Abstract
High-resolution amplicon melting is a simple method for genotyping that uses only generic PCR primers and a saturating DNA dye. Multiplex amplicon genotyping has previously been reported in a single color, but two instruments were required: a carousel-based rapid cycler and a high-resolution melting instrument for capillaries. Manual transfer of capillaries between instruments and sequential melting of each capillary at 0.1°C/s seriously limited the throughput. In this report, a single instrument that combines rapid-cycle real-time PCR with high-resolution melting [LightScanner-32 (LS-32), Idaho Technology, Salt Lake City, UT] was used for multiplex amplicon genotyping. The four most common mutations associated with thrombophilia, F5 (factor V Leiden 1691G>A), F2 (prothrombin 20210G>A), and methylenetetrahydrofolate reductase (MTHFR; 1298A>C and 677C>T) were genotyped in a single homogeneous assay with internal controls to adjust for minor chemistry and instrument variation. Forty temperature cycles required 9.2 min, and each capillary required 2.2 min by melting at 0.3°C/s, 3× the prior rate. Sample volume was reduced from 20 μl to 10 μl. In a blinded study of 109 samples (436 genotypes), complete concordance with standard assays was obtained. In addition, the rare variant MTHFR 1317T>C was genotyped correctly when present. The LS-32 simplifies more complex high-resolution melting assays by reducing hands-on manipulation, total time of analysis, and reagent cost while maintaining the resolution necessary for multiplex amplicon genotyping.
Keywords: Thrombophilia, F5, F2, MTHFR, PCR, SNP, LightScanner-32
INTRODUCTION
Genotyping by high-resolution amplicon melting uses only two PCR primers/locus and a generic, saturating DNA dye that detects heteroduplexes as well as homoduplexes. Heterozygous genotypes have a characteristic melting curve shape and a broader width than homozygous genotypes, which are assigned based on small melting temperature (Tm) differences that are usually around 1.0°C but can be less depending on the base change and the length of the amplicon.1,2 Genotyping accuracy depends on the resolution of the melting instrument and appropriate software for analysis.3–5 The discrimination of different homozygous genotypes by Tm is affected by chemistry and instrument variance and can be corrected by the inclusion of temperature correction controls within each PCR.6,7
Multiplex genotyping by amplicon melting is also possible. Multiple amplicons targeting different loci may be separated naturally in Tm based on their guanine-cytosine content and sequence. If the Tms of different loci are not separated naturally, adequate separation can often be achieved by modifying the length of the amplicons or by selectively adding Tm-shifting primer tails.8,9 The ability to genotype up to four loci in the same reaction using amplicon melting has been reported using rapid-cycle PCR for amplification in one instrument and high-resolution melting in separate instruments.9 However, individual capillary samples had to be transferred manually between instruments, and the melting rates were slow (0.1°C/s), resulting in limited throughput.
Here, we report use of a new instrument that combines rapid-cycle PCR with high-resolution melting for multiplex amplicon genotyping to eliminate manual sample handling. Associated advantages include a threefold increase in melting rate to 0.3°C/s, which triples throughput, and a twofold reduction in reaction volume that halves reagent costs. Genotyping accuracy is demonstrated using a four-plex thrombophilia amplicon melting assay for the F5 (factor V Leiden, 1691G>A), F2 (prothrombin 20210G>A), and methylenetetrahydrofolate reductase (MTHFR; 1298A>C and 677C>T) mutations and also discriminates MTHFR 1317T>C successfully.
MATERIALS AND METHODS
LightScanner-32 (LS-32)
LS-32 (Idaho Technology, Salt Lake City, UT) is a new instrument that combines LightCycler® (Roche Applied Systems, Indianapolis, IN) and HR-1™ (Idaho Technology) technologies (Fig. 1). The LS-32 eliminates the manual transfer of individual capillaries from the LightCycler® to the HR-1™. This transfer is replaced by the automatic rotation of the LS-32 carousel into position over the high-resolution melting ingot (Fig. 1B). The ingot is elevated to enclose each capillary in turn to perform the high-resolution melting analysis. When one tube is finished, the ingot drops, and the carousel rotates the next sample into position. This greatly reduces the hands-on time associated with the original method. The LS-32 can perform rapid-cycle PCR, acquire real-time fluorescence, and provide low- and high-resolution melting analysis on 10 μl samples.
FIGURE 1.
Photo (A) and schematic (B) of LS-32. (A) The instrument. (B) The internal design of LS-32 showing the 32-sample carousel and the high-resolution melting ingot with light source/fluorescence-detection housing.
Study Samples and DNA Extraction
Whole blood samples were submitted to ARUP (Salt Lake City, UT) for F5 (1691G>A), MTHFR (1298A>C and 677C>T), or F2 (20210G>A) genotyping. DNA was extracted with the Roche Applied Systems MagNA Pure LC system (Roche Applied Systems), resulting in concentrations of 10–67 ng/μl by absorbance at 260 nm, which were diluted to a uniform concentration of 10 ng/μl prior to four-plex amplification. Samples were genotyped at all loci using HybProbe (MTHFR)7 or unlabeled probe (F5 and F2) assays.10 A total of 109 samples was selected to enrich rare genotypes at each locus. The genotypes of these samples were 89 wild-type, 10 heterozygous, and 10 homozygous at F5 Leiden; 70 wild-type, 24 heterozygous, and 15 homozygous at MTHFR 1298; 59 wild-type, 33 heterozygous, and 17 homozygous at MTHFR 677; and 90 wild-type, 15 heterozygous, and 4 homozygous at F2 20210. At MTHFR 1317, 103 of these samples were wild-type, five heterozygous, and one homozygous. The samples were de-identified according to a global ARUP protocol (IRB #7275) after blinding and analyzed by the thrombophilia multiplex amplicon melting assay.
Thrombophilia Multiplex High-Resolution Amplicon Melting Assay
Oligonucleotide sequences for the primers as well as the internal controls have been published previously.9 PCR was performed in 10 μl vol with 1× LightCycler® FastStart DNA Master HybProbe (Roche Applied Systems), 0.5 μM each F5 primers, 0.15 μM each MTHFR 1298 and 677 primers, 0.16 μM each F2 primers, 0.06 μM low-temperature correction control and 0.08 μM high-temperature correction control, 3.5 mM MgCl2 (including 1 mM MgCl2 contributed by the LightCycler® FastStart DNA Master HybProbe solution), 0.01 U/reaction heat-labile uracil-DNA glycosylase (Roche Applied Systems), 1× LCGreen® Plus (Idaho Technology), and 20 ng template DNA. All oligonucleotides were mixed together and stored as a 20× stock solution.
PCR and high-resolution melting were done on the LS-32 (Idaho Technology). PCR included an initial hold of 95°C for 10 min, followed by 15 cycles of 95°C for 2 s, 56°C for 1 s, and 72°C for 1 s, and 25 cycles of 95°C for 2 s, 58°C for 1 s, and 72°C for 4 s. During amplification, no fluorescence acquisition was performed to avoid prolonging the temperature cycles. All heating and cooling steps during PCR were done with ramp rates programmed at 20°C/s. After PCR, samples were cooled (10°C/s) from 95°C to 40°C and melting curves generated with continuous fluorescence acquisition from 55°C to 95°C at 0.3°C/s. Data processing included normalization of fluorescence, exponential background removal,11 and display of derivative melting curves, which were adjusted by identifying the maxima of the temperature correction control peaks and aligning curves by shifting and linear-scaling using custom software.7 Heterozygotes were identified by melting-peak width and shape. Homozygotes were assigned genotypes by visual inspection based on Tm (melting-peak maxima). Predicted Tms based on nearest-neighbor parameters were calculated as described previously.1,2
RESULTS
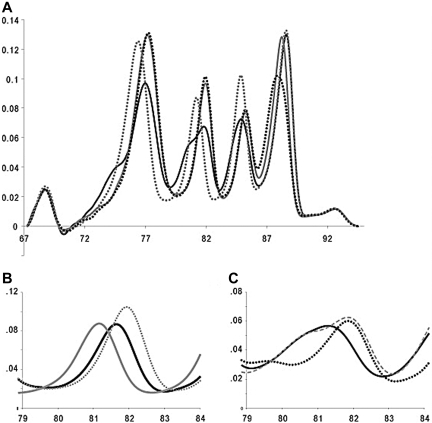
Representative multiplex genotyping results are shown in Figure 2A as derivative melting curves after normalization, exponential background subtraction, and correction with internal controls. The melting transitions for genotyping are spread over an 18°C temperature range with the controls bracketing this range by an additional 5°C. For each locus, heterozygous genotypes result in an altered melting-curve shape with a wider transition, and homozygous genotypes can be distinguished by Tm.
FIGURE 2.
Derivative melting plots of the multiplex thrombophilia melting assay. (A) Four representative melting profiles are shown, which in combination, contain examples of all genotypes at each locus. The melting plots are shown as a solid black line (F5 1691GA, MTHFR 1298AC and 677CT, and F2 20210GG), solid gray line (F5 1691GG, MTHFR 1298CC and 677CC, and F2 20210AA), dotted black line (F5 1691GG, MTHFR 1298CC and 677CC, and F2 20210GA), and dotted gray line (F5 1691AA, MTHFR 1298AA and 677TT, and F2 20210GG). The derivative melting plot includes all four thrombophilia loci and high and low 50 bp complimentary oligonucleotide temperature correction controls. (B and C) Atypical derivative melting plots from the MTHFR 1298A>C amplicon. The gray lines represent characteristic MTHFR 1298 melting curves for AA (solid line), CC (dotted line), and AC (dashed line) genotypes. (B) Derivative melting curves of a MTHFR 1298 AA sample that was homozygous MTHFR 1317T>C (solid black). (C) A representative derivative melting curve of a MTHFR 1298 AA sample that was heterozygous MTHFR 1317T>C (solid black). The dotted black line represents the derivative melting curve of a sample heterozygous for MTHFR 1298A>C and 1317T>C.
All 109 blinded F5, MTHFR 677C>T, MTHFR 1298A>C, and F2 variants were genotyped correctly. Furthermore, variation at the 1317T>C locus that is internal to the MTHFR 1298 amplicon was also detected and genotyped correctly in six samples, which included four 1298AA, 1317TC heterozygotes, one 1298AC, 1317TC double heterozygote, and one 1298AA, 1317CC homozygote. The 1298AA, 1317CC homozygote had a Tm of 81.6°C between the 1298AA, 1317TT (81.1°C) and 1298CC, 1317TT (81.9°C) homozygotes (Fig. 2B). Nearest-neighbor modeling predicted the same order of homozygote stabilities with separations of 0.4°C between 1298AA, 1317TT and 1298AA, 1317CC and 0.3°C between 1298AA, 1317CC and 1298CC, 1317TT. Four 1298AA, 1317TC heterozygotes followed a melting-curve cluster distinctly different from 1298AC, 1317TT heterozygotes (Fig. 2C). The double heterozygote (1298 AC and 1317TC) correlated with a distinct heteroduplex peak between 79°C and 80°C (Fig. 2C).
The Tms and SDs of all homozygotes, with and without correction by internal controls, are shown in Table 1. The Tms of alternative homozygotes were separated by 0.60°C (F5), 0.76°C (MTHFR 1298), 0.39°C (MTHFR 677), and 0.10°C (F2) without correction and 0.80°C (F5), 0.73°C (MTHFR 1298), 0.40°C (MTHFR 677), and 0.30°C (F2) after correction. Internal controls decreased the Tm SD by an average of 67%. Internal controls decreased the average homozygous Tm SD from 0.13°C to 0.04°C, a variation low enough to allow accurate genotyping of all homozygotes. Without internal control correction, 14 of the possible 436 genotypes were assigned incorrectly. Six of the 14 incorrectly assigned genotypes were MTHFR 1298 samples confounded by the presence of the 1317 variant. The other samples included one F5 1691GG assigned as 1691AA, one F2 20210GG and two 20210AA called 20210AA and GG, respectively, and one MTHFR 677CC assigned as 677TT.
TABLE 1.
Tms and SDs of Homozygous Genotypes
| Wild-typea |
Wild-typeb |
Varianta |
Variantb |
|||||
|---|---|---|---|---|---|---|---|---|
| Tm | SD | Tm | SD | Tm | SD | Tm | SD | |
| F5 | 77.20 | 0.121 | 77.27 | 0.052 | 76.60 | 0.067 | 76.47 | 0.048 |
| MTHFR 1298 | 81.09 | 0.121 | 81.16 | 0.050 | 81.85 | 0.155 | 81.89 | 0.026 |
| MTHFR 677 | 85.03 | 0.133 | 85.10 | 0.018 | 84.64 | 0.123 | 84.70 | 0.039 |
| F2 | 88.28 | 0.146 | 88.33 | 0.047 | 88.18 | 0.150 | 88.03 | 0.050 |
Data without internal controls
Internal control temperature adjusted data.
DISCUSSION
Deep venous thrombosis is dependent on many factors including heredity and acquired lifestyle risk factors.12 Mutations at several well-defined loci in genes coding for proteins involved in coagulation, fibrinolysis, and homocysteine metabolism contribute to the development of deep venous thrombosis.13 The potential cooperative interaction of these mutations in the development of thrombophilia suggests multiplex genotyping. High-resolution melting of amplicons is an attractive method for genotyping because of its simplicity and has been reviewed recently.14–16
Up to four different single-base variants were amplified and genotyped previously by high-resolution melting in one reaction using a single fluorescence color.9 However, two different instruments, one for rapid-cycle PCR and the other for high-resolution melting, were required.9 Furthermore, manual transfer of individual sample capillaries was necessary. Each 20 μl sample was melted at 0.1°C/s requiring 6.7 min on an instrument that only melted one sample at a time. Although the data quality was excellent, the throughput was low and required significant hands-on time. Internal controls decreased the SD within a genotype by 38% to an average weighted SD of 0.06°C. In this prior study, internal controls were not necessary for correct genotyping.
With minimal adjustments of oligonucleotide concentrations, the same quadraplex assay was adapted to the LS-32 platform. The LS-32 integrates rapid-cycle PCR and high-resolution melting on one instrument so that no manual transfers are necessary. Temperature cycling for PCR required 9.2 min, similar to the time of 10.1 min for the LightCycler® 1.5. We found that the LS-32 generated high-resolution melting profiles similar to the HR-1™ at 3× the rate (0.3°C/s), lowering the analysis time of each sample to 2.2 min. In addition, the sample volume was reduced from 20 μl to 10 μl. Internal controls were required for correct genotyping. When internal control adjustments were not made, 14 of the possible 336 genotypes were assigned incorrectly. The Tm SD after internal control correction was decreased by 67% to an average weighted SD of 0.04°C, even lower than similar data reported on the HR-1™.
The MTHFR 1298 primers bracket position 1317, where a known variant is observed in <5% of Caucasians and >40% of Blacks.17 In our prior study, the 1317 variants were unexpected and resulted in atypical MTHFR 1298 melting profiles that could not be assigned a genotype. Sequencing confirmed that the variations were correlated with 1317 variants. In the current study, similar variants also occurred, but the patterns were expected, and by prior experience, all melting curves were genotyped correctly at 1298 and 1317. The 1317 variant can confound MTHFR 1298 genotyping with hydrolysis probe and restriction fragment-length polymorphism assays.18 In contrast, all common genotypes with 1317 variants (1298AC/1317TC, 1298AA/1317TC, and 1298AA/1317CC) were clearly distinct by melting analysis (Fig. 2B and C). Furthermore, we expect the more rare genotypes 1298CC/1317CC, 1298AC/1317CC, and 1298CC/1317TC to be distinct also. For example, by nearest-neighbor predictions, the Tm of the 1298CC/1317CC homozygote should be 0.4°C higher than the 1298CC/1317TT homozygote and should be easily identifed. In addition, different single heterozygotes within the same amplicon are usually distinct.19,20
These studies were performed to evaluate the LS-32 as a rapid-cycle real-time PCR instrument with high-resolution melting. The LS-32 required 9.2 min for 40 cycles compared with 10.1 min on a carousel LightCycler®. High-resolution melting on the LS-32 easily discriminated homozygous from heterozygous melting profiles as well as different heterozygous profiles from each other. The Tm precision of the LS-32 with internal controls appears superior to the HR-1™, previously reported as the instrument with the highest melting precision.3 The LS-32 is more convenient than the LightCycler®/HR-1™ pair by eliminating manual transfer of capillaries between instruments. The LS-32 should be able to run all carousel LightCycler® assays with the added advantage of high-resolution melting.
Rapid cycling and high-resolution melting appear necessary for multiplex amplicon genotyping.9 We have not been able to reproduce this work on instruments other than the LS-32 or the carousel LightCycler®/HR-1™ combination, presumably because of limits in cycling speed and/or melting resolution of other instruments. Access to exponential background subtraction methods21 may be another reason why multiplex amplicon genotyping has only appeared on these platforms. Further work will focus on automatic genotype analysis in software to provide objective results not requiring expert evaluation.
Footnotes
Statement of Competing Interests: Aspects of high-resolution melting and rapid-cycle PCR are licensed by the University of Utah to Idaho Technology and from Idaho Technology to Roche Applied Systems. C. T. W. holds equity interest in Idaho Technology.
REFERENCES
- 1.Liew M, Pryor R, Palais R, et al. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem 2004; 50: 1156– 1164 [DOI] [PubMed] [Google Scholar]
- 2.Palais RA, Liew MA, Wittwer CT.Quantitative heteroduplex analysis for single nucleotide polymorphism genotyping. Anal Biochem 2005; 346: 167– 175 [DOI] [PubMed] [Google Scholar]
- 3.Herrmann MG, Durtschi JD, Bromley LK, Wittwer CT, Voelkerding KV.Amplicon DNA melting analysis for mutation scanning and genotyping: cross-platform comparison of instruments and dyes. Clin Chem 2006; 52: 494– 503 [DOI] [PubMed] [Google Scholar]
- 4.Herrmann MG, Durtschi JD, Bromley LK, Wittwer CT, Voelkerding KV.Instrument comparison for heterozygote scanning of single and double heterozygotes: a correction and extension of Herrmann et al.. Clin Chem 2006; 52: 494– 503Clin Chem 2007;53:150–152 [DOI] [PubMed] [Google Scholar]
- 5.Herrmann MG, Durtschi JD, Wittwer CT, Voelkerding KV.Expanded instrument comparison of amplicon DNA melting analysis for mutation scanning and genotyping. Clin Chem 2007; 53: 1544– 1548 [DOI] [PubMed] [Google Scholar]
- 6.Liew M, Seipp M, Durtschi J, et al. Closed-tube SNP genotyping without labeled probes/a comparison between unlabeled probe and amplicon melting. Am J Clin Pathol 2007; 127: 341– 348 [DOI] [PubMed] [Google Scholar]
- 7.Seipp MT, Durtschi JD, Liew MA, et al. Unlabeled oligonucleotides as internal temperature controls for genotyping by amplicon melting. J Mol Diagn 2007; 9: 284– 289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erali M, Pounder JI, Woods GL, Petti CA, Wittwer CT.Multiplex single-color PCR with amplicon melting analysis for identification of Aspergillus species. Clin Chem 2006; 52: 1443– 1445 [DOI] [PubMed] [Google Scholar]
- 9.Seipp MT, Pattison D, Durtschi JD, Jama M, Voelkerding KV, Wittwer CT.Quadruplex genotyping of F5, F2, and MTHFR variants in a single closed tube by high-resolution amplicon melting. Clin Chem 2008; 54: 108– 115 [DOI] [PubMed] [Google Scholar]
- 10.Chou LS, Meadows C, Wittwer CT, Lyon E.Unlabeled oligonucleotide probes modified with locked nucleic acids for improved mismatch discrimination in genotyping by melting analysis. Biotechniques 2005; 39: 644– 648– 646 [DOI] [PubMed] [Google Scholar]
- 11.Erali M, Palais R, Wittwer CT.SNP genotyping by unlabeled probe melting analysis. Seitz O, Marx A. (eds.): Methods in Molecular Biology (Clifton, N.J.)2008; 429: 199– 206 [DOI] [PubMed] [Google Scholar]
- 12.Dahlback B.Blood coagulation. Lancet 2000; 355: 1627– 1632 [DOI] [PubMed] [Google Scholar]
- 13.Key NS, McGlennen RC.Hyperhomocyst(e) inemia and thrombophilia. Arch Pathol Lab Med 2002; 126: 1367– 1375 [DOI] [PubMed] [Google Scholar]
- 14.Erali M, Voelkerding KV, Wittwer CT.High resolution melting applications for clinical laboratory medicine. Exp Mol Pathol 2008; 85: 50– 58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farrar JS, Reed GH, Wittwer CT.High Resolution Melting Curve Analysis for Molecular Diagnostics, 2nd ed. Burlington, MA: Elsevier, 2009 [Google Scholar]
- 16.Reed GH, Kent JO, Wittwer CT.High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics 2007; 8: 597– 608 [DOI] [PubMed] [Google Scholar]
- 17.Pegoraro RJ, Chikosi A, Rom L, Roberts C, Moodley J.Methylenetetrahydrofolate reductase gene polymorphisms in black South Africans and the association with preeclampsia. Acta Obstet Gynecol Scand 2004; 83: 449– 454 [DOI] [PubMed] [Google Scholar]
- 18.Allen RA, Gatalica Z, Knezetic J, Hatcher L, Vogel JS, Dunn ST.A common 1317TC polymorphism in MTHFR can lead to erroneous 1298AC genotyping by PCR-RE and TaqMan probe assays. Genet Test 2007; 11: 167– 173 [DOI] [PubMed] [Google Scholar]
- 19.Graham R, Liew M, Meadows C, Lyon E, Wittwer CT.Distinguishing different DNA heterozygotes by high-resolution melting. Clin Chem 2005; 51: 1295– 1298 [DOI] [PubMed] [Google Scholar]
- 20.Montgomery J, Wittwer CT, Kent JO, Zhou L.Scanning the cystic fibrosis transmembrane conductance regulator gene using high-resolution DNA melting analysis. Clin Chem 2007; 53: 1891– 1898 [DOI] [PubMed] [Google Scholar]
- 21.Erali M, Palais R, Wittwer C.SNP genotyping by unlabeled probe melting analysis. Methods Mol Biol 2008; 429: 199– 206 [DOI] [PubMed] [Google Scholar]