Abstract
AIM: To study the association of the frequency and pattern of KIT and PDGFRA mutations and clinicopathological factors in a group of patients with gastrointestinal stromal tumors (GIST).
METHODS: Thirty patients with GIST were examined. Exons 9, 11, 13, and 17 of the KIT and exons 12 and 18 of the PDGFRA gene were analyzed for the presence of mutations by PCR amplification and direct sequencing.
RESULTS: KIT or PDGFRA mutations were detected in 21 of the 30 patients (70%). Sixteen patients had mutations within KIT exon 11, three within KIT exon 9, and two within PDGFRA exon 18. GISTs with KIT exon 9 mutations were predominantly located in the small intestine, showed a spindle cell phenotype, and were assessed as potentially malignant. GISTs with KIT exon 11 mutations were located in the stomach and intestine, showed mainly a spindle cell phenotype, and were scored as potentially malignant (P < 0.05). Tumors with KIT exon 11 codon 557/558 deletion/insertion mutations were found to be associated with a potentially malignant clinical behaviour (P < 0.003). GISTs with PDGFRA mutations located in stomach showed a mixed cell phenotype and were classified as of very low or low moderate malignant potential.
CONCLUSION: Determination of KIT and PDGFRA mutations should be additional parameters for the better prediction of GISTs clinical behaviour. Tumors with deletion/insertion mutations affecting codons 557/558 of the KIT gene seem to represent a distinct subset of malignant GISTs.
Keywords: Gastrointestinal stromal tumors, KIT gene, Platelet derived growth factor receptor alpha, Mutations, Malignant
INTRODUCTION
Gastrointestinal stromal tumors (GISTs) are the most common primary mesenchymal tumors of the gastrointestinal tract. Their biological behaviour is difficult to predict. GIST prognosis is largely dependent on the size, mitotic index, and presence or absence of metastases[1–3]. We now know that GISTs may have either a well-developed or an incomplete myoid, neural, autonomic nerve, or mixed phenotype, or may remain undifferentiated[2]. Typically, GISTs are immunohistochemical positive for KIT tyrosine kinase receptor which is perhaps their single best defining feature[4,5].
Some GISTs are benign tumors and most of these are found incidentally. Other GISTs metastasize to liver or disseminate in the peritoneal cavity. Most of the latter type GISTs do not respond to chemotherapy and ultimately kill the hosts. The pharmaceutical development and therapeutic implications of protein tyrosine kinase inhibitors has refocused the attention on GIST. Until now, the treatment with selective tyrosine kinase inhibitors, such as imatinib mesylate, for patients with GISTs has hinged on the KIT positive immunostaining tumors[6]. Although the KIT positivity by immunohistochemistry becomes invaluable in the diagnosis of GISTs, some authors believe that a small subgroup of these tumors fulfill the clinical and morphological criteria of GISTs, and lack KIT expression[7,8]. Studies in the last decade have established that activating mutations of KIT are present in 40% to 92% of GISTs and likely play an essential role in the development of these tumors[9]. The subset of GISTs that lack detectable mutations could be divided into a group that has activating mutations in the related tyrosine kinase platelet-derived growth factor receptor alpha (PDGFRA) and a group without identified kinase mutations[10,11].
A proportion of GISTs shows mutations in the regulatory juxtamembrane domain of the c-kit gene. These KIT mutations have been shown to represent gain-of-function mutations leading to ligand independent activation of the tyrosine kinase and the phosphorylation cascade that leads into mitogenic activation[12,13]. Benign and malignant GISTs carry mutations in KIT and PDGFRA gene, but although these mutations vary among GISTs the definitive genotype/phenotype correlations are still under consideration[9–11,14]. Moreover, currently it is not clear whether mutations are independent prognostic factors[15–18].
In this study we examined a series of 30 patients with primary GISTs defined by different types of KIT and PDGFRA mutations, to investigate whether mutations’ type and distribution are associated with GISTs clinical behaviour.
MATERIALS AND METHODS
Clinical and pathological data
Using the database of Surgery and Pathology Departments of Areteion University Hospital and Evangelismos General Hospital, we collected records with a pathologic diagnosis of stromal tumor within the GI tract. Thirty patients’ records with the diagnosis of GIST between 2001 and 2005 were reviewed. Patients’ age and gender, clinical manifestations, tumor size, pathological characteristics including cell type, cellularity, nuclear atypia, the number of mitoses, the presence of necrosis, or hemorrhage were recorded.
Formalin-fixed and paraffin-embedded samples were used for immunohistochemical examination. Tumors were divided according to their morphologic profile into four histological categories: epithelioid (Ep), spindle cell (Sp), mixed spindle cell with focal epithelioid component (mixed type 1), and tumors with mixed epithelioid and focal spindle cell component (mixed type 2). Mitoses were counted in 5 separate groups of 50 HPFs, a total area of 5 mm2, and the highest of these 5 counts was recorded. Mitotic activity was scored as low (< 5/50 HPF), intermediate (5-10/50 HPF), or high (> 10/50 HPF). Tumors were assigned to risk assessment categories based on size, mitotic index, and location according to published criteria[17].
Immunohistochemistry
Immunohistochemical staining was performed using the following primary antibodies: KIT (CD 117 antigen, polyclonal, Dako, USA; 1:50 dilution), PDGFRA (polyclonal, Santa Cruz, California, USA, 1:400), a-smooth-muscle actin (clone asm-1, Dako; 1:200), desmin (clone DE-R-11, NovocastraLabs; 1:100), S-100 (clone S1/61/69, Novocastra Labs; 1:40), CD34 (clone QBEnd/10, Novocastra Labs; 1:50), Ki-67 (clone MM1, Novocastra Labs; 1:200) by a standard three step immunoperoxidase procedure (APAAP, DAKO, Glostroup, Danemark). Appropriate positive controls were run in parallel for all antibodies tested. According to the proportion of tumor cells showing an immunopositive reaction, tumors were classified as negative (< 10%) or positive (> 10%).
DNA sequencing
Exons 9, 11, 13, and 17 of the KIT and exons 12 and 18 of the PDGFRA gene were evaluated for the presence of mutations by PCR amplification and direct sequencing. DNA was extracted from formalin-fixed, paraffin embedded tissue samples using the PUREGEN DNA Purification System (Gentra Systems, Minneapolis, USA). The primer pairs used for PCR amplification and direct sequencing are given in Table 1.
Table 1.
Primer sequences used for KIT and PDGFRA PCR
| Kit exon 9F tttggaaagctagtggttca | Kit exon 9R atggtagacagagcctaaac |
| Kit exon11F ctatttttccctttctcccc | Kit exon11R tacccaaaaaggtgacatgg |
| Kit exon 13F cttgacatcagtttgccag | Kit exon 13R aaaggcagcttggacacggcttta |
| Kit exon 17F tttctcctccaacctaatag | Kit exon 17R cctttgcaggactgtcaagc |
| PDGFRA exon 12aF ccagttacctgtcctggtcat | PDGFRA exon 12aR tggaaactcccatcttgagtc |
| PDGFRA exon 12bF aaattcgctggagggtcatt | PDGFRA exon 12bR ggaggttaccccatggaagt |
| PDGFR exon 18F agtgtgtccaccgtgatctg | PDGFRA exon 18R gtgtgggaagtgtggaggta |
Annealing temperature for the KIT exon 9 and 13 and for the PDGFRA primer sets was 53°C and for the KIT exon 13 and 17 primer sets 56°C, respectively. Amplification products were separated by 2% agarose ethidium bromide gel electrophoresis to confirm correct amplification. Products were purified with NucleoSpin Extract II kit (Macherey-Nagel, Duren, Germany) and applied to bi-directional sequencing on an ABI130 sequencer using the Big Dye Terminator v.1.1 KIT (Applied Biosystems, Foster City, CA).
Statistical analysis
Data were analyzed using Statistical software SPSS Version 13.0. Fisher's Exact Test of Independence was used to evaluate the statistical significance of associations in two-way contingency tables to determine whether KIT and PDGFRA gene mutations are independent of clinicopathological parameters. Statistical significance would be inferred at a two-tailed P < 0.05.
RESULTS
Types and distribution of KIT and PDGFRA gene mutations among GISTs
All detected mutations, irrespective of whether involving single nucleotide substitutions or insertion/deletions, preserved the open reading frame. KIT or PDGFRA mutations were detected in 21/30 GIST patients (70%). Nine patients had no detectable mutations with the methods used. Sixteen patients had mutations within KIT exon 11 (Figure 1), three patients within KIT exon 9, and two patients within PDGFRA exon 18. No mutations were found in KIT exons 13, 17, or PDGFRA exon 12. The mutations within KIT exon 11 were heterogeneous and consisted of 10 simple deletions, 4 point mutations, and 2 insertions (Figures 1 and 2A). Codons 557 and 558 deletion/insertion mutations were found in 8 samples (50%) of the KIT exon 11 mutations followed by codon 560 (3 cases) and codon 559 (2 cases). Six of the mutations affecting codons 557/558 were deletions of various sizes and two were 3nt insertions.
Figure 1.
Kit exon 11 wild type and mutated amino acid sequences in 30 GISTs analyzed in this study.
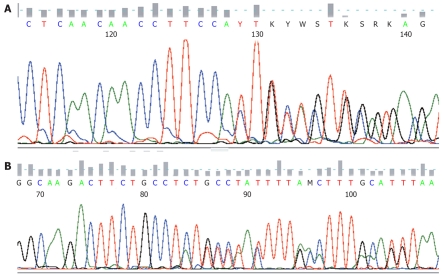
Figure 2.
The sequencing data from a GIST. A: Showing KIT exon 11 deletion (pP551-Q556del); B: Showing KIT exon 9 insertion (p.A502-Y503insAS).
KIT exon 9 mutations were all 6 bp insertions resulting in tandem duplication of the amino acids 502Ala-503Tyr (2 out of 3) (Figure 2B) or the amino acids 501Ser-502Ala. PDGFRA mutations affecting exon 18 consisted of a D842V substitution and a simple 12 bp deletion.
Clinicopathologic profile of patients with GISTs
The clinicopathological and molecular findings of the GIST patients are summarized in Table 2. Sixteen of the patients (53.3%) were male and 14 (46.7%) were female. Patients with KIT mutation exon 9 GISTs were exclusively male. Patient age ranged from 46 to 82 years with a median of 63.4 years. Sixteen tumors (53.3%) were located in the stomach, 9 (30%) in small intestine, and 5 (16.7%) in large intestine. Two out of three GISTs with KIT exon 9 mutations were located within the small intestine. GISTs with KIT exon11 mutations were located in the stomach (6/16) and within the small and the large intestine (10/16). GISTs with PDGFRA mutations were exclusively located in the stomach (2/2). The majority of GISTs with no detectable mutations in KIT or PDGFRA (77.8%) were located in stomach.
Table 2.
Clinicopathological and molecular characteristics of GIST patients
| Case No. | Sex | Age | Site | Size (cm) | Mitoses (/50 HPF) | Differentiation | CD117 | Diagnosis | KIT mutations | PDGFRA mutations |
| 1 | F | 72 | LI | 9 | 6 | SP | N | Malignant potential | wt | wt |
| 2 | M | 65 | SI | 2.8 | 10 | M1 | P | Malignant potential | p.A502-Y503insA | wt |
| 3 | M | 79 | S | 16 | 9 | EP | P | Malignant potential | p.K558>NP | wt |
| 4 | F | 60 | SI | 17 | 3 | EP | P | Malignant potential | p.K558>NP | wt |
| 5 | F | 59 | SI | 5 | 2 | SP | P | Low malignant potential | p.W557-K558del | wt |
| 6 | M | 72 | SI | 7.8 | 2 | SP | P | Malignant potential | p.W557-K558del | wt |
| 7 | M | 82 | S | 5.5 | 5 | SP | P | Very low malignant potential | p.D579del | wt |
| 8 | M | 52 | SI | 4 | 3 | SP | P | Low malignant potential | p.V559A | wt |
| 9 | F | 63 | S | 8 | 7 | SP | P | Malignant potential | p.V559A | wt |
| 10 | F | 70 | SI | 6 | 1 | M1 | P | Malignant potential | p.W557-K558del | wt |
| 11 | F | 60 | S | 9 | 4 | M1 | P | Very low malignant potential | wt | p.D842V |
| 12 | M | 61 | S | 13 | 2 | M2 | P | Low-moderate malignant potential | wt | p.I 843-D846del |
| 13 | M | 64 | S | 9 | 3 | SP | P | Very low malignant potential | wt | wt |
| 14 | F | 70 | S | 1.5 | 1 | SP | P | Benign | p.P551-Q556del | wt |
| 15 | M | 66 | S | 10 | 20 | SP | P | Malignant potential | wt | wt |
| 16 | M | 60 | SI | 12 | 4 | SP | P | Malignant potential | p.Y503-F504insAY | wt |
| 17 | F | 62 | SI | 3.5 | 1 | SP | P | Low malignant potential | pV560D | wt |
| 18 | M | 56 | SI | 4 | 1 | SP | P | Low malignant potential | p.K550-E554del | wt |
| 19 | F | 67 | S | 19 | 22 | M1 | P | Malignant potential | p.K550-K558del | wt |
| 20 | F | 46 | S | 19 | 10 | EP | P | Malignant potential | wt | wt |
| 21 | F | 61 | S | 3.5 | 3 | SP | P | Very low malignant potential | wt | wt |
| 22 | M | 57 | LI | 8 | 5 | SP | P | Malignant potential | pV560D | wt |
| 23 | M | 58 | S | 0.7 | 0 | SP | P | Benign | p.V559-G565del | wt |
| 24 | M | 60 | S | 7.5 | 3 | M2 | P | Very low malignant potential | wt | wt |
| 25 | M | 67 | S | 8 | 1 | M2 | P | Very low malignant potential | wt | wt |
| 26 | M | 72 | S | 5.5 | 1 | SP | P | Very low malignant potential | p.Y503-F504insAY | wt |
| 27 | F | 70 | LI | 17 | 9 | SP | P | Malignant potential | p.W557-K558del | wt |
| 28 | M | 61 | LI | 2.6 | 6 | SP | P | Malignant potential | p.P551-K558del | wt |
| 29 | F | 53 | S | 5 | 0 | SP | N | Very low malignant potential | wt | wt |
| 30 | F | 57 | LI | 2.5 | 0 | SP | P | Low malignant potential | wt | wt |
S: Stomach; SI: Small intestine; LI: Large intestine; SP: Spindle cell; EP: Epithelioid; M1: Mixed type 1; M2: Mixed type 2; P: Positive; N: Negative.
Tumor size ranged from 0.7 cm to 19 cm (mean: 8, std: 5.2). The mean tumor size of GISTs with KIT exon 9 mutations was 6.7 cm (std: 8.1), and that of GISTs with KIT exon 11 mutations 7.8 cm (std: 8.2). The mean tumor’s size of GISTs with KIT exon 11 codon 557/558 mutations was 11.3 cm, while 4.4 cm in GISTs without 557/558 mutations (P = 0.025) (Table 3). The mean size of GISTs with PDFGRA mutations was 11 cm (std: 7.8).
Table 3.
Clinicopathological data of GIST patients according to the presence of codon 557/558 deletion/insertion mutations
| Variable | Wt (%) | 557/558 mutations (%) | Non 557/558 mutations (%) | P |
| Age (yr) | 61.7 | 67.2 | 62.5 | NS |
| Size (cm) | 8.2 | 11.3 | 4.4 | 0.025 |
| Mitoses | ||||
| Low (≤ 5/50 HPF) | 71.4 | 50 | 87.5 | NS |
| Intermediate | 21.4 | 37.5 | 12.5 | |
| 5-10/50 HPF | ||||
| High (> 10/50 HPF) | 7.1 | 12.5 | ||
| Differentiation | ||||
| Epithelioid | 7.1 | 25 | NS | |
| Spindle cell | 57.1 | 50 | 100 | |
| Mixed type 1 | 14.3 | 25 | ||
| Mixed type 2 | 21.4 | |||
| Risk assessment | ||||
| Benign | 25 | 0.003 | ||
| Very low malignant potential | 57.1 | 12.5 | ||
| Low malignant potential | 7.1 | 12.5 | 37.5 | |
| Low moderate malignant potential | 14.3 | |||
| Malignant potential | 21.4 | 87.5 | 25 |
Grossly, most tumors were presented as circumscribed or lobulated masses. Cystic change was recognized in several cases. Histologically, the majority of tumors (66.7%) showed spindle cell phenotype, another 10% epithelioid, 13.3% mixed type 1, and 10% mixed type 2 phenotypes.
Deletions in KIT exon 11 were frequently found to be associated with a spindle cell phenotype, substitutions in KIT exon 11 were exclusively found to be associated with a mixed type 1 phenotype, while insertions in KIT exon 11 that affect codons 557/558 exclusively showed an epithelioid phenotype (P < 0.05) (Table 4). The majority of GISTs with KIT exon 9 mutations showed a spindle cell phenotype, whereas GISTs with PDGFRA exon 18 mutations were both of mixed type morphology. The mitotic count ranged from 0 to 22/50 HPF (× 400) (mean: 4.8). The majority of tumors (70%) contained less than 5/50 HPF mitoses, 23.3% of tumors contained mitoses between 5 and 10/50 HPF, while only 6.7% of tumors contained mitoses > 10/50 HPF. GISTs with KIT exon 9 and exon 11 mutations exhibited a mean value of 5/50 HPF mitoses (std: 4.7) and 4.8/50 HPF mitoses (std: 4.7), respectively. In GISTs with PDGFRA mutations, mitotic activity was low with 2 to 4/50 HPF.
Table 4.
GIST phenotypes according to the presence and type of KIT mutations
|
Differentiation |
|||||
| Epithelioid (%) | Spindle cells (%) | Mixed type 1 (%) | Mixed type 2 (%) | P | |
| KIT mutation | |||||
| Positive | 10.5 | 73.3 | 15.8 | NS | |
| Negative | 9.1 | 54.5 | 9.1 | 27.3 | |
| Exon 9 | 66.7 | 33.3 | NS | ||
| Exon 11 | 12.5 | 75 | 12.5 | ||
| Insertions | 100 | 0.03 | |||
| Deletions | 80.0 | 20.0 | |||
| Substitutions | 100 | ||||
Cellularity was high in 40% of the tumors, moderate in 36.7%, and mild in 23.3%. Cellularity was high in the majority (86%) of GISTs with KIT exon 9 mutations, moderate in the majority GISTs with exon 11 mutations, and moderate or high in all GISTs with PDGFRA mutations. The majority (73%) of the tumors showed no necrosis. Absence of necrosis was present in 2/3 of GISTs with KIT exon 9-11 mutations, and in all GIST patients with PDGFRA mutations. A high proportion of GISTs with codon 557/558 mutations (50%) was found to be necrotic. The majority (66.6%) of the tumors showed no hemorrhage. Hemorrhage was found in all cases of GISTs with PDGFRA mutations (P = 0.038), but to be absent in most of the GISTs with KIT mutations. The majority (80%) of the tumors showed no ulceration. All the tumors with ulceration were positive for KIT exon 9 or 11 mutations.
Immunohistochemically, 28 tumors were positive for KIT expression. All GISTs with KIT and PDGFRA mutations showed weak to strongly and diffuse positive staining for the KIT gene. PDGFRA expression was immunohistochemically detected in 18 cases (60%). Alpha-smooth muscle actin, desmin, and S-100 protein were positive in 12 (40%), 3 (10%), and 4 (13.3%) cases, respectively. A-SMA was positive in 47.6% of GISTs with KIT mutations. CD34 expression was positive in 20 (67%) cases. Ki-67 expression was strongly positive (5%) in 13 cases (43%). Applying the clinical behaviour (risk assessment) of primary tumors according to Miettinen and Lasota (2006) 6.7% of GISTs were benign, 30% of very low malignant, 16.7% of low malignant, 6.7% of low moderate malignant, and 40% of malignant potential. 57.9% of GISTs with KIT mutations were associated with malignant potential (P = 0.003) whereas none of the GISTs with PDGFRA mutations was assessed as of malignant potential. 66.7% of the GISTs with exon 9 KIT mutations and 56.3% of those with exon 11 KIT mutations were assessed as of malignant potential (P = 0.036) (Tables 4 and 5). Tumors with KIT exon 11 codon 557/558 mutations showed a statistically significant correlation with malignant potential scoring (87.5% vs 25% of the non 557/558, P = 0.003).
Table 5.
Risk assessment according to the KIT/PDGFRA mutations
|
Risk assessment |
||||||
| Benign (%) | Very low malignant potential (%) | Low malignant potential (%) | Low moderate malignant potential (%) | Malignant (%) | P | |
| KIT mutation | ||||||
| Positive | 10.5 | 10.5 | 21.1 | 57.9 | 0.003 | |
| Negative | 63.6 | 9.1 | 18.2 | 9.1 | ||
| Exon 9 | 33.3 | 66.7 | 0.036 | |||
| Exon 11 | 12.5 | 6.3 | 25 | 56.3 | ||
| PDGFRA mutation | ||||||
| Positive | NS | |||||
| Negative | 50 | 50 | ||||
DISCUSSION
In the late 1990s it was shown that GISTs share morphological, immunohistochemical, and genetic characteristics with the interstitial cells of Cajal (ICCs). Most GISTs express strongly and specific the tyrosine kinase KIT oncoprotein that it was claimed to be required for the diagnosis[4,5]. It is now clear that a small but significant group of GISTs (5%-10%) are KIT negative[7,8]. It seems that GISTs probably do not constitute a homogenous group of tumors. Until now the prediction of their biological behaviour depended on classic clinicopathological characteristics as size and mitotic activity or location. Moreover, in the last years a significant correlation has been suggested between these pathological parameters and molecular alterations[11].
It has been suggested that tumors’ location were associated with mutations. GISTs with KIT exon 9 mutations were predominantly located in the small intestine whereas GISTs with PDGFRA mutations represent gastric tumors[10,15,17]. Other studies failed to find a significant association between KIT exon 11 mutation status and tumor location[18,19].
With regard to the primary tumor location, our results indicated that KIT exon 9 mutations were almost always detected in GISTs of intestinal origin, whereas PDGFRA mutations were associated with GISTs of gastric origin. The incidence of exon 11 KIT mutations did not appear to be related to tumor location.
The mutation type in GISTs has been reported to be associated with the phenotype[20,21]. With respect to histological phenotype, KIT exon 11 mutations were strongly associated with spindle cell phenotype GISTs. Deletions in KIT exon 11 have been mostly associated with spindle cell phenotype, substitutions in KIT exon 11 were associated exclusively with mixed cell phenotype, whereas insertions in KIT exon 11 affecting codons 557/558 were exclusively associated with epithelioid cell phenotype GISTs. PDGFRA mutations, in this study, were exclusively associated with mixed type phenotype and low risk assessment GISTs.
An association between the occurrence of KIT exon 9 and 11 mutations in GISTs and malignancy was suggested by previous studies[14,22]. Our data support this notion by showing a significant association between KIT exon 11 and exon 9 mutations and malignant GISTs. Moreover, in our data this association remains significant when only the exon 11 mutations affecting codons 557/558 were analysed. This observation is in agreement with previous studies that indicate an association of 557/558 deletions with poor prognosis and metastatic behaviour[23]. In addition to deletions it is possible that insertions affecting the 557/558 codon, although rare, are also associated with malignant phenotype as suggested by previously published data[24–26]. Codons 557/558 have been also found to represent significant a/a residues either for inhibitory role in the control of the receptor tyrosine kinase activity (Tryp557) or in constitutive receptor phosphorylation (Lys558)[27,28].
In conclusion, in this study we have analyzed the frequency and pattern of KIT and PDGFRA mutations in a group of GISTs, and we presented evidence that tumors defined by KIT codon 557/558 deletion/insertion mutations represent a subgroup of GISTs with malignant clinical behaviour. These findings underline the need for a new classification system that would integrate specific molecular alterations to the pathological criteria.
COMMENTS
Background
In the late 1990s it was shown that most gastrointestinal stromal tumors (GISTs) share morphological, immunohistochemical, and genetic characteristics with the interstitial cells of Cajal (ICCs). Most GISTs express strongly and specifically the tyrosine kinase KIT oncoprotein that it was claimed to be required for the diagnosis. GISTs are the most common primary mesenchymal tumors of the gastrointestinal tract. Their biological behaviour is difficult to predict. GISTs prognosis is largely dependent on the size, mitotic index, and presence or absence of metastases. We now know that GISTs may have either a well-developed or an incomplete myoid, neural, autonomic nerve, or mixed phenotype, or may remain undifferentiated. Typically, GISTs are immunohistochemical positive for KIT tyrosine kinase receptor which is perhaps their single best defining feature. Studies in the last decade have established that activating mutations of KIT are present in 40% to 92% of GISTs and likely play an essential role in the development of these tumors. The subset of GISTs that lack detectable mutations could be divided into a group that has activating mutations in the related tyrosine kinase platelet-derived growth factor receptor alpha (PDGFRA) and a group without identified kinase mutations. A proportion of GISTs shows mutations in the regulatory juxtamembrane domain of the c-kit gene. These KIT mutations have been shown to represent gain-of-function mutations leading to ligand independent activation of the tyrosine kinase and the phosphorylation cascade that leads into mitogenic activation.
Reasearch frontiers
Benign and malignant GISTs carry mutations in KIT and PDGFRA gene, but although these mutations vary among GISTs the definitive genotype/phenotype correlations are still under consideration. Moreover, currently it is not clear whether mutations are independent prognostic factors. It has been suggested that tumors’ location was associated with mutations in GISTs. KIT-MT exon 9 GISTs were predominantly located in small intestine whereas GISTs with PDGFRA mutations represent gastric tumors. Others studies failed to find a significant association between KIT exon 11 mutation status and tumor location. The mutation type in GISTs has been reported to be associated with the phenotype. With respect to histological phenotype, KIT exon 11 mutations were strongly associated with spindle cell phenotype GISTs. Deletions in KIT exon 11 have been mostly associated with spindle cell phenotype, substitutions in KIT exon 11 were associated exclusively with mixed cell phenotype, whereas insertions in KIT exon 11 affecting codons 557/558 were exclusively associated with epithelioid cell phenotype GISTs.
Innovations and breakthroughs
In addition to deletions it is possible that insertions affecting the 557/558 codon, although rare, are also associated with malignant phenotype as suggested by previously published data. Codons 557/558 have been also found to represent significant a/a residues either for inhibitory role in the control of the receptor tyrosine kinase activity (Tryp557) or in constitutive receptor phosphorylation (Lys558). We presented evidence that tumors defined by KIT codon 557/558 deletion/insertion mutations represent a subgroup of GISTs with malignant clinical behaviour.
Applications
These findings underline the need for a new classification system that would integrate specific molecular alterations to the pathological criteria.
Peer review
This is a nice study which analysed the frequency and pattern of KIT and PDGFRA mutations in a group of patients with GISTs and the association of these mutations with other clinicopathological factors.
Acknowledgments
We are grateful to Eugenia Tsompanaki, MSs, Department of Statistics, Athens University, School of Economics and Business, for her invaluable help in data management.
Supported by The Hellenic State Scholarship Foundation, Dept of Science Promotion, 2005 Grant for Scientific Research. No. 19366/2005
Peer reviewer: Yoshiharu Motoo, MD, PhD, FACP, FACG, Professor and Chairman, Department of Medical Oncology, Kanazawa Medical University,1-1 Daigaku, Uchinada, Ishikawa 920-0293, Japan
S- Editor Yang RH L- Editor Mihm S E- Editor Lu W
References
- 1.Miettinen M, El-Rifai W, H L Sobin L, Lasota J. Evaluation of malignancy and prognosis of gastrointestinal stromal tumors: a review. Hum Pathol. 2002;33:478–483. doi: 10.1053/hupa.2002.124123. [DOI] [PubMed] [Google Scholar]
- 2.Fletcher CD, Berman JJ, Corless C, Gorstein F, Lasota J, Longley BJ, Miettinen M, O'Leary TJ, Remotti H, Rubin BP, et al. Diagnosis of gastrointestinal stromal tumors: A consensus approach. Hum Pathol. 2002;33:459–465. doi: 10.1053/hupa.2002.123545. [DOI] [PubMed] [Google Scholar]
- 3.Kontogianni K, Demonakou M, Kavantzas N, Lazaris ACh, Lariou K, Vourlakou C, Davaris P. Prognostic predictors of gastrointestinal stromal tumors: a multi-institutional analysis of 102 patients with definition of a prognostic index. Eur J Surg Oncol. 2003;29:548–556. doi: 10.1016/s0748-7983(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 4.Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 5.Kindblom LG, Remotti HE, Aldenborg F, Meis-Kindblom JM. Gastrointestinal pacemaker cell tumor (GIPACT): gastrointestinal stromal tumors show phenotypic characteristics of the interstitial cells of Cajal. Am J Pathol. 1998;152:1259–1269. [PMC free article] [PubMed] [Google Scholar]
- 6.Demetri GD. Identification and treatment of chemoresistant inoperable or metastatic GIST: experience with the selective tyrosine kinase inhibitor imatinib mesylate (STI571) Eur J Cancer. 2002;38 Suppl 5:S52–S59. doi: 10.1016/s0959-8049(02)80603-7. [DOI] [PubMed] [Google Scholar]
- 7.Debiec-Rychter M, Wasag B, Stul M, De Wever I, Van Oosterom A, Hagemeijer A, Sciot R. Gastrointestinal stromal tumours (GISTs) negative for KIT (CD117 antigen) immunoreactivity. J Pathol. 2004;202:430–438. doi: 10.1002/path.1546. [DOI] [PubMed] [Google Scholar]
- 8.Kontogianni-Katsarou K, Lariou C, Tsompanaki E, Vourlakou C, Kairi-Vassilatou E, Mastoris C, Pantazi G, Kondi-Pafiti A. KIT-negative gastrointestinal stromal tumors with a long term follow-up: a new subgroup does exist. World J Gastroenterol. 2007;13:1098–1102. doi: 10.3748/wjg.v13.i7.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taniguchi M, Nishida T, Hirota S, Isozaki K, Ito T, Nomura T, Matsuda H, Kitamura Y. Effect of c-kit mutation on prognosis of gastrointestinal stromal tumors. Cancer Res. 1999;59:4297–4300. [PubMed] [Google Scholar]
- 10.Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen CJ, Joseph N, Singer S, Griffith DJ, Haley A, Town A, et al. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- 11.Lasota J, Wozniak A, Sarlomo-Rikala M, Rys J, Kordek R, Nassar A, Sobin LH, Miettinen M. Mutations in exons 9 and 13 of KIT gene are rare events in gastrointestinal stromal tumors. A study of 200 cases. Am J Pathol. 2000;157:1091–1095. doi: 10.1016/S0002-9440(10)64623-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yarden Y, Kuang WJ, Yang-Feng T, Coussens L, Munemitsu S, Dull TJ, Chen E, Schlessinger J, Francke U, Ullrich A. Human proto-oncogene c-kit: a new cell surface receptor tyrosine kinase for an unidentified ligand. EMBO J. 1987;6:3341–3351. doi: 10.1002/j.1460-2075.1987.tb02655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitamura Y, Hirota S, Nishida T. Molecular pathology of c-kit proto-oncogene and development of gastrointestinal stromal tumors. Ann Chir Gynaecol. 1998;87:282–286. [PubMed] [Google Scholar]
- 14.Lasota J, Jasinski M, Sarlomo-Rikala M, Miettinen M. Mutations in exon 11 of c-Kit occur preferentially in malignant versus benign gastrointestinal stromal tumors and do not occur in leiomyomas or leiomyosarcomas. Am J Pathol. 1999;154:53–60. doi: 10.1016/S0002-9440(10)65250-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubin BP. Gastrointestinal stromal tumours: an update. Histopathology. 2006;48:83–96. doi: 10.1111/j.1365-2559.2005.02291.x. [DOI] [PubMed] [Google Scholar]
- 16.Kim TW, Lee H, Kang YK, Choe MS, Ryu MH, Chang HM, Kim JS, Yook JH, Kim BS, Lee JS. Prognostic significance of c-kit mutation in localized gastrointestinal stromal tumors. Clin Cancer Res. 2004;10:3076–3081. doi: 10.1158/1078-0432.ccr-03-0581. [DOI] [PubMed] [Google Scholar]
- 17.Lasota J, Dansonka-Mieszkowska A, Sobin LH, Miettinen M. A great majority of GISTs with PDGFRA mutations represent gastric tumors of low or no malignant potential. Lab Invest. 2004;84:874–883. doi: 10.1038/labinvest.3700122. [DOI] [PubMed] [Google Scholar]
- 18.Tornillo L, Terracciano LM. An update on molecular genetics of gastrointestinal stromal tumours. J Clin Pathol. 2006;59:557–563. doi: 10.1136/jcp.2005.031112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antonescu CR, Viale A, Sarran L, Tschernyavsky SJ, Gonen M, Segal NH, Maki RG, Socci ND, DeMatteo RP, Besmer P. Gene expression in gastrointestinal stromal tumors is distinguished by KIT genotype and anatomic site. Clin Cancer Res. 2004;10:3282–3290. doi: 10.1158/1078-0432.CCR-03-0715. [DOI] [PubMed] [Google Scholar]
- 20.Koon N, Schneider-Stock R, Sarlomo-Rikala M, Lasota J, Smolkin M, Petroni G, Zaika A, Boltze C, Meyer F, Andersson L, et al. Molecular targets for tumour progression in gastrointestinal stromal tumours. Gut. 2004;53:235–240. doi: 10.1136/gut.2003.021238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou YY, Tan YS, Sun MH, Wei YK, Xu JF, Lu SH, A-Ke-Su SJ, Zhou YN, Gao F, Zheng AH, et al. C-kit gene mutation in human gastrointestinal stromal tumors. World J Gastroenterol. 2004;10:1310–1314. doi: 10.3748/wjg.v10.i9.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersson J, Bumming P, Meis-Kindblom JM, Sihto H, Nupponen N, Joensuu H, Oden A, Gustavsson B, Kindblom LG, Nilsson B. Gastrointestinal stromal tumors with KIT exon 11 deletions are associated with poor prognosis. Gastroenterology. 2006;130:1573–1581. doi: 10.1053/j.gastro.2006.01.043. [DOI] [PubMed] [Google Scholar]
- 23.Related Articles, LinksMartín J, Poveda A, Llombart-Bosch A, Ramos R, López-Guerrero JA, García del Muro J, Maurel J, Calabuig S, Gutierrez A, et al. Deletions affecting codons 557-558 of the c-KIT gene indicate a poor prognosis in patients with completely resected gastrointestinal stromal tumors: a study by the Spanish Group for Sarcoma Research (GEIS) J Clin Oncol. 2005;23:6190–6198. doi: 10.1200/JCO.2005.19.554. [DOI] [PubMed] [Google Scholar]
- 24.Ma Y, Cunningham ME, Wang X, Ghosh I, Regan L, Longley BJ. Inhibition of spontaneous receptor phosphorylation by residues in a putative alpha-helix in the KIT intracellular juxtamembrane region. J Biol Chem. 1999;274:13399–13402. doi: 10.1074/jbc.274.19.13399. [DOI] [PubMed] [Google Scholar]
- 25.Miettinen M, Lasota J. Gastrointestinal stromal tumors: pathology and prognosis at different sites. Semin Diagn Pathol. 2006;23:70–83. doi: 10.1053/j.semdp.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 26.Antonescu CR, Sommer G, Sarran L, Tschernyavsky SJ, Riedel E, Woodruff JM, Robson M, Maki R, Brennan MF, Ladanyi M, et al. Association of KIT exon 9 mutations with nongastric primary site and aggressive behavior: KIT mutation analysis and clinical correlates of 120 gastrointestinal stromal tumors. Clin Cancer Res. 2003;9:3329–3337. [PubMed] [Google Scholar]
- 27.Wardelmann E, Losen I, Hans V, Neidt I, Speidel N, Bierhoff E, Heinicke T, Pietsch T, Buttner R, Merkelbach-Bruse S. Deletion of Trp-557 and Lys-558 in the juxtamembrane domain of the c-kit protooncogene is associated with metastatic behavior of gastrointestinal stromal tumors. Int J Cancer. 2003;106:887–895. doi: 10.1002/ijc.11323. [DOI] [PubMed] [Google Scholar]
- 28.Andersson J, Sjogren H, Meis-Kindblom JM, Stenman G, Aman P, Kindblom LG. The complexity of KIT gene mutations and chromosome rearrangements and their clinical correlation in gastrointestinal stromal (pacemaker cell) tumors. Am J Pathol. 2002;160:15–22. doi: 10.1016/S0002-9440(10)64343-X. [DOI] [PMC free article] [PubMed] [Google Scholar]