Abstract
Soluble N-ethyl-maleimide sensitive fusion protein attachment protein receptors (SNAREs) are hypothesized to trigger membrane fusion by complexing in trans through their membrane-distal N termini and zippering toward their membrane-embedded C termini, which in turn drives the two membranes together. In this study, we use a set of truncated SNAREs to trap kinetically stable, partially zipped trans-SNARE complexes on intact organelles in the absence of hemifusion and content mixing. We show that the C-terminal zippering of SNARE cytoplasmic domains controls the onset of lipid mixing but not the subsequent transition from hemifusion to full fusion. Moreover, we find that a partially zipped nonfusogenic trans-complex is rescued by Sec17, a universal SNARE cochaperone. Rescue occurs independently of the Sec17-binding partner Sec18, and it exhibits steep cooperativity, indicating that Sec17 engages multiple stalled trans-complexes to drive fusion. These experiments delineate distinct functions within the trans-complex, provide a straightforward method to trap and study prefusion complexes on native membranes, and reveal that Sec17 can rescue a stalled, partially zipped trans-complex.
Introduction
Fusion is the terminal step of most membrane trafficking processes in the secretory and endocytic systems. It requires that two bilayers be brought into close proximity and disrupted in a controlled manner, facilitating the local rearrangement of lipids into a single topologically continuous bilayer. SNARE proteins are thought to catalyze fusion by assembling into tight membrane-bridging (trans-) complexes (Hanson et al., 1997a; Jahn and Scheller, 2006; Rizo and Rosenmund, 2008). Analyses of membrane-free SNARE core complexes revealed that four SNARE domains pack into a parallel coiled-coil bundle with the integral membrane anchors held together at the bundle’s C-terminal end (Hanson et al., 1997b; Sutton et al., 1998; Xiao et al., 2001; Antonin et al., 2002). These observations suggested that complete folding of a trans-SNARE complex should be possible only when the docked membranes are in extremely close proximity and prompted the suggestion that “zippering” of trans-SNARE complexes catalyzes fusion (Hanson et al., 1997a,b; Weber et al., 1998). In this model, trans-SNARE complexes nucleate at the membrane-distal SNARE N termini and zipper toward the membrane-embedded C termini. Energy liberated by a concerted oligomerization and folding reaction is thereby coupled to the mechanical work of pulling the membranes into tight apposition.
The zippering hypothesis was supported by experiments with secretory cell preparations and with purified SNAREs. These studies demonstrated distinct outcomes for N- versus C-terminal disruptions of SNARE complexes (Matos et al., 2003; Pobbati et al., 2006; Sorensen et al., 2006; Li et al., 2007). In experiments with reconstituted proteoliposomes and living cells, the insertion of flexible linkers between SNARE domains and transmembrane anchors diminished fusion, presumably by allowing trans-complexes to zipper fully without bringing the membranes into close apposition. (McNew et al., 1999; Wang et al., 2001; Kesavan et al., 2007). However, major questions about the precise relationship between SNARE zippering and fusion are unanswered.
Trans-SNARE complex assembly and fusion occur spontaneously in reconstituted systems, supporting the notion that SNAREs represent a minimal membrane fusion machinery (Weber et al., 1998; Giraudo et al., 2005). However, in living cells, fusion is subject to stringent spatiotemporal regulation by Rab GTPases, tethering proteins, SNARE-binding proteins, and other factors that conspire to control SNARE availability, trans-SNARE complex assembly, and fusion. A major challenge is to discover how these factors guide, regulate, and respond to the assembly of prefusion trans-complexes. Under normal conditions, trans-SNARE complexes are evanescent, neither long lived nor abundant at steady state. Moreover, it is notoriously difficult to distinguish prefusion trans-SNARE complexes from postfusion cis-SNARE complexes. Consequently, the molecular compositions and configurations of prefusion complexes (containing trans-complexed SNAREs and the associated accessory factors) remain largely undefined, as do the biochemical pathways leading to assembly of these complexes.
Ca2+-stimulated exocytosis can occur on a submillisecond time scale, which is likely faster than trans-complex nucleation and assembly. Experiments with exocytic and synaptic model systems implied that docked vesicles attach to target membranes through “loose” (i.e., partially zipped) trans-SNARE complexes (Xu et al., 1998; Hua and Charlton, 1999; Xu et al., 1999; Lagow et al., 2007). Thus, at least in these systems, the fusion pathway might traverse a metastable, partially zipped intermediate that undergoes a triggered transition from loose (partial) to “tight” (complete) zippering. It is unknown whether vesicles docked through such a mechanism would have partially completed the fusion reaction (as in a hemifusion intermediate; Schaub et al., 2006; Zampighi et al., 2006; Wong et al., 2007). It is also unknown whether the tightening of partially zipped SNARE complexes could be sufficient, or even necessary, to trigger fusion.
In this study, we dissect the zippering of trans-SNARE complexes on an intact organelle, the Saccharomyces cerevisiae vacuolar lysosome. The cell-free assay of homotypic vacuole fusion offers unique advantages. It allows us to measure trans-SNARE complex assembly, lipid mixing, and content mixing in a preparation of intact organelles replete with native fusion-regulatory factors (Haas, 1995; Merz and Wickner, 2004a; Reese and Mayer, 2005; Collins and Wickner, 2007; Jun and Wickner, 2007). Vacuolar trans-SNARE complex assembly and fusion are stringently controlled by the Rab GTPase Ypt7 and its cognate tethering complex, Vps-C–HOPS (homotypic fusion and vacuole protein sorting complex; Seals et al., 2000). HOPS contains at least six subunits, including the Ypt7 activator Vps39 and the Ypt7 effector Vps41 (Wurmser et al., 2000; Brett et al., 2008). HOPS also contains Vps33, a Sec1/Munc-18 family protein that is thought to catalyze trans-complex assembly or to regulate SNARE function (Collins et al., 2005).
In this study, a set of purified, truncated SNAREs provides precise control over the extent of C-terminal (membrane proximal) SNARE complex zippering. These truncated SNAREs are added acutely to purified vacuoles, precluding indirect pleiotropic effects on membrane traffic, a bane of conventional genetic approaches. We capture a stalled assembly intermediate in which kinetically stable trans-SNARE complexes have formed but neither lipid nor content mixing has initiated. We show that C-terminal zippering beyond that needed for trans-complex assembly initiates lipid mixing but that downstream stages of the reaction are not directly controlled by C-terminal zippering of the SNARE cytoplasmic domains. Finally, we demonstrate that a stalled, partially zipped trans-complex is rendered fusogenic by a universal SNARE complex–binding protein, Sec17 (yeast α-SNAP).
Results
Experimental perturbation of SNARE C-terminal zippering
In all systems studied to date, four SNARE domains, designated R, Qa, Qb, and Qc are required for trans-complex assembly and fusion catalysis (Sutton et al., 1998; Jahn and Scheller, 2006). Freshly isolated yeast vacuoles bear free R, Qa, and Qb (but not Qc) SNAREs; they also bear stable cis-complexes that contain R, Qa, and Qb along with the Qc Vam7 (Ungermann et al., 1998a; Thorngren et al., 2004; Collins et al., 2005). Vam7 is soluble and is targeted to the membrane by an N-terminal phox homology (PX) domain (Cheever et al., 2001; Boeddinghaus et al., 2002). Because the Vam7 on isolated vacuoles is sequestered within cis-complexes, it is functionally inert. This native Vam7 cannot participate in fusion unless liberated through the ATP-dependent priming activity of Sec17 and Sec18 (Boeddinghaus et al., 2002). Purified Vam7 potently stimulates homotypic vacuole fusion, completely bypassing the requirement for ATP, Sec17, and Sec18-dependent priming (Merz and Wickner, 2004a,b; Thorngren et al., 2004). The priming bypass fusion reaction is on pathway, as it requires GTP-bound Ypt7 (the vacuolar Rab), the Vps-C–HOPS effector complex, and unpaired Qa, Qb, and R SNAREs (Thorngren et al., 2004).
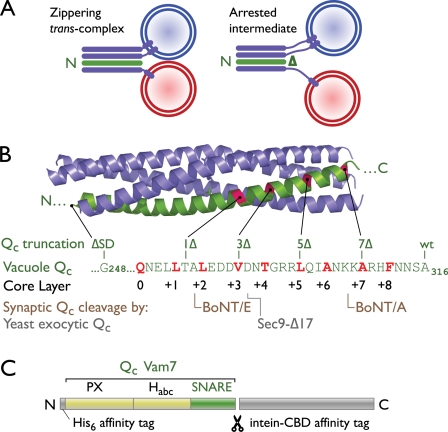
We hypothesized that the removal of C-terminal residues from Vam7 should facilitate partial trans-SNARE complex assembly with zippering arrested at the C-terminal core layer residue of the truncated protein (Fig. 1 A). We therefore prepared a set of C-terminally truncated rVam7 (Qc) mutants (Fig. 1, B and C). These proteins are designated QcXΔ, where X denotes the most C-terminal (membrane proximal) core-packing layer to which the mutant’s Qc-SNARE domain can contribute. We also prepared a mutant lacking the entire SNARE domain (QcΔSD).
Figure 1.
Strategy to arrest trans-SNARE complex zippering. (A) A schematic of rationale is shown. N- to C-terminal zippering of trans-SNARE complexes (purple and green) drives membranes into close apposition. A C-terminally truncated Qc-SNARE is predicted to permit only partial zippering. (B) Experimental constructs are shown. The structure of an endosomal SNARE complex (Protein Data Bank accession no. 1GL2; Antonin et al., 2002) highlights the Qc chain in green. In the aligned Vam7 (vacuolar Qc) amino acid sequence, core-packing layer residues are highlighted in red. The C-terminal residue present in each Qc construct is indicated. Also shown are locations of truncation after BoNT A and E cleavage of the neuronal Qc, SNAP-25 (Binz et al., 1994), and the location of the Qc truncation in a dominant-negative Saccharomyces cerevisiae exocytic SNARE mutant (Sec9-Δ17; Rossi et al., 1997). (C) rVam7 (Qc) domain structure and expression constructs. A two-tag purification strategy was used to obtain homogenous preparations of each Qc protein. The C-terminal intein/chitin-binding domain (CBD) affinity tag was removed during purification to yield the His6-Vam7 (Qc) products used in this study. wt, wild type.
C-terminal SNARE zippering controls fusion
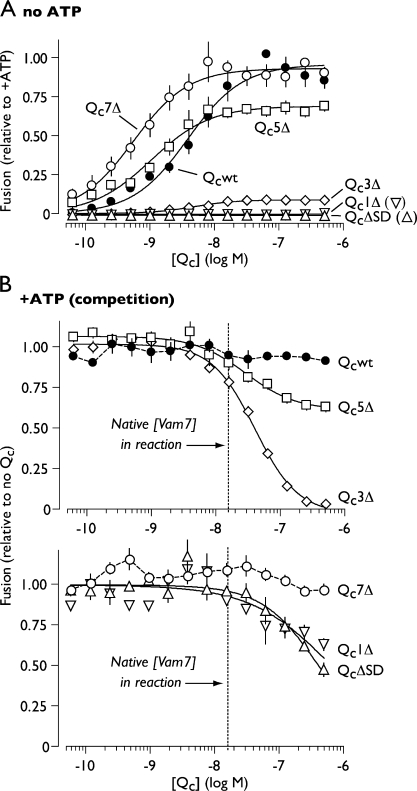
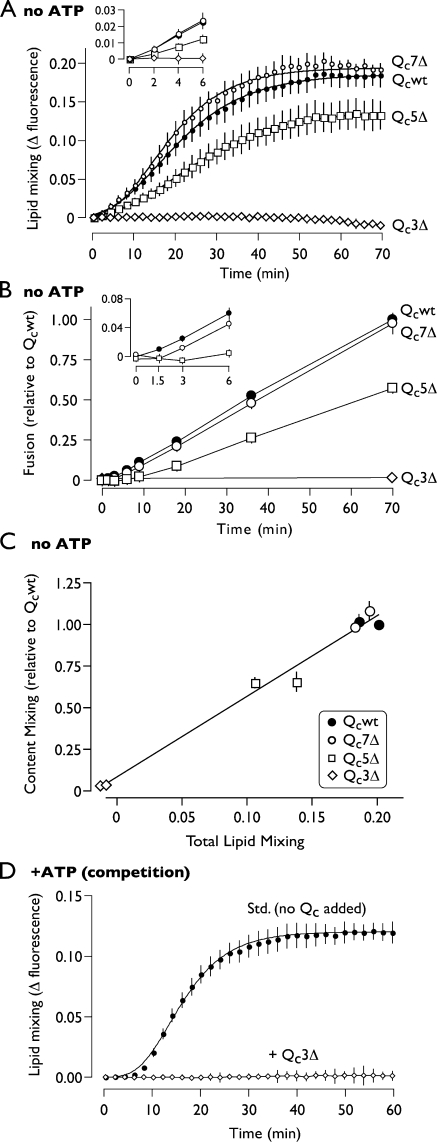
In priming bypass fusion assays, purified Qcwt (Qc wild type) stimulated robust fusion (content mixing) with a 50% effective concentration (EC50) of ∼4 nM (Fig. 2 A and Table I). C-terminal truncation had two consequences. First, the mild truncation mutants Qc7Δ and Qc5Δ were three to seven times more potent than Qcwt, eliciting substantial fusion at subnanomolar concentrations. Thus, the extreme C terminus of Vam7 negatively regulates fusion. Second, truncation beyond the +7 core layer resulted in the progressive loss of fusion activity. At saturating concentrations, Qc5Δ stimulated ∼65% as much fusion as Qcwt. In contrast, Qc3Δ was nearly inactive, eliciting ∼5% as much fusion as Qcwt. Qc1Δ and QcΔSD were completely inactive (Fig. 2 A).
Figure 2.
Fusion driven by truncated Qc-SNAREs. (A) Homotypic vacuole fusion driven by truncated Qc proteins in priming bypass reactions lacking ATP. Solid lines show best-fit sigmoidal dose-response curves. Error bars indicate mean ± SEM (n ≥ 4 experiments). (B) Competitive inhibition by truncated Qc-SNAREs. Vacuole fusion was driven by ATP, Sec17, and Sec18 in the presence of Qc proteins. Solid lines represent best-fit dose-inhibition curves. The concentration of native Qc (Vam7) in a standard in vitro fusion reaction is indicated (Thorngren et al., 2004). Extended dose-response curves for Qc1Δ and QcΔSD are shown in Fig. S1. Fit parameters are presented in Table I. Relative fusion value of 1 equals 3.3 fusion units. Error bars span ±1 SEM (n ≥ 4 experiments).
Table I.
Curve fit parameters for selected Qc and Sec17 dose-response relationships
| Protein | Figure | Experimental condition | EC50 | IC50 | Hill coefficient | r2 |
| nM | nM | |||||
| Qcwt | 2 A | No ATP (priming bypass) | 3.74 ± 0.66 | NA | 1.04 ± 0.15 | 0.979 |
| Qc7Δ | 2 A | No ATP (priming bypass) | 0.55 ± 0.07 | NA | 1.03 ± 0.11 | 0.982 |
| Qc5Δ | 2 A | No ATP (priming bypass) | 1.05 ± 0.15 | NA | 0.96 ± 0.11 | 0.982 |
| Qc3Δ | 2 A | No ATP (priming bypass) | 4.75 ± 0.53 | NA | 1.34 ± 0.16 | 0.989 |
| Qc5Δ | 2 C | +ATP (competition with wt) | NA | 28.9 ± 7.8 | −1.10 ± 0.25 | 0.947 |
| Qc3Δ | 2 C | +ATP (competition with wt) | NA | 40.6 ± 3.4 | −1.30 ± 0.12 | 0.996 |
| Qc1Δ | 2 C and S1 | +ATP (competition with wt) | NA | 656 ± 110 | −0.70 ± 0.08 | 0.733 |
| QcΔSD | 2 C and S1 | +ATP (competition with wt) | NA | 417 ± 45 | −0.99 ± 0.10 | 0.841 |
| Qc GST-PX | S1 | +ATP (competition with wt) | NA | 2,010 ± 247 | −0.89 ± 0.09 | 0.892 |
| Qcwt | 6 A | No ATP, +94 nM Sec17 | 34.3 ± 4.8 | NA | 0.94 ± 0.08 | 0.997 |
| Qc7Δ | 6 A | No ATP, +94 nM Sec17 | 8.6 ± 1.1 | NA | 1.13 ± 0.16 | 0.990 |
| Qc5Δ | 6 A | No ATP, +94 nM Sec17 | 11.7 ± 1.2 | NA | 1.33 ± 0.13 | 0.996 |
| Qc3Δ | 6 A | No ATP, +94 nM Sec17 | 19.9 ± 1.6 | NA | 1.29 ± 0.07 | 0.999 |
| Sec17 wt | 6 C | No ATP, +75 nM Qc3Δ | 125 ± 3.0 | NA | 4.97 ± 0.46 | 0.995 |
| Sec17-LALA | 6 C | No ATP, +75 nM Qc3Δ | 158 ± 5.0 | NA | 4.30 ± 0.45 | 0.992 |
| His6-Sec17 | S1 | No ATP, +75 nM Qc3Δ | 41.0 ± 3.6 | NA | 5.71 ± 2.23 | 0.933 |
IC50, 50% inhibitory concentration; NA, not applicable; wt, wild type. Errors indicate ±SEM for fits to three or more independently obtained data sets.
The compromised fusion activity of QcΔ truncation mutants could result either from reduced recruitment into prefusion complexes (loss of function) or from functional defects manifesting after QcΔ entry into prefusion complexes (gain of inhibitory function). To distinguish between these mechanisms, we assayed the activity of each recombinant Qc protein in competition with native Qc (Vam7). When fusion reactions are run in the presence of ATP, vacuolar cis-SNARE complexes are disassembled by Sec17 and Sec18, liberating native Vam7 (∼15 nM) to compete with the added Qc (Ungermann et al., 1998a; Boeddinghaus et al., 2002). Qcwt and Qc7Δ, both of which support efficient fusion in the absence of ATP, did not strongly influence ATP-driven fusion. Both Qc5Δ and Qc3Δ inhibited ATP-driven fusion at concentrations consistent with competition against native Vam7, suggesting that Qc5Δ and Qc3Δ efficiently enter prefusion complexes. Qc5Δ was a potent but incomplete fusion inhibitor. This partial competitive inhibition together with the finding that Qc5Δ is an incomplete fusogen in the absence of competitor (Fig. 2 A) defines Qc5Δ as a partial agonist of fusion. Qc3Δ was a potent and nearly complete fusion inhibitor. Together, these findings indicate that Qc5Δ and Qc3Δ are gain of function mutants that assemble into partially or completely defective prefusion complexes.
Qc1Δ and QcΔSD also inhibited fusion but did so at least an order of magnitude less potently than Qc5Δ or Qc3Δ (Fig. 2 B; Table I; and Fig. S1). Importantly, Qc1Δ and QcΔSD inhibited fusion with nearly identical potency, indicating that the partial SNARE domain of Qc1Δ, unlike that of Qc3Δ, is not itself inhibitory. The remaining inhibitory activity of these mutants is probably a result of the N-terminal PX domain, which by itself inhibits fusion at low micromolar concentrations (Fig. S1; Boeddinghaus et al., 2002; Merz and Wickner, 2004b). Together, these experiments suggest a working model in which entry of the Qc-SNARE into a prefusion complex requires a region of the Qc-SNARE domain falling between the N terminus and the +3 core-packing layer, whereas fusion catalysis requires zippering beyond layer +3.
Stable trans-complex assembly requires zippering beyond layer +1
To evaluate whether the Qc truncation mutants stimulate on-pathway (i.e., Rab dependent) assembly of kinetically stable trans-SNARE complexes, we used a direct biochemical assay of trans-complex formation (Collins and Wickner, 2007). In this assay, the Qa-SNARE Vam3::CBP (Vam3 bearing an internal calmodulin-binding affinity tag) and the R-SNARE Nyv1 are initially present on opposite membranes (Fig. 3 A). After Rab-mediated vacuole docking, the membranes are dissolved in nonionic detergent. Trans-complexes are then identified by coprecipitation of Nyv1 (R) with Vam3::CBP (Qa) on calmodulin-agarose resin. In the absence of fusion, the Qa- and R-SNAREs are on different membranes and can complex only in trans. When fusion occurs, both prefusion trans-complexes and postfusion cis-complexes that originally assembled in trans are detected. As described in subsequent sections of the Results, multiple lines of evidence indicate that under the assay conditions used in the this study, all or nearly all of the complexes recovered formed in trans, and few or none formed in cis after fusion.
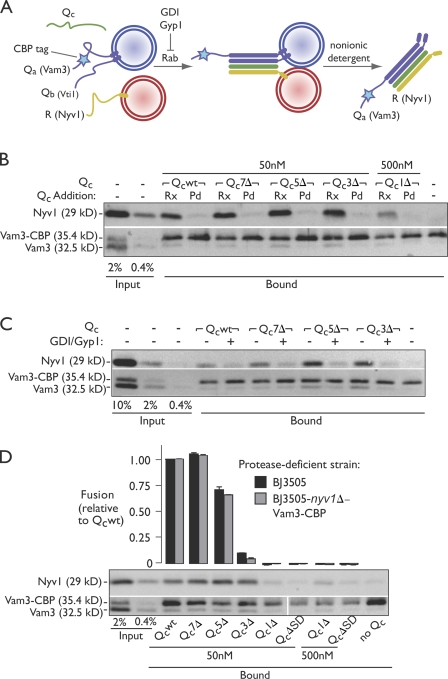
Figure 3.
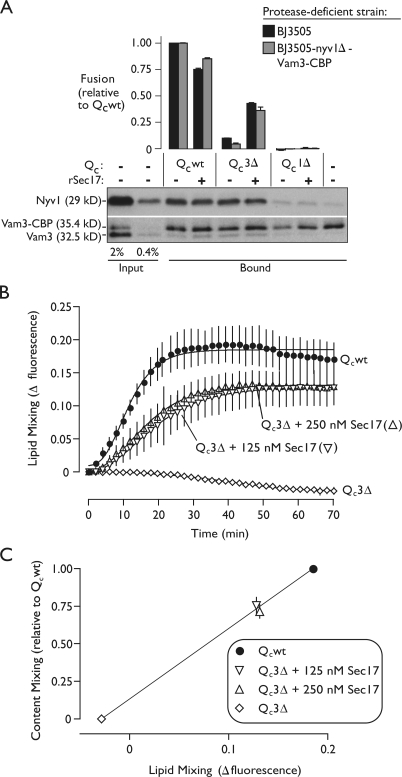
Truncated Qc proteins nucleate trans-SNARE complexes. (A) A schematic of the assay is shown. Trans-SNARE complex assembly was monitored by assaying physical association of Vam3::CBP and Nyv1, which were initially present on separate vacuoles. Pull-down reactions contained only protease-deficient vacuoles (equal mixture of vacuoles from BJ3505 and BJ3505 VAM3::CBP nyv1Δ cells). (B) Vacuolar trans-SNARE complex assembly driven by Qc proteins. Priming bypass reactions lacked ATP and contained Qc proteins as indicated. Qc proteins were added either at the start of in vitro fusion reactions (Rx) or to vacuole detergent extracts before Vam3::CBP pull-down (Pd). (C) Qc-driven trans-SNARE complex assembly requires the activated vacuolar Rab, Ypt7. Reactions were run in the absence or presence of Rab inhibitors 2.4 µM GDI and 16 µM Gyp1-46. (D) Qc-driven trans-SNARE complex assembly and fusion assayed in parallel. Trans-SNARE complex assays are representative of three independent experiments. Relative fusion value of 1 equals 1.9 fusion units. Error bars for fusion span ±1 SEM (n = 3 experiments).
Using this assay (Fig. 3, B–D), we found that Qcwt, Qc7Δ, Qc5Δ, and Qc3Δ potently stimulated trans-SNARE complex formation. In contrast, Qc1Δ and QcΔSD were unable to stimulate substantial complex formation even when added at 10-fold higher concentrations. Qc-stimulated complexes were assembled through the physiological docking pathway; Qc proteins did not stimulate complex formation when added to postreaction detergent lysates (Fig. 3 B), and complex formation was abolished when docking was blocked by Rab GTPase inhibitors (guanine nucleotide dissociation inhibitor [GDI] and Gyp1; Fig. 3 C; Ungermann et al., 1998b; Collins and Wickner, 2007). The tagged R-SNARE–deficient vacuoles used for the SNARE complex assembly assay exhibited fusion responses to the QcΔ mutants identical to responses obtained using vacuoles bearing wild-type SNAREs (Fig. 3 D).
The most important conclusions obtained with this assay are for the Qc3Δ mutant. Qc3Δ drove efficient SNARE complex assembly in the near absence of fusion and, as will be shown, in the complete absence of detectable lipid mixing. Thus, Qc3Δ-SNARE complexes are unambiguously captured in the trans-configuration. In contrast, Qcwt, Qc7Δ, and Qc5Δ all drove fusion. The assay results obtained for these Qc’s therefore reflect the sum of complexes formed in trans- plus any cis-complexes that might have formed after fusion. If substantial numbers of cis-complexes did form after fusion, we would expect to retrieve the largest amount of SNARE complex with Qcwt and Qc7Δ, less with Qc5Δ (which drives less fusion), and least of all with Qc3Δ (which does not drive fusion). But this is not the result obtained. Instead, we always retrieved as much or more SNARE complex with Qc5Δ and Qc3Δ as with Qcwt and Qc7Δ. In other words, the less opportunity there was for postfusion cis-SNARE complexes to form, the more SNARE complexes we recovered. Because ATP is not present to drive complex disassembly, the obvious inference is that the Qcwt-, Qc7Δ-, and Qc5Δ-SNARE complexes recovered in these assays, like the Qc3Δ complexes, were assembled in trans.
Collectively, our fusion experiments (Figs. 2 and 3) and biochemical results (Fig. 3) indicate that zippering beyond the +1 core-packing layer is necessary for the assembly of stable trans-SNARE complexes, whereas efficient fusion catalysis requires zippering beyond layer +3. Our results further indicate that Qc5Δ and Qc3Δ inhibit fusion by sequestering unpaired Qa-, Qb-, and R-SNAREs within partially (Qc5Δ) or completely (Qc3Δ) nonfusogenic trans-complexes. Consistent with this interpretation, we frequently observed increased trans-SNARE complex abundance for Qc3Δ and Qc5Δ compared with Qcwt and Qc7Δ (Fig. 3, B–D). This implies that Qc3Δ- and Qc5Δ-mediated trans-complexes may be even less efficient at catalyzing fusion relative to Qcwt than the raw fusion data suggest, and it indicates that trans-SNARE complexes continue to assemble and accumulate between membranes when fusion is blocked at a late stage.
Partially zipped complexes exhibit late-stage fusion defects
To further examine how incomplete zippering impairs fusion, we used stage-specific fusion inhibitors (Fig. 4 A). Because Qc3Δ competitively inhibits the standard ATP-driven reaction, we examined the kinetics with which standard reactions acquire resistance to Qc3Δ-mediated competitive inhibition. A large-scale ATP-driven fusion reaction was initiated. At intervals, 1× reaction aliquots were withdrawn and combined with the indicated inhibitor for the remainder of the incubation. GDI (a Rab inhibitor) and affinity-purified Vam3 antibody (αVam3; an inhibitor of the Qa-SNARE) block fusion at the docking stage, during which trans-SNARE complexes form. We found that ATP-driven reactions became resistant to inhibition by Qc3Δ with kinetics indistinguishable from GDI and αVam3 (Fig. 4 B). Qc3Δ therefore must be present during trans-complex assembly to function as a competitive inhibitor, again supporting the interpretation that Qc3Δ nucleates unproductive trans-complexes.
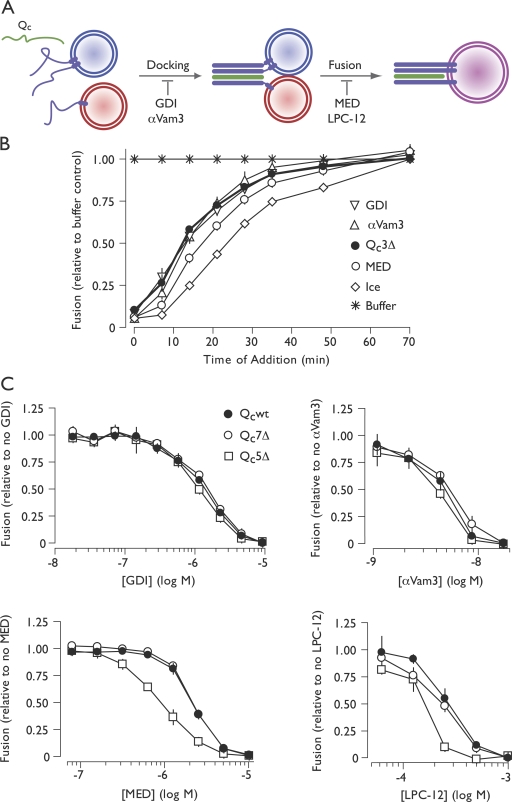
Figure 4.
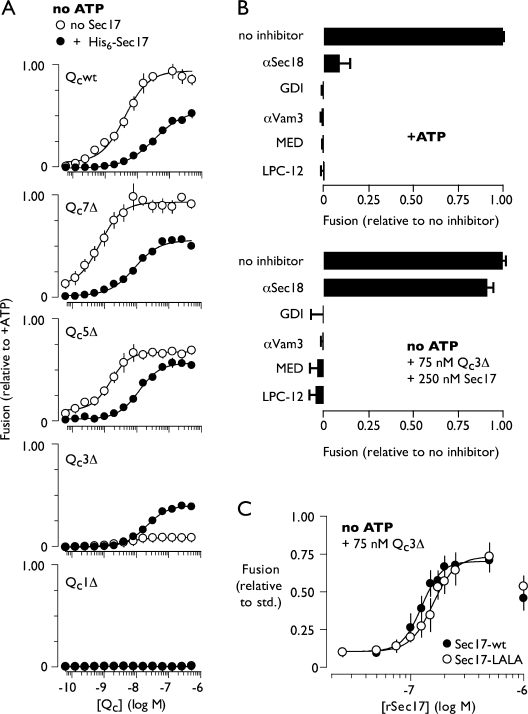
C-terminal trans-SNARE complex zipping controls a late, lipid-dependent stage of fusion. (A) Stages of vacuole fusion and stage-specific inhibitors. (B) Staging of Qc3Δ-mediated competitive inhibition. 10× scale ATP-driven reactions were initiated at t = 0. At the indicated times, 1× reaction aliquots were withdrawn and mixed with inhibitor or buffer or transferred to ice. Inhibitors were used at the following final concentrations: GDI, 2.4 µM; αVam3, 18 nM; Qc3Δ, 500 nM; MED, 10 µM. (C) Dosage sensitivity of priming bypass fusion driven by Qc proteins (40 nM Qc, no ATP) to stage-specific reaction inhibitors. Note that all values in this experiment are normalized to the no inhibitor condition for each Qc-SNARE. (B and C) Relative fusion value of 1 equals 2.7 fusion units (B), 3.1 fusion units for Qcwt and Qc7Δ (C), and 2.3 fusion units for Qc5Δ (C). Error bars span ±1 SEM (n = 3 experiments).
We next screened fusion reactions driven by Qcwt, Qc7Δ, or the partial agonist Qc5Δ for altered dosage sensitivity to stage-specific fusion inhibitors (Fig. 4 C). Fusion reactions driven by each of these Qc proteins exhibited similar dosage sensitivities to the Rab and SNARE inhibitors GDI and αVam3. This result suggests that decreasing the extent of C-terminal SNARE complex zippering does not influence the docking phase of the reaction. However, a major difference was found with two different late-stage bilayer-perturbing inhibitors, myristoylated alanine-rich C kinase substrate effector domain (MED; Fratti et al., 2004) and LPC-12 (Reese and Mayer, 2005). Both MED and LPC-12 (1-lauroyl-2-hydroxy-sn-glycero-3-phosphocholine) inhibited fusion driven by Qc5Δ more potently than fusion driven by Qcwt or Qc7Δ (Fig. 4 C). These synthetic interactions between Qc5Δ and late-stage bilayer-perturbing inhibitors implicate zippering between layers +3 and +7 in postdocking lipid-dependent fusion processes.
Zippering controls initiation of outer leaflet lipid mixing
SNARE-mediated fusion is hypothesized to involve a hemifusion intermediate (Chernomordik and Kozlov, 2008). In this intermediate state, outer (cytoplasmic) leaflet lipids intermix, but inner (lumenal) leaflets and lumenal contents remain unmixed. If the hemifusion hypothesis is valid, at least two distinct subreactions must occur: initiation of hemifusion and resolution of the hemifused intermediate to yield a fully fused product. In principle, either or both of these subreactions could be controlled by SNARE zippering.
In one model, N-terminal SNARE assembly drives hemifusion, and C-terminal zippering promotes conversion of the hemifused intermediate to a fully fused product. If this model is correct, C-terminal Qc truncations should cause accumulation of hemifused intermediates, and we should observe efficient lipid mixing with attenuated or absent content mixing. In an alternative model, C-terminal zippering controls entry into the hemifused state, but other factors (e.g., SNARE transmembrane domains) control conversion of the hemifused intermediate to a fully fused product. If this alternative model is correct, C-terminal Qc truncations should not cause accumulation of hemifused intermediates, and outer leaflet lipid mixing and content mixing should be attenuated to the same extent. To discriminate between these models, we monitored lipid and content mixing in parallel.
To assay outer leaflet lipid mixing, we incorporated rhodamine-phosphatidylethanolamine (R-PE) into donor vacuoles at a self-quenching concentration and then mixed the R-PE–labeled donor vacuoles with unlabeled acceptors. Lipid mixing between donors and acceptors results in R-PE dilution and fluorescence dequenching. Because intact vacuoles are labeled with R-PE, this assay predominantly measures mixing between proximal (cytoplasmic) bilayer leaflets. As reported previously (Reese and Mayer, 2005; Jun and Wickner, 2007), a slow fusion-independent dequenching signal is observed (Fig. S2), but when fusion is allowed to proceed, robust and highly reproducible dequenching is observed above this background. This lipid-mixing signal depends on Rab and SNARE function, and its kinetics map between docking and content mixing (Reese and Mayer, 2005; Jun and Wickner, 2007). Because the transition from lipid mixing (i.e., hemifusion) to content mixing is the rate-limiting, postdocking subreaction in vacuole fusion (Merz and Wickner, 2004a; Reese and Mayer, 2005; Jun and Wickner, 2007), any delay in the conversion of hemifused to fully fused reaction products should be readily detected when the lipid- and content-mixing signals are compared.
Lipid mixing driven by saturating concentrations of each Qc was assayed under priming bypass conditions (Fig. 5 A and Fig. S2). The results show that zippering past layer +3 up to layer +7 is required for the onset of lipid mixing and presumably for hemifusion. This is illustrated by the partial defect in lipid mixing exhibited by Qc5Δ and by the complete defect exhibited by Qc3Δ (Fig. 5 A). When lipid and content mixing were compared (Fig. 5, A and B), lipid mixing in each case preceded content mixing, and the rates of lipid and content mixing were proportional for each Qc. The final extents of lipid and content mixing were also closely correlated for each mutant (Fig. 5 C). Collectively, these results indicate that kinetically stable, partially zipped trans-SNARE complexes can assemble on native membranes in the absence of lipid or content mixing. Zippering beyond layer +3 is required to initiate lipid mixing with efficiency increasing as zippering proceeds to layer +7. This conclusion is further buttressed by the finding that Qc3Δ, in competition with native Vam7, totally suppressed SNARE-dependent lipid mixing (Fig. 5 D). The failure of Qc5Δ-driven reactions to disproportionately accumulate hemifused intermediates indicates that once a hemifused intermediate is established, the rate of the transition to full fusion is not substantially controlled by the extent of C-terminal zippering of the SNARE cytoplasmic domains.
Figure 5.
C-terminal trans-SNARE complex zippering initiates lipid mixing. (A) Lipid mixing in priming bypass reactions driven by saturating concentrations (∼2 × EC90) of the indicated Qc proteins (Qcwt, 64 nM; Qc7Δ, 16 nM; Qc5Δ, 16 nM; Qc3Δ, 64 nM). Plotted values are normalized to the fluorescence at infinite probe dilution (a standard reaction including 0.33% Triton X-100) and are corrected for background (SNARE independent) dequenching by subtracting the fluorescence change measured from reactions inhibited by 17.6 nM anti-Vam3. Uncorrected traces are presented in Fig. S2. Best-fit curves (Gompertz function) are also shown. (B) Content mixing (fusion) in priming bypass reactions driven by saturating concentrations (∼2 × EC90) of the indicated Qc proteins (Qcwt, 50 nM; Qc7Δ, 10 nM; Qc5Δ, 21 nM). (C) Correlation of endpoint lipid and content-mixing values obtained from fusion reactions driven by the indicated Qc proteins. Data for lipid and content mixing were acquired from the same reactions, with content-mixing endpoints quantified after collection of lipid-mixing data. The total extent of lipid mixing was obtained from the Gompertz fit to the aggregate time series for each Qc protein. Individual data points and error bars result from reactions driven by each Qc at either 16 nM or 64 nM. (D) Lipid mixing in reactions incubated with an ATP regenerating system in the absence or presence of 500 nM Qc3Δ. Data for D are normalized and baseline subtracted as for A; uncorrected traces are presented in Fig. S2. Relative fusion value of 1 equals 2.0 fusion units in B and C. Insets shows early time points. Error bars span ±1 SEM (n = 3 experiments).
Sec17-mediated rescue of Qc3Δ trans-complexes
To gain additional insight into why partially zipped Qc3Δ-mediated trans-SNARE complexes are fusion deficient, we sought conditions that might rescue the catalytic function of these complexes. Fortuitously, we discovered that addition of the universal SNARE cochaperone Sec17 rescues fusion in Qc3Δ-driven reactions lacking ATP (Fig. 6 A). This was completely unexpected because the best-characterized function of Sec17 is in postfusion cis-SNARE recycling, not fusion catalysis (Mayer et al., 1996; Ungermann et al., 1998a; Thorngren et al., 2004). The fusion-promoting activity of recombinant Sec17 was observed only with Qc3Δ. Sec17 partially inhibited fusion driven by Qcwt, Qc7Δ, and Qc5Δ and completely failed to rescue the assembly defective mutant Qc1Δ (Fig. 6 A). Partially zipped trans-SNARE complexes are therefore a prerequisite for Sec17-mediated rescue. Rescue was sensitive to Rab, SNARE, and bilayer-perturbing inhibitors (Fig. 6 B), indicating that Sec17 rescues fusion without fundamentally altering the reaction pathway.
Figure 6.
Qc3Δ function is rescued by excess Sec17. (A) Selective rescue of Qc3Δ by Sec17. Fusion driven by C-terminal Qc truncation mutants in the absence of ATP and in the presence of Sec17 (94 nM). (B) Sec17-mediated rescue occurs through the normal fusion pathway and does not involve Sec18. Sensitivity of Qc3Δ + Sec17-driven fusion to docking and fusion inhibitors. Reagents were included at the following final concentrations: Qc3Δ, 75 nM; Sec17, 94 nM; αSec18, 2.0 µM (total IgG); GDI, 2.4 µM; αVam3, 17.6 nM; MED, 10 µM; LPC-12, 500 µM. (C) Rescue of Qc3Δ by a Sec17 mutant that cannot interact with Sec18. Priming bypass reactions lacked ATP and contained Qc3Δ (75 nM) along with the indicated concentrations of Sec17 or Sec17-LALA. Sec17 used in A was purified as a His6-tagged protein and included at 94 nM (Fig. S3); both forms of Sec17 used in B and C were untagged. Fit parameters for A and C are presented in Table I. Relative fusion value of 1 equals 3.5 fusion units for the +ATP condition and 1.6 fusion units for Sec17 rescue condition (B, bottom). Error bars indicate ±1 SEM (n = 3 experiments).
Sec17 mediates binding of the AAA ATPase Sec18 (yeast NSF) to SNARE complexes, allowing Sec18 to power SNARE complex disassembly. Because the rescue reaction lacks ATP, it is unlikely that Sec18 is required for Sec17-mediated rescue. The absence of a Sec18 requirement was confirmed by two additional experiments. First, Sec18 antibodies, which inhibit the standard ATP-driven, Sec18-dependent fusion reaction (Haas and Wickner, 1996), did not significantly inhibit the rescue reaction (Fig. 6 B). Second, a Sec17 mutant with two residues essential for stimulation of Sec18 ATP ase activity (Barnard et al., 1996) changed from Leu- to Ala (Sec17-LALA)-rescued Qc3Δ reactions to the same extent as wild-type Sec17 (Table I; Fig. 6 C; and Figs. S3 and S4). Thus, Sec17 selectively rescues Qc3Δ trans-complexes without assistance from Sec18.
Mechanism of Sec17-mediated rescue
How does Sec17 rescue Qc3Δ trans-SNARE complexes? We found that dose-response curves for Sec17 in Qc3Δ rescue reactions were steeply sigmoidal, indicating a cooperative mechanism of Sec17 action (Fig. 6 C; Table I; and Fig. S3). In dose-response experiments with untagged Sec17, the Hill coefficient was 5.0 ± 0.46 (mean ± SEM; 95% confidence interval 3.9–6.1). Indistinguishable Hill coefficients (Table I) were obtained independently for Sec17-LALA and for hexahistidine-tagged Sec17 (note that the extent but not the cooperativity of rescue was reduced when an N-terminal His6 tag was present). These large Hill coefficients for Sec17-mediated rescue diverged dramatically from the coefficients obtained for the various Qc proteins, all of which were close to unity (Table I). Biochemical and structural studies from several groups indicate that Sec17 binds SNARE core complexes with 3:1 stoichiometry (Hohl et al., 1998; Rice and Brunger, 1999; Wimmer et al., 2001; Marz et al., 2003). Moreover, the Qc3Δ trans-complex, when zipped to its full potential, should present a complete Sec17-binding surface (Fig. S5; Marz et al., 2003). The most straightforward interpretation of these findings (see Discussion) is that a minimum of four to six Sec17 molecules sequentially engages a minimum of two Qc3Δ trans-SNARE complexes to rescue fusion.
Sec17 binding to Qc3Δ trans-complexes might stimulate fusion by stabilizing preformed, partially zipped complexes in a fusion-competent conformation. Alternatively, Sec17 addition might increase the number of trans-complexes spanning docked membranes, allowing a larger number of weaker, partially zipped complexes to overcome the same energy barrier as a smaller number of wild-type complexes. To evaluate these models, we measured the amount of Qc3Δ trans-SNARE complex formed in the absence or presence of added Sec17. There was no change in the steady-state quantity of Qc3Δ-stimulated trans-complex (Fig. 7 A), indicating that Sec17 rescues fusion by augmenting the function of existing Qc3Δ trans-complexes, not by increasing the total number of Qc3Δ trans-complexes. Note that this experiment also provides further evidence that our biochemical assay of trans-SNARE pairing does not recover cis-complexes that form after fusion. If such complexes formed, we would expect to retrieve more SNARE complexes with Qc3Δ and Sec17 addition (where there is substantial fusion) than with Qc3Δ alone (where there is very little). But in each case, we retrieve similar numbers of complexes, indicating that our assay retrieves few or no complexes assembled in cis after fusion has occurred.
Figure 7.
Sec17 rescues Qc3Δ by interacting with preassembled, partially zipped trans-complexes and driving outer leaflet lipid mixing. (A) Trans-SNARE complex assembly and fusion stimulated by the indicated Qc proteins were monitored in parallel, as in Fig. 3 D, in the absence or presence of Sec17. Priming bypass reactions included 50 nM of the indicated Qc protein and 94 nM His6-Sec17. The blot is representative of three independent experiments. (B) Lipid mixing in priming bypass reactions driven by Qcwt (64 nM) or Qc3Δ (75 nM) with 0, 125, or 250 nM Sec17. Plotted values are normalized to the fluorescence at infinite probe dilution (a standard reaction including 0.33% Triton X-100) and are corrected for background (SNARE independent) dequenching by subtracting the fluorescence change measured from reactions lacking a fusion stimulus. Best-fit curves (Gompertz) are also shown. (C) Correlation of endpoint lipid and content-mixing values obtained from fusion reactions driven by the indicated Qc proteins. Data for lipid and content mixing were acquired from the same reactions, with content-mixing endpoints quantified after collection of lipid-mixing data. The final extent of lipid mixing was obtained from the Gompertz fit to the aggregate time series for each condition. Relative fusion value of 1 equals 1.9 fusion units for A and 5.5 fusion units for C. Error bars span ±1 SEM (n = 3 experiments).
To further characterize the rescue reaction, we monitored outer leaflet lipid mixing and content mixing in parallel in reactions driven by Qc3Δ and Sec17 (Fig. 7, B and C). Under conditions of maximum rescue, we found that lipid- and content-mixing signals were correlated just as in reactions containing Qcwt, Qc7Δ, and Qc5Δ. Thus, by several criteria, Sec17 binding augments the function of Qc3Δ trans-complexes, rescuing fusion through an overall pathway that closely resembles the normal zippering-mediated fusion mechanism.
Discussion
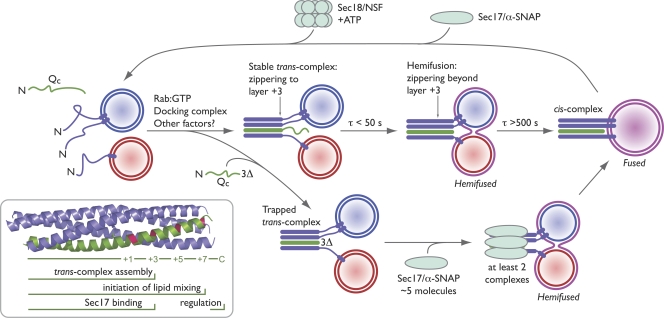
It is clear that SNARE proteins are core components of the fusion machinery and that trans-SNARE complex assembly is a pivotal step in the fusion pathway. However, surprisingly little is understood about the molecular mechanisms governing trans-SNARE zippering and the mechanistic role of zippering in driving fusion. This is due in part to technical challenges. Chemically defined systems provide crucial information about the capabilities of the components in the system, but proteoliposomes at best approximate biological membranes, and in any case, observations with synthetic systems must be validated with biological preparations. Genetic approaches permit us to study physiology in situ but are often complicated by off-pathway pleiotropic effects. Progress has also been impeded by the difficulty of discriminating between cis- and trans-complexes and by a paucity of methods for kinetic trapping and accumulation of on-pathway fusion complex assembly intermediates. To sidestep these limitations, we combined a well-characterized in vitro docking and fusion system with acute administration of engineered recombinant SNARE proteins. The major results of our experiments are summarized in Fig. 8. Arresting zippering at layer +3 traps an on-pathway, trans-paired prefusion intermediate with little or no fusion; assembly beyond layer +3 up to layer +7 controls the onset of lipid mixing but does not appear to control the efficiency or kinetics of the subsequent transition from hemifusion to content mixing. Moreover, the SNARE cochaperone Sec17 rescues an arrested, partially zipped trans-SNARE complex, raising anew the possibility that at least in certain contexts Sec17 functions before as well as after fusion.
Figure 8.
Working model of trans-SNARE zippering in vacuolar docking and fusion. The time constants for posttethering trans-SNARE assembly, lipid mixing, and content mixing are derived from previous studies (Merz and Wickner, 2004a; Jun and Wickner, 2007). (inset) The crystal structure of the endosomal SNARE complex (Protein Data Bank accession no. 1GL2; Antonin et al., 2002) is shaded as in Fig. 1 B. Horizontal bars indicate the region of the SNARE complex implicated in each function. See Discussion for further details.
Generality of zippering in SNARE-mediated fusion
There are strong parallels between SNARE zippering during exocytosis at the metazoan neuronal synapse, exocytosis in yeast, and homotypic fusion of the yeast vacuolar lysosome. Botulinum neurotoxins (BoNTs) E and A are site-selective proteases that cleave SNAP-25, a mammalian exocytic SNARE (Montecucco et al., 2005; Sakaba et al., 2005). BoNT E cleavage of SNAP-25 after layer +1 blocks SNARE complex formation altogether, whereas on the yeast vacuole, we find that a Qc1Δ mutant is unable to support trans-SNARE complex assembly. BoNT A cleaves after layer +6 of the SNAP-25 Qc-SNARE domain (Fig. 1 B) and raises the Ca2+ threshold for exocytosis, a result consistent with our finding that the Qc5Δ mutant exhibits a partial defect in fusion manifesting after trans-complex assembly. No neurotoxin is known to cleave a Qc-SNARE at layer +3, but truncation of the yeast SNAP-25 paralogue Sec9 after layer +3 (Fig. 1 B) resulted in a protein that could assemble into cis-SNARE complexes in vitro and conferred a dominant lethal phenotype attributed to defective exocytosis (Rossi et al., 1997). These findings are consistent with our results for the Qc3Δ mutant. It appears that not only the structures of SNARE core complexes but also the zippering requirements for trans-complex assembly and fusion will be broadly conserved across phyla and between different transport pathways.
Zippering, hemifusion, and content mixing
Theoretical considerations and accreting experimental evidence suggest that passage through a hemifused intermediate state should be an obligate step in many membrane fusion reactions (Chernomordik and Kozlov, 2008; Jackson and Chapman, 2008). However, the relationship between SNARE zippering and lipid and content mixing has not been thoroughly analyzed, and in native biological membrane systems is almost completely unexplored. Insertion of linkers between the R-SNARE domain and the transmembrane anchor of an exocytic SNARE indicated that the stability of fusion pores and the dynamics of pore opening are perturbed as SNARE zippering and membrane proximity are progressively uncoupled (Kesavan et al., 2007), but lipid mixing was not examined in these experiments. In experiments with synthetic liposomes, linker insertion inhibited lipid mixing, but content mixing was not monitored in this study (McNew et al., 1999). Electron tomography, fluorescence studies, and experiments with liposomes suggest that docked secretory vesicles, which might attach through partially zipped SNARE complexes, exist in a hemifused state (Schaub et al., 2006; Zampighi et al., 2006; Wong et al., 2007).
Our experiments show that partial zippering of a native trans-SNARE complex does not necessarily trigger hemifusion. Yeast vacuoles tether in a Rab-dependent reaction before trans-SNARE complex assembly (Ungermann et al., 1998b). Upon Qc addition to predocked vacuoles, trans-SNARE complex assembly and lipid mixing occur within seconds (Fig. 8, τ [time constant] < 50 s). Content mixing occurs more slowly (Fig. 8, τ > 500 s), indicating that egress from the trans-SNARE paired, hemifused intermediate is rate limiting (Merz and Wickner, 2004a; Reese and Mayer, 2005; Jun and Wickner, 2007). This study shows that stable on-pathway, partially zipped (to layer +3) trans-SNARE complexes can form between docked vacuoles without lipid mixing and thus in the absence of hemifusion.
Zippering beyond layer +3 triggers lipid and content mixing with increasing and correlated efficiency up to layer +7 (Fig. 5). The involvement of core layers +5 to +7 in initiation of lipid rearrangements is underscored by experiments with two inhibitors, MED and LPC-12 (Fig. 4). These late-stage inhibitors perturb bilayer structure and allow at least some trans-complex assembly to occur but inhibit lipid mixing (Reese and Mayer, 2005; Melia et al., 2006; Collins and Wickner, 2007). We found that fusion reactions driven by the partial agonist Qc5Δ are hypersensitive to MED and LPC-12. This synthetic defect strengthens the interpretation that layer +5 to +7 zippering, although not essential for fusion, increases the efficiency of the initial lipid-mixing step or prolongs the lifetime of a reversibly hemifused intermediate. In contrast, the subsequent conversion of the putatively hemifused intermediate to a content-mixed final fusion product does not appear to depend on complete C-terminal SNARE zippering (Fig. 5).
If SNARE domain zippering does not control the resolution of hemifusion, what does? Pioneering experiments with hemagglutinin, the influenza fusogen, indicated that the transmembrane anchor has a crucial role in egress from a hemifused intermediate (Kemble et al., 1994; Melikyan et al., 1995; Armstrong et al., 2000). Similarly, experiments with proteoliposomes and mutational analyses suggest that SNARE transmembrane anchors have important functions in late steps of fusion (McNew et al., 2000; Langosch et al., 2001; Han et al., 2004; Xu et al., 2005; Roy et al., 2006; Lu et al., 2008). Nevertheless, the possibility remains that SNAREs do not act alone in the final subreactions of fusion and that other proteins play supporting roles.
Sec17-mediated rescue of arrested trans-complexes: implications for triggered fusion
Rapid Ca2+-evoked fusion events have been hypothesized to involve triggering of metastable, partially zipped SNARE complexes (Xu et al., 1998; Hua and Charlton, 1999; Xu et al., 1999; Lagow et al., 2007). Synaptotagmins are major Ca2+ sensors thought to synchronize or trigger exocytosis in neurons and secretory cells, but the mechanisms of synaptotagmin function are complex (Chapman, 2008; Rizo and Rosenmund, 2008). Synaptotagmins bind acidic phospholipids, SNARE complexes, and other proteins in a Ca2+-dependent manner. In addition, synaptotagmins functionally interact with the SNARE-binding complexin proteins. It has been proposed that synaptotagmin binding stabilizes partially zipped trans-SNARE complexes to trigger fusion, but it has been challenging to disentangle the various functions of the synaptotagmins to rigorously test this idea (Chapman, 2008; Rizo and Rosenmund, 2008). Synaptotagmin and Sec17/α-SNAP bind to overlapping sites on the SNARE core complex, but Sec17/α-SNAP does not strongly interact with membranes, and yeast cells lack both synaptotagmin and complexin paralogues. Our discovery that Sec17 rescues partially zipped Qc3Δ trans-complexes provides synaptotagmin- and complexin-independent evidence that an extrinsic binding factor can render a partially zipped, nonfusogenic trans-SNARE complex fusogenic. This presumably occurs through Sec17-mediated stabilization of unzipped, unstructured, or partially structured C-terminal portions of the trans-SNARE complex.
Sec17/α-SNAP and Sec18/NSF were originally hypothesized to act during fusion as part of the core fusion machinery (Sollner et al., 1993). Subsequent studies demonstrated that the major function of Sec17 and Sec18 is to disassemble postfusion cis-SNARE complexes (Mayer et al., 1996; Hanson et al., 1997b). However, Ungermann et al. (1998b) suggested that Sec17 and Sec18 bind and disassemble trans- as well as cis-complexes, although this idea has been controversial (Weber et al., 2000). This study provides new evidence that Sec17 can functionally interact with trans-SNARE complexes.
These effects are quite selective: Sec17 rescues fusion mediated by Qc3Δ but inhibits fusion driven by Qcwt and Qc7Δ (Fig. 6 A). Moreover, Sec17 decreases the potency of the partial agonist Qc5Δ but has little effect on the overall efficiency of Qc5Δ-mediated fusion. Sec17 rescues Qc3Δ complexes in the absence of Sec18 activity (Fig. 6, B and C). We speculate that, more generally, Sec17 binding might rescue a subset of defective, partially zipped trans-complexes and that Sec18 then engages in kinetic proofreading to disassemble any remaining stalled prefusion complexes. Experimental tests of this hypothesis are under way in our laboratory.
Sec17-mediated rescue of vacuolar Qc3Δ complexes, like evoked exocytosis, displays strong biochemical cooperativity. Ca2+-evoked fusion requires binding of at least four to five Ca2+ ions (Dodge and Rahamimoff, 1967; Lando and Zucker, 1994) and is proposed to require at least three SNARE complexes (Hua and Scheller, 2001). Our experiments show that Sec17 rescue of Qc3Δ-mediated fusion is also highly cooperative, with a Hill coefficient of 5.0 ± 1.1 (95% confidence interval) for the Sec17 dose-response curve. This indicates that rescue requires the concerted action of at least four to six Sec17 molecules. As three molecules of Sec17 bind to each SNARE complex, rescue involves binding of Sec17 to a minimum of two trans-SNARE complexes. It is important to note that each of these cooperativity coefficients may be interpreted as lower-bound estimates of the minimum number of complexes required to mediate a single fusion event. However, cooperativity coefficients tend to systematically underestimate the number of sites or complexes involved in a biochemical process (Weiss, 1997). Consequently, a firm upper-bound estimate of the minimum number of SNARE complexes needed for fusion (i.e., the number of complexes sufficient for fusion) is not yet available (Montecucco et al., 2005). We are now exploiting the extraordinary sensitivity of Qc7Δ-mediated fusion (ED50 < 600 pM) to address this question.
Materials and methods
Yeast strains
BY4742 pho8Δ (MATα ura3Δ leu2Δ his3Δ lys2Δ pho8Δ::neo) and BY4742 pep4Δ (PHO8 pep4Δ::neo) were used for all experiments except the trans-SNARE complex assays (Collins and Wickner, 2007). In those experiments, DKY6281 (MATα pho8::TRP1 leu2–3 leu 2–112 ura3–52 his3–200 trp1–901 lys2–801 suc2–9), BJ3505 (MATα pep4::HIS3 prb1-1.6R his3–200 lys2–801 trp1101 [gal3] ura3–52 gal2 can1), and the VAM3::CBP nyv1Δ derivative of BJ3505 (vam3::kanMX6-pVAM3-2-CaMBP-VAM3 nyv1Δ::natMX6) were used.
Plasmid construction
Vam7 (Qc) expression constructs.
DNA encoding the multiple cloning site and intein/chitin-binding domain tag from the vector pTYB3 (New England Biolabs, Inc.) were isolated by digestion with XbaI and PstI and ligated into the pET41(a) (Invitrogen) backbone. A secondary (nonmultiple cloning site) SapI restriction site in the resultant vector was then removed by site-directed mutagenesis. DNA encoding a His6 affinity tag and a tobacco etch virus protease recognition sequence was isolated from the vector p-His-Parallel1 by digestion with XbaI and EcoRI and ligated into the pET41(a)-derived fusion vector, resulting in pMS101. The VAM7 coding sequence (lacking appropriate portions of the 3′ sequence in the case of truncation mutants) was amplified from BY4741 genomic DNA with primer-introduced 5′-NcoI and 3′-SapI restriction sites, which were used to clone the VAM7 DNA into pMS101.
Sec17 expression constructs.
The SEC17 coding sequence was amplified from pQE3-Sec17 DNA (Haas and Wickner, 1996) with primer-introduced 5′-NdeI and 3′-XhoI restriction sites, which were used to clone the SEC17 DNA into the vector pTYB12 (New England Biolabs, Inc.). To generate Sec17-LALA, mutations were embedded in the reverse primer.
Vam7 (Qc) expression and purification
BL21(DE3) pRP Escherichia coli were grown in terrific broth to OD600 = 1.0–2.0, and protein expression was induced with 350 µM IPTG at 30°C for 4 h. Cells were disrupted by sonication on ice in buffer A (20 mM Tris-Cl, pH 7.9, 500 mM KCl, 1 mM EDTA, and 10% glycerol) supplemented with protease inhibitors (PMSF, pepstatin, and leupeptin) and DNase I. Purification by chitin-affinity chromatography was conducted as described by the manufacturer (New England Biolabs, Inc.) with the addition of a high salt wash (buffer A with 1.5 M KCl) before on-column cleavage. Intein-mediated cleavage (Fig. 1 C) was performed in buffer A supplemented with 50 mM DTT at ∼22°C overnight. Cleaved eluate protein was exchanged into buffer B (20 mM NaPO4, pH 7.4, 500 mM KCl, 20 mM imidazole, and 10% glycerol, pH 7.4) by repeated dilution and concentration in a centrifugal concentrator. Proteins in the cleaved eluate bearing an intact N terminus were selected by nickel nitrilotriacetic acid affinity chromatography on Ni-Sepharose 6 resin (GE Healthcare) and exchanged into storage buffer (20 mM NaPO4, 250 mM KCl, and 10% glycerol, pH 7.4) by repeated dilution and concentration in a centrifugal concentrator. Single-use aliquots of purified Qc at 5–50 µM were frozen over liquid nitrogen and stored at −80°C. Protein concentrations were determined by Coomassie plus assay (Thermo Fisher Scientific) against BSA standards.
Sec17 expression and purification
BL21(DE3) pRP E. coli were grown in terrific broth to OD600 = 1.0–2.0, and protein expression was induced with 350 µM IPTG at 18°C for 16 h. The proteins were purified by chitin-affinity chromatography following the same general procedure as for Vam7, but glycerol was omitted from all buffers, and the high salt column wash was omitted. Proteins were exchanged into PS buffer (20 mM Pipes-KOH, pH 6.8, and 200 mM sorbitol) with 125 mM KCl by dialysis, and aliquots were frozen over liquid nitrogen and stored at −80°C.
Fusion inhibitors
His6-Sec17, His6-Gyp1-46, affinity-purified αVam3 (rabbit polyclonal), and MED were prepared as described by Fratti et al. (2004). GDI was prepared as described by Starai et al. (2007), and LPC-12 (Avanti Polar Lipids, Inc.) was prepared as described by Reese and Mayer (2005).
Content-mixing assay
Vacuoles were purified, and standard fusion reactions were assembled as described previously (Merz and Wickner, 2004a,b; Brett and Merz, 2008). 1× reactions (30 µl) contained 6 µg vacuoles (3 µg each from BY4742 pep4Δ and pho8Δ or DKY6281 and either BJ3505 or BJ3505 VAM3::CBP nyv1Δ) in 20 mM Pipes-KOH, pH 6.8, 200 mM sorbitol, 125 mM KCl, 5 mM MgCl2, 10 uM coenzyme A (Sigma-Aldrich), 3.2 µg/ml Pbi2 (IB2), and 10 mg/ml BSA (Thermo Fisher Scientific; Thorngren et al., 2004). Where indicated, an ATP regenerating system of 1 mM ATP, 1 mg/ml creatine kinase, and 29 mM creatine phosphate was included (all components obtained from Roche). Reactions were incubated at 27°C for 70 min and were assayed for alkaline phosphatase activity (Merz and Wickner, 2004a). Raw values were corrected by subtracting the background signal (measured value of an ATP- or Qcwt-driven reaction held on ice; typically ≤0.05 of the standard condition value) and normalized to a background-corrected standard condition. The standard condition was either a standard +ATP reaction or the Qcwt bypass reaction within the experiment. Absolute fusion values for the standard conditions in each figure were calculated as previously described (Merz and Wickner, 2004a).
Lipid-mixing assay
Lipid-mixing assays were conducted as described previously (Reese and Mayer, 2005; Jun and Wickner, 2007) with the following modifications: 6× scale reactions contained 4 µg BY4742 pep4Δ vacuoles labeled with R-PE (Invitrogen), 14 µg unlabeled BY4742 pep4Δ vacuoles, and 18 µg BY4742 pho8Δ vacuoles. BSA was omitted from all lipid-mixing reactions; in its place, 1 mg/ml creatine kinase was included in bypass reactions to serve as a carrier for Vam7. Fluorescence was monitored in a microplate reader (Victor3; PerkinElmer) using black conical bottom microplates. After measurement of lipid mixing, 1× reaction aliquots were withdrawn from each well and assayed for content mixing by the alkaline phosphatase activity assay.
Trans-SNARE complex assay
Trans-SNARE complex assays were conducted as described previously (Collins and Wickner, 2007; Jun and Wickner, 2007) with the following modifications: pull-downs were conducted from 15× scale reactions containing 45 µg vacuoles from each of BJ3505 and BJ3505 VAM3::CBP nyv1Δ cells. Content mixing was measured from parallel 1× scale reactions containing vacuoles from DKY6281 and one of the two protease-deficient strains. 15× scale pull-down reactions were incubated at 27°C for 45 min before vacuole solubilization, and parallel content-mixing reactions were developed after 70 min at 27°C. The solubilization buffer was modified to 20 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.5% NP-40, 10% glycerol, and 1× protease inhibitor cocktail (0.46 µg/ml leupeptin, 3.5 µg/ml pepstatin, and 1 mM PMSF). Samples were brought to 2 mM CaCl2 before incubation with CaM-agarose resin (Agilent Technologies). Vacuole extracts were incubated with resin for 1 h at 4°C, washed five times with 1 ml of solubilization buffer containing 2 mM CaCl2, and eluted as described previously (Collins and Wickner, 2007).
Data analysis
Prism software (version 4.03; GraphPad Software, Inc.) was used for statistical analyses and to prepare graphs. Dose-response curves were obtained by nonlinear least squares fitting of the sigmoidal Hill equation to experimental log-transformed dosages and mean fusion values normalized as specified in Figs. 2, 6, S1, and S3. Lipid-mixing curves were obtained by nonlinear least squares fitting of the Gompertz equation, which is particularly well suited for description of irreversible, time-dependent ensemble processes. Immunoblot images were globally adjusted for brightness and contrast using ImageJ (version 1.36b; National Institutes of Health). Figures were prepared in Prism and Illustrator (CS3; Adobe).
Online supplemental material
Fig. S1 shows the complete dose-inhibition curves for Qc1Δ, QcΔSD, and GST-PX. Fig. S2 shows the lipid-mixing data from Fig. 5 before subtraction of the background (SNARE independent) signal. Fig. S3 shows the dose-response relationship for rescue of Qc3Δ-mediated fusion by His-tagged Sec17. Fig. S4 shows the dose-inhibition curves for inhibition of ATP-driven reactions by untagged Sec17-wt and Sec17-LALA. Fig. S5 depicts the putative α-SNAP (mammalian Sec17)–binding surface on the synaptic SNARE complex as determined by Marz et al. (2003). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200811082/DC1.
Acknowledgments
We thank Drs. Bertil Hille, Justin Taraska, Dawn Wenzel, and members of our group for critical discussions, Phyllis Hanson for suggesting the Sec17-LALA mutant, and Bill Wickner for sharing reagents.
This study is supported by the National Institutes of Health grants to A.J. Merz (RO1 GM077349) and M.L. Schwartz (T32 GM07270).
Footnotes
Abbreviations used in this paper: BoNT, botulinum neurotoxin; EC, effective concentration; GDI, guanine nucleotide dissociation inhibitor; MED, myristoylated alanine-rich C kinase substrate effector domain; PX, phox homology; R-PE, rhodamine-phosphatidylethanolamine.
References
- Antonin W., Fasshauer D., Becker S., Jahn R., Schneider T.R. 2002. Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs.Nat. Struct. Biol. 9:107–111 [DOI] [PubMed] [Google Scholar]
- Armstrong R.T., Kushnir A.S., White J.M. 2000. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition.J. Cell Biol. 151:425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard R.J., Morgan A., Burgoyne R.D. 1996. Domains of alpha-SNAP required for the stimulation of exocytosis and for N-ethylmalemide-sensitive fusion protein (NSF) binding and activation.Mol. Biol. Cell. 7:693–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz T., Blasi J., Yamasaki S., Baumeister A., Link E., Sudhof T.C., Jahn R., Niemann H. 1994. Proteolysis of SNAP-25 by types E and A botulinal neurotoxins.J. Biol. Chem. 269:1617–1620 [PubMed] [Google Scholar]
- Boeddinghaus C., Merz A.J., Laage R., Ungermann C. 2002. A cycle of Vam7p release from and PtdIns 3-P–dependent rebinding to the yeast vacuole is required for homotypic vacuole fusion.J. Cell Biol. 157:79–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett C.L., Merz A.J. 2008. Osmotic regulation of Rab-mediated organelle docking.Curr. Biol. 18:1072–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett C.L., Plemel R.L., Lobinger B.T., Vignali M., Fields S., Merz A.J. 2008. Efficient termination of vacuolar Rab GTPase signaling requires coordinated action by a GAP and a protein kinase.J. Cell Biol. 182:1141–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman E.R. 2008. How does synaptotagmin trigger neurotransmitter release? Annu. Rev. Biochem. 77:615–641 [DOI] [PubMed] [Google Scholar]
- Cheever M.L., Sato T.K., de Beer T., Kutateladze T.G., Emr S.D., Overduin M. 2001. Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes.Nat. Cell Biol. 3:613–618 [DOI] [PubMed] [Google Scholar]
- Chernomordik L.V., Kozlov M.M. 2008. Mechanics of membrane fusion.Nat. Struct. Mol. Biol. 15:675–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K.M., Wickner W.T. 2007. Trans-SNARE complex assembly and yeast vacuole membrane fusion.Proc. Natl. Acad. Sci. USA. 104:8755–8760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K.M., Thorngren N.L., Fratti R.A., Wickner W.T. 2005. Sec17p and HOPS, in distinct SNARE complexes, mediate SNARE complex disruption or assembly for fusion.EMBO J. 24:1775–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge F.A.J., Rahamimoff R. 1967. Co-operative action a calcium ions in transmitter release at the neuromuscular junction.J. Physiol. 193:419–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratti R.A., Jun Y., Merz A.J., Margolis N., Wickner W. 2004. Interdependent assembly of specific regulatory lipids and membrane fusion proteins into the vertex ring domain of docked vacuoles.J. Cell Biol. 167:1087–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudo C.G., Hu C., You D., Slovic A.M., Mosharov E.V., Sulzer D., Melia T.J., Rothman J.E. 2005. SNAREs can promote complete fusion and hemifusion as alternative outcomes.J. Cell Biol. 170:249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A. 1995. A quantitative assay to measure homotypic vacuole fusion in vitro.Methods Cell Sci. 17:283–294 [Google Scholar]
- Haas A., Wickner W. 1996. Homotypic vacuole fusion requires Sec17p (yeast a-SNAP) and Sec18p (yeast NSF).EMBO J. 15:3296–3305 [PMC free article] [PubMed] [Google Scholar]
- Han X., Wang C.T., Bai J., Chapman E.R., Jackson M.B. 2004. Transmembrane segments of syntaxin line the fusion pore of Ca2+-triggered exocytosis.Science. 304:289–292 [DOI] [PubMed] [Google Scholar]
- Hanson P.I., Heuser J.E., Jahn R. 1997a. Neurotransmitter release - four years of SNARE complexes.Curr. Opin. Neurobiol. 7:310–315 [DOI] [PubMed] [Google Scholar]
- Hanson P.I., Roth R., Morisaki H., Jahn R., Heuser J.E. 1997b. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy.Cell. 90:523–535 [DOI] [PubMed] [Google Scholar]
- Hohl T.M., Parlati F., Wimmer C., Rothman J.E., Sollner T.H., Engelhardt H. 1998. Arrangement of subunits in 20 S particles consisting of NSF, SNAPs, and SNARE complexes.Mol. Cell. 2:539–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S.Y., Charlton M.P. 1999. Activity-dependent changes in partial VAMP complexes during neurotransmitter release.Nat. Neurosci. 2:1078–1083 [DOI] [PubMed] [Google Scholar]
- Hua Y., Scheller R.H. 2001. Three SNARE complexes cooperate to mediate membrane fusion.Proc. Natl. Acad. Sci. USA. 98:8065–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M.B., Chapman E.R. 2008. The fusion pores of Ca(2+)-triggered exocytosis.Nat. Struct. Mol. Biol. 15:684–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R., Scheller R.H. 2006. SNAREs–engines for membrane fusion.Nat. Rev. Mol. Cell Biol. 7:631–643 [DOI] [PubMed] [Google Scholar]
- Jun Y., Wickner W. 2007. Assays of vacuole fusion resolve the stages of docking, lipid mixing, and content mixing.Proc. Natl. Acad. Sci. USA. 104:13010–13015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemble G.W., Danieli T., White J.M. 1994. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion.Cell. 76:383–391 [DOI] [PubMed] [Google Scholar]
- Kesavan J., Borisovska M., Bruns D. 2007. v-SNARE actions during Ca(2+)-triggered exocytosis.Cell. 131:351–363 [DOI] [PubMed] [Google Scholar]
- Lagow R.D., Bao H., Cohen E.N., Daniels R.W., Zuzek A., Williams W.H., Macleod G.T., Sutton R.B., Zhang B. 2007. Modification of a hydrophobic layer by a point mutation in syntaxin 1A regulates the rate of synaptic vesicle fusion.PLoS Biol. 5:e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando L., Zucker R.S. 1994. Ca2+ cooperativity in neurosecretion measured using photolabile Ca2+ chelators.J. Neurophysiol. 72:825–830 [DOI] [PubMed] [Google Scholar]
- Langosch D., Crane J.M., Brosig B., Hellwig A., Tamm L.K., Reed J. 2001. Peptide mimics of SNARE transmembrane segments drive membrane fusion depending on their conformational plasticity.J. Mol. Biol. 311:709–721 [DOI] [PubMed] [Google Scholar]
- Li F., Pincet F., Perez E., Eng W.S., Melia T.J., Rothman J.E., Tareste D. 2007. Energetics and dynamics of SNAREpin folding across lipid bilayers.Nat. Struct. Mol. Biol. 14:890–896 [DOI] [PubMed] [Google Scholar]
- Lu X., Zhang Y., Shin Y.K. 2008. Supramolecular SNARE assembly precedes hemifusion in SNARE-mediated membrane fusion.Nat. Struct. Mol. Biol. 15:700–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marz K.E., Lauer J.M., Hanson P.I. 2003. Defining the SNARE complex binding surface of alpha-SNAP: implications for SNARE complex disassembly.J. Biol. Chem. 278:27000–27008 [DOI] [PubMed] [Google Scholar]
- Matos M.F., Mukherjee K., Chen X., Rizo J., Sudhof T.C. 2003. Evidence for SNARE zippering during Ca2+-triggered exocytosis in PC12 cells.Neuropharmacology. 45:777–786 [DOI] [PubMed] [Google Scholar]
- Mayer A., Wickner W., Haas A. 1996. Sec18p (NSF)-driven release of Sec17p (a-SNAP) can precede docking and fusion of yeast vacuoles.Cell. 85:83–94 [DOI] [PubMed] [Google Scholar]
- McNew J.A., Weber T., Engelman D.M., Sollner T.H., Rothman J.E. 1999. The length of the flexible SNAREpin juxtamembrane region is a critical determinant of SNARE-dependent fusion.Mol. Cell. 4:415–421 [DOI] [PubMed] [Google Scholar]
- McNew J.A., Weber T., Parlati F., Johnston R.J., Melia T.J., Sollner T.H., Rothman J.E. 2000. Close is not enough: SNARE-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors.J. Cell Biol. 150:105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melia T.J., You D., Tareste D.C., Rothman J.E. 2006. Lipidic antagonists to SNARE-mediated fusion.J. Biol. Chem. 281:29597–29605 [DOI] [PubMed] [Google Scholar]
- Melikyan G.B., White J.M., Cohen F.S. 1995. GPI-anchored influenza hemagglutinin induces hemifusion to both red blood cell and planar bilayer membranes.J. Cell Biol. 131:679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz A.J., Wickner W.T. 2004a. Resolution of organelle docking and fusion kinetics in a cell-free assay.Proc. Natl. Acad. Sci. USA. 101:11548–11553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz A.J., Wickner W.T. 2004b. Trans-SNARE interactions elicit Ca2+ efflux from the yeast vacuole lumen.J. Cell Biol. 164:195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco C., Schiavo G., Pantano S. 2005. SNARE complexes and neuroexocytosis: how many, how close? Trends Biochem. Sci. 30:367–372 [DOI] [PubMed] [Google Scholar]
- Pobbati A.V., Stein A., Fasshauer D. 2006. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion.Science. 313:673–676 [DOI] [PubMed] [Google Scholar]
- Reese C., Mayer A. 2005. Transition from hemifusion to pore opening is rate limiting for vacuole membrane fusion.J. Cell Biol. 171:981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice L.M., Brunger A.T. 1999. Crystal structure of the vesicular transport protein Sec17: implications for SNAP function in SNARE complex disassembly.Mol. Cell. 4:85–95 [DOI] [PubMed] [Google Scholar]
- Rizo J., Rosenmund C. 2008. Synaptic vesicle fusion.Nat. Struct. Mol. Biol. 15:665–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi G., Salminen A., Rice L.M., Brunger A.T., Brennwald P. 1997. Analysis of a yeast SNARE complex reveals remarkable similarity to the neuronal SNARE complex and a novel function for the C terminus of the SNAP-25 homolog, Sec9.J. Biol. Chem. 272:16610–16617 [DOI] [PubMed] [Google Scholar]
- Roy R., Peplowska K., Rohde J., Ungermann C., Langosch D. 2006. Role of the Vam3p transmembrane segment in homodimerization and SNARE complex formation.Biochemistry. 45:7654–7660 [DOI] [PubMed] [Google Scholar]
- Sakaba T., Stein A., Jahn R., Neher E. 2005. Distinct kinetic changes in neurotransmitter release after SNARE protein cleavage.Science. 309:491–494 [DOI] [PubMed] [Google Scholar]
- Schaub J.R., Lu X., Doneske B., Shin Y.K., McNew J.A. 2006. Hemifusion arrest by complexin is relieved by Ca2+-synaptotagmin I.Nat. Struct. Mol. Biol. 13:748–750 [DOI] [PubMed] [Google Scholar]
- Seals D.F., Eitzen G., Margolis N., Wickner W.T., Price A. 2000. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion.Proc. Natl. Acad. Sci. USA. 97:9402–9407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner T., Bennett M.K., Whiteheart S.W., Scheller R.H., Rothman J.E. 1993. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion.Cell. 75:409–418 [DOI] [PubMed] [Google Scholar]
- Sorensen J.B., Wiederhold K., Muller E.M., Milosevic I., Nagy G., de Groot B.L., Grubmuller H., Fasshauer D. 2006. Sequential N- to C-terminal SNARE complex assembly drives priming and fusion of secretory vesicles.EMBO J. 25:955–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai V.J., Jun Y., Wickner W. 2007. Excess vacuolar SNAREs drive lysis and Rab bypass fusion.Proc. Natl. Acad. Sci. USA. 104:13551–13558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton R.B., Fasshauer D., Jahn R., Brunger A.T. 1998. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution.Nature. 395:347–353 [DOI] [PubMed] [Google Scholar]
- Thorngren N., Collins K.M., Fratti R.A., Wickner W., Merz A.J. 2004. A soluble SNARE drives rapid docking, bypassing ATP and Sec17/18p for vacuole fusion.EMBO J. 23:2765–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermann C., Nichols B.J., Pelham H.R., Wickner W. 1998a. A vacuolar v-t-SNARE complex, the predominant form in vivo and on isolated vacuoles, is disassembled and activated for docking and fusion.J. Cell Biol. 140:61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungermann C., Sato K., Wickner W. 1998b. Defining the functions of trans-SNARE pairs.Nature. 396:543–548 [DOI] [PubMed] [Google Scholar]
- Wang Y., Dulubova I., Rizo J., Sudhof T.C. 2001. Functional analysis of conserved structural elements in yeast syntaxin Vam3p.J. Biol. Chem. 276:28598–28605 [DOI] [PubMed] [Google Scholar]
- Weber T., Zemelman B.V., McNew J.A., Westermann B., Gmachl M., Parlati F., Sollner T.H., Rothman J.E. 1998. SNAREpins: minimal machinery for membrane fusion.Cell. 92:759–772 [DOI] [PubMed] [Google Scholar]
- Weber T., Parlati F., McNew J.A., Johnston R.J., Westermann B., Sollner T.H., Rothman J.E. 2000. SNAREpins are functionally resistant to disruption by NSF and αSNAP.J. Cell Biol. 149:1063–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J.N. 1997. The Hill equation revisited: uses and misuses.FASEB J. 11:835–841 [PubMed] [Google Scholar]
- Wimmer C., Hohl T.M., Hughes C.A., Muller S.A., Sollner T.H., Engel A., Rothman J.E. 2001. Molecular mass, stoichiometry, and assembly of 20 S particles.J. Biol. Chem. 276:29091–29097 [DOI] [PubMed] [Google Scholar]
- Wong J.L., Koppel D.E., Cowan A.E., Wessel G.M. 2007. Membrane hemifusion is a stable intermediate of exocytosis.Dev. Cell. 12:653–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser A.E., Sato T.K., Emr S.D. 2000. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion.J. Cell Biol. 151:551–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Poirier M.A., Bennett M.K., Shin Y.K. 2001. The neuronal t-SNARE complex is a parallel four-helix bundle.Nat. Struct. Biol. 8:308–311 [DOI] [PubMed] [Google Scholar]
- Xu T., Binz T., Niemann H., Neher E. 1998. Multiple kinetic components of exocytosis distinguished by neurotoxin sensitivity.Nat. Neurosci. 1:192–200 [DOI] [PubMed] [Google Scholar]
- Xu T., Rammner B., Margittai M., Artalejo A.R., Neher E., Jahn R. 1999. Inhibition of SNARE complex assembly differentially affects kinetic components of exocytosis.Cell. 99:713–722 [DOI] [PubMed] [Google Scholar]
- Xu Y., Zhang F., Su Z., McNew J.A., Shin Y.K. 2005. Hemifusion in SNARE-mediated membrane fusion.Nat. Struct. Mol. Biol. 12:417–422 [DOI] [PubMed] [Google Scholar]
- Zampighi G.A., Zampighi L.M., Fain N., Lanzavecchia S., Simon S.A., Wright E.M. 2006. Conical electron tomography of a chemical synapse: vesicles docked to the active zone are hemi-fused.Biophys. J. 91:2910–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]