Abstract
We have established that two homologous nucleoporins, Nup170p and Nup157p, play an essential role in the formation of nuclear pore complexes (NPCs) in Saccharomyces cerevisiae. By regulating their synthesis, we showed that the loss of these nucleoporins triggers a decrease in NPCs caused by a halt in new NPC assembly. Preexisting NPCs are ultimately lost by dilution as cells grow, causing the inhibition of nuclear transport and the loss of viability. Significantly, the loss of Nup170p/Nup157p had distinct effects on the assembly of different architectural components of the NPC. Nucleoporins (nups) positioned on the cytoplasmic face of the NPC rapidly accumulated in cytoplasmic foci. These nup complexes could be recruited into new NPCs after reinitiation of Nup170p synthesis, and may represent a physiological intermediate. Loss of Nup170p/Nup157p also caused core and nucleoplasmically positioned nups to accumulate in NPC-like structures adjacent to the inner nuclear membrane, which suggests that these nucleoporins are required for formation of the pore membrane and the incorporation of cytoplasmic nups into forming NPCs.
Introduction
Molecules travel across the nuclear envelope (NE) through nuclear pore complexes (NPCs). The NPC (∼66 MD in yeast) possesses eightfold rotational symmetry perpendicular to the plane of the NE and can be divided in three parts: a central core or annulus, the nuclear basket, and cytoplasmic filaments. Structural analyses have revealed that the central core consists of distinct cytoplasmic, luminal spokes, and nucleoplasmic rings (Yang et al., 1998; Beck et al., 2004; Beck et al., 2007). Movement of macromolecules through the NPC is controlled by nuclear transport receptors, which bind to nuclear localization signals (NLSs) or nuclear export signals on these molecules. Karyopherins (kaps) are a family of structurally related proteins, of which there are 14 in yeast, that bind to different signals and function as importers (importins) or exporters (exportins; Wozniak et al., 1998). Kaps interact with a subset of NPC proteins (termed nucleoporins [nups]) containing phenylalanine-glycine repeats (termed FG-nups) that populate the translocation channel. Distinct models suggest that the FG-nups form a hydrophobic barrier (hydrogel; Frey et al., 2006; Frey and Gorlich, 2007) or, by the rapid Brownian motion of unstructured FG domains, a virtual gate (Rout et al., 2003). By binding FG-nups, kaps overcome this barrier and progress through the channel.
The FG-nups are attached to the symmetrical core that forms the architectural framework of the NPC. The core is composed largely of nups that lack FG repeats (Hetzer et al., 2005; Tran and Wente, 2006). Several of these are transmembrane proteins (termed poms), which probably function by anchoring the NPC to the NE. The structural organization of nups that contribute to the rings and spokes of the NPC has been investigated using various biochemical, genetic, and cytological techniques (for review see Suntharalingam and Wente, 2003). Seminal contributions have included the isolation and characterization of subcomplexes. In yeast, these include the heptameric Nup84p subcomplex, the Nup188p–Nic96p–Pom152p subcomplex, and the Nup170p–Nup53p–Nup59p subcomplex (Nehrbass et al., 1996; Marelli et al., 1998; Lutzmann et al., 2002; Lutzmann et al., 2005). Recently, through an elegant biochemical and computational study, Alber et al. (2007a,b) proposed a structure of the NPC that provides a view of how nups are likely organized within the NPC. A growing body of crystal structures of nups, among them the structure of a Sec13–Nup145 complex (Hsia et al., 2007), have also led to predictions for the structure of the NPC. These varied approaches are likely to provide a high-resolution map of the NPC core in the near future.
The assembly of subcomplexes likely plays a role in the formation of mature NPCs. This process occurs both when the NE is intact and during NE reassembly in the final stages of mitosis. Postmitotic NPC assembly occurs in organisms that undergo an open mitosis, and at the end of mitosis, when membrane vesicles and NPC components are recruited back to the chromatin surface. Studies using in vitro Xenopus NE assembly assays and tissue culture cells show that NPC formation occurs through the ordered assembly of nup subcomplexes on the chromatin surface together with coordinated interactions with the NE membrane (for reviews see Hetzer et al., 2005; Tran and Wente, 2006). First, the Nup107–160 subcomplex (vertebrate equivalent of the yeast Nup84p subcomplex) is recruited to chromatin through its interactions with ELYS/Mel28 (Rasala et al., 2006, 2008; Franz et al., 2007; Gillespie et al., 2007). These prepores act as binding sites for membrane vesicles containing Ndc1 and Pom121 (Antonin et al., 2005; Rasala et al., 2008). This step is functionally linked to the incorporation of Nup155 and Nup53 (Franz et al., 2005; Hawryluk-Gara et al., 2008) and precedes a cascade of events that ultimately leads to the formation of mature NPCs.
NPCs are also formed across an intact NE. In higher eukaryotic cells, the number of NPCs appears to double during S phase (Maul et al., 1972). Accordingly, NPC assembly has been observed across an intact NE of nuclei reconstituted in vitro using Xenopus egg extracts and in vivo using HeLa cells (D'Angelo et al., 2006). The assembly of NPCs across the NE is likely the only mechanism functioning in unicellular eukaryotes that undergo closed mitoses (i.e., the NE remains intact during mitosis). In Saccharomyces cerevisiae, the number of NPCs increases throughout the cell cycle (Winey et al., 1997), allowing NPC numbers to be maintained in both the mother and daughter. A recent study suggests that the process may be more complicated, with new NPC formation being the primary source of NPCs in the daughter nucleus (Shcheprova et al., 2008).
A requisite step in the formation of NPCs across the NE is the fusion of the inner and outer nuclear membranes to form transcisternal nuclear pores. Poms and nups located close to the membrane are likely to play a role in this and accompanying steps of NPC assembly. In higher eukaryotes, three poms—gp210, Pom121, and Ndc1—have each been implicated in NPC assembly (Antonin et al., 2005; Mansfeld et al., 2006; Stavru et al., 2006a,b; Funakoshi et al., 2007). Three S. cerevisiae poms—Pom152p, Pom34p, and Ndc1p—also appear to share a redundant function in NPC assembly (Chial et al., 1998; Lau et al., 2004; Madrid et al., 2006; Miao et al., 2006). In addition to poms, analyses of several yeast nups have lead to the conclusion that they are required for NPC assembly. Mutants in NIC96, NUP192, or NSP1 cause a decrease in NPC numbers (Mutvei et al., 1992; Zabel et al., 1996; Kosova et al., 1999; Gomez-Ospina et al., 2000). Studies using Xenopus egg extracts have also implicated nuclear and cytoplasmic pools of the Nup107–160 subcomplex in forming NPCs within intact nuclei (D'Angelo et al., 2006). Intriguingly, NPC assembly also employs soluble components of the nuclear transport machinery including Ran, Kap121p, and Kap95p (Lusk et al., 2002; Ryan et al., 2003, 2007; D'Angelo et al., 2006).
The NUP170 gene family is conserved among eukaryotes and is essential for growth in various species (Gigliotti et al., 1998; Kiger et al., 1999; Galy et al., 2003). The S. cerevisiae genome encodes two homologous genes—NUP170 and NUP157—one or the other of which is required for growth (Aitchison et al., 1995). Both genetic (Aitchison et al., 1995; Tcheperegine et al., 1999; Miao et al., 2006) and structural studies (Alber et al., 2007a,b) suggest that Nup170p and Nup157p are located close to the pore membrane, where they could interact with poms and the membrane. Nup170p and Nup157p are predicted to contain β-propeller and α-solenoid domains that are reminiscent of membrane coat proteins such as clathrin (Devos et al., 2004, 2006). To uncover their functions, we constructed a conditional lethal strain that allowed repression of NUP170 in the absence of NUP157. We found that loss of these proteins caused a decrease in NPC density due to a defect in new NPC assembly. In the absence of Nup170p and Nup157p, newly synthesized nups accumulated in cytoplasmic foci and inner nuclear membrane-associated structures, which is suggestive of stalled NPC assembly intermediates. Importantly, reinitiation of Nup170p synthesis induces the dissociation of the cytoplasmic foci and their incorporation into the NE. We conclude that Nup170p and Nup157p are required for the assembly of subcomplexes within the forming NPC at a step that likely coincides with the fusion of the outer and inner nuclear membranes and the formation of the nuclear pore.
Results
Loss of Nup170p and Nup157p causes a decrease in the number of the NPCs
One of the major lineages that comprise the S. cerevisiae complex shares a common ancestor that underwent a whole genome duplication (Scannell et al., 2007). The duplicated S. cerevisiae genome retains ∼551 duplicate pairs, including the nup genes NUP170 and NUP157. Nup170p and Nup157p have both redundant and distinct functions (Aitchison et al., 1995; Kerscher et al., 2001). Neither gene is essential for growth, but deletion of the pair is lethal, which suggests that they share essential functions.
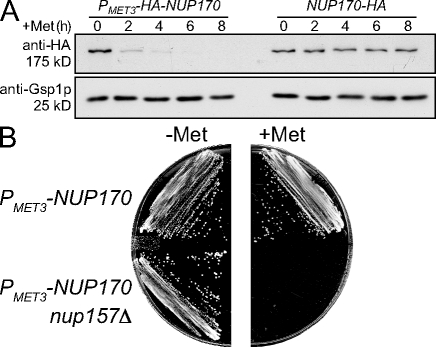
To investigate the molecular basis underlying the synthetic lethality of nup170 and nup157 null alleles, the endogenous NUP170 promoter was replaced by a repressible MET3 promoter in backgrounds containing or lacking NUP157 (PMET3-NUP170 NUP157 and PMET3-NUP170 nup157Δ). In the absence of methionine, the MET3 promoter is active, leading to levels of Nup170p that are similar to those of wild-type (WT) cells (Fig. 1 A). Upon addition of methionine, the promoter is repressed and levels of Nup170p decrease. By 4 h after methionine addition, Nup170p levels are barely detectable (Fig. 1 A). In strains containing Nup157p (PMET3-NUP170 NUP157), depletion of Nup170p had little effect on growth. However, strains lacking Nup157p and depleted of Nup170p (which we will refer to as depleted of Nup170p [nup157Δ]) failed to form colonies on plates (Fig. 1 B).
Figure 1.
Depletion of Nup170p in the nup157Δ background causes a defect in cell growth. (A) The strains TMY1098 (PMET3-HA-NUP170) and TMY1269 (NUP170-HA) were grown in medium lacking methionine. Methionine was added to induce NUP170 shut-off and incubated for the indicated times. Samples were analyzed by Western blotting using anti-HA and anti-Gsp1p (load control) antibodies. (B) The yeast strains TMY1098 (PMET3-HA-NUP170) and TMY1126 (PMET3-HA-NUP170 nup157Δ) were streaked on CM (−Met) and CM (+Met) plates, and grown for 2 d at 30°C.
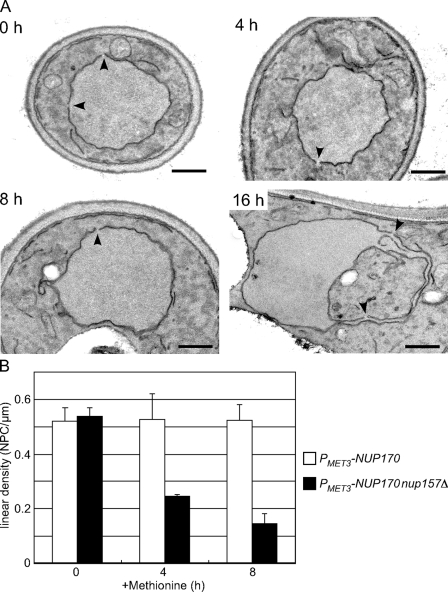
To understand the functional basis for the growth inhibition observed in the absence of Nup170p and Nup157p, we examined the morphology of the NE after repression of NUP170 in the PMET3-NUP170 nup157Δ strain. Samples were harvested at various time points after the addition of methionine, postfixed with potassium permanganate, and analyzed by thin-section transmission EM (TEM). With this method, membranes are darkly stained while proteinaceous structures are not highlighted. Thus, the NE membrane is clearly visible and nuclear pores appear as gaps of defined size in the continuity of the NE. WT or cells depleted of only Nup170p (PMET3-NUP170 NUP157) contained largely spherical nuclei with visible nuclear pores (unpublished data). Similarly, PMET3-NUP170 nup157Δ cells showed no striking changes in the shape or size of their nuclei through 8 h after the addition of methionine (Fig. 2 A). However, these nuclei contained fewer pores. We quantified the number of NPCs by counting the number of pores per micrometer of linear distance along the NE in multiple sections. A linear density of 0.54 ± 0.03 pores/µm was determined for the PMET3-NUP170 nup157Δ strain at 0 h, which was similar to that of the PMET3-NUP170 NUP157 strain (0.52 ± 0.05 pores/µm). As shown in Fig. 2 B, by 4 and 8 h after repressing NUP170 (nup157Δ), we detected a progressive decrease in NPC density to 0.24 ± 0.01 pores/µm and 0.15 ± 0.03 pores/µm, respectively. By later time points (16 h), gross morphological changes were observed in the nuclei, including the formation of NE projections and invaginations (Fig. 2 A). We speculate that these changes occur in response to decreased levels of NPCs.
Figure 2.
Depletion of Nup170p (nup157Δ) leads to a progressive decrease in the density of NPCs. The strain TMY1126 was grown in media lacking methionine. Methionine was added to repress NUP170 expression, and cells were processed for examination by TEM. (A) Typical TEM images of TMY1126 at the indicated times after methionine addition. Arrowheads indicate gaps in the NE of a size consistent with NPCs. Note that by 16 h, the NE becomes distorted, with this section showing a large invagination of the cytoplasm. Bars, 0.5 µm. (B) The number of NPCs was counted in each cell section and divided by the length of the NE to calculate a linear density of NPCs. A minimum of 30 cells was examined in each experiment, and standard deviations were estimated from three independent experiments. White bar, TMY1098; shaded bar, TMY1126.
Depletion of Nup170p and Nup157p is accompanied by a progressive loss of nuclear transport
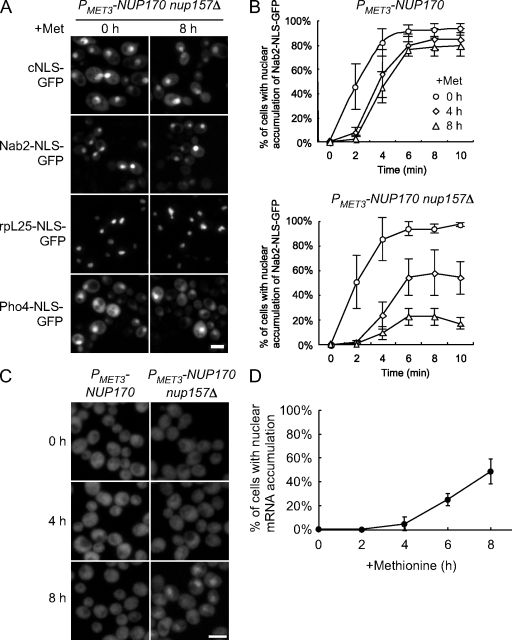
As described in the previous section, by 4 and 8 h after repression of NUP170 (nup157Δ), the numbers of NPCs are reduced to one half (4 h) and one quarter (8 h) those observed in WT strains. To investigate the effects of depletion on nuclear transport, we examined the localization of reporter proteins imported by Kap121p (Pho4-NLS-GFP; Kaffman et al., 1998), Kap123p (rpL25-NLS-GFP; Timney et al., 2006), Kap104p (Nab2-NLS-GFP; Lee and Aitchison, 1999), and the Kap60p/Kap95p heterodimer (cNLS-GFP; Shulga et al., 1996). All continued to accumulate within nuclei 6 h after methionine addition (Fig. 3 A). Nuclear import rates are generally much faster than needed to achieve good steady-state nuclear localization. Therefore, we used an assay described by Shulga et al. (1996) that provides a relative measure of in vivo import rates. As shown in Fig. 3 B, the rates of Nab2-NLS-GFP import slowed between 0 and 8 h after Nup170p (nup157Δ) depletion. Interestingly, the relative rates of import decreased by ∼50% and ∼75% after 4 and 8 h of depletion, which mirrors the decrease in NPCs observed at these time points. In contrast, depletion of Nup170p alone did not significantly alter the import rate of the Nab2-NLS-GFP reporter, although some lag was seen at early time points (Fig. 3 B). The lag may be related to the increased permeability of nup170Δ NPCs to the relatively small Nab2-NLS-GFP (Shulga et al., 2000).
Figure 3.
Decrease in NPC density caused by the depletion of Nup170p (nup157Δ) is accompanied by a decrease in the rate of protein import and mRNA export. (A) TMY1126 cells harboring plasmids encoding the cNLS-GFP, Nab2-NLS-GFP, rpL25-NLS-GFP, and Pho4-NLS-GFP reporters were transferred to media containing methionine for the indicated times to repress NUP170 expression, then examined using an epifluorescence microscope. Bar, 5 µm. (B) Analysis of Nab2-NLS–mediated import. Strains of TMY1098 and TMY1126 harboring a Nab2-NLS-GFP plasmid were grown in media containing methionine for the indicated times. A relative measure of in vivo import rates of the Nab2-NLS-GFP reporter was examined. Standard deviations estimated from at least four independent measurements are shown as error bars. (C) NUP170 expression was repressed in the TMY1098 and TMY1126 strains, and, at the indicated times, the localization of poly-A mRNA was examined by FISH analysis using an oligo (dT50) probe. Bar, 5 µm. (D) For TMY1126 (PMET3-NUP170 nup157Δ), the number of cells that showed a nuclear accumulation of poly-A mRNA at each time point was determined. A minimum of 200 cells was counted in each experiment and standard deviations were estimated from two independent experiments.
We also examined the effects of the loss of Nup170p and Nup157p on mRNA export. FISH analysis using an oligo (dT50) probe showed that the number of cells that accumulated poly-A mRNA in their nuclei increased upon repression of NUP170 (nup157Δ) but not NUP170 alone (Fig. 3 C). Over 50% of the NUP170 (nup157Δ) cells showed nuclear mRNA accumulation after 8 h of NUP170 repression (Fig. 3 D). We conclude that the defects in mRNA export and protein import observed upon loss of Nup170p and Nup157p occur as a consequence of NPC depletion.
The loss of Nup170p and Nup157p causes the mislocalization of other nups
The reduction in NPCs detected upon depletion of Nup170p and Nup157p is likely to arise from an inhibition of new NPC assembly, either exclusively or in conjunction with the destabilization of existing NPCs. To more closely evaluate the role of these nups in NPC assembly, we examined the consequences of depleting Nup170p and Nup157p on the association of individual nups with the NE. To visualize each nup, a GFP tag was introduced at the 3′ end of its open reading frame (ORF). In most cases, the NUP-GFP allele did not affect cell growth. However, strains containing the GLE2-GFP, NUP192-GFP, NUP57-GFP, NUP49-GFP, NSP1-GFP, and NIC96-GFP alleles exhibited severe growth defects upon depletion of Nup170p even in the presence of Nup157p (unpublished data), which suggests that the C-terminal GFP compromises the function of these nups and causes a synthetically lethal interaction with reduced NUP170 expression. As a consequence, these nups were omitted from further analysis.
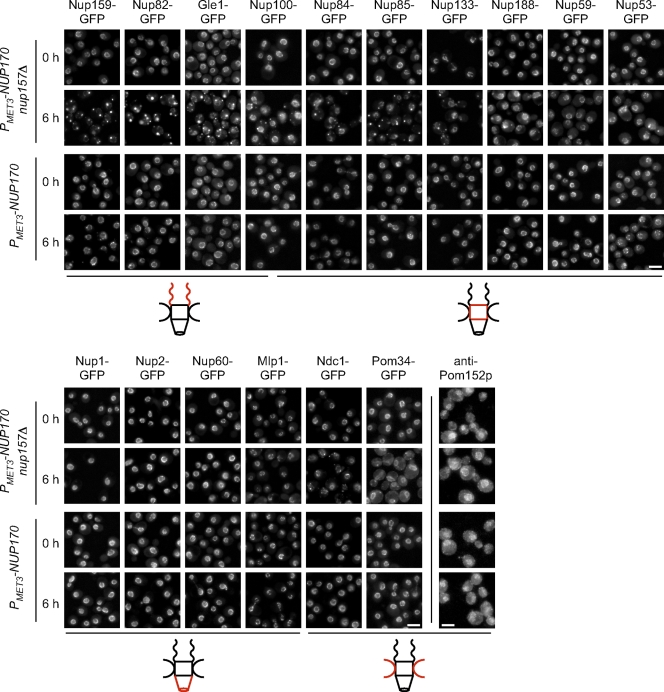
The localization of each nup-GFP fusion was examined 6 h after methionine-induced repression of NUP170. Nups belonging to various subdomains of the NPC were examined, including those positioned on the cytoplasmic or nucleoplasmic face (here referred to as cytoplasmic or nucleoplasmic nups), and within the core, integral membrane proteins. The most dramatic effects of Nup170p (nup157Δ) depletion were on nups positioned exclusively on the cytoplasmic face of the NPC, including Nup82-GFP and Nup159-GFP. Upon depletion of Nup170p (nup157Δ; 6 h), the NE localization of Nup82-GFP and Nup159-GFP was largely lost, and the proteins were found in foci located in the cytoplasm and occasionally adjacent the NE (Fig. 4). Generally, 3–7 foci were visible per cell. In contrast, the nucleoplasmic nups Nup1-GFP, Nup2-GFP, Nup60-GFP, and Mlp1-GFP, and the membrane protein Pom152p retained their nuclear rim localization over the same time period (Fig. 4). Finally, nups that contribute to the central core of the NPC showed a mixed phenotype upon depletion of Nup170p (nup157Δ). For example, components of the Nup84p complex (Nup84-GFP, Nup85-GFP, and Nup133-GFP), Gle1-GFP, Nup100-GFP, and Ndc1-GFP were still visible at the NE but generally exhibited a weaker signal than seen at the time of 0 h. In addition, these nups were also detected in punctate cytoplasmic structures (Fig. 4). Cytoplasmic foci were also visible, but less abundant, when we examined a group of more centrally located core nups and poms including Nup188-GFP, Nup59-GFP, Nup53-GFP, and Pom34-GFP (Fig. 4). Each of these retained an NE signal at 6 h after Nup170p (nup157Δ) depletion.
Figure 4.
The loss of Nup170p and Nup157p has distinct effects on the localization of NPC components. (A) The endogenous gene encoding the indicated nup was tagged with GFP in the TMY1098 and TMY1126 strains, and the fusion proteins were examined at the indicated times after repression of NUP170 expression using an epifluorescence microscope. Pom152p localization was analyzed by indirect immunofluorescence using an anti-Pom152p antibody (mAb118C3). Note, the brighter Ndc1-GFP foci of seen at the NE at 0 h likely reflect the shared spindle pole body association of Ndc1p (Chial et al., 1998). Diagrams adjacent to the images depict the general localization of each nup within the NPC (red). Nup100p is present on both faces of the NPC but is biased to the cytoplasm. Bars, 5 µm.
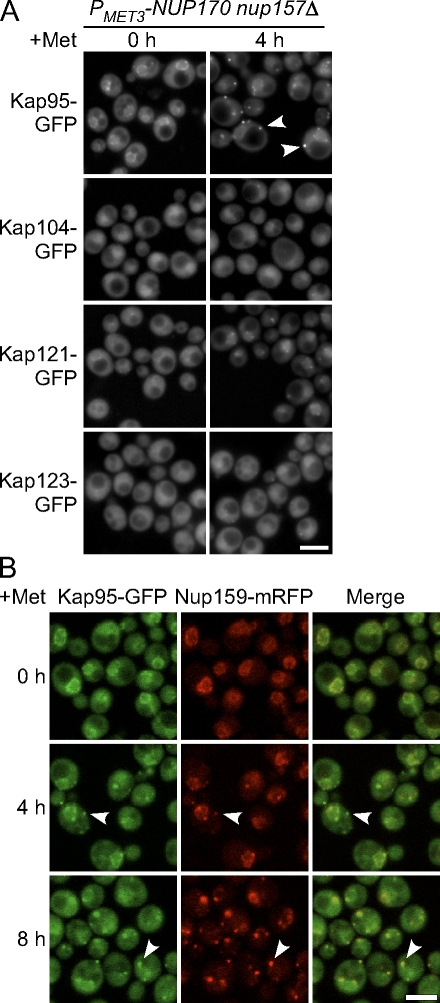
To determine whether the mislocalized nups coassembled into foci with other nups, we examined the localization of Nup159–monomeric RFP (mRFP) in cells coproducing Nup188-GFP, Nup82-GFP, Nup2-GFP, or Ndc1-GFP. All of the double-tagged strains grew normally when either Nup157p or Nup170p was present, which suggests that the tags did not alter the functions of these nups. 4 h after addition of methionine, both Nup159-mRFP and Nup82-GFP colocalized to the same cytoplasmic foci adjacent the NE (Fig. 5). In contrast, Nup188-GFP, Nup2-GFP, and Ndc1-GFP were generally restricted to the NE at the 4-h time point (Fig. 5). By 8 h, Nup188-GFP and Ndc1-GFP began to accumulate in cytoplasmic foci (Fig. 5). The Nup188-GFP cytoplasmic foci generally colocalized with Nup159-mRFP, which suggests that Nup188-GFP can associate with structures containing the cytoplasmic nups. Similarly, overlapping Ndc1-GFP and Nup159-mRFP spots were detected. However, distinct Ndc1-GFP and Nup159-mRFP foci were also seen: the Ndc1-GFP–specific foci that likely represent spindle pole body–associated protein, and the Nup159-mRFP–specific foci perhaps arising from their positioning away from membrane containing Ndc1-GFP. Finally, little or no colocalization of Nup2-GFP and Nup159-mRFP was detected in cells at 4 or 8 h after depletion (Fig. 5). These results suggest that the depletion of Nup170p (nup157Δ) causes the mislocalization of cytoplasmically positioned nups into foci, which at later time points also contain detectable amounts of core nups and certain poms.
Figure 5.
Cytoplasmic foci contain multiple nups. Strains expressing Nup159-mRFP and either Nup188-GFP, Nup2-GFP, or Ndc1-GFP within the PMET3-NUP170 nup157Δ background were examined at the indicated times after repression of NUP170 expression using a confocal fluorescence microscope. It should be noted that the foci of Ndc1-GFP and Nup188-GFP generally colocalize with those of Nup159-mRFP (arrows) but not vice versa (arrowheads). Bars, 5 µm.
The distributions of certain importins were also affected by Nup170p (nup157Δ) depletion (Fig. 6). By 4 h after methionine addition, Kap95-GFP was visible primarily in cytoplasmic foci and its normal NE association was lost, a pattern largely similar to that observed in the cytoplasmic nups. Consistent with this, we observed that the Kap95-GFP signal colocalized with Nup159-mRFP (Fig. 6 B). Kap121-GFP and Kap123-GFP were less frequently seen in cytoplasmic foci and remained concentrated at the nuclear periphery at the 4-h time point. This distribution pattern appeared to have more similarity to that seen with core nups such as Nup53 (Fig. 4). Finally, cellular distribution of Kap104-GFP appeared unaffected by depletion of Nup170p (nup157Δ).
Figure 6.
Kap95p binds cytoplasmic foci in cells lacking Nup170p and Nup157p. (A) Strains producing GFP-tagged Kap95p, Kap104p, Kap121p, or Kap123p in the PMET3-NUP170 nup157Δ background were transferred to media containing methionine to repress NUP170 expression. Kap-GFP localization was examined using an epifluorescence microscope at the indicated times. At 4 h after NUP170 repression (nup157Δ), Kap95-GFP was visible at cytoplasmic foci (arrowheads). (B) A PMET3-NUP170 nup157Δ strain producing Kap95-GFP and Nup159-mRFP was grown in media containing methionine for the indicated times. Images were acquired using a confocal microscope. As indicated by the arrowheads, the Nup159-mRFP foci overlap with those of the Kap95-GFP. Bars, 5 µm.
The decrease in NPC density upon loss of Nup170p and Nup157p is caused by a defect in NPC assembly
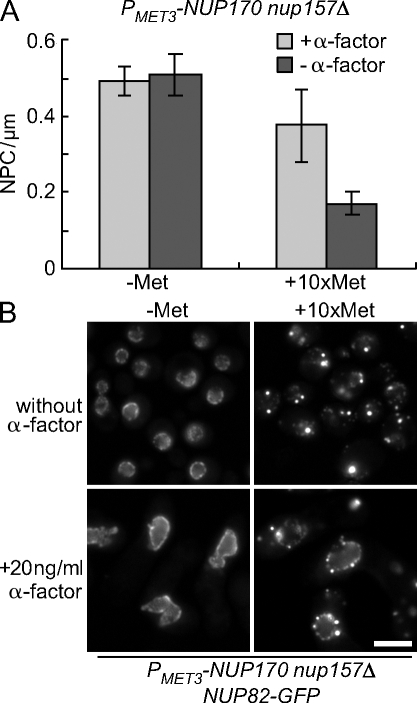
Our data are consistent with a model in which the loss of Nup170p and Nup157p inhibits new NPC assembly, and preexisting NPCs, being largely stable, are diluted out between mother and daughter cells during repeated cell cycles. Based on this model, we predict that the inhibition of the cell cycle would prevent the loss of preexisting NPCs. To test this idea, we inhibited cell cycle progression using α-factor to arrest cells in G1 phase, and TEM analysis was performed to determine NPC density (Fig. 7 A). When Nup170p was present in the nup157Δ background, the linear density of NPCs in the NEs of cells arrested with α-factor for 6 h was estimated at 0.49 NPC/µm, which was similar to cycling cells (0.51 NPC/µm; Fig. 7 A). In cells incubated for 6 h in the presence of α-factor and methionine, in order to inhibit Nup170p synthesis, NPC density decreased slightly (to 0.37 NPC/µm) when compared with cells expressing NUP170 but was significantly higher than that seen in cycling cells depleted of Nup170p (nup157Δ) for 6 h (0.17 NPC/µm). Consistent with these data, Nup82-GFP remained visible at the NE in α-factor–arrested cells after 6 h of Nup170p (nup157Δ) depletion, in contrast to what is seen in cycling cells (Fig. 7 B). Moreover, Nup82-GFP also accumulated in cytoplasmic foci, which is consistent with the inhibition of new NPC biogenesis. These foci were not detected in arrested cells expressing NUP170 (Fig. 7 B).
Figure 7.
Cell cycle arrest induced by α-factor inhibits the decrease in NE-associated Nup82-GFP after the repression of NUP170 (nup157Δ). (A) The strain TMY1203 (PMET3-NUP170 nup157Δ) was transferred to media lacking (dark gray bars) or containing (light gray bars) α-factor in the presence (+Met) or absence (−Met) of methionine for 6 h. The linear density of NPCs was calculated as in Fig. 2. A minimum of 30 cells was examined in each experiment, and the standard deviation was estimated from two independent experiments. (B) Nup82-GFP localization. The localization of Nup82-GFP in TMY1209 (PMET3-NUP170 nup157Δ NUP82-GFP) cells was examined by epifluorescence microscopy after 6 h with or without α-factor and in the presence (+Met) or absence (−Met) of methionine. Bar, 5 µm.
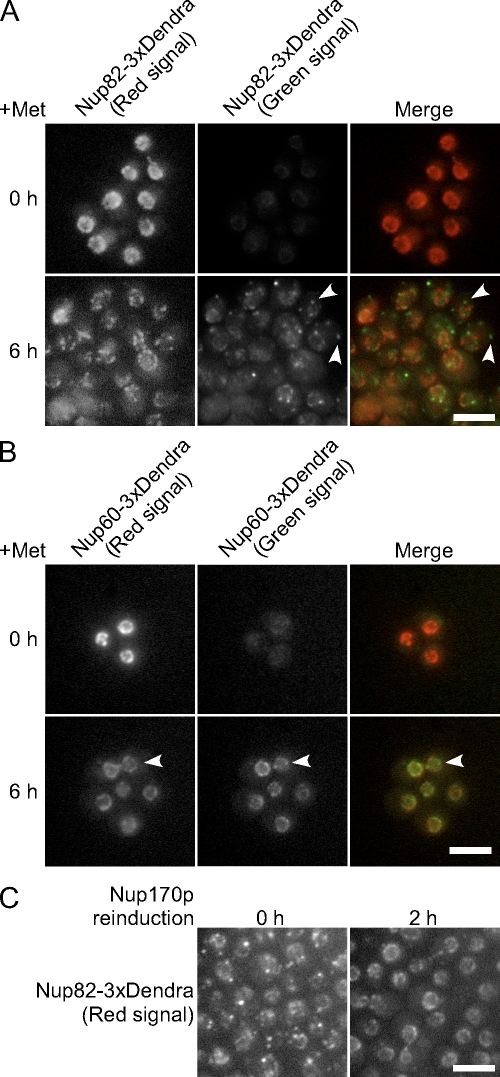
The fates of existing NPCs and the formation of new ones in the absence of Nup170p and Nup157p was also examined using nups tagged with the fluorescent protein Dendra. Dendra undergoes a photoconversion from green to red fluorescent states in response to visible blue or UV light (Gurskaya et al., 2006). This allows one to distinguish the pool of Dendra-tagged protein present at the point of photoconversion from that synthesized after exposure. We used this tag to follow the fates of existing and newly synthesized Nup82-Dendra and Nup60-Dendra after depletion of Nup170p (nup157Δ). These nups were selected as they represent different NPC substructures, and they exhibited different localization patterns after depletion of Nup170p (nup157Δ). After addition of methionine, cells were flashed with UV light to induce conversion of Nup-Dendra-green to Nup-Dendra-red. After conversion, no Nup82-Dendra-green signal was visible, and Nup82-Dendra-red appeared along the NE in an NPC-like pattern (Fig. 8 A). By 6 h after repression of NUP170 (nup157Δ), the Nup82-Dendra-red signal showed a striking decrease (Fig. 8 A), which is consistent with our observed decrease in NPCs (Fig. 2 B). In most cells, the remaining Nup82-Dendra-red signal was largely localized in a roughly spherical pattern, which is consistent with its continued presence in the dwindling number of NPCs. In contrast, newly synthesized Nup82-Dendra-green appeared in cytoplasmic foci with occasional foci adjacent to the NE (Fig. 8 A). These foci were generally distinct from the Nup82-Dendra-red containing NPCs, which suggests that the Nup82-Dendra-green failed to incorporate into existing or new NPCs.
Figure 8.
Nup170p and Nup157p are required for the incorporation of newly synthesized nups into NPCs. The strains KWY2216 (NUP82-Dendra PMET3-NUP170 nup157Δ; A) and KWY2215 (NUP60-Dendra PMET3-NUP170 nup157Δ; B) were transferred to media containing methionine to repress NUP170 expression. At the same time, UV light was applied to the cells to induce photoconversion of Dendra (green to red). Cells were imaged at 0 and 6 h after photoconversion. Nup82-Dendra produced after photoconversion (green) appears at foci generally distinct from existing NPCs (A, arrowheads). NE-associated Nup60-Dendra-green foci that were distinct from Nup60-Dendra-red were also detected (B, arrowheads). (C) KWY2216 cells were switched to media containing methionine; then, at 5 h after NUP170 repression, UV light was applied to the cells to induce photoconversion of Dendra to the red fluorophore. Cells were transferred to the media lacking methionine for reinduction of NUP170. At 0 and 2 h after induction, images of the Nup82-Dendra (red) were captured using an epifluorescence microscope. Bars, 5 µm.
Nup60p is one of the nups whose association with the NE was unaffected after 6 h of Nup170p (nup157Δ) depletion (Fig. 4). Because NPC formation is inhibited under these conditions, we were interested in comparing existing Nup60-Dendra to that synthesized after depletion of Nup170p (nup157Δ). 6 h after photoconversion and repression of NUP170 (nup157Δ), newly synthesized Nup60-Dendra-green was visible along the NE in foci that were, in many cases, distinct from Nup60-Dendra-red in preexisting NPCs (Fig. 8 B). Because few NPCs are formed in the absence of Nup170p and Nup157p, the nature of these deposits was of interest.
Loss of Nup170p and Nup157p induces the formation of proteinaceous structures adjacent to the inner nuclear membrane
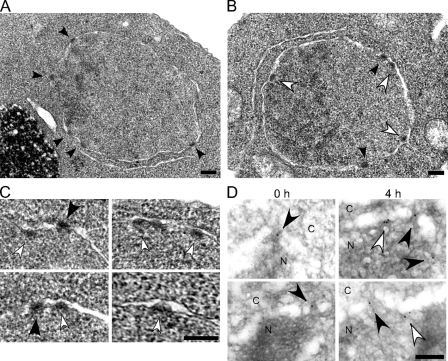
As the loss of Nup170p and Nup157p inhibits NPC formation, we were perplexed by the continued localization of core and nucleoplasmic nups at the NE after 6 h of Nup170p (nup157Δ) depletion. To further investigate the basis for these observations, we examined Nup170p (nup157Δ)-depleted cells using TEM techniques (osmium tetroxide fixation and uranyl acetate staining) that highlight NPCs. In addition to NPCs, we observed electron-dense structures adjacent to the nucleoplasmic face of the inner nuclear membrane at 4 h after repression of NUP170 (nup157Δ; Fig. 9, B and C, white arrowheads). The inner and outer NE membranes were clearly distinguishable at these sites, with no connecting pore membrane domain. Measurements of the width and thickness of these structures (71 ± 7 nm wide and 34 ± 9 nm thick; n = 32) revealed that they are similar in dimension to those of native NPCs (75 ± 7 nm wide and 33 ± 4 nm thick; n = 32). Similar structures were not detected before repression of NUP170 (nup157Δ; 0 h) or in strains lacking only Nup170p or Nup157p (Fig. 9 A and not depicted). We propose that these structures arise from core and nucleoplasmic nups forming NPC assembly intermediates at the inner membrane. To further test this conclusion, we performed immunoelectron microscopy analysis using mAb414, a monoclonal antibody that binds to several XFXFG repeats containing nups but mainly the core and nucleoplasmic nups Nsp1p and Nup2p (Kenna et al., 1996). We detected mAb414 binding to NPCs, as defined by discontinuities in the lumen of the NE (Fig. 9 D, black arrowheads). After Nup170p (nup157Δ) depletion, however, a significant number of gold particles were also observed at sites apart from NPCs (Fig. 9 D, white arrowheads; and Table I) and adjacent to the inner nuclear membrane. These results support the conclusion that core and nucleoplasmic nups contribute to the proteinaceous structures accumulating along the inner nuclear membrane.
Figure 9.
NPC-like structures accumulate at the inner nuclear membrane after the loss of Nup170p and Nup157p. (A–C) The strains TMY1098 (PMET3-NUP170; A) and TMY1126 (PMET3-NUP170 nup157Δ; B and C) were harvested at 4 h after NUP170 repression. The cells were then examined by TEM. Black arrowheads indicate the mature NPCs. White arrowheads indicate structures attached on the inner nuclear membrane with similar size and staining properties of NPCs. (D) TMY1126 cells were grown in media containing methionine for the indicated time points. Cells were then harvested and analyzed by immunoelectron microscopy using the mAb414 antibody. The nucleoplasm (N) and the cytoplasm (C) are indicated. Black arrowheads indicate gold particles bound to NPCs as defined by discontinuities in NE lumen. White arrowheads indicate gold particles attached to the inner nuclear membrane. Bars, 0.2 µm.
Table I.
Quantification of mAb414 immunogold labeling of Nup170p (nup157Δ)-depleted cells
| +Met (h) | Total number of gold particles | Gold particles at NPCs | Gold particles at the inner nuclear membrane |
| 0 | 98 | 82 (84%) | 16 (16%) |
| 4 | 110 | 54 (49%) | 56 (51%) |
The +Met (h) column shows time after addition of methionine to the TMY1126 strain. Gold particles present at discontinuities in NE lumen of a size consistent with NPCs were scored as NPC associated. Gold particles at the inner nuclear membrane but distinct from NPCs were scored as inner membrane associated. Percentages of the total number of particles counted are given in parentheses.
Reinitiation of Nup170p synthesis induces targeting to the NE of cytoplasmic pools of mislocalized nups
If the structures observed upon depletion of Nup170p (nup157Δ) represent an accumulation of intermediates in NPC assembly, it is possible that these nups could be used in new NPC formation after reinitiation of Nup170p synthesis. To test this idea, Nup82-Dendra-green was induced to accumulate in cytoplasmic foci by a 5-h depletion of Nup170p (nup157Δ), then converted to Nup82-Dendra-red. Cells were then switched to medium lacking methionine to induce Nup170p synthesis. As shown in Fig. 8 C, Nup170p production induced both the disassembly of the cytoplasmic foci and the relocation of Nup82-Dendra-red to the NE in a pattern consistent with its localization to NPCs, which suggests that nups in the cytoplasmic foci remain competent to assemble into NPCs.
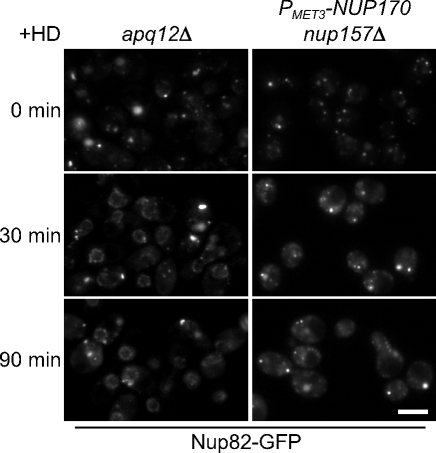
Structures similar to the cytoplasmic foci seen in Nup170p (nup157Δ)-depleted cells have also been observed in strains lacking Apq12p (Scarcelli et al., 2007). This ER membrane protein has been proposed to play a role in the biophysical properties of the nuclear membrane and the formation of NPCs. An apq12Δ strain at 23°C accumulates cytoplasmic foci containing nups, including Nup82-GFP. The addition of benzyl alcohol to the medium disperses these nups and promotes their association with the NE and incorporation into NPCs (Scarcelli et al., 2007). Similarly, we observed that the incubation of apq12Δ cells with another alcohol, 3% hexane-1, 6-diol, also induced the dispersion of Nup82-GFP–containing cytoplasmic foci and localization to the nuclear periphery (Fig. 10). This alcohol has been demonstrated to disrupt nup–nup interactions in various contexts, including dispersing cytoplasmic aggregates of FG-nups (Patel et al., 2007). Treatment with 3% hexane-1, 6-diol also partially dispersed Nup82-GFP–containing foci formed upon loss of Nup170p and Nup157p, but Nup82-GFP failed to relocalize to the nuclear rim (Fig. 10) and remained diffusely distributed throughout the cytoplasm. Thus, although hexane-1, 6-diol can bypass the requirement for Apq12p and induce reassembly of cytoplasmic nups onto the NE, this latter process requires the presence of Nup170p and Nup157p.
Figure 10.
Hexane-1, 6-diol suppresses the NPC assembly defect of the apq12Δ strain but not that of cells lacking Nup170p and Nup157p. apq12Δ (TMY1290) and PMET3-NUP170 nup157Δ (TMY1135) strains expressing Nup82-GFP were grown in YPD overnight at 23°C, and for 6 h in media containing methionine to induce NPC assembly defects and accumulation of Nup82-GFP foci. Hexane-1, 6-diol (+HD; final 3% wt/vol) was then added to the media, and samples were taken at the indicated times. Images of Nup82-GFP were acquired using an epifluorescence microscope. Bar, 5 µm.
Discussion
The assembly of NPCs across an intact NE requires the fusion of the inner and outer NE and the assembly of nups within the newly forming NPC. Here, we show that the related nups Nup170p and Nup157p are required for the assembly of subcomplexes within the forming NPC at a step before or coinciding with the membrane fusion and pore formation. Depletion of these nups does not appear to affect existing NPCs, which is consistent with the predicted stability of these structures (Daigle et al., 2001; Rabut et al., 2004a,b). However, the loss of these nups leads to the arrest of NPC formation and small foci composed of distinct groups of nups accumulated in either the cytoplasm or the nucleus adjacent to the inner nuclear membrane. We demonstrate that the cytoplasmic foci represent pools of nups that remain competent to assemble into NPCs after reinitiation of NUP170 expression.
Other nup mutants also show a decrease in NPC numbers, including nic96-1 and nup192-15 (Zabel et al., 1996; Kosova et al., 1999). In these mutants, the overall NE structure, protein import, and mRNA export are not significantly altered at early times after transfer into a nonpermissive condition; however, the number of the NPCs is significantly reduced (Zabel et al., 1996; Kosova et al., 1999; Gomez-Ospina et al., 2000). Thus, much like cells lacking Nup170p and Nup157p, these mutations are likely to affect NPC assembly but not the function of intact NPCs. This difference is not always discernible, and, in the case of several other nup mutants, it has been difficult to discriminate between those mutations that affect the integrity or function of the NPC versus those inhibiting assembly. Among the most interesting phenotypes observed are those of nup mutants that induce NE herniations that seal over NPCs (e.g., Wente and Blobel, 1993). It is not clear, however, whether this phenotype arises as a consequence of assembly defects or represents a cellular response to damaged NPCs.
Of the various nup mutants that have now been examined, only cells lacking both Nup170p and Nup157p exhibit the unique feature of accumulating putative NPC assembly intermediates. These intermediates were revealed by the inspection of individual nups after loss of Nup170p and Nup157p. Nups that contribute to defined NPC subcomplexes or are positioned at a common location in the NPC (e.g., the nucleoplasmic face of the NPC) exhibited one of three distinct localization patterns. For example, the cytoplasmic nups Nup82p and its binding partner Nup159p (Belgareh et al., 1998) rapidly accumulated in cytoplasmic foci after loss of Nup170p and Nup157p (Fig. 4). In a second phenotype, nups associated with the symmetrical core of the NPC showed a mixed distribution between the cytoplasmic foci and structures along the NE. Several nups thought to be closely associated with Nup82p in the NPC, including Gle1p, Nup100p, and members of the Nup84p complex (Alber et al., 2007a,b), showed a clear association with the cytoplasmic foci as well as the NE. Other core nups including Nup188p and Nup53p exhibited a weaker, but detectable, signal in cytoplasm foci. Similarly, Ndc1p and Pom34p were seen in a subset of the cytoplasm foci, presumably those at or near the NE or ER membrane. Each of these core nups and poms also exhibited a significant punctate NE localization (Fig. 4). In contrast, nucleoplasmic nups were found exclusively at the NE after the loss of Nup170p and Nup157p (6 h; Fig. 4). These results, coupled with the results of the Nup60-Dendra and the immunoelectron microscopy experiments (Fig. 8 and 9), have lead us to conclude that association of the core and nucleoplasmic nups with the NE reflected their presences in the inner membrane–associated, NPC-like structures.
The nup localization patterns seen after the loss of Nup170p and Nup157p provide important insights into their role in NPC assembly. We conclude that the removal of Nup170p and Nup157p does not inhibit the targeting of core and nucleoplasmic nups to the nuclear periphery or their assembly into structural precursors of the NPC, which are what we interpret the inner nuclear membrane–associated structures to represent. In contrast, Nup170p and Nup157p appear essential for the incorporation of the cytoplasmically positioned nups into NPCs. Although displaced from the NE, these nups appear to associate with known binding partners (e.g., the interaction of the Nup82p/Nup159p with the Nup84p complex) within cytoplasmic foci. These foci are likely composed of assembly intermediates, not nonspecific aggregates, as their nups remain competent to target to the NE after derepression of NUP170, and appear to assemble into NPCs (Fig. 8 C).
We propose that Nup170p and Nup157p function at the NE membrane, where they facilitate insertion of the core and cytoplasmic structures into the forming nuclear pore, potentially by assisting in the fusion of the inner and outer nuclear membranes. Alternatively, these factors could be required for the bending and stabilization of the incipient nuclear pore, which may form but collapse in their absence. These activities would necessitate the positioning of these proteins at or near the membrane. A growing number of observations support these ideas; among these are synthetic lethal interactions between mutations in NUP170 and POM152 or POM34 (Aitchison et al., 1995; Tcheperegine et al., 1999; Miao et al., 2006). These interactions appear to be based, in part, on physical associations. Nup170p binds directly to the pore-exposed region of Pom152p (Fig. S1) and is detected in subcomplexes with Ndc1p, Pom34p, and Pom152p; Nup157p is detected bound to Pom34p (Alber et al., 2007b; see Onischenko et al. on p. 475 of this issue). Also, based on their analysis, Alber et al. (2007a,b) have positioned Nup170p and Nup157p adjacent to the membrane in their proposed architecture of the NPC (Alber et al., 2007a,b).
The placement of Nup170p at or near the pore is also consistent with functions suggested by its structural features. Like members of the Nup84p complex, Nup170p and Nup157p are predicted to contain an N-terminal β-propeller and a C-terminal α-solenoid domain, features they also share with membrane vesicle coatomer proteins such as clathrin (Devos et al., 2004, 2006). By analogy to clathrin, these regions of Nup170p and Nup157p provide surfaces for interactions with membrane proteins and perhaps a means for these proteins to influence the curvature of the pore membrane. In support of this idea, near the N-terminal end of the α-solenoid region of Nup170p lies a predicted membrane-binding, amphipathic α-helix, or ALPS, domain (Drin et al., 2007). An ALPS domain has also been detected in Nup133p, and its ability to preferentially bind curved membranes has been documented in vitro (Drin et al., 2007).
In addition to their own membrane binding potential, Nup170p and Nup157p also directly interact with Nup53p (Fig. S1; Lusk et al., 2002; Alber et al., 2007a,b; Onischenko et al., 2009; unpublished data). Nup53p contains a C-terminal amphipathic helix that mediates membrane binding and is required for the formation of inner membrane–derived tubular vesicular structures induced by the overexpression of Nup53p (Marelli et al., 2001; Patel and Rexach, 2008). We envisage that the complexes formed between Nup53p–Nup170p–Nup157p are likely capable of interacting with the membrane at multiple interfaces and by various mechanisms, which are properties consistent with a role for these proteins in steps that promote membrane curvature and fusion during NPC assembly.
It remains to be determined whether the mammalian counterpart of Nup170p and Nup157p, Nup155, would play a similar role in interphase NPC assembly. However, Nup155 is essential for postmitotic NPC assembly, and its function has been linked to events at the membrane (Franz et al., 2005). Depletion of Nup155 from Xenopus extracts inhibits NPC and NE assembly at a step after vesicle binding to chromatin. This phenotype is similar to that observed upon depletion of the membrane proteins POM121 or NDC1 (Antonin et al., 2005; Mansfeld et al., 2006) and the Nup155 binding partner Nup53 (Hawryluk-Gara et al., 2008), but is distinct from phenotypes seen after depletion of other nups, including members of the Nup107–Nup160 complex (Harel et al., 2003; Walther et al., 2003). These results have been interpreted to suggest that the functions of Nup155 and Nup53 in NPC assembly are tightly linked to those of POM121 or NDC1, which is consistent with a potential role for these proteins in influencing pore membrane structure (Hawryluk-Gara et al., 2008).
The steps that lead to pore formation in yeast, including changes in membrane structure and fusion, are also likely to be controlled by additional factors, including non-NPC proteins and lipids (Schneiter et al., 1996). In this regard, mutations in Kap95p and Apq12p have been described that exhibit the accumulation of NPC intermediates similar to those we have documented in the Nup170p- and Nup157p-depleted cells. The phenotype of the kap95 mutant may be particularly relevant to our observations, as it disrupts the localization of Nup170p, thus raising the possibility that the phenotype of the kap mutant is, at least in part, caused by a defect in the targeting of Nup170p to its site of function in pore formation. Importantly, the kap95 mutant also exhibits a synthetic sick phenotype in combination with a functionally compromised nup170-GFP allele (Ryan et al., 2007). We have previously proposed the idea that kaps play an important role in NPC assembly through their targeting of nups to the inner nuclear membrane, where assembly intermediates would be formed (Marelli et al., 2001). In these studies, Kap121p was shown to assist in the targeting of Nup53p to NPCs, and it was required for the inner membrane proliferation induced by overexpression of NUP53. In light of the phenotypic similarities between the kap95 mutant and the Nup170p- and Nup157p-depleted cells, a similar role for Kap95p in targeting Nup170p and possibly other nups, including Nup2p (Dilworth et al., 2001), to the nucleoplasm seems reasonable.
In addition to contributing a nuclear targeting function, it has been suggested that Kap95p may play a role in the assembly of the cytoplasmic face of the NPC, functioning here to regulate the fusion of putative nup-containing vesicles to the outer nuclear membrane (Ryan et al., 2007). Consistent with a cytoplasmic role for Kap95p, we also detected Kap95-GFP in association with the cytoplasmic nup foci formed after the loss of Nup170p and Nup157p (Fig. 6). The time course of the appearance of Kap95-GFP foci after depletion paralleled that of cytoplasmic nups, which suggests that the association of Kap95 with these foci is linked to this group of nups. Whether or not these cytoplasmic foci are membrane associated remains to be addressed.
Null mutants lacking the integral ER and NE membrane protein Apq12p also contain mislocalized cytoplasmic nups and inner nuclear membrane–associated, NPC-like structures when grown at a semipermissive temperature (Scarcelli et al., 2007). Moreover, combinations of apq12Δ and mutations in several NPC components, including Pom34p, Ndc1p, and Nup170p, exhibit synthetic growth defects, with an apq12Δ nup170Δ mutant showing especially strong defects in nuclear membrane structure. Although the role of Apq12p in NPC assembly remains to be clearly established, it has been proposed that it contributes to membrane fluidity by modulating the lipid composition of the NE (Scarcelli et al., 2007). This conclusion is based, in part, on the observation that benzyl alcohol treatment of apq12Δ cells can induce the dispersion of the cytoplasmic nup foci and the recruitment of their nups to the NE, presumably stimulating their assembly into NPCs. We observed a similar effect using hexane-1, 6-diol treatment of apq12Δ cells. Thus, alcohols can suppress the defects caused by the loss of Apq12p. However, hexane-1, 6-diol treatment of cells depleted of Nup170p and Nup157p, although capable of dispersing cytoplasmic nup foci, did not induce their incorporation into the NE. Based on the phenotypic similarities caused by the loss of either Apq12p or Nup170p and Nup157p, we conclude that they likely function at a similar point in NPC assembly, but that their different responses to alcohol suggest they contribute to distinct aspects of the process. The relationships between these proteins are again consistent with a role for Nup170p and Nup157p in mediating structural changes in the NE membrane that drive formation of the pore membrane and contribute to its stability. Although they required for these essential processes, Nup170p and Nup157p are expected to function in concert with pore membrane proteins, Apq12p, and other nups to guide these events. Understanding the physical relationships between these proteins and the pore membrane will be a key next step in further understanding the mechanisms of nuclear pore formation.
Materials and methods
Yeast strains, media, and plasmids
Yeast strains used in this study are shown in Table S1. All strains were grown in YPD (1% yeast extract, 2% bactopeptone, and 2% glucose) or synthetic media containing 0.17% yeast nitrogen base (without amino acids and ammonium sulfate), 0.5% ammonium sulfate, 2% glucose, and the appropriate supplements at 30°C. Cell transformations were performed using a lithium acetate method (Gietz and Schiestl, 2007) and a PCR-based, one-step gene modification method (Longtine et al., 1998). The plasmid used for the PMET3-NUP170 integration, pTM1046, was constructed by the replacement of the BglII–PacI region of pFA6a-kanMX6-PGAL1-3HA (Longtine et al., 1998) with a BglII–PacI fragment containing a promoter region of the MET3 gene, which was made by PCR using BY4742 genomic DNA as a template and the primers 5′-GCGAGATCTTTTAGTACTAACAGAGAC-3′ and 5′-GCGTTAATTAAAGACATGTTAATTATACTTTATTC-3′. The following NLS reporter plasmids were used: pGAD-GFP (cNLS-GFP; Shulga et al., 1996), pEB0836 (Pho4-NLS-GFP; Kaffman et al., 1998), pNab2(200–249)-GFP (rg-NLS-GFP; Lee and Aitchison, 1999), and pBT008 (rpL25-NLS-GFP; Timney et al., 2006). The following plasmids were used for protein expression in Escherichia coli: pGST-170 (full-length NUP170 ORF cloned in pGEX-4T1), pGST-152N (coding region POM152 corresponding to amino acid residues 1–111 in pGEX-6P1), and pET53 (full-length NUP53 ORF cloned into pET9d).
For repression of MET3-NUP170, cells were grown in the supplemented minimal media lacking methionine to a mid-logarithmic growth phase. Methionine (from the 20-mg/ml stock) was then added into the media to a final concentration of 200 µg/ml and incubated at 30°C for the indicated times. For α-factor arrest experiments, α-factor was added into the media to a final concentration of 20 ng/ml at the same time as the methionine addition and incubated for 6 h.
Fluorescence microscopy
Yeast strains synthesizing various GFP and mRFP fusion proteins were taken from cultures at various time points after methionine addition. The locations of GFP and mRFP fusion proteins in live cells were visualized using either an epifluorescence or confocal microscope. Epifluorescence images were obtained with either a microscope (BX50; Olympus) using a UPlanS-Apochromat 100×/1.40 NA oil objective lens (Olympus) and a charge-coupled device (CCD) camera (AxioCam HRm; Carl Zeiss, Inc.) controlled by AxioVision software (Carl Zeiss, Inc.), or with a microscope (IX80; Olympus) using a UPlanS-Apochromat 100×/1.40 NA oil objective lens (Olympus) with a CCD camera (CoolSNAP HQ; Photometrics) controlled by InVivo software (Media Cybernetics). Confocal images were obtained with a microscope (Axiovert 200M; Carl Zeiss, Inc.) equipped with a confocal scanning system (LSM 510 META; Carl Zeiss, Inc.) using a Plan-Apochromat 63×/1.4 NA oil objective lens (Carl Zeiss, Inc.). Image processing (cropping, background subtraction, and linear normalization of images) was performed with the ImageJ software (National Institutes of Health).
Immunofluorescence microscopy using anti-Pom152p (mAb118C3 at 1:20 dilution; Strambio-de-Castillia et al., 1995) and Alexa Fluor 594–labeled anti–mouse IgG (1:200 dilution; Invitrogen) was performed as described previously (Nishikawa et al., 2003). FISH analysis was performed as described previously (Amberg et al., 1992; Cole et al., 2002) using Texas red–labeled oligo (dT50) as a probe.
Kinetic nuclear import assay of NLS reporter proteins
For the kinetic nuclear import assay, the yeast strains expressing a Nab2-NLS-GFP reporter protein were harvested from cultures at various time points after methionine addition. Nucleoplasmic equilibration of the reporter protein was achieved by energy depletion using sodium azide and deoxyglucose. Nuclear import of the reporter was initiated by removal of the metabolic poison and addition of media containing glucose, and cells were examined at the indicated times as described previously (Shulga et al., 1996).
Photoconversion of Dendra fusion proteins
We performed two sets of photoconversion experiments using Dendra fusion proteins. In the first set, we examined the localization of Dendra fusion proteins after loss of Nup170p and Nup157p. Yeast cells expressing Dendra fusion proteins were harvested from cultures cells grown in media lacking methionine. Methionine (from the 20 mg/ml stock) was then added to repress NUP170 expression, and cells were immediately placed on a slide-mounted agarose pad (2% low-melt agarose containing the supplemented minimal medium and 200 µg/ml methionine) and then on the fluorescence microscope (BX61 with a UPlanFl 100×/1.30 oil objective; Olympus). Images were acquired with a digital camera (CA742-98; Hamamatsu Photonics) controlled by the Metamorph software program (MDS Analytical Technologies). Dendra fusion proteins were then converted by 4 × 20-ms pulses of UV light, and the Dendra-red signals were imaged just after photoconversion. Cells were then allowed to grow on the agarose pad for 6 h at 30°C. Dendra-red (proteins present at the point of photoconversion) and Dendra-green (proteins synthesized after photoconversion) signals were imaged after the incubation. The second set of experiments examined the localization change of Dendra fusion proteins after derepression of NUP170. A PMET3-NUP170 nup157Δ yeast strain expressing NUP82-3xDendra or NUP60-3xDendra was grown in media lacking methionine. Methionine was then added, and the cells were incubated for 5 h to induce cytoplasmic foci of Nup82-3xDendra or Nup60-3xDendra. The cells were washed with media lacking methionine and then placed on an agarose pad (2% low-melt agarose containing the supplemented minimal medium lacking methionine) to allow Nup170p production. Nup82-3xDendra or Nup60-3xDendra was photoconverted by 4 × 20-ms pulses of UV light, and the Dendra-red signals were imaged just after photoconversion. Cells were allowed to grow on the agarose pad for 2 h at 30°C, and images of the Dendra-red signal were captured.
EM
TEM for visualizing membrane structures using potassium permanganate fixation, and visualizing membranous and proteinous structures using osmium tetroxide fixation and uranyl acetate was performed as described previously (Marelli et al., 2001). For potassium permanganate fixation experiments, the cells were harvested from cultures at various times after methionine addition, washed briefly with water, and fixed with 3% potassium permanganate. For osmium tetroxide fixation experiments, the strains were harvested from cultures 4 h after methionine addition. Cells were incubated in the supplemental media with 200 µg/ml methionine for first 2 h and then in YPD plus 200 µg/ml methionine for another 2 h. The latter step has been suggested to lead to improved image contrast (Wright and Rine, 1989). Glutaraldehyde fixation, cell wall removal, and osmium tetroxide postfixation were performed as described previously (Marelli et al., 2001). Samples were examined with using an electron microscope (410; Philips), and images were collected with a CCD camera (Megaview III; Soft Imaging System) controlled by AnalySIS software (Soft Imaging System). Image processing (cropping, rotation, and linear normalization of images) and measurement of the length of the NE were performed with the ImageJ software.
Immunoelectron microscopy was performed as described previously (Wente et al., 1992), with modifications. Spheroplasts were incubated in 0.6 M sorbitol and 100 mM potassium phosphate, pH 6.5 (buffer A), for 15 min on ice ,and then fixed with 4% formaldehyde and 0.05% glutaraldehyde in buffer A for 1 h on ice. The samples were dehydrated twice in 50% ethanol for 10 min, twice in 80% ethanol for 10 min at room temperature, and then incubated with LR white resin for infiltration at 4°C overnight. The resin was polymerized using UV light for 23 h at 4°C. Thin sections were collected on nickel grids. The samples were blocked with 1% BSA in TBS-T and then incubated with mAb414 (Covance) diluted 1:2,000 in 1% BSA in TBS-T at room temperature for 1 h. After washing with 1% BSA in TBS-T, the grids were incubated with 12 nm of colloidal gold-conjugated goat anti–mouse IgG (Jackson ImmunoResearch Laboratories) diluted 1:30 in 1% BSA in TBS-T. After final washes, the samples were contrasted with 2% aqueous uranyl acetate for 5 min.
Isolation of recombinant proteins expressed in bacteria and in vitro binding assays
An E. coli strain BL21 (DE3) containing plasmids expressing GST, Nup53p, GST-Nup170p, and GST-Pom152p1−111 was grown to an OD of 0.6–1.0 at 37°C, and induced with 1 mM IPTG for 2 h at 37°C for GST, Nup53p, and GST- Pom152p1–111, and overnight at room temperature for GST-Nup170p. GST and GST fusion proteins were purified on glutathione Sepharose 4B beads (GE Healthcare), according to the manufacturer's instructions, in lysis buffer (50 mM Tris HCl, pH 7.5, 300 mM NaCl, 150 mM potassium acetate, 2 mM MgCl2, 0.1% tergitol, 10% glycerol, 1 mM DTT, and complete protease inhibitor cocktail [Roche]). Nup170p was separated from GST and released from the beads by incubation with 0.6 units of thrombin (Sigma-Aldrich) per 10 µl of beads for 4 h at room temperature.
For in vitro binding assays examining the interactions of Nup170p and GST- Pom152p1–111, 25 µl of purified Nup170p in buffer B (50 mM Tris HCl, pH 7.5, 150 mM potassium acetate, 2 mM MgCl2, 0.1% tergitol, 10% glycerol, 1 mM DTT, and complete protease inhibitor cocktail) was incubated with 25 µl of glutathione Sepharose 4B beads preloaded with either purified GST or purified GST-Pom152p1−111 for 2 h at 4°C. The unbound fraction was obtained from the supernatant after the incubation. After washing, proteins were eluted from the beads with SDS-PAGE sample buffer and analyzed by SDS-PAGE and Coomassie blue staining.
For in vitro binding assays examining the interactions of Nup53p and GST-Nup170p, 15 ml of bacterial lysate containing Nup53p in lysis buffer was supplemented with 2 mM ATP and 10 mM MgSO4, and then incubated with 100 µl of glutathione Sepharose 4B beads preloaded with purified GST-Nup170p for 20 min at 4°C. The unbound fraction was obtained from the supernatant just after the incubation. After washing, proteins were released from the beads by incubation with 0.6 units of thrombin. Proteins in the various fractions were analyzed by SDS-PAGE and Coomassie blue staining.
Online supplemental material
Fig. S1 shows that recombinant Nup170p directly binds an N-terminal fragment of Pom152p and Nup53p. Table S1 lists the strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200810029/DC1.
Acknowledgments
We would like to thank Dr. Ed Hurt and Dr. Patrick Lusk for sharing unpublished results, Honey Chan for assistance in the EM experiments, and members of Wozniak laboratory for helpful discussions and sharing reagents.
Funds for this work were provided to R.W. Wozniak by the Canadian Institutes of Health Research, the Alberta Heritage Foundation for Medical Research, and the Howard Hughes Medical Institute; to D.S. Goldfarb by the National Science Foundation (grant MCB-072064); and to K. Weis by the National Institutes of Health (grant GM058065). T. Makio was partially supported by a postdoctoral fellowship from the Uehara Memorial Foundation.
Footnotes
Abbreviations used in this paper: CCD, charge-coupled device; kap, karyopherin; mRFP, monomeric red fluorescent protein; NE, nuclear envelope; NLS, nuclear localization signal; NPC, nuclear pore complex; nup, nucleoporin; ORF, open reading frame; TEM, transmission electron microscopy; WT, wild type.
References
- Aitchison J.D., Rout M.P., Marelli M., Blobel G., Wozniak R.W. 1995. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p.J. Cell Biol. 131:1133–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T., et al. 2007a. Determining the architectures of macromolecular assemblies.Nature. 450:683–694 [DOI] [PubMed] [Google Scholar]
- Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T., et al. 2007b. The molecular architecture of the nuclear pore complex.Nature. 450:695–701 [DOI] [PubMed] [Google Scholar]
- Amberg D.C., Goldstein A.L., Cole C.N. 1992. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA.Genes Dev. 6:1173–1189 [DOI] [PubMed] [Google Scholar]
- Antonin W., Franz C., Haselmann U., Antony C., Mattaj I.W. 2005. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation.Mol. Cell. 17:83–92 [DOI] [PubMed] [Google Scholar]
- Beck M., Forster F., Ecke M., Plitzko J.M., Melchior F., Gerisch G., Baumeister W., Medalia O. 2004. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography.Science. 306:1387–1390 [DOI] [PubMed] [Google Scholar]
- Beck M., Lucic V., Forster F., Baumeister W., Medalia O. 2007. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography.Nature. 449:611–615 [DOI] [PubMed] [Google Scholar]
- Belgareh N., Snay-Hodge C., Pasteau F., Dagher S., Cole C.N., Doye V. 1998. Functional characterization of a Nup159p-containing nuclear pore subcomplex.Mol. Biol. Cell. 9:3475–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chial H.J., Rout M.P., Giddings T.H., Winey M. 1998. Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies.J. Cell Biol. 143:1789–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C.N., Heath C.V., Hodge C.A., Hammell C.M., Amberg D.C. 2002. Analysis of RNA export.Methods Enzymol. 351:568–587 [DOI] [PubMed] [Google Scholar]
- D'Angelo M.A., Anderson D.J., Richard E., Hetzer M.W. 2006. Nuclear pores form de novo from both sides of the nuclear envelope.Science. 312:440–443 [DOI] [PubMed] [Google Scholar]
- Daigle N., Beaudouin J., Hartnell L., Imreh G., Hallberg E., Lippincott-Schwartz J., Ellenberg J. 2001. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells.J. Cell Biol. 154:71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D., Dokudovskaya S., Alber F., Williams R., Chait B.T., Sali A., Rout M.P. 2004. Components of coated vesicles and nuclear pore complexes share a common molecular architecture.PLoS Biol. 2:e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D., Dokudovskaya S., Williams R., Alber F., Eswar N., Chait B.T., Rout M.P., Sali A. 2006. Simple fold composition and modular architecture of the nuclear pore complex.Proc. Natl. Acad. Sci. USA. 103:2172–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth D.J., Suprapto A., Padovan J.C., Chait B.T., Wozniak R.W., Rout M.P., Aitchison J.D. 2001. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex.J. Cell Biol. 153:1465–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G., Casella J.F., Gautier R., Boehmer T., Schwartz T.U., Antonny B. 2007. A general amphipathic alpha-helical motif for sensing membrane curvature.Nat. Struct. Mol. Biol. 14:138–146 [DOI] [PubMed] [Google Scholar]
- Franz C., Askjaer P., Antonin W., Iglesias C.L., Haselmann U., Schelder M., de Marco A., Wilm M., Antony C., Mattaj I.W. 2005. Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates.EMBO J. 24:3519–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C., Walczak R., Yavuz S., Santarella R., Gentzel M., Askjaer P., Galy V., Hetzer M., Mattaj I.W., Antonin W. 2007. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly.EMBO Rep. 8:165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S., Gorlich D. 2007. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes.Cell. 130:512–523 [DOI] [PubMed] [Google Scholar]
- Frey S., Richter R.P., Gorlich D. 2006. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties.Science. 314:815–817 [DOI] [PubMed] [Google Scholar]
- Funakoshi T., Maeshima K., Yahata K., Sugano S., Imamoto F., Imamoto N. 2007. Two distinct human POM121 genes: requirement for the formation of nuclear pore complexes.FEBS Lett. 581:4910–4916 [DOI] [PubMed] [Google Scholar]
- Galy V., Mattaj I.W., Askjaer P. 2003. Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo.Mol. Biol. Cell. 14:5104–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R.D., Schiestl R.H. 2007. Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method.Nat. Protoc. 2:35–37 [DOI] [PubMed] [Google Scholar]
- Gigliotti S., Callaini G., Andone S., Riparbelli M.G., Pernas-Alonso R., Hoffmann G., Graziani F., Malva C. 1998. Nup154, a new Drosophila gene essential for male and female gametogenesis is related to the nup155 vertebrate nucleoporin gene.J. Cell Biol. 142:1195–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie P.J., Khoudoli G.A., Stewart G., Swedlow J.R., Blow J.J. 2007. ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing.Curr. Biol. 17:1657–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Ospina N., Morgan G., Giddings T.H., Jr., Kosova B., Hurt E., Winey M. 2000. Yeast nuclear pore complex assembly defects determined by nuclear envelope reconstruction.J. Struct. Biol. 132:1–5 [DOI] [PubMed] [Google Scholar]
- Gurskaya N.G., Verkhusha V.V., Shcheglov A.S., Staroverov D.B., Chepurnykh T.V., Fradkov A.F., Lukyanov S., Lukyanov K.A. 2006. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light.Nat. Biotechnol. 24:461–465 [DOI] [PubMed] [Google Scholar]
- Harel A., Orjalo A.V., Vincent T., Lachish-Zalait A., Vasu S., Shah S., Zimmerman E., Elbaum M., Forbes D.J. 2003. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores.Mol. Cell. 11:853–864 [DOI] [PubMed] [Google Scholar]
- Hawryluk-Gara L.A., Platani M., Santarella R., Wozniak R.W., Mattaj I.W. 2008. Nup53 is required for nuclear envelope and nuclear pore complex assembly.Mol. Biol. Cell. 19:1753–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M.W., Walther T.C., Mattaj I.W. 2005. Pushing the envelope: structure, function, and dynamics of the nuclear periphery.Annu. Rev. Cell Dev. Biol. 21:347–380 [DOI] [PubMed] [Google Scholar]
- Hsia K.C., Stavropoulos P., Blobel G., Hoelz A. 2007. Architecture of a coat for the nuclear pore membrane.Cell. 131:1313–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A., Rank N.M., O'Shea E.K. 1998. Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121.Genes Dev. 12:2673–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna M.A., Petranka J.G., Reilly J.L., Davis L.I. 1996. Yeast Nle3p/Nup170p is required for normal stoichiometry of FG nucleoporins within the nuclear pore complex.Mol. Cell. Biol. 16:2025–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O., Hieter P., Winey M., Basrai M.A. 2001. Novel role for a Saccharomyces cerevisiae nucleoporin, Nup170p, in chromosome segregation.Genetics. 157:1543–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiger A.A., Gigliotti S., Fuller M.T. 1999. Developmental genetics of the essential Drosophila nucleoporin nup154: allelic differences due to an outward-directed promoter in the P-element 3′ end.Genetics. 153:799–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosova B., Pante N., Rollenhagen C., Hurt E. 1999. Nup192p is a conserved nucleoporin with a preferential location at the inner site of the nuclear membrane.J. Biol. Chem. 274:22646–22651 [DOI] [PubMed] [Google Scholar]
- Lau C.K., Giddings T.H., Jr., Winey M. 2004. A novel allele of Saccharomyces cerevisiae NDC1 reveals a potential role for the spindle pole body component Ndc1p in nuclear pore assembly.Eukaryot. Cell. 3:447–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.C., Aitchison J.D. 1999. Kap104p-mediated nuclear import. Nuclear localization signals in mRNA-binding proteins and the role of Ran and Rna.J. Biol. Chem. 274:29031–29037 [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae.Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Lusk C.P., Makhnevych T., Marelli M., Aitchison J.D., Wozniak R.W. 2002. Karyopherins in nuclear pore biogenesis: a role for Kap121p in the assembly of Nup53p into nuclear pore complexes.J. Cell Biol. 159:267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M., Kunze R., Buerer A., Aebi U., Hurt E. 2002. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins.EMBO J. 21:387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M., Kunze R., Stangl K., Stelter P., Toth K.F., Bottcher B., Hurt E. 2005. Reconstitution of Nup157 and Nup145N into the Nup84 complex.J. Biol. Chem. 280:18442–18451 [DOI] [PubMed] [Google Scholar]
- Madrid A.S., Mancuso J., Cande W.Z., Weis K. 2006. The role of the integral membrane nucleoporins Ndc1p and Pom152p in nuclear pore complex assembly and function.J. Cell Biol. 173:361–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeld J., Guttinger S., Hawryluk-Gara L.A., Pante N., Mall M., Galy V., Haselmann U., Muhlhausser P., Wozniak R.W., Mattaj I.W., et al. 2006. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells.Mol. Cell. 22:93–103 [DOI] [PubMed] [Google Scholar]
- Marelli M., Aitchison J.D., Wozniak R.W. 1998. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p.J. Cell Biol. 143:1813–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli M., Lusk C.P., Chan H., Aitchison J.D., Wozniak R.W. 2001. A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope.J. Cell Biol. 153:709–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul G.G., Maul H.M., Scogna J.E., Lieberman M.W., Stein G.S., Hsu B.Y., Borun T.W. 1972. Time sequence of nuclear pore formation in phytohemagglutinin-stimulated lymphocytes and in HeLa cells during the cell cycle.J. Cell Biol. 55:433–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao M., Ryan K.J., Wente S.R. 2006. The integral membrane protein Pom34p functionally links nucleoporin subcomplexes.Genetics. 172:1441–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutvei A., Dihlmann S., Herth W., Hurt E.C. 1992. NSP1 depletion in yeast affects nuclear pore formation and nuclear accumulation.Eur. J. Cell Biol. 59:280–295 [PubMed] [Google Scholar]
- Nehrbass U., Rout M.P., Maguire S., Blobel G., Wozniak R.W. 1996. The yeast nucleoporin Nup188p interacts genetically and physically with the core structures of the nuclear pore complex.J. Cell Biol. 133:1153–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S., Terazawa Y., Nakayama T., Hirata A., Makio T., Endo T. 2003. Nep98p is a component of the yeast spindle pole body and essential for nuclear division and fusion.J. Biol. Chem. 278:9938–9943 [DOI] [PubMed] [Google Scholar]
- Onischenko E., Stanton L.H., Madrid A.S., Kieselbach T., Weis K. 2009. Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance.J. Cell Biol. 185:475–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S.S., Rexach M.F. 2008. Discovering novel interactions at the nuclear pore complex using bead halo: a rapid method for detecting molecular interactions of high and low affinity at equilibrium.Mol. Cell. Proteomics. 7:121–131 [DOI] [PubMed] [Google Scholar]
- Patel S.S., Belmont B.J., Sante J.M., Rexach M.F. 2007. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex.Cell. 129:83–96 [DOI] [PubMed] [Google Scholar]
- Rabut G., Doye V., Ellenberg J. 2004a. Mapping the dynamic organization of the nuclear pore complex inside single living cells.Nat. Cell Biol. 6:1114–1121 [DOI] [PubMed] [Google Scholar]
- Rabut G., Lenart P., Ellenberg J. 2004b. Dynamics of nuclear pore complex organization through the cell cycle.Curr. Opin. Cell Biol. 16:314–321 [DOI] [PubMed] [Google Scholar]
- Rasala B.A., Orjalo A.V., Shen Z., Briggs S., Forbes D.J. 2006. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division.Proc. Natl. Acad. Sci. USA. 103:17801–17806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala B.A., Ramos C., Harel A., Forbes D.J. 2008. Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly.Mol. Biol. Cell. 19:3982–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M.P., Aitchison J.D., Magnasco M.O., Chait B.T. 2003. Virtual gating and nuclear transport: the hole picture.Trends Cell Biol. 13:622–628 [DOI] [PubMed] [Google Scholar]
- Ryan K.J., McCaffery J.M., Wente S.R. 2003. The Ran GTPase cycle is required for yeast nuclear pore complex assembly.J. Cell Biol. 160:1041–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K.J., Zhou Y., Wente S.R. 2007. The karyopherin Kap95 regulates nuclear pore complex assembly into intact nuclear envelopes in vivo.Mol. Biol. Cell. 18:886–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell D.R., Butler G., Wolfe K.H. 2007. Yeast genome evolution–the origin of the species.Yeast. 24:929–942 [DOI] [PubMed] [Google Scholar]
- Scarcelli J.J., Hodge C.A., Cole C.N. 2007. The yeast integral membrane protein Apq12 potentially links membrane dynamics to assembly of nuclear pore complexes.J. Cell Biol. 178:799–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R., Hitomi M., Ivessa A.S., Fasch E.V., Kohlwein S.D., Tartakoff A.M. 1996. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex.Mol. Cell. Biol. 16:7161–7172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcheprova Z., Baldi S., Frei S.B., Gonnet G., Barral Y. 2008. A mechanism for asymmetric segregation of age during yeast budding.Nature. 454:728–734 [DOI] [PubMed] [Google Scholar]
- Shulga N., Roberts P., Gu Z., Spitz L., Tabb M.M., Nomura M., Goldfarb D.S. 1996. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation.J. Cell Biol. 135:329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N., Mosammaparast N., Wozniak R., Goldfarb D.S. 2000. Yeast nucleoporins involved in passive nuclear envelope permeability.J. Cell Biol. 149:1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavru F., Hulsmann B.B., Spang A., Hartmann E., Cordes V.C., Gorlich D. 2006a. NDC1: a crucial membrane-integral nucleoporin of metazoan nuclear pore complexes.J. Cell Biol. 173:509–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavru F., Nautrup-Pedersen G., Cordes V.C., Gorlich D. 2006b. Nuclear pore complex assembly and maintenance in POM121- and gp210-deficient cells.J. Cell Biol. 173:477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia C., Blobel G., Rout M.P. 1995. Isolation and characterization of nuclear envelopes from the yeast Saccharomyces.J. Cell Biol. 131:19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam M., Wente S.R. 2003. Peering through the pore: nuclear pore complex structure, assembly, and function.Dev. Cell. 4:775–789 [DOI] [PubMed] [Google Scholar]
- Tcheperegine S.E., Marelli M., Wozniak R.W. 1999. Topology and functional domains of the yeast pore membrane protein Pom152p.J. Biol. Chem. 274:5252–5258 [DOI] [PubMed] [Google Scholar]
- Timney B.L., Tetenbaum-Novatt J., Agate D.S., Williams R., Zhang W., Chait B.T., Rout M.P. 2006. Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo.J. Cell Biol. 175:579–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E.J., Wente S.R. 2006. Dynamic nuclear pore complexes: life on the edge.Cell. 125:1041–1053 [DOI] [PubMed] [Google Scholar]
- Walther T.C., Alves A., Pickersgill H., Loiodice I., Hetzer M., Galy V., Hulsmann B.B., Kocher T., Wilm M., Allen T., et al. 2003. The conserved Nup107-160 complex is critical for nuclear pore complex assembly.Cell. 113:195–206 [DOI] [PubMed] [Google Scholar]
- Wente S.R., Blobel G. 1993. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic.J. Cell Biol. 123:275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente S.R., Rout M.P., Blobel G. 1992. A new family of yeast nuclear pore complex proteins.J. Cell Biol. 119:705–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Yarar D., Giddings T.H., Jr., Mastronarde D.N. 1997. Nuclear pore complex number and distribution throughout the Saccharomyces cerevisiae cell cycle by three-dimensional reconstruction from electron micrographs of nuclear envelopes.Mol. Biol. Cell. 8:2119–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R.W., Rout M.P., Aitchison J.D. 1998. Karyopherins and kissing cousins.Trends Cell Biol. 8:184–188 [DOI] [PubMed] [Google Scholar]
- Wright R., Rine J. 1989. Transmission electron microscopy and immunocytochemical studies of yeast: analysis of HMG-CoA reductase overproduction by electron microscopy.Methods Cell Biol. 31:473–512 [DOI] [PubMed] [Google Scholar]
- Yang Q., Rout M.P., Akey C.W. 1998. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications.Mol. Cell. 1:223–234 [DOI] [PubMed] [Google Scholar]
- Zabel U., Doye V., Tekotte H., Wepf R., Grandi P., Hurt E.C. 1996. Nic96p is required for nuclear pore formation and functionally interacts with a novel nucleoporin, Nup188p.J. Cell Biol. 133:1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]