Abstract
Quantitative RT-PCR can be carried out as a one- or a two-step reaction. However, the choice of method raises controversy from the perspective of the researcher and manufacturer, because of advantages and disadvantages with both systems. We therefore hypothesize that running the RNA-to-CT™ 2-Step kit [(Applied Biosystems (AB), Foster City, CA] using a one-step protocol (as recommended) is not appropriate for quantitation of gene expression levels and should not be performed. Consequently, we ran comparative studies of the two suggested methods to evaluate their efficiency, sensitivity, and accuracy. To ensure precession, two different PCR machines were used: the StepOnePlus system and Chromo4. In addition, the RNA-to-CT™ 1-Step kit (recently launched by AB) was also used to compare its efficiency with these methods. Efficiency, sensitivity, and linearity were determined by standard curves generated using RNA isolated from C2 myoblasts to amplify the housekeeping gene GAPDH. When the RNA-to-CT™ 2-Step kit was run as a two-step reaction on the Chromo4 or StepOnePlus, respectively, not only did the efficiency increase (100±1.5% and 99.7±0.95%) but also the sensitivity (comparative threshold cycle for the lowest standard: 33.2±0.5 and 32.5±0.7) and linearity (0.997±0.001 and 0.993±0.006) compared with RNA-to-CT™ 2-Step run as one-step and RNA-to-CT™ 1-Step kit. This is the first study to demonstrate that the RNA-to-CT™ 2-Step kit is not reliable to be performed as a one-step reaction but as a two-step reaction, is even more sensitive than the newly launched RNA-to-CT™ 1-Step kit.
Keywords: linearity, real time PCR, methodology validation
RT-PCR reaction converts and amplifies a ssRNA template to yield abundant dsDNA product. This technique has gained renewed importance since the development of quantitative real-time RT-PCR,1 which is a powerful tool for analysis of gene expression and quantitation and for characterization of RNA splice variants.2,3 These techniques rely on measuring the amplification of a fluorescence signal generated during the log linear phase of the reaction, where the sensitivity and efficiencies of the reaction for the target gene are normalized against a housekeeping gene.1,4 Ideally, the qRT-PCR is validated and optimized by standard curve linearity, amplification efficiency, and sensitivity. As the real-time quantitation is based on the relation between the initial template amount and the comparative threshold cycle (CT), at which fluorescence signal crosses the threshold line during the exponential phase of the amplification, validation, and optimization of qRT-PCR is highly essential for accurate and reproducible quantitation of samples.5 By definition, qRT-PCR efficiency is the doubling of DNA concentration in each amplification cycle, which can be calculated from the slope of the standard curve (generated using tenfold serial dilutions) using the following formula: Efficiency (E) = 10(−1/slope).6 Sensitivity of qRT-PCR refers to the minimum quantity of target that can be detected above the background noise of the system. Linearity of qRT-PCR refers to a measure of variability across assay replicates and whether the amplification efficiency is the same for different starting template copy numbers. Linearity can be measured by the correlation coefficient of the line (R2), which should be >0.985 to validate the data.5,7 The RT-PCR is generally carried out as a one-step or two-step reaction. In the one-step reaction, all reagents are combined in one single tube, and RT and PCR are performed sequentially in the tube. For the two-step reaction, the RT reaction is first performed, and the resultant template subsequently undergoes PCR in a second step.
The choice of method is potentially somewhat controversial between the researchers and the manufacturers, as there are advantages and disadvantages for both systems. The purpose behind running RT and PCR reactions in a single tube (one-step) is that the method is less time-consuming and requires no user intervention, therefore minimizing the chance of pipetting errors and cross-contamination. However, several groups have determined that the one-step reaction is intrinsically variable and significantly less sensitive than the two-step method.8,9 One major disadvantage to using the one-step reaction is that it is not possible to store cDNA for any further applications. In contrast, the two-step method offers the opportunity to generate and store cDNA for any subsequent investigations. In the one-step reaction method, cDNA can only be generated by a gene-specific primer, and in the two-step method, a variety of priming options can be used to generate cDNA (gene-specific, poly-dT, and random hexamer primers), which may provide greater flexibility and optimization.
Applied Biosystems (AB; Foster City, CA) advertises that its two-step reagents (Power SYBR® Green RNA-to-CT™ 2-Step kit; P/N #4399449) may be used to perform a two-step or a one-step reaction. Therefore, the purpose of this study was to examine the efficiency, linearity, and sensitivity of the RNA-to-CT™ 2-Step (carried out as one-step) against the RNA-to-CT™ 2-Step (carried out as two-step) and newly launched RNA-to-CT™ 1-Step (Power SYBR® Green RNA-to-CT™ 1-Step kit; P/N #4389986) using two different PCR machines [AB and Bio-Rad (BR), Hercules, CA] and the universal housekeeping gene GAPDH. Our data show clearly that when the RNA-to-CT™ 2-Step kit is carried out as a two-step reaction, the resultant data are highly efficient, sensitive, and linear compared with data generated using the RNA-to-CT™ 2-Step as a one-step method, as advertised as being plausible by the manufacturers. Moreover, efficiency, sensitivity, and linearity obtained by the RNA-to-CT™ 2-Step kit (carried out as a two-step reaction) were superior to those produced by the RNA-to-CT™ 1-Step. These data were reproducible and accurate and confirmed using two different PCR machines.
MATERIALS AND METHODS
Cell Culture
C2 mouse skeletal myoblasts10 were grown to ∼80% cofluency in a humidified 5% CO2 atmosphere at 37°C in growth medium (GM) composed of DMEM with Glutamax supplemented with 10% FBS, 10% newborn calf serum, and penstrep and L-glutamine at final concentrations of 10,000 U/ml and 2 mM, respectively. Six-well plates were precoated with 0.2% gelatine for 5 min at room temperature, and cells were plated at 1 × 105 cells/ml in GM for next-day confluency. The following day, differentiation was initiated following washing with PBS by transferring cultured cells to low serum-containing differentiation medium [DMEM plus glutamax, supplemented with 2% human serum, penstrep, and L-glutamine (supplemented as above)].
Primer Design and Synthesis
Optimal primer designs are essential to ensure that only a single PCR product is amplified in particular when using real-time PCR-based SYBR Green methodology. We therefore used web-based software from Invitrogen (Carlsbad, CA) to design our primers (F: GC CT TCCGTGTT CT TACC; R: GC CT G CT TCACCAC CT TC), which were analyzed further by Sigma-Genosys (Haverhill, UK) software. The primers were designed to yield products spanning exon-intron boundaries to prevent any possible genomic DNA contaminations from total RNA isolation. Sequence homology searches against the database of GenBank showed that these primers matched only the sequence to which they were designed.
qRT-PCR and Data Analysis
RNA was extracted using the TRIzol method, according to the manufacturer's instructions (Invitrogen), followed by DNase digestion to minimize genomic contamination.11 For the RNA-to-CT™ 1-Step reaction (Power SYBR® Green RNA-to-CT™ 1-Step kit; P/N #4389986), the standard curve was generated by a fivefold serial dilution using total RNA at concentrations of 500 ng/reaction to 3.4 pg/reaction to amplify the housekeeping gene GAPDH. For the RNA-to-CT™ 2-Step reaction, the same RNA concentration was used for RT reaction, and then the cDNA was diluted using fivefold serial dilutions (as above) to generate the standard curve. Real-time PCR amplifications were carried out using the Power SYBR® Green RNA-to-CT™ 1-Step and RNA-to-CT™ 2-Step kits on the Chromo4 PCR (BR) and the StepOnePlus (AB) machines, according to the manufacturers' instructions. In brief, for RNA-to-CT™ 1-Step, real-time PCR was performed using the following cycles: 48°C for 30 min (for cDNA synthesis), 95°C for 10 min (transcriptase inactivation), followed by the following cycling parameters: 95°C for 15 s and 60°C for 1 min for 40 cycles. For the RNA-to-CT™ 2-Step reaction, the RNA-to-CT™ 1-Step protocol was used with the addition of one extra step at the beginning for the RT reaction (cDNA generation), which was as follows: 25°C for 15 min, 37°C for 60 min, and 95°C for 5 min. The resultant cDNA reaction (5 μl) was used to perform the real-time PCR. At the conclusion of the 40 cycles, dissociation curve (melting curve) analyses were performed using the following protocol: hot start at 60°C for 15 s, and then measure the fluorescence every 0.5°C until 95°C to confirm specific amplification. The Step-One software v2.0.1 (AB) and Opticon Monitor software, version 3.1 (BR), were used to analyze the data of the real-time PCR. Using data from samples designated as standard points with assigned concentrations, an arbitrary threshold level was set, and CT values for all PCR samples were calculated, allowing generation of standard curves.
Statistical Analysis
All experiments were repeated at least twice independently, and the measurements in each experiment were run in triplicate. Statistical analysis and the significance of the data were determined using SPSS, UK (Surry, UK) software, version 12.0. Results are presented as the mean ± SD. Statistical significance was determined using one-way ANOVA analysis of variance with multiple post hoc analyses. Results were considered statistically significant with P < 0.05 versus appropriate controls.
RESULTS
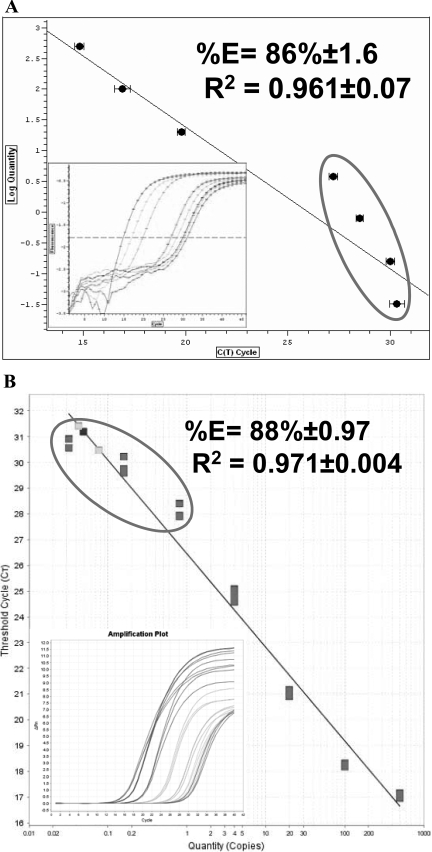
To address whether the original Power SYBR® Green RNA-to-CT™ 2-step and the newly launched RNA-to-CT™ 1-Step show comparable efficiency and sensitivity for the expression levels of the GAPDH housekeeping gene, fivefold dilutions of total RNA extracted from differentiated C2 myoblast cells were analyzed using AB and BR PCR machines. Representative real-time PCR standard curve linearity (log concentration versus CT) is shown in Figures 1–3, and inset graphs of amplification plots (Δ reaction vs. CT) are also presented in Figures 1–3. Each real-time RT-PCR was run in triplicate to generate standard curves on a log scale graph of RNA concentration versus CT. The slope of each standard curve was used to determine reaction efficiency.5 Initially, we sought to evaluate the efficiency and sensitivity of the RNA-to-CT™ 2-Step using the one-step protocol on the BR machine (Fig. 1A). It demonstrates lower-than-acceptable efficiency and sensitivity. The efficiency was 86 ± 1.6%, and the sensitivity was inaccurate in the quantitation of low RNA concentrations (0.032, 0.16, 0.8, 4.0 ng/reaction). The CT values for these standards were 30.0, 30, 28.7, and 27, respectively, and were not separated as we expected. Moreover, the linearity of the standard curve was low, R2 = 0.961 ± 0.07 (Fig. 1A). To elucidate whether the low efficiency and poor sensitivity at low concentrations, obtained from running RNA-to-CT™ 2-Step using a one-step protocol on a BR machine, were not occurring as a result of using a non-AB PCR machine, we tested the RNA-to-CT™ 2-Step using the one-step protocol on an AB machine. Similarly, the efficiency and standard curve linearity were comparable with data obtained on the BR machine, as 88 ± 0.97% and R2 = 0.971 ± 0.004, respectively. Compared with the BR machine, sensitivity was lost at the bottom end of the curve on the AB machine, with CT values of 30.7, 29.8, 28.2, and 24.8 (Fig. 1B).
FIGURE 1.
Efficiency and linearity of the Power SYBR® Green RNA-to-CT™ 2-Step kit run as one-step reaction. PCR amplification of standard curves generated using GAPDH as a reference gene on BR (A) and AB machines (B) using total RNA in amounts ranging from 500 ng to 3.2 pg (fivefold serial dilutions). (Insets) Graphs show the amplification plot of the standard curves, which were run on BR and on AB, respectively.
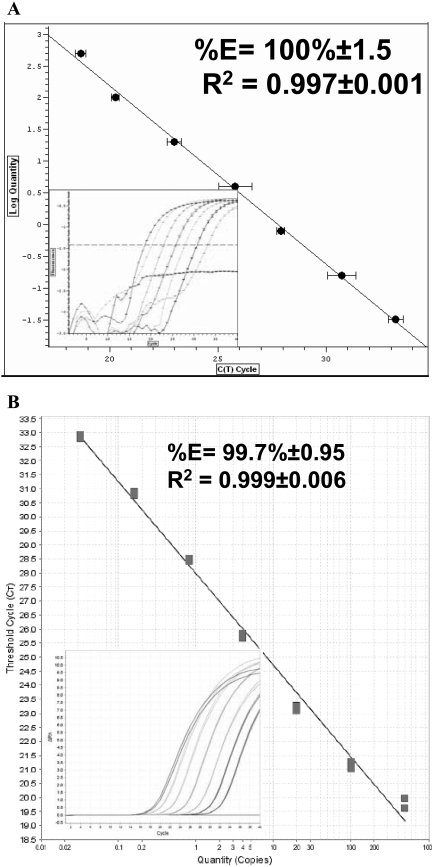
FIGURE 2.
Efficiency and linearity of the Power SYBR® Green RNA-to-CT™ 2-Step kit run as a two-step reaction. Standard curves were generated using GAPDH as a reference gene on BR (A) and AB machines (B), as mentioned in Figure 1. (Insets) Graphs show the amplification plot of the standard curves, which were run on BR and on AB, respectively.
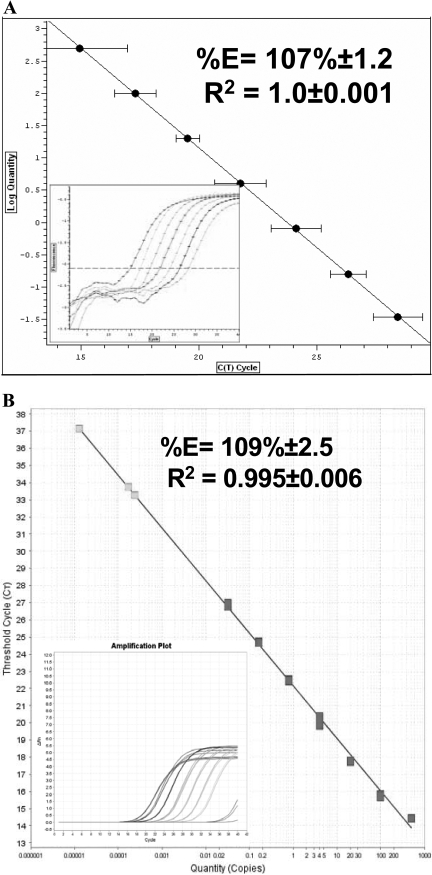
FIGURE 3.
Efficiency and linearity of Power SYBR® Green RNA-to-CT™ 1-Step kit. (A and B) The linearity of standard curves generated using GAPDH as a reference gene. Seven samples containing fivefold serial dilutions of GAPDH in amounts ranging from 500 ng to 3.2 pg were assayed on BR (A) and AB (B) PCR machines. (Insets) Graphs show the amplification plot of the standard curves, which were performed on BR and on AB, respectively.
To provide additional lines of evidence that the RNA-to-CT™ 2-Step is not accurate and appropriate to be run using the one-step protocol, the RNA-to-CT™ 2-Step was performed using two-step protocol on both machines (Fig. 2A and B). The efficiency was increased significantly up to 100 ± 1.5% on the BR and 99.7% ± 0.95 on the AB machine, and the standard curve linearity (R2) was also good on both machines: 0.997 ± 0.001 and 0.993 ± 0.006 for BR and AB, respectively (Fig. 2A and B). The sensitivity of both PCR machines was high and much closer to our expectation. The CT values of the lowest standard RNA concentration detected on the BR and AB machine were 33.2 ± 0.5 and 32.5 ± 0.7, respectively.
Finally, we carried out the newly launched RNA-to-CT™ 1-Step (Power SYBR® Green RNA-to-CT™ 1-Step kit; P/N #4389986) on both machines, and the efficiency, sensitivity, and linearity were determined (Fig. 3A and B). The data analysis using the BR and AB software show that the efficiency was 107 ± 1.2% and 109.5 ± 2.5%, and linearity was 1 ± 0.001 and 0.995 ± 0.006, respectively. The CT values of the lowest standard RNA concentration, as detected on the BR and AB machines, were 28.4 ± 0.9 and 27.2 ± 0.3, respectively.
DISCUSSION
The real-time, fluorescence-based qRT-PCR is a major development of PCR technology that enables reliable detection and measurement of the target-specific products amplified during each cycle of the PCR process.1,2 RT-PCR can be performed by a one-step method, in which the cDNA synthesis (RT reaction) and PCR are carried out in one tube as a single reaction, or by a two-step reaction, in which the RT reaction is run first, followed by the PCR reaction in a separate tube. Although the two-step RT-PCR reaction provides greater flexibility and better optimization, one-step protocols are powerful, as they minimize handling and therefore, reduce chances of pipetting errors and cross-contamination.12–14 Therefore, the preferred method of choice (one- or two-step) for RT-PCR has become somewhat controversial.
AB, one of the leading commercial companies in DNA/RNA quantitation and molecular biology techniques, produces a Power SYBR® Green RNA-to-CT™ 2-Step kit, which advocates one-step or two-step protocols for performing RT and cDNA quantitation by real-time PCR. Recently, AB launched a new Power SYBR® Green RNA-to-CT™ 1-Step kit that enables RT of RNA into cDNA, which is then quantitated by SYBR Green real-time PCR in a one-step protocol. Therefore, it was our aim to test whether the results of these two different protocols were comparable in terms of efficiency, sensitivity, and reliability.
In the last decade, it has been well established that validated qRT-PCR assays should show a good linear standard curve with the expected R2 as >0.980 and elicit a high amplification efficiency (90–110%) using: E = 10(−1/slope), where the slope is 2n = dilution factor of the standard curve, and n = the number of CT between curves.5,9,15 For example, in our system with a fivefold serial dilution of RNA, 2n = 5. Therefore, n = 2.35, and the CT values should be separated by 2.23 cycles.
Any variability in the efficiency, linearity, and sensitivity in real-time RT-PCR reactions could be a result of one or more of the following reasons: using different PCR machines, stability and quality of isolated RNA, and cDNA synthesis efficiency. In our current study, as the same RNA samples, reagents, and PCR machines were used, these may be excluded from any contribution to variability in efficiency and linearity of real-time RT-PCR. Therefore, the remaining reason that may explain the variability in our data could be the cDNA synthesis step. The RT (or cDNA) synthesis step is considered the main source of variability in a qRT-PCR experiment, so an optimal RT is essential for reliable and successful qRT-PCR.15,16 Generally, the cDNA, which is used in quantitation real-time assays, can be generated using target gene-specific primers, oligo-dT, or random hexamer primers. In real-time PCR, target-specific primers are used in ∼20% of experiments to generate cDNA for quantitation assays.12,15 However, ∼30% and ∼40% of cDNA, which are used in qRT-PCR experiments, are synthesized using random hexamer and oligo-dT primers, respectively.15 Theoretically, random hexamer and oligo-dT primers convert all mRNAs into cDNA (nonspecifically), and target gene-specific primers prime only target-gene mRNA, resulting in the most-specific cDNA, which in turn provides the highest sensitivity for quantitation assays.12,16 Oligo-dT primers are more specific than random primers and could be more favorable over the target gene-specific primers, as higher cDNA yielded in two-step reaction (1 μg RNA) compared with one-step reaction (50 ng RNA/reaction).15 In our study, the one-step qRT-PCR was carried out using target-specific primers, and the two-step qRT-PCR was performed using oligo-dT primers. These variables in the above studies may not only explain the variability in the efficiency, linearity, and sensitivity between RNA-to-CT™ 2-Step, which was run as a one- and two-step reaction, but also between RNA-to-CT™ 1-Step and RNA-to-CT™ 2-Step assays. Thus, our data are consistent with the results of a previous study, which demonstrated that the two-step RT-PCR method is more efficient and sensitive when compared with the one-step method.17 Theoretically, a one-step method should have higher or at least the same sensitivity of a two-step method because of using gene-specific primers, but this was not the case here or in studies by Battaglia et al.8 This variability in data becomes crucial in, for example, viral research-diagnostic laboratories, where the sensitivity is a vital issue.18–20 In agreement with these findings, our data demonstrated that RNA-to-CT™ 1-Step is less sensitive (where the CT value of the lowest RNA concentration was 27) than RNA-to-CT 2-step as a two-step assay (where the CT value of the lowest RNA concentration was 33). The sensitivity of RNA-to-CT™ 1-Step could be limited by the relative nonspecificity of the RT step.21 Indeed, the manufacturers may recommend carrying out RNA-to-CT™ 1-Step and RNA-to-CT™ 2-Step as a one-step method to facilitate research for the scientists by minimizing contamination, pipetting inaccuracies, and carryover. Therefore, each laboratory must decide what is more important relative to their outcomes, sensitivity, or speed of the assay protocol. Nevertheless, it is highly recommended not to carry out the RNA-to-CT™ 2-Step using a one-step protocol nor to replace the RNA-to-CT™ 2-Step with the newly launched RNA-to-CT™ 1-Step kit, particularly in viral diagnostic research laboratories.
ACKNOWLEDGMENT
Funding for these studies was provided by Institute for Biomedical Research into Human Movement and Health, Manchester Metropolitan University, UK.
REFERENCES
- 1.Gibson UE, Heid CA, Williams PM.A novel method for real time quantitative RT-PCR. Genome Res 1996; 6: 995– 1001 [DOI] [PubMed] [Google Scholar]
- 2.Orlando C, Pinzani P, Pazzagli M.Developments in quantitative PCR. Clin Chem Lab Med 1998; 36: 255– 269 [DOI] [PubMed] [Google Scholar]
- 3.Lockey C, Otto E, Long Z.Real-time fluorescence detection of a single DNA molecule. Biotechniques 1998; 24: 744– 746 [DOI] [PubMed] [Google Scholar]
- 4.Walker NJ.Tech.Sight. A technique whose time has come. Science 2002; 296: 557– 559 [DOI] [PubMed] [Google Scholar]
- 5.Pfaffl MW.A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tichopad A, Dilger M, Schwarz G, Pfaffl MW.Standardized determination of real-time PCR efficiency from a single reaction set-up. Nucleic Acids Res 2003; 31: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong ML, Medrano JF.Real-time PCR for mRNA quantitation. Biotechniques 2005; 39: 75– 85 [DOI] [PubMed] [Google Scholar]
- 8.Battaglia M, Pedrazzoli P, Palermo B, et al. Epithelial tumor cell detection and the unsolved problems of nested RT-PCR: a new sensitive one step method without false positive results. Bone Marrow Transplant 1998; 22: 693– 698 [DOI] [PubMed] [Google Scholar]
- 9.Bustin SA.Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 2000; 25: 169– 193 [DOI] [PubMed] [Google Scholar]
- 10.Yaffe D, Saxel O.Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 1977; 270: 725– 727 [DOI] [PubMed] [Google Scholar]
- 11.Overbergh L, Valckx D, Waer M, Mathieu C.Quantification of murine cytokine mRNAs using real time quantitative reverse transcriptase PCR. Cytokine 1999; 11: 305– 312 [DOI] [PubMed] [Google Scholar]
- 12.Lekanne Deprez RH, Fijnvandraat AC, Ruijter JM, Moorman AF.Sensitivity and accuracy of quantitative real-time polymerase chain reaction using SYBR Green I depends on cDNA synthesis conditions. Anal Biochem 2002; 307: 63– 69 [DOI] [PubMed] [Google Scholar]
- 13.Stahlberg A, Hakansson J, Xian X, Semb H, Kubista M.Properties of the reverse transcription reaction in mRNA quantification. Clin Chem 2004; 50: 509– 515 [DOI] [PubMed] [Google Scholar]
- 14.Raja S, Luketich JD, Kelly LA, Ruff DW, Godfrey TE.Increased sensitivity of one-tube, quantitative RT-PCR. Biotechniques 2000; 29: 702– 704, 706 [DOI] [PubMed] [Google Scholar]
- 15.Bustin SA, Benes V, Nolan T, Pfaffl MW.Quantitative real-time RT-PCR—a perspective. J Mol Endocrinol 2005; 34: 597– 601 [DOI] [PubMed] [Google Scholar]
- 16.Wacker MJ, Godard MP.Analysis of one-step and two-step real-time RT-PCR using SuperScript III. J Biomol Tech 2005; 16: 266– 271 [PMC free article] [PubMed] [Google Scholar]
- 17.Leutenegger CM, Mislin CN, Sigrist B, Ehrengruber MU, Hofmann-Lehmann R, Lutz H.Quantitative real-time PCR for the measurement of feline cytokine mRNA. Vet Immunol Immunopathol 1999; 71: 291– 305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mentel R, Wegner U, Bruns R, Gurtler L.Real-time PCR to improve the diagnosis of respiratory syncytial virus infection. J Med Microbiol 2003; 52: 893– 896 [DOI] [PubMed] [Google Scholar]
- 19.Corless CE, Guiver M, Borrow R, et al. Development and evaluation of a “real-time” RT-PCR for the detection of enterovirus and parechovirus RNA in CSF and throat swab samples. J Med Virol 2002; 67: 555– 562 [DOI] [PubMed] [Google Scholar]
- 20.Whiley DM, Syrmis MW, Mackay IM, Sloots TP.Detection of human respiratory syncytial virus in respiratory samples by LightCycler reverse transcriptase PCR. J Clin Microbiol 2002; 40: 4418– 4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters IR, Helps CR, Hall EJ, Day MJ.Real-time RT-PCR: considerations for efficient and sensitive assay design. J Immunol Methods 2004; 286: 203– 217 [DOI] [PubMed] [Google Scholar]