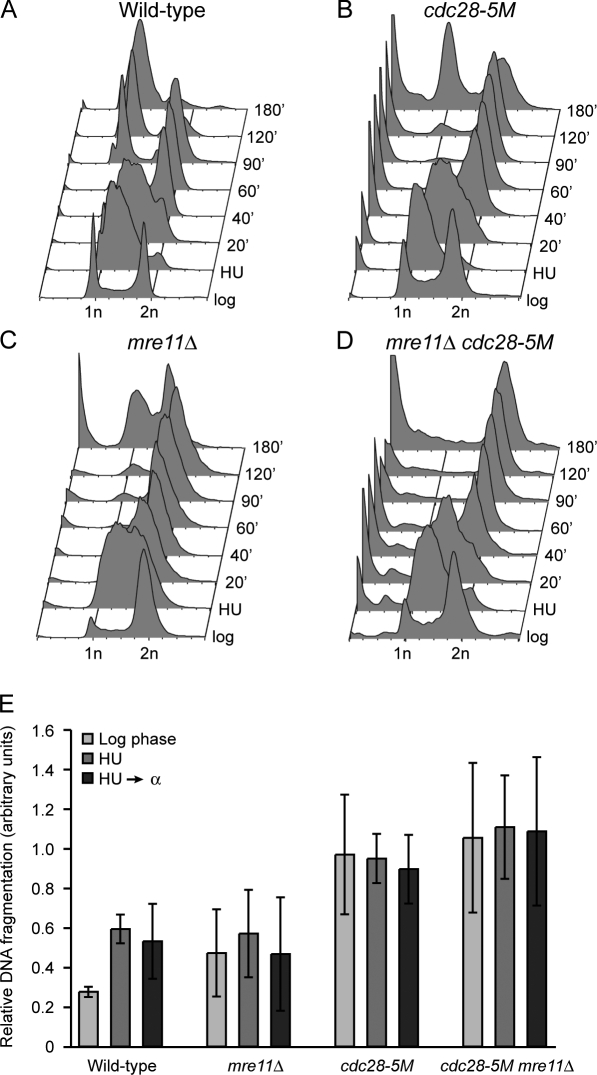
Although Cdc28 by itself may not be required to recover from DNA replication arrest, it might cooperate with certain DNA repair pathways for recovery and resumption of the cell cycle. Indeed, we found that CDC28 genetically interacts with many pathways involved in the maintenance of genome stability, including HR (see next section). To explore the possibility that Cdc28 cooperates with HR in recovery from DNA replication arrest, we made use of FACS analysis to test the ability of WT cells, cdc28-5M and mre11Δ single mutants, and cdc28-5M mre11Δ double mutants to recover from S-phase arrest and to go through M phase by treating them with HU and releasing them into medium containing α factor. Mutants that do not recover from replication arrest should be unable to complete M phase or to exit from M phase. We made use of the cdc28-5M allele because Cdc28-5M has strongly reduced kinase activity even at permissive temperatures (Li and Cai, 1997) while retaining sufficient kinase activity to allow cells to go through M phase (inhibition of Cdc28 activity using 1-NM-PP1 treatment of cdc28-as1 mutants results in complete M-phase arrest [Bishop et al., 2000] and, therefore, is not suitable for studying exit from M phase). As shown in Fig. 4 A, after 40 min, WT cells had recovered from HU arrest and completed bulk DNA synthesis. All cells had exited from M phase and accumulated in G1 phase after 180 min. The cdc28-5M single mutant also completed DNA synthesis after 40 min but exited from M phase slower than WT, accumulating in G1 phase after 180 min (Fig. 4 B). Interestingly, a portion of cdc28-5M mutants accumulated with sub-G1 DNA content, indicating a certain degree of mitotic catastrophe, which is consistent with a previous study implicating Cdc28 in prevention of mitotic catastrophe (Kitazono and Kron, 2002). mre11Δ mutants were delayed in passing through M phase (Fig. 4 C), although a relatively large number of cells ultimately accumulated in G1 phase. However, the mre11Δ cdc28-5M double mutant had a strong defect in recovering from DNA replication arrest, as the majority of cells were unable to progress through M phase (Fig. 4 D). In addition, those cells that did manage to exit from M phase appeared to have <1 N DNA content, as indicated by the absence of a distinct G1-phase peak and the presence of a large sub–G1 phase population instead. We also made use of pulsed-field gel electrophoresis (PFGE) to monitor chromosomal integrity. We found that log-phase WT cells had very low levels of chromosome fragmentation, which increased after treatment with HU (Fig. 4 E and Fig. S2). mre11Δ mutants had levels of chromosome fragmentation that were comparable with that of WT cells. Interestingly, log-phase cdc28-5M mutants showed increased chromosome fragmentation (Fig. 4 E), although this was not further increased by HU treatment. Furthermore, mre11Δ cdc28-5M double mutants did not have higher levels of chromosome fragmentation than cdc28-5M single mutants. Collectively, these results indicate that Cdc28 cooperates with Mre11 to prevent catastrophic M-phase progression after DNA replication stress.
Figure 4.
Cdc28 cooperates with Mre11 to maintain genome stability. Cdc28 cooperates with Mre11 to prevent mitotic catastrophe after DNA replication arrest. (A–D) WT cells (A), cdc28-5M mutants (B), mre11Δ mutants (C), and mre11Δ cdc28-5M double mutants (D) were arrested in HU for 3 h before being released into YPD supplemented with α factor at 30°C. Samples were taken at the indicated time intervals and analyzed using FACS analysis. (E) Cdc28 prevents chromosome fragmentation. WT cells and mre11Δ, cdc28-5M, and mre11Δ cdc28-5M mutants were treated as indicated, and whole chromosomes were analyzed by PFGE. The relative amount of chromosome fragmentation (at 30°C because incubation at 37°C did not further increase fragmentation) was calculated as described in Materials and methods. Error bars represent the standard error of the mean.
Cdc28 is required for the formation of GCRs
To further characterize the effect of Cdc28 on the maintenance of genome stability, we analyzed its effect on GCR formation. First, we measured GCR rates in strains with increased Cdc28 activity generated by deleting genes encoding endogenous inhibitors of Cdc28. Mutants lacking the CKI Sic1 (the functional homologue of mammalian p27KIP1 [Barberis et al., 2005]), which prevents precocious entry into S phase by inhibiting cyclin B–Cdc28 complexes (Schwob et al., 1994), had a 17-fold increase in GCR rate (Table I). This is significantly lower than the ∼600–fold increase that has been published previously (Lengronne and Schwob, 2002) but consistent with the 19-fold increase in GCR rates obtained with overexpression of CLN2-1, which encodes a stabilized form of Cln2 that also induces precocious entry into S phase by elevating the activity of Cdc28 (Tanaka and Diffley, 2002). In addition, deletion of FAR1, a CKI that directly inhibits cyclin–Cdc28 complexes to prevent entry into S phase (Peter and Herskowitz, 1994), resulted in a similar increase in GCR rates (Table I), whereas simultaneous deletion of SIC1 and FAR1 appeared to result in an additive effect, leading to a 33-fold increase in GCR rates (Table I). Mutants lacking Swe1, the budding yeast homologue of mammalian Wee1 that directly phosphorylates and inhibits Cdc28 to inhibit entry into M phase when certain aspects of cytoskeletal function or bud formation are impaired (Lew, 2003), did not have increased GCR rates (Table I). Therefore, consistent with previous findings (Lengronne and Schwob, 2002; Tanaka and Diffley, 2002), unscheduled activation of Cdc28 during G1–S phase resulted in increased GCR rates, reiterating the belief that most GCRs stem from processes related to DNA replication (for review see Kolodner et al., 2002).
Table I.
Mutations in CDC28 suppress GCR rates
| Relevant genotype | Mutation rate (Canr − 5FOAr) (×10−10) | ||
| CDC28 | cdc28-as1 | cdc28-5M | |
| WT | 3.50 (1)a | <12.5 (<3.6)b | <128 (<37)b |
| CKIs | |||
| sic1Δ | 59.5 (17) | NT | NT |
| far1Δ | 71.1 (20) | NT | NT |
| sic1Δ far1Δ | 115.9 (33) | NT | NT |
| swe1Δ | <5.69 (<1.6)b | NT | NT |
| HR | |||
| rad50Δ | 2,300 (657)a | 148 (42) | NT |
| mre11Δ | 2,490 (711) | 128 (37) | <327 (93)b |
| mre11Δ in nocodazoleg | 5,700 (1,622) | NT | NT |
| mre11Δ in SC, RTh | 1,890 (543) | NT | NT |
| mre11Δ yku80Δ | 6,156 (1,759) | 542 (155) | NT |
| rad52Δ | 350 (100)c | 290 (83) | <446 (127)b |
| sae2Δ | 177 (51) | <67 (<19)b | NT |
| sae2Δ mre11Δ | 2,798 (797) | NT | NT |
| DNA damage and replication checkpoint | |||
| mec1Δ | 680 (194)d | NT | <33 (<9)b |
| mec1Δ tel1Δ | 45,000 (12,857)d | NT | 2,390 (683) |
| rad53Δ | 95 (27)d | <9.6 (<2.7)b | NT |
| pds1Δ | 670 (190)d | <64 (<18)b | NT |
| rfc5-1Δ | 660 (189)d | <5.1 (<1.5)b | NT |
| rad17Δ | 30 (9)d | <14 (<4)b | NT |
| rad24Δ | 40 (11)d | <26 (<7.4)b | NT |
| Others | |||
| sgs1Δ | 77 (22)e | 60 (17) | <74 (<21)b |
| rfa1-t11 (CEN) | 420 (120)a | <69 (<20)b | NT |
| rad27Δ | 4,400 (1,257)e | NT | <277 (<79)b |
| tsa1Δ | 173 (49)f | NT | <84 (24)b |
| pif1Δ | 3,530 (1,008)c | 519 (148) | 200 (57) |
NT, not tested. Numbers in parentheses indicate fold increase over WT. The mutation rates are shown as events per generation.
Data from Chen and Kolodner (1999).
GCR rates could not be accurately calculated because GCRs were not detected in a large enough proportion of the cultures.
Data from Myung et al. (2001a).
Data from Myung et al. (2001c).
Data from Myung et al. (2001b).
Data from Huang and Kolodner (2005).
mre11Δ mutants were grown in 10 µM nocodazole until stationary phase to test the effect of delaying the cell cycle on GCR rates.
mre11Δ mutants were grown in synthetic complete medium (SC) at RT until stationary phase to test the effect of delaying the cell cycle on GCR rates.
Because increased Cdc28 activity resulted in increased GCR rates, we next asked what the effect would be of reduced Cdc28 activity on GCR rates. Typically, mutations in genes that are involved in the DNA damage response lead to increased GCR rates, presumably because damaged chromosomes are not faithfully repaired (for review see Kolodner et al., 2002). Therefore, because we found that Cdc28 is involved in the response to DNA damage (Fig. 1 and Fig. S1 A), we expected cdc28 mutants to have increased GCR rates. However, we did not observe a single GCR event in cultures of either cdc28-as1 or cdc28-5M mutants and, therefore, could only calculate an upper limit for the GCR rate of these strains (Table I); the actual rate is likely to be lower. We next tested whether cdc28 mutations can suppress the increased GCR rates caused by well-known mutator mutations such as mre11Δ and rad50Δ (Myung et al., 2001a). As shown in Table I, mre11Δ and rad50Δ single mutants have GCR rates that are ∼700-fold higher than WT. However, even in the absence of 1-NM-PP1, mre11Δ cdc28-as1 and rad50Δ cdc28-as1 double mutants only had ∼40-fold increased GCR rates. Thus, a modest reduction in Cdc28 activity was sufficient to largely suppress GCR rates of mre11Δ and rad50Δ mutants. In addition, cultures of mre11Δ cdc28-5M double mutants did not form even a single GCR, and, therefore, accurate GCR rates could not be calculated for this strain. The increased GCR rate of mutants lacking Rad52 was also suppressed by cdc28 mutations (Table I).
One might argue that the suppressive effect of cdc28 mutations on GCR rates might be caused by the reduced speed with which the mutants traverse the cell cycle. We tested that possibility by artificially reducing growth rates by culturing mre11 mutants at RT in synthetic complete media. These conditions substantially reduced growth rates of this mutant (unpublished data) but had basically no effect on the GCR rate, which was 543-fold over WT (Table I). In addition, we cultured mre11 mutants in YPD supplemented with a sublethal dose (10 µM) of nocodazole, which strongly reduced the growth rate of this mutant, taking up to 5 d to reach stationary phase (unpublished data). However, this treatment resulted in increased rather than decreased GCR rates of mre11 mutants (∼1,622-fold over WT; Table I). Therefore, the suppression of GCR formation by hypomorphic cdc28 alleles is not a result of the reduced speed with which these mutants pass through the cell cycle. Alternatively, the suppressive effect of cdc28-as1 and cdc28-5M on formation of GCRs in mre11Δ mutants could also be a result of the reduced activity of the nuclease Sae2. Cdc28 has recently been shown to phosphorylate and thereby stimulate the activity of Sae2, resulting in the resection of DSBs to expose ssDNA, which is the first step in HR (Huertas et al., 2008). One could argue that mutants harboring cdc28 alleles suppress the GCRs that arise in HR-defective mre11Δ mutants because Cdc28 is unable to fully activate Sae2, thereby preventing resection and futile attempts to perform HR that might otherwise lead to GCRs. The DSBs might be channeled into the NHEJ pathway instead, resulting in an apparent reduction in GCR rates. If this model were correct, one would predict that depleting Sae2 would result in suppression of the GCR rates of mre11Δ mutants. Conversely, inactivation of the NHEJ pathway by deleting YKU80 in an mre11Δ cdc28-as1 background should then restore GCR rates. However, we found that a sae2Δ single mutant had a GCR rate of ∼50-fold over WT, and a sae2Δ mre11Δ double mutant had a GCR rate of ∼797-fold over WT, which is identical to that of mre11Δ single mutants (Table I). Furthermore, the GCR rate of sae2Δ mutants was suppressed by the cdc28-as1 allele. Finally, yku80Δ mre11Δ cdc28-as1 triple mutants had a GCR rate of 155-fold over WT, compared with 1,759-fold for yku80Δ mre11Δ double mutants (Table I). We conclude that channeling of DSBs into the NHEJ pathway is not involved in cdc28-mediated suppression of GCRs because (a) deletion of SAE2 does not result in suppression of GCRs and therefore failure to fully activate Sae2 cannot explain the suppressive effect of cdc28 mutations on formation of GCRs, and (b) deletion of YKU80 does not restore the suppressed GCR rates of mre11Δ cdc28-as1 mutants.
We also determined the effect of cdc28-as1 and cdc28-5M mutations on the GCR rates of cells with deletions in various DNA damage and replication checkpoint genes, including RAD17, RAD24, RAD53, MEC1, and a combination of MEC1 and TEL1. In all cases, mutations in CDC28 suppressed the increased GCR rates (Table I), and in most cases, no GCR events could be detected in mutants in the cdc28-5M background. The one exception was the mec1Δ tel1Δ cdc28-5M triple mutant, which had a GCR rate that was increased ∼700-fold over the WT rate; however, because mec1Δ tel1Δ double mutants have a GCR rate that is increased 12,000-fold over the WT rate, the cdc28-5M mutation was still a potent suppressor of GCRs. The cdc28-as1 and cdc28-5M mutations also suppressed GCR rates of rfa1-t11, rfc5-1, tsa1Δ, pds1Δ, asf1Δ, sgs1Δ, rad27Δ, and tsa1Δ mutants and at least partially suppressed the increased GCR rate of a pif1Δ mutant, which acquires GCRs that are exclusively de novo telomere additions. In conclusion, these data show that mutations in CDC28 have a broad suppressive effect on the increased accumulation of GCRs in many mutant backgrounds, indicating a general requirement for Cdc28 activity in formation of GCRs.
Cdc28 activity does not suppress the increased mutation rate caused by an msh2 mutation
To determine whether Cdc28 also affects the accumulation of mutations like base substitutions and frameshifts, we made use of three mutator assays: the CAN1 forward mutation assay, which scores for mutations that inactivate the CAN1 gene, the lys2-Bgl reversion assay, which detects reversion of a +4 insertion in the LYS2 gene, and the hom3-10 reversion assay, which detects reversion of a +1 insertion in the HOM3 gene. The lys2-Bgl and the hom3-10 assays are particularly sensitive for detecting defects in mismatch repair, whereas the CAN1 assay detects a broad range of mutator phenotypes (Marsischky et al., 1996; Greene and Jinks-Robertson, 1997; Tishkoff et al., 1997; Tran et al., 1997; Umezu et al., 1998). As shown in Table II, the cdc28-5M mutation did not affect mutation rates in any of these assays. Furthermore, the cdc28-5M mutation did not suppress the high mutation rates of mutants lacking the Msh2 mismatch repair protein. Therefore, it seems likely that Cdc28 might specifically affect the processing or the stability of broken chromosomes that underlie the formation of GCRs.
Table II.
Reduced Cdc28 activity does not affect non-GCR mutation rates
| Relevant genotype | Hom+ rate (×10−8) | Lys+ rate (×10−8) | Canr rate (×10−7) |
| WT | 1.4 (1.2–1.5) | 1.9 (1.4–2.7) | 3.6 (3.1–5.2) |
| cdc28-5M | 1.2 (0.8–1.6) | 1.8 (1.6–2.5) | 2.3 (1.9–5.8) |
| msh2Δ | 670.2 (405.9–832.8) | 62.1 (50.8–99.0) | 34.6 (16.6–48.8) |
| msh2Δ cdc28-5M | 967.7 (505.4–1,730.9) | 55.3 (45.1–73.6) | 39.3 (31.4–67.6) |
Numbers in parentheses indicate confidence intervals. The mutation rates are shown as events per generation.
CDC28 is part of a genetic network that preserves chromosomal stability
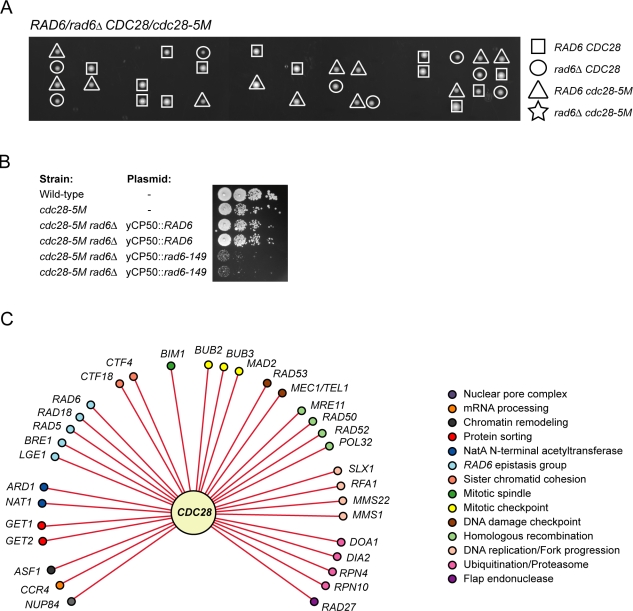
We next wanted to gain insight into the genetic network that involves CDC28 and that promotes cell survival. We made use of a directed chemical–genetics approach to test for genetic interactions between cdc28-as1 and defects in the major pathways involved in maintenance of genome stability. We spotted double mutants on either YPD plates supplemented with DMSO or YPD plates containing 200 nM 1-NM-PP1, which is a sublethal concentration that still permits growth of cdc28-as1 single mutants. We found that a variety of genetic pathways function with CDC28 to maintain cell viability (Table III and Fig. S3 A). For instance, CDC28 genetically interacted with genes involved in HR (MRE11, RAD50, RAD52, and POL32), sister chromatid cohesion (CTF4 and CTF18), the DNA replication and DNA damage checkpoints (RAD53 and a combination of MEC1 and TEL1), the spindle checkpoint and mitotic exit network (BUB3, MAD2, and BUB2), the RNA component of telomerase (TLC1), DNA replication (RFA1), flap endonuclease (RAD27), and chromatin remodeling (ASF1). CDC28 also showed strong genetic interactions with components of the postreplication repair pathway, particularly RAD6, and also with RAD18 and RAD5. Deletion of RAD6 resulted in synthetic lethality in a cdc28-5M background (Fig. 5 A), which was rescued by a plasmid harboring WT RAD6 (Fig. 5 B) but not by a plasmid encoding the catalytically inactive Rad6-C88A mutant (not depicted) and was partially rescued by a plasmid harboring rad6-149, which encodes a mutant of Rad6 that lacks the acidic C terminus and which is particularly deficient in ubiquitination of histones (Sung et al., 1988; Robzyk et al., 2000). We also found genetic interactions between CDC28 and BRE1 and LGE1, whose gene products function in complex with Rad6 in H2B ubiquitination (Hwang et al., 2003; Wood et al., 2003). All of the genetic interactions with CDC28 that we observed in this study were seen under conditions of chronic but not acute inhibition of Cdc28; a brief treatment of each of the double mutants with 1-NM-PP1 resulted in little or no loss of cell viability (Fig. S3 B). The fact that a brief inhibition of Cdc28 in these mutants is reversible indicates that these double mutants may have to go through several rounds of the cell cycle under conditions of reduced Cdc28 activity before losing cell viability, which could potentially be the result of chromosome loss events. Finally, using a bioinformatics approach (see Materials and methods), we identified additional genetic interactions between CDC28 and genes previously implicated in the maintenance of genome stability (Table S1; Pan et al., 2006). Although we did not confirm every predicted genetic interaction, we found that 14 out of 18 (78%) genes tested genetically interacted with CDC28. Therefore, our methodology for prediction of genetic interactions should be widely applicable. An overview of the CDC28 genetic network involved in maintenance of genome stability is shown in Fig. 5 C; CDC28 genetically interacts with a wide range of pathways involved in genome stability, thus underscoring the importance of Cdc28 in this process.
Table III.
Genetic interactions with CDC28
| Relevant genotype | Sensitivity to 200 nM 1-NM-PP1 | |
| CDC28 | cdc28-as1 | |
| WT | − | − |
| Recombination | ||
| mre11Δ | − | ++++ |
| rad50Δ | − | ++++ |
| rad51Δ | − | + |
| rad52Δ | − | +++ |
| rad54Δ | − | − |
| pol32Δ | − | ++++ |
| sae2Δ | − | + |
| RPA complex | ||
| rfa1-t6 | − | ++++ |
| rfa1-t11 | − | ++++ |
| rfa1-t19 | − | ++++ |
| rfa1-t48 | − | ++++ |
| NHEJ | ||
| yku80Δ | − | + |
| lig4Δ | − | − |
| Replication fork progression | ||
| top3Δ | − | + |
| sgs1Δ | − | + |
| rrm3Δ | − | − |
| srs2Δ | − | − |
| mus81Δ | − | − |
| mus81Δ mms4Δ | − | − |
| slx1Δ | − | +++ |
| Nucleotide excision repair | ||
| rad10Δ | − | − |
| Okazaki fragments | ||
| rnh201Δ rnh203Δ | − | − |
| rad27Δ | − | ++++ |
| Postreplication repair | ||
| rad5Δ | − | ++ |
| rad6Δ | − | ++++ |
| rad18Δ | − | ++ |
| pol30-119 | − | + |
| DNA damage and replication checkpoints | ||
| rad17Δ | − | − |
| rad24Δ | − | − |
| rfc5-1 | − | + |
| elg1Δ | − | + |
| tof1Δ | − | + |
| csm3Δ | − | ++ |
| mec1Δ | − | − |
| tel1Δ | − | − |
| mec1Δ tel1Δ | − | ++ |
| rad53Δ | − | ++ |
| chk1Δ | − | − |
| Spindle checkpoint/ mitotic exit | ||
| bub3Δ | − | ++++ |
| mad2Δ | − | +++ |
| bub2Δ | − | +++ |
| Sister chromatid cohesion | ||
| ctf4Δ | − | ++++ |
| ctf18Δ | − | ++++ |
| Chromatin organization | ||
| asf1Δ | − | +++ |
| cac1Δ | − | − |
| Telomeres | ||
| pif1Δ | − | − |
| tlc1Δ | − | ++++ |
| Oxidative stress response | ||
| tsa1Δ | − | − |
| Cdc28 regulation | ||
| sic1Δ | − | − |
−, not more sensitive/equally sensitive to cdc28-as1; +, slightly slower growth compared with cdc28-as1 single mutant; ++, slow growth compared with cdc28-as1 single mutant; +++, severely reduced growth compared with cdc28-as1 single mutant; ++++, near death or dead compared with cdc28-as1 single mutant.
Figure 5.
CDC28 genetically interacts with pathways involved in maintenance of genome stability. (A) Synthetic lethal interaction between cdc28-5M and rad6Δ. CDC28/cdc25-5M RAD6/rad6Δ diploids were sporulated, and tetrads were dissected. All combinations except the rad6 cdc28-5M double mutant were observed. (B) Expression of rad6-149 partially rescues the synthetic lethality of the cdc28-5M rad6Δ mutant. 10-fold dilutions of log-phase cell cultures were spotted on YPD plates and incubated until colonies were visible. (C) CDC28 genetic interaction map.
Discussion
CDC28 is required for survival of chronic but not acute DNA damage
The role of Cdc28 in DNA damage–induced checkpoint activation is currently unclear. It has been shown that Cdc28 is required for checkpoint activation after induction of a homothallic switching (HO) endonuclease break during the G2 phase but not the G1 phase of the cell cycle (Ira et al., 2004). Furthermore, artificial recruitment of Ddc1-lacI (which recruits the Ddc1–Mec1 complex) and Ddc2-lacI (which recruits the 9-1-1 complex that consists of Ddc2, Mec3, and Rad17) chimaeras to a chromosomal array of lac operators has been shown to induce Rad53 phosphorylation even in the absence of DNA damage, and this was dependent on Cdc28-mediated phosphorylation of Rad9 (Bonilla et al., 2008). In another study, it was reported that Cdc28 was important for Ddc2 foci formation but, by itself, is not required for ionizing radiation–induced Rad53 phosphorylation (Barlow et al., 2008); rather, Cdc28 had a redundant role with RPA in the activation of Rad53 (Barlow et al., 2008). In this study, we have shown that Cdc28 functions in the maintenance of cell viability in the presence of various types of DNA damage, including HU-induced replication fork stalling, alkylation of DNA (MMS), cross-linked nucleotides (UV) and free radical–mediated DNA damage (phleomycin). However, Cdc28 by itself did not seem to have a major effect on either checkpoint activation or cell survival upon acute exposure to most of these types of DNA-damaging agents, indicating that Cdc28 is not a key regulator of checkpoint responses to most types of DNA damage. We only observed a partial effect of Cdc28 on Rad53 phosphorylation after phleomycin treatment, which is thought to induce DSBs (Sleigh, 1976). Because phleomycin-induced formation of Ddc2-GFP foci in cdc28-as1 mutants was not affected by 1-NM-PP1, Cdc28 appears to have a function downstream of Ddc2 but upstream of Rad53, which is in accordance with a previous study showing that Cdc28 might be important for Rad9 activity (Bonilla et al., 2008). Apart from this, our finding that Cdc28 by itself was not a major player in checkpoint activation seems at variance with previous studies that implicate Cdc28 in checkpoint activation (Ira et al., 2004; Barlow et al., 2008; Bonilla et al., 2008). One explanation might be that Cdc28 is involved in DNA damage checkpoint activation by DSBs that are induced by HO breaks (Ira et al., 2004), γ irradiation (Barlow et al., 2008), and, to a certain extent, by phleomycin (this study), but Cdc28 might not be required for checkpoint activation after DNA-damaging treatments that mainly result in replication fork stalling such as MMS and HU. Furthermore, the effect of Cdc28 on DNA damage checkpoint activation has thus far mainly been studied in cells that were arrested in either G1 phase or in G2/M phase (Li and Cai, 1997; Ira et al., 2004; Barlow et al., 2008; Bonilla et al., 2008), whereas we studied the effect of DNA damage in S phase. It was recently shown that DNA damage during S phase results in much more potent checkpoint activation than during either G1 phase or G2/M phase as a result of replication fork stalling (Zierhut and Diffley, 2008). Because we used the replication fork stalling agents MMS and HU, it is possible that checkpoint activation by stalled replication forks may either not require Cdc28 or it might result in such a strong signal that the requirement for Cdc28 is overridden.
Although Cdc28 activity alone did not appear to be involved in recovery from replication checkpoint arrest, we found that it cooperates with Mre11 to prevent mitotic catastrophe after replication arrest. Cdc28 has previously been shown to be required for the resection step of HR (Aylon et al., 2004; Ira et al., 2004; Henderson et al., 2006), but our finding that CDC28 is not epistatic to MRE11 and RAD52 (Tables I and III) indicates that Cdc28 likely has additional functions as well. We currently do not know the nature of this function, but based on the results of our GCR assays (see next section), we speculate that Cdc28 may somehow help prevent loss of damaged chromosomes, as has been suggested previously (Kitazono and Kron, 2002; Kitazono et al., 2003).
Cdc28 activity is required for formation of GCRs
We evaluated the function of CDC28 in formation of GCRs and found that Cdc28 activity directly correlated with the rate of GCR formation; augmented Cdc28 activity led to elevated GCR rates, whereas a reduction in Cdc28 activity resulted in suppression of GCRs. Reduced Cdc28 activity did not affect the rate of accumulation of mutations in the CAN1, hom3-10, or lys2-Bgl assays and did not suppress the mutator phenotype caused by an msh2 mutation, indicating that Cdc28 specifically affects the formation of GCRs. Mutations that cause defects in mitotic checkpoint functions were previously shown to suppress the formation of GCRs (Myung et al., 2004), indicating that mitotic checkpoint functions are required for the formation of GCRs, possibly by preventing loss of damaged chromosomes. We do not know the exact mechanism of suppression of GCR rates by the hypomorphic cdc28-as1 and cdc28-5M alleles, although we can exclude a role for the Cdc28 target Sae2 because deletion of SAE2 did not suppress the GCR rate of mre11Δ mutants. Although we found that Cdc28 activity by itself was not required for recovery from DNA replication arrest, it was essential when Mre11-dependent DSB repair mechanisms were defective (Fig. 4). Therefore, one explanation for the suppression of GCRs is that Cdc28 has a redundant function in the repair of damaged chromosomes and cooperates with different DNA repair pathways to promote cell viability by repairing DSBs or by healing broken chromosomes in a GCR-prone manner. For instance, Cdc28 has been found to be required for HR (Ira et al., 2004), which is error free, but in the absence of HR, the cell deploys alternative chromosome healing pathways that can give rise to GCRs (for review see Kolodner et al., 2002). A major pathway involved in the formation of GCRs is de novo telomere addition (Pennaneach et al., 2006). Interestingly, Cdc28 has several functions in the processing of telomeres (Frank et al., 2006; Vodenicharov and Wellinger, 2006; Li et al., 2009) and in telomeric recombination in telomerase-deficient cells (Grandin and Charbonneau, 2003). Consequently, inhibiting Cdc28 activity could reduce the activity of several of the major pathways that repair and heal damaged chromosomes. The damaged chromosomes might then be lost during the next cell cycle, resulting in cell death and an apparent suppression of GCR rates. A similar model has been proposed previously to explain suppression of GCRs by mutations in mitotic checkpoint genes (Myung et al., 2004). Suppression of GCR rates through loss of damaged chromosomes resulting in cell death is supported by several of our other findings, including (a) hypomorphic cdc28 alleles show strong growth defects when combined with mutations like mre11Δ and rad27Δ, which are known to cause high levels of spontaneous DSBs and increased GCR rates, (b) cdc28-5M mre11Δ double mutants have increased mitotic catastrophe after treatment with HU, and (c) mre11Δ cdc28-5M and rad27Δ cdc28-5M double mutants had defects in maintenance of minichromosomes (unpublished data), indicating that chromosomes might indeed be lost after being damaged in these mutants.
There may be additional explanations for suppression of GCRs by cdc28 alleles. For example, initiation of DNA replication is controlled by Cdc28 (Diffley, 2004), and deletion of CDC28 or overexpression of CLN2-1 induces unscheduled entry into S phase, resulting in increased GCR rates (Lengronne and Schwob, 2002; Tanaka and Diffley, 2002). Introducing seven extra copies of the autonomously replicating sequence ARSH4 between CAN1 and URA3 suppressed the GCR rates induced by CLN2-1 overexpression, and, therefore, GCRs that arise by precocious S-phase entry could be a result of poor assembly of prereplicative complexes (Tanaka and Diffley, 2002). Therefore, we hypothesized that suppression of GCRs by hypomorphic cdc28 alleles might be a result of delayed entry into S phase, allowing cells to more efficiently assemble prereplicative complexes. Interestingly, we found that the introduction of extra copies of ARSH4 into a WT strain did not reduce GCR rates (Table IV) but, in fact, resulted in a fourfold increase in the GCR rate. Extra copies of ARSH4 also increased the GCR rate of mre11Δ mutants by about fourfold (Table IV). Therefore, suppression of GCR rates by hypomorphic cdc28 alleles does not appear to be the result of improved assembly of prereplicative complexes. An alternative explanation for suppression of GCRs in strains with hypomorphic cdc28 alleles could be that these mutants fire their origins of replication less efficiently, resulting in a lower total number of replication forks and therefore a smaller chance of replication fork collapse and thus less damage that could lead to GCRs. However, this is not likely to be the mechanism of GCR suppression because, in that case, one would expect to see fewer Ddc2 foci during S phase in mutants with hypomorphic cdc28 alleles, but this is not the case (Fig. 3, A and B). Furthermore, hypomorphic cdc28 alleles did not reduce the number of Ddc2 foci in mre11Δ mutants during S phase, indicating that the amount of damage in these cells is not reduced, and, therefore, it is unlikely that suppression of GCR rates by cdc28 alleles is caused by a lower amount of DNA damage.
Table IV.
GCR rates in mutants with seven copies of ARSH4
| Strain | Relevant genotype | GCR rates (×10−10) |
| RDK3615 | WT | 3.5 (1)a |
| RDK6307 | sit1∷control DNA | <4.0 (1)b |
| RDK6308 | sit1∷7xARSH4 | 13.9 (4) |
| RDK6311 | mre11Δ sit1∷control DNA | 2,963 (847) |
| RDK6312 | mre11Δ sit1∷7xARSH4 | 12,539 (3,580) |
Numbers in parentheses indicate fold increase over WT. The GCR rates are shown as events per generation.
Data from Chen and Kolodner (1999).
GCR rate could not be accurately calculated because GCRs were not detected in a large enough proportion of the cultures.
The CDC28 genetic network
We identified genetic interactions between CDC28 and genes involved in a plethora of pathways that function in DNA damage responses and in the maintenance of genome stability. These include HR, sister chromatid cohesion, the spindle checkpoint, postreplication repair, telomere maintenance, and chromatin remodeling, underscoring the importance of Cdc28 in maintenance of genome stability. Given that Cdc28 controls a wide variety of cell cycle–regulated processes, we expect that the CDC28 genetic network is much larger than presented in this study. The finding that CDC28 genetically interacted with factors involved in HR was unexpected because Cdc28 has previously been shown to be required for HR (Aylon et al., 2004; Ira et al., 2004), and, therefore, one might have expected an epistatic relationship between Cdc28 and factors involved in HR. Our data indicate that although Cdc28 might have an important function in HR, it must also have additional functions in the maintenance of genome stability. Indeed, we found that Cdc28 may cooperate with Mre11 to prevent mitotic catastrophe after DNA damage during S phase, which is in line with a previous study that showed that Cdc28 prevents chromosome loss during mitosis (Kitazono and Kron, 2002). Our finding that Cdc28 appears to be required for formation of GCRs supports the idea that it prevents loss of damaged chromosomes, although the mechanism and the relevant targets of Cdc28 in this process still need to be revealed.
Cdks and genome stability in cancer
Evasion of antigrowth signals to allow unrestricted entry into S phase is a characteristic of cancer, as is genome instability (Hanahan and Weinberg, 2000), and the types of genome rearrangements that are seen in cancer parallel those of GCRs in S. cerevisiae (Putnam et al., 2005). Although a single Cdk (Cdc28/Cdk1) controls the cell cycle in S. cerevisiae, in mammalian cells, five Cdks related to Cdc28 (Cdk1, Cdk2, Cdk4, and Cdk6) have been implicated in driving the cell cycle. Recent genetic evidence identified Cdk1 as the main player, and although, under normal conditions, Cdk2, Cdk4, and Cdk6 may be more important for cycling of specialized cells, they appear to play a role in driving the cell cycle in tumor cells (Malumbres, 2005; Malumbres and Barbacid, 2005, 2009). Interestingly, aberrant Cdk activity induces DNA damage in mammalian cells and may contribute to genome rearrangements that are observed in cancer (Bartkova et al., 2005; Enders and Maude, 2006; Malumbres and Barbacid, 2009). Indeed, overexpression of cyclin E leads to chromosomal instability in immortalized rat embryo fibroblasts and human breast epithelial cells (Spruck et al., 1999). Our data show that aberrant Cdk activity is not just sufficient but actually also required for formation of genome rearrangements. We also found that cells with reduced Cdk activity are greatly sensitized to DNA-damaging agents, and, furthermore, they depend heavily on pathways that are involved in DNA repair such as the Me11 DSB repair pathway and the Rad6 pathway but also on intact M-phase checkpoints. Therefore, the efficacy of current cancer treatments based on γ irradiation, DNA damage–based chemotherapy, or paclitaxel-based drugs that target microtubules might be improved by combining them with broad-range CKIs. Indeed, several ongoing clinical trials are focused on Cdks and involve combination therapies (Shapiro, 2006; Malumbres and Barbacid, 2009).
In conclusion, CDC28 functions in a genetic network that preserves genome integrity. It cooperates with the MRE11 pathway in recovery from DNA replication arrest by preventing mitotic catastrophe during mitosis; however, Cdc28 is also required for the formation of GCRs. Therefore, Cdc28-mediated maintenance of cell viability during DNA damage may come at a cost: genome rearrangements.
Materials and methods
Yeast strains and plasmids
S. cerevisiae strains were grown in standard YPD medium. Strains were directly derived from the S288c strain RDKY3615 using either standard gene replacement methods or intercrossing (Table S2). To construct the cdc28-as1 strain, the coding region of cdc28-as1 was amplified from pRS306–cdc28-as1 (gift from K. Shokat, University of California, San Francisco, San Francisco, CA) and subcloned into pRS303 and pRS304. Subsequently, a PCR product spanning the coding sequences of cdc28-as1 and either HIS3 or TRP1 was used to transform RDK3615. Positive clones were identified by sensitivity to 1-NM-PP1. Sequencing revealed that two sites differed from the published sequence: A24C and T874C (resulting in Thr8Pro and Ile291Thr). These mutations did not affect sensitivity to 1-NM-PP1 (Fig. 1 and not depicted). To construct the cdc28-5M strain, a region spanning the coding sequences of cdc28-5M and TRP1 was PCR amplified from pRS304–cdc28-5M (gift from M. Cai, National University of Singapore, Singapore) and then used to transform RDK3615 to Trp+. Plasmids pRDK1293 and pRDK1294 were constructed by inserting a PCR product containing the nourseothricin resistance marker NAT-NT1 into the BamHI site of yCP50-derived plasmids harboring RAD6 and rad6-149, respectively, which were provided by M.A. Osley (University of New Mexico, Albuquerque, NM). To construct strains RDK6307 and RDK6308, the SIT1 ORF was replaced with a PCR product spanning either part of plasmid YIplac204, including the TRP1 marker or part of plasmid YIplac204–7xARSH4 linked to TRP1, respectively. Both plasmids were provided by J.D. Diffley (Cancer Research UK London Research Institute, Hertfordshire, England, UK).
Sensitivity to chronic exposure to DNA-damaging agents and genetic interaction screen
10-fold dilutions of log-phase cell cultures were spotted on YPD supplemented with drugs, as indicated in the figures and figure legends, and incubated at 30°C, and pictures were taken when colonies were visible. UV irradiation was performed using a UV cross-linker (model 2400; Stratalinker). Genetic interactions with cdc28-as1 were determined by spotting mutants on YPD or on YPD containing the indicated concentrations of 1-NM-PP1.
Cell survival after short treatments with drugs
Cells from log-phase cultures were arrested in 10 µg/ml α factor for 3 h and released into YPD for 30 min to allow cells to pass Start and enter S phase, which is a Cdc28-dependent event. As indicated, 10 µM 1-NM-PP1 (experiments involving cdc28-as1 mutants) was added for 5 min before cells were treated with 200 mM HU, 0.05% MMS, or 0.1, 0.3, or 1 µg/ml phleomycin for 2 h, as indicated; alternatively, in experiments involving cdc28-5M, cells were incubated in a 42°C water bath for 5 min to rapidly inactivate Cdc28-5M before incubating at 37°C in the presence of drugs. Cells were then washed, plated on YPD plates, and incubated at 30°C until colonies appeared. Colonies were counted, and cell survival was calculated as the percentage of the colony-forming units of untreated WT cells. To determine cell survival of 1-NM-PP1–mediated cell cycle arrest, log-phase cell cultures were arrested with 10 µM 1-NM-PP1 for 3 or 8 h before being washed and plated on YPD. Cell survival was then calculated as described above.
Determination of Ddc2 foci
Log-phase cells were arrested in 10 µg/ml α factor for 3 h and released into YPD supplemented with 15 µg/ml nocodazole (to prevent untreated cells from entering the next cell cycle) for 30 min to allow cells to enter S phase. 10 µM 1-NM-PP1 was then added for 5 min, and cells were either left untreated or treated with 1 µg/ml phleomycin for 2 h. Cells were transferred to ice until imaged. Ddc2-GFP foci in living cells were visualized in YPD at RT with an inverted microscope (Eclipse TE300; Nikon) equipped with a 100× NA 1.40 Plan-Apochromat objective lens (Nikon), using a charge-coupled device camera (Orca-ER; Hamamatsu Photonics) and MetaMorph software (MDS Analytical Technologies). Images were processed using Photoshop (Adobe) and Illustrator (Adobe). At least 100 cells were counted, and the number of cells with at least one Ddc2-GFP focus was calculated as the percentage of the total cell population of that specific sample.
GCR assays and fluctuation assays
GCR rates and fluctuation rates using the CAN1, lys2-Bgl, and the hom3-10 assays were determined as described previously (Marsischky et al., 1996; Greene and Jinks-Robertson, 1997; Tishkoff et al., 1997; Tran et al., 1997; Umezu et al., 1998; Chen and Kolodner, 1999).
Bioinformatics
Identification of components of the CDC28 genetic network involved in the preservation of genome stability was performed as described previously (Huang and Kolodner, 2005), with some modifications. In brief, a Saccharomyces Genome Database search was conducted with a query set of genes that showed the strongest genetic interactions with CDC28 (RAD53, MRE11, RAD50, RAD52, BUB2, BUB3, MAD2, CTF4, CTF18, CSM3, RAD6, RAD18, RAD5, ASF1, POL32, RAD27, and TLC1) followed by data sorting with Excel (Microsoft). This dataset was then filtered by discarding all genes having less than five genetic interactions in common with the set of query genes to remove false interactions. Predictions of genetic interactions that we already tested in our initial screen were discarded. Finally, the remaining genes were grouped according to gene ontology annotations, and those gene ontology groups that contained genes that tested negative in our initial screen for genetic interaction with CDC28 were discarded.
Western blotting
Cell pellets were resuspended in 20% TCA and disrupted by vortexing at 4°C for 15 min in the presence of glass beads. Lysates were centrifuged, pellets were washed with 70% ethanol and resuspended in Laemmli sample buffer, and the pH was neutralized using Tris base. Lysates were boiled, centrifuged, and resolved on SDS-PAGE followed by Western blotting using HRP-coupled Flag antibodies (Sigma-Aldrich).
FACS analysis
Cells were fixed in 70% ethanol for 1 h at RT, harvested by centrifugation, and resuspended in 50 mM sodium citrate buffer, pH 7.0. Cells were sonicated (three pulses of 1 s each), centrifuged, resuspended in sodium citrate buffer containing 250 µg/ml RNase A and 1 mg/ml proteinase K, and incubated overnight at 37°C. The cells were then harvested by centrifugation, resuspended in 1 ml of sodium citrate buffer containing 1 µM Sytox green (Invitrogen), and incubated at RT for at least 1 h before being analyzed by FACS.
PFGE
Overnight cultures of the indicated strains were diluted in fresh YPD and allowed to resume growth at 30°C to reach ∼107 cells/ml (hemocytometer readings). 10 ml from each culture was centrifuged, and cells were fixed with 70% ethanol for at least 1 h at RT. The remaining culture of each strain was synchronized by the addition of HU (Sigma-Aldrich) to a final concentration of 0.2 M. After incubation at 30°C for 2 h, the majority of the cells (>90%) was arrested in S phase with large buds. Aliquots were taken and fixed in 70% ethanol. The remaining culture was washed to remove HU and resuspended in fresh YPD containing 10 µg/ml α factor (Sigma-Aldrich). After incubation at 30°C (or 37°C as indicated) for 3 h, samples were collected, and cells were fixed in 70% ethanol as above. Preparation of agarose-embedded yeast DNA was performed with contour-clamped homogeneous electric field (CHEF) genomic DNA plug kit (Bio-Rad Laboratories) according to the manufacturer's instructions. 6 × 107 fixed cells were washed and set into each agarose plug. All plugs were subsequently treated with lyticase and proteinase K before loading onto an agarose separation gel (1% Megabase agarose; Bio-Rad Laboratories). PFGE was run in CHEF Mapper equipment (Bio-Rad Laboratories) in 0.5× Tris-borate-EDTA buffer at 14°C with an angle of 120°, a voltage gradient of 6 V/cm, and switch times of 60 s for 15 h and 90 s for 7 h. The gel was stained with ethidium bromide before being photographed. The relative amount of DNA fragmentation was determined as follows. Part of the fragmented DNA of subchromosomal size (Fig. S2) was quantified using the histogram tool of Photoshop and corrected for background noise. This was then normalized against the amount of DNA of chromosome XI (quantified and corrected in the same way). The mean and standard error of the mean were then calculated using data from three independent experiments. Only treatments at 30°C were analyzed because incubation at 37°C did not further increase chromosome fragmentation.
Online supplemental material
Fig. S1 shows that Cdc28 functions in the DNA damage response but is not important for survival of acute DNA damage. Fig. S2 shows an example of a PFGE gel. Fig. S3 quantifies the genetic interactions of several genes with CDC28. Table S1 displays the predicted as well as confirmed genetic interactions with CDC28 using a bioinformatics approach. Table S2 shows the yeast strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200811083/DC1.
Acknowledgments
We thank Jason Chan (Ludwig Institute for Cancer Research, University of California, San Diego School of Medicine, La Jolla, CA) for help with bioinformatics and construction of strains and Christopher Putnam, Tikvah Hayes, Stephanie Soltero, and Sandra Martinez (Ludwig Institute for Cancer Research, University of California, San Diego School of Medicine) for the construction of strains. We are grateful to Ismail Iraqui (Institut Curie, Centre Universitaire, Orsay, France) for technical assistance with PFGE.
This work was supported by a postdoctoral fellowship from the Koningin Wilhelmina Fonds Dutch Cancer Society (to J.M. Enserink), a Yngre, Fremragende Forskere young excellent researcher award from the Norwegian Research Council (to J.M. Enserink), and National Institutes of Health grant GM26017 (to R.D. Kolodner).
Footnotes
Abbreviations used in this paper: CKI, Cdk inhibitor; CPT, camptothecin; DSB, DNA double-strand break; GCR, gross chromosomal rearrangement; HR, homologous recombination; HU, hydroxyurea; MMS, methylmethanesulfonate; NHEJ, nonhomologous end joining; PFGE, pulsed-field gel electrophoresis; RPA, replication protein A; ssDNA, single-stranded DNA; WT, wild type; YPD, yeast extract/peptone/dextrose.
References
- Amon A., Surana U., Muroff I., Nasmyth K. 1992. Regulation of p34CDC28 tyrosine phosphorylation is not required for entry into mitosis in S. cerevisiae.Nature. 355:368–371 [DOI] [PubMed] [Google Scholar]
- Aylon Y., Liefshitz B., Kupiec M. 2004. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle.EMBO J. 23:4868–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberis M., De Gioia L., Ruzzene M., Sarno S., Coccetti P., Fantucci P., Vanoni M., Alberghina L. 2005. The yeast cyclin-dependent kinase inhibitor Sic1 and mammalian p27Kip1 are functional homologues with a structurally conserved inhibitory domain.Biochem. J. 387:639–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow J.H., Lisby M., Rothstein R. 2008. Differential regulation of the cellular response to DNA double-strand breaks in G1.Mol. Cell. 30:73–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J., Horejsi Z., Koed K., Kramer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J.M., Lukas C., et al. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis.Nature. 434:864–870 [DOI] [PubMed] [Google Scholar]
- Bishop A.C., Ubersax J.A., Petsch D.T., Matheos D.P., Gray N.S., Blethrow J., Shimizu E., Tsien J.Z., Schultz P.G., Rose M.D., et al. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase.Nature. 407:395–401 [DOI] [PubMed] [Google Scholar]
- Bloom J., Cross F.R. 2007. Multiple levels of cyclin specificity in cell-cycle control.Nat. Rev. Mol. Cell Biol. 8:149–160 [DOI] [PubMed] [Google Scholar]
- Bonilla C.Y., Melo J.A., Toczyski D.P. 2008. Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage.Mol. Cell. 30:267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Kolodner R.D. 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants.Nat. Genet. 23:81–85 [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O., Koshland D. 1997. The anaphase inhibitor of Saccharomyces cerevisiae Pds1p is a target of the DNA damage checkpoint pathway.Proc. Natl. Acad. Sci. USA. 94:14361–14366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix O., Koshland D. 1999. Pds1p of budding yeast has dual roles: inhibition of anaphase initiation and regulation of mitotic exit.Genes Dev. 13:1950–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J.F. 2004. Regulation of early events in chromosome replication.Curr. Biol. 14:R778–R786 [DOI] [PubMed] [Google Scholar]
- Dotiwala F., Haase J., Arbel-Eden A., Bloom K., Haber J.E. 2007. The yeast DNA damage checkpoint proteins control a cytoplasmic response to DNA damage.Proc. Natl. Acad. Sci. USA. 104:11358–11363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enders G.H., Maude S.L. 2006. Traffic safety for the cell: influence of cyclin-dependent kinase activity on genomic stability.Gene. 371:1–6 [DOI] [PubMed] [Google Scholar]
- Enserink J.M., Smolka M.B., Zhou H., Kolodner R.D. 2006. Checkpoint proteins control morphogenetic events during DNA replication stress in Saccharomyces cerevisiae.J. Cell Biol. 175:729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank C.J., Hyde M., Greider C.W. 2006. Regulation of telomere elongation by the cyclin-dependent kinase CDK1.Mol. Cell. 24:423–432 [DOI] [PubMed] [Google Scholar]
- Gibson D.G., Aparicio J.G., Hu F., Aparicio O.M. 2004. Diminished S-phase cyclin-dependent kinase function elicits vital Rad53-dependent checkpoint responses in Saccharomyces cerevisiae.Mol. Cell. Biol. 24:10208–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandin N., Charbonneau M. 2003. Mitotic cyclins regulate telomeric recombination in telomerase-deficient yeast cells.Mol. Cell. Biol. 23:9162–9177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S., Chapman J.R., Magill C., Jackson S.P. 2008. DNA helicases Sgs1 and BLM promote DNA double-strand break resection.Genes Dev. 22:2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene C.N., Jinks-Robertson S. 1997. Frameshift intermediates in homopolymer runs are removed efficiently by yeast mismatch repair proteins.Mol. Cell. Biol. 17:2844–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. 2000. The hallmarks of cancer.Cell. 100:57–70 [DOI] [PubMed] [Google Scholar]
- Hartwell L.H., Weinert T.A. 1989. Checkpoints: controls that ensure the order of cell cycle events.Science. 246:629–634 [DOI] [PubMed] [Google Scholar]
- Hasty P., Campisi J., Hoeijmakers J., van Steeg H., Vijg J. 2003. Aging and genome maintenance: lessons from the mouse? Science. 299:1355–1359 [DOI] [PubMed] [Google Scholar]
- Henderson K.A., Kee K., Maleki S., Santini P.A., Keeney S. 2006. Cyclin-dependent kinase directly regulates initiation of meiotic recombination.Cell. 125:1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M.E., Kolodner R.D. 2005. A biological network in Saccharomyces cerevisiae prevents the deleterious effects of endogenous oxidative DNA damage.Mol. Cell. 17:709–720 [DOI] [PubMed] [Google Scholar]
- Huertas P., Cortes-Ledesma F., Sartori A.A., Aguilera A., Jackson S.P. 2008. CDK targets Sae2 to control DNA-end resection and homologous recombination.Nature. 455:689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W.W., Venkatasubrahmanyam S., Ianculescu A.G., Tong A., Boone C., Madhani H.D. 2003. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control.Mol. Cell. 11:261–266 [DOI] [PubMed] [Google Scholar]
- Ira G., Pellicioli A., Balijja A., Wang X., Fiorani S., Carotenuto W., Liberi G., Bressan D., Wan L., Hollingsworth N.M., et al. 2004. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1.Nature. 431:1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg D.R. 2003. Wee1-dependent mechanisms required for coordination of cell growth and cell division.J. Cell Sci. 116:4883–4890 [DOI] [PubMed] [Google Scholar]
- Kitazono A.A., Kron S.J. 2002. An essential function of yeast cyclin-dependent kinase Cdc28 maintains chromosome stability.J. Biol. Chem. 277:48627–48634 [DOI] [PubMed] [Google Scholar]
- Kitazono A.A., Garza D.A., Kron S.J. 2003. Mutations in the yeast cyclin-dependent kinase Cdc28 reveal a role in the spindle assembly checkpoint.Mol. Genet. Genomics. 269:672–684 [DOI] [PubMed] [Google Scholar]
- Kolodner R.D., Putnam C.D., Myung K. 2002. Maintenance of genome stability in Saccharomyces cerevisiae.Science. 297:552–557 [DOI] [PubMed] [Google Scholar]
- Krishnan V., Nirantar S., Crasta K., Cheng A.Y., Surana U. 2004. DNA replication checkpoint prevents precocious chromosome segregation by regulating spindle behavior.Mol. Cell. 16:687–700 [DOI] [PubMed] [Google Scholar]
- Lengronne A., Schwob E. 2002. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1).Mol. Cell. 9:1067–1078 [DOI] [PubMed] [Google Scholar]
- Leroy C., Lee S.E., Vaze M.B., Ochsenbien F., Guerois R., Haber J.E., Marsolier-Kergoat M.C. 2003. PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break.Mol. Cell. 11:827–835 [DOI] [PubMed] [Google Scholar]
- Lew D.J. 2003. The morphogenesis checkpoint: how yeast cells watch their figures.Curr. Opin. Cell Biol. 15:648–653 [DOI] [PubMed] [Google Scholar]
- Li S., Makovets S., Matsuguchi T., Blethrow J.D., Shokat K.M., Blackburn E.H. 2009. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression.Cell. 136:50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Cai M. 1997. Inactivation of the cyclin-dependent kinase Cdc28 abrogates cell cycle arrest induced by DNA damage and disassembly of mitotic spindles in Saccharomyces cerevisiae.Mol. Cell. Biol. 17:2723–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberi G., Chiolo I., Pellicioli A., Lopes M., Plevani P., Muzi-Falconi M., Foiani M. 2000. Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and Cdk1 activity.EMBO J. 19:5027–5038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M., Barlow J.H., Burgess R.C., Rothstein R. 2004. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins.Cell. 118:699–713 [DOI] [PubMed] [Google Scholar]
- Lopes M., Cotta-Ramusino C., Pellicioli A., Liberi G., Plevani P., Muzi-Falconi M., Newlon C.S., Foiani M. 2001. The DNA replication checkpoint response stabilizes stalled replication forks.Nature. 412:557–561 [DOI] [PubMed] [Google Scholar]
- Majka J., Niedziela-Majka A., Burgers P.M. 2006. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint.Mol. Cell. 24:891–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M. 2005. Revisiting the “Cdk-centric” view of the mammalian cell cycle.Cell Cycle. 4:206–210 [DOI] [PubMed] [Google Scholar]
- Malumbres M., Barbacid M. 2005. Mammalian cyclin-dependent kinases.Trends Biochem. Sci. 30:630–641 [DOI] [PubMed] [Google Scholar]
- Malumbres M., Barbacid M. 2009. Cell cycle, CDKs and cancer: a changing paradigm.Nat. Rev. Cancer. 9:153–166 [DOI] [PubMed] [Google Scholar]
- Marsischky G.T., Filosi N., Kane M.F., Kolodner R. 1996. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair.Genes Dev. 10:407–420 [DOI] [PubMed] [Google Scholar]
- Mimitou E.P., Symington L.S. 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing.Nature. 455:770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K., Kolodner R.D. 2002. Suppression of genome instability by redundant S-phase checkpoint pathways in Saccharomyces cerevisiae.Proc. Natl. Acad. Sci. USA. 99:4500–4507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K., Chen C., Kolodner R.D. 2001a. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae.Nature. 411:1073–1076 [DOI] [PubMed] [Google Scholar]
- Myung K., Datta A., Chen C., Kolodner R.D. 2001b. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination.Nat. Genet. 27:113–116 [DOI] [PubMed] [Google Scholar]
- Myung K., Datta A., Kolodner R.D. 2001c. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae.Cell. 104:397–408 [DOI] [PubMed] [Google Scholar]
- Myung K., Pennaneach V., Kats E.S., Kolodner R.D. 2003. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability.Proc. Natl. Acad. Sci. USA. 100:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K., Smith S., Kolodner R.D. 2004. Mitotic checkpoint function in the formation of gross chromosomal rearrangements in Saccharomyces cerevisiae.Proc. Natl. Acad. Sci. USA. 101:15980–15985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill B.M., Szyjka S.J., Lis E.T., Bailey A.O., Yates J.R., III, Aparicio O.M., Romesberg F.E. 2007. Pph3-Psy2 is a phosphatase complex required for Rad53 dephosphorylation and replication fork restart during recovery from DNA damage.Proc. Natl. Acad. Sci. USA. 104:9290–9295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Ye P., Yuan D.S., Wang X., Bader J.S., Boeke J.D. 2006. A DNA integrity network in the yeast Saccharomyces cerevisiae.Cell. 124:1069–1081 [DOI] [PubMed] [Google Scholar]
- Paulovich A.G., Margulies R.U., Garvik B.M., Hartwell L.H. 1997. RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage.Genetics. 145:45–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C.E., Edamura K.N., Cleary J.D. 2005. Repeat instability: mechanisms of dynamic mutations.Nat. Rev. Genet. 6:729–742 [DOI] [PubMed] [Google Scholar]
- Pennaneach V., Kolodner R.D. 2004. Recombination and the Tel1 and Mec1 checkpoints differentially effect genome rearrangements driven by telomere dysfunction in yeast.Nat. Genet. 36:612–617 [DOI] [PubMed] [Google Scholar]
- Pennaneach V., Putnam C.D., Kolodner R.D. 2006. Chromosome healing by de novo telomere addition in Saccharomyces cerevisiae.Mol. Microbiol. 59:1357–1368 [DOI] [PubMed] [Google Scholar]
- Peter M., Herskowitz I. 1994. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1.Science. 265:1228–1231 [DOI] [PubMed] [Google Scholar]
- Putnam C.D., Pennaneach V., Kolodner R.D. 2005. Saccharomyces cerevisiae as a model system to define the chromosomal instability phenotype.Mol. Cell. Biol. 25:7226–7238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robzyk K., Recht J., Osley M.A. 2000. Rad6-dependent ubiquitination of histone H2B in yeast.Science. 287:501–504 [DOI] [PubMed] [Google Scholar]
- Sanchez Y., Bachant J., Wang H., Hu F., Liu D., Tetzlaff M., Elledge S.J. 1999. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms.Science. 286:1166–1171 [DOI] [PubMed] [Google Scholar]
- Santocanale C., Diffley J.F. 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication.Nature. 395:615–618 [DOI] [PubMed] [Google Scholar]
- Schwob E., Bohm T., Mendenhall M.D., Nasmyth K. 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae.Cell. 79:233–244 [DOI] [PubMed] [Google Scholar]
- Shapiro G.I. 2006. Cyclin-dependent kinase pathways as targets for cancer treatment.J. Clin. Oncol. 24:1770–1783 [DOI] [PubMed] [Google Scholar]
- Sleigh M.J. 1976. The mechanism of DNA breakage by phleomycin in vitro.Nucleic Acids Res. 3:891–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka M.B., Chen S.H., Maddox P.S., Enserink J.M., Albuquerque C.P., Wei X.X., Desai A., Kolodner R.D., Zhou H. 2006. An FHA domain-mediated protein interaction network of Rad53 reveals its role in polarized cell growth.J. Cell Biol. 175:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger P.K., Murray A.W. 1992. S-phase feedback control in budding yeast independent of tyrosine phosphorylation of p34cdc28.Nature. 355:365–368 [DOI] [PubMed] [Google Scholar]
- Spruck C.H., Won K.A., Reed S.I. 1999. Deregulated cyclin E induces chromosome instability.Nature. 401:297–300 [DOI] [PubMed] [Google Scholar]
- Sung P., Prakash S., Prakash L. 1988. The RAD6 protein of Saccharomyces cerevisiae polyubiquitinates histones, and its acidic domain mediates this activity.Genes Dev. 2:1476–1485 [DOI] [PubMed] [Google Scholar]
- Symington L.S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair.Microbiol. Mol. Biol. Rev. 66:630–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Diffley J.F. 2002. Deregulated G1-cyclin expression induces genomic instability by preventing efficient pre-RC formation.Genes Dev. 16:2639–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff D.X., Filosi N., Gaida G.M., Kolodner R.D. 1997. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair.Cell. 88:253–263 [DOI] [PubMed] [Google Scholar]
- Tran H.T., Keen J.D., Kricker M., Resnick M.A., Gordenin D.A. 1997. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants.Mol. Cell. Biol. 17:2859–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu K., Sugawara N., Chen C., Haber J.E., Kolodner R.D. 1998. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism.Genetics. 148:989–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodenicharov M.D., Wellinger R.J. 2006. DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase.Mol. Cell. 24:127–137 [DOI] [PubMed] [Google Scholar]
- Wood A., Krogan N.J., Dover J., Schneider J., Heidt J., Boateng M.A., Dean K., Golshani A., Zhang Y., Greenblatt J.F., et al. 2003. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter.Mol. Cell. 11:267–274 [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Guacci V., Koshland D. 1996a. Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae.J. Cell Biol. 133:85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Guacci V., Koshland D. 1996b. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s).J. Cell Biol. 133:99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Chung W.H., Shim E.Y., Lee S.E., Ira G. 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends.Cell. 134:981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut C., Diffley J.F. 2008. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response.EMBO J. 27:1875–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]