Abstract
MicroRNAs (miRNAs) are highly conserved small RNAs that act as translational regulators of gene expression, exerting their influence by selectively targeting mRNAs bearing complementary sequence elements. These RNAs function in diverse aspects of animal development and physiology. Because of an ability to act as rapid responders at the level of translation, miRNAs may also influence stress response. In this study, we show that the miR-8 family of miRNAs regulates osmoregulation in zebrafish embryos. Ionocytes, which are a specialized cell type scattered throughout the epidermis, are responsible for pH and ion homeostasis during early development before gill formation. The highly conserved miR-8 family is expressed in ionocytes and enables precise control of ion transport by modulating the expression of Nherf1, which is a regulator of apical trafficking of transmembrane ion transporters. Ultimately, disruption of miR-8 family member function leads to an inability to respond to osmotic stress and blocks the ability to properly traffic and/or cluster transmembrane glycoproteins at the apical surface of ionocytes.
Introduction
MicroRNAs (miRNAs) are a class of small (∼22 nt) noncoding RNAs that negatively regulate gene expression (Reinhart et al., 2000; Lagos-Quintana et al., 2001). Functional miRNAs are derived from larger precursors that mature through sequential nuclear and cytoplasmic cleavages performed by the RNase III enzymes Drosha and Dicer, respectively (Bernstein et al., 2001; Ketting et al., 2001; Lee et al., 2002, 2003). The longer primary miRNA transcripts contain hairpin folds that are recognized and excised by a Drosha-containing complex and are required for nuclear export and final maturation by Dicer in the cytoplasm (Lee et al., 2003). Normally, one strand of the fully processed 22-nt double-stranded miRNA is incorporated into the RNA-induced silencing complex, a multisubunit complex that associates with polyribosomes and is responsible for inhibiting translation of associated mRNAs (Tuschl et al., 1999; Zamore et al., 2000; Ishizuka et al., 2002; Okamura et al., 2004).
miRNAs target specific mRNAs for down-regulation, usually by pairing imperfectly to miRNA recognition elements (MREs) in 3′ untranslated regions (UTRs; Lai, 2002; Enright et al., 2003; Lewis et al., 2003; Brennecke et al., 2005). Higher eukaryotic genomes encode anywhere from hundreds to thousands of miRNAs to enable precise control of gene expression (Kloosterman and Plasterk, 2006). Understanding and identifying the exact genes regulated by specific miRNAs remain a difficult problem. The prediction of miRNA targets through genome-wide analysis of 3′ UTR sequences is complicated by imperfect complementarity between most miRNAs and their targets. Therefore, reporter assays and direct functional tests are required to verify prediction algorithms.
The expression patterns of multiple miRNAs have been described in different organisms, tissues, and developmental time points (Miska et al., 2004; Sempere et al., 2004; Giraldez et al., 2005; Thatcher et al., 2007). In vertebrate embryos, particularly zebrafish, temporal expression patterns have been complemented by in situ localization using locked nucleic acid (LNA) oligonucleotides to hybridize to mature miRNA sequences (Wienholds et al., 2005; Kloosterman et al., 2006a,b). These analyses have revealed a striking variety of expression patterns of different miRNAs during early vertebrate development. The sequences of many miRNAs are conserved, showing similar expression patterns, genomic organization, and copy numbers, suggesting that the use of genetically tractable organisms such as zebrafish could yield insight into the role of miRNAs in humans and their potential role in physiology and disease.
One such conserved family of miRNAs is the miR-8 family, which has five members in vertebrates. These miRNAs (miR-200a, miR-200b, miR-200c, miR-141, and miR-429) are very similar in sequence, particularly at their 5′ ends, and appear to have descended from miR-8 in insects (Ambros, 2003; Griffiths-Jones, 2004; Griffiths-Jones et al., 2006). All vertebrates encode miR-8 homologues arranged identically in two polycistrons and, at least in zebrafish, show identical tissue specificity in nasal epithelia, neuromasts, the pronephros, and a subset of epidermal cells (Wienholds et al., 2005).
Although the aforementioned tissues may seem quite distinct, they all are composed of cells that can be readily stained with dyes that are reported to mostly target mitochondria-rich cells (MRCs). In this study, we focused on ionocytes, which are cells that are interspersed among keratinocytes in the skin of developing zebrafish embryos. Functionally, these cells mimic intercalated cells in the mammalian distal nephron and collecting duct that function to regulate ion flux (Hsiao et al., 2007; Janicke et al., 2007). We show that in zebrafish, these cells express miR-8 family miRNAs that participate in osmoregulation through the targeting of nherf1. Nherf1 was originally shown to regulate the activity of NHE3 in renal brush border cells (Weinman et al., 2000), but it also controls apical presentation and trafficking of membrane proteins such as ion transporters and receptors (Lin et al., 2006; Hsiao et al., 2007; Janicke et al., 2007). Disruption of miR-8 miRNAs results in zebrafish embryos deficient in responding to osmotic stress and incapable of properly maintaining ion and acid base homeostasis.
Results
miR-8 family miRNAs are expressed in ionocytes
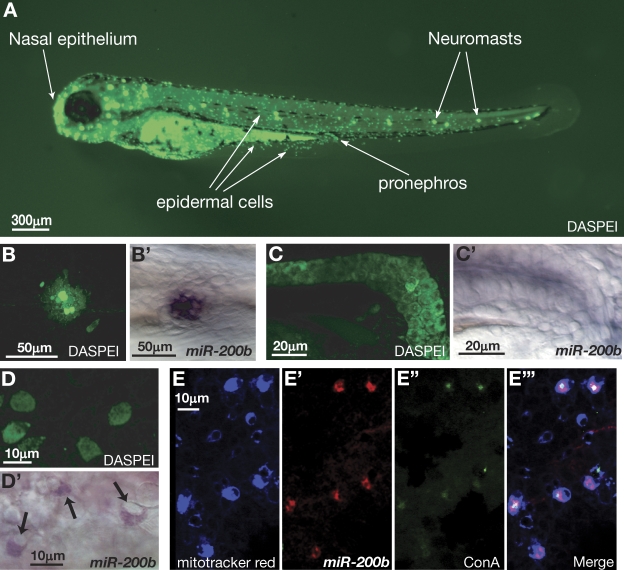
In situ hybridization experiments using LNA probes complementary to miR-200a and miR-200b have shown that these miRNAs are expressed in several tissues in zebrafish embryos, including nasal epithelium, neuromasts, the pronephros, and scattered epithelial cells (Wienholds et al., 2005). Interestingly, these same tissues can be stained with fluorescent dyes that are thought to preferentially target MRCs (Jonz and Nurse, 2006). One of these stains is the styryl dye DASPEI, which is cell permeable, accumulates in mitochondria, and allows staining and visualization in live embryos (Fig. 1 A; Harris et al., 2003). Structures stained by DASPEI that also show an accumulation of miR-200b include neuromasts, the pronephros, and dispersed epithelial cells (Fig. 1, B–D′). The dispersed epidermal cells that express miR-200b resemble ionocytes based on their ovoid cell morphology and their location in the epidermis (Fig. 1, D and D′; Jonz and Nurse, 2006; Lin et al., 2006). There are at least two different populations of ionocytes present in the skin of zebrafish embryos that can be differentiated based on the expression of H+ ATPases (H+ pump–rich cells [HRCs]) or Na+-K+ ATPases (Na+-K+ pump–rich cells [NRCs]; Lin et al., 2006; Esaki et al., 2007). HRCs can be differentiated from NRCs by their strong affinity to the lectin Con A (Lin et al., 2006). HRCs are responsible for the accumulation of Na+, whereas NRCs are thought to participate in regulating appropriate levels of K+ and Na+, with a subset responsible for the uptake of Ca2+ (Esaki et al., 2007; Janicke et al., 2007). To determine whether the epidermal cells in zebrafish skin that express miR-200b are ionocytes and, if so, which subclass of ionocytes, we localized miR-200b by fluorescent in situ hybridization in embryos stained with both MitoTracker red and Con A before fixation. MitoTracker red behaves similarly to DASPEI, accumulating as a fluorescent marker of mitochondria that can be visualized in living embryos (Esaki et al., 2007). However, unlike DASPEI, the dye exhibits a much narrower emission spectra and becomes covalently attached to mitochondrial proteins through thiol conjugation. Thus, MitoTracker red staining persists after fixation of embryos, allowing colabeling experiments. Triple labeling demonstrated that miR-200b is expressed in MitoTracker red–positive, Con A–positive cells, indicating that these cells are ionocytes of the HRC subtype (Fig. 1, E–E‴).
Figure 1.
miR-200 is expressed in MRCs. (A) Live imaging of 3-dpf zebrafish embryos incubated in the mitochondrial stain DASPEI. DASPEI preferentially labels the nasal epithelium, neuromasts, epidermal cells, and the pronephros, as indicated. (B) DASPEI staining of a single neuromast. (B′) In situ hybridization was performed on zebrafish embryos at 36 hpf with LNA antisense oligonucleotides complementary to miR-200b. Localization was performed using NBT/BCIP color development. (C) DASPEI staining of the pronephros. (C′) miR-200b in situ localization shows expression in the pronephros. (D) DASPEI staining in epidermal cells overlying the yolk in 36-hpf zebrafish embryos. (D′) miR-200b in situ localization in epidermal cells. Arrows point to miR-200b expression. (E) MitoTracker red staining of epidermal cells. (E′) Fluorescence signal derived from Cy3-labeled antidigoxigenin antibodies bound to LNA probes hybridized to miR-200b. (E″) Signal from FITC-conjugated Con A bound to glycoproteins on the apical membrane of HRC ionocytes. (E‴) Merge of MitoTracker red, miR-200b in situ, and FITC–Con A signals.
Morpholino knockdown of miR-8 family members
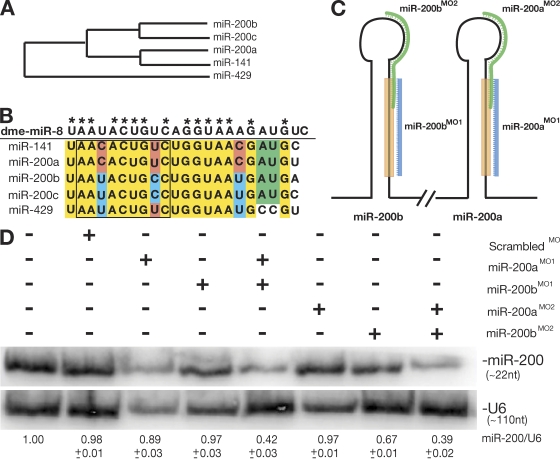
miR-200b is a member of a larger family of miRNAs named for the founder miRNA in Drosophila melanogaster, miR-8 (Aravin et al., 2003; Chen et al., 2005b). Although all members of the miR-8 family share a high degree of sequence similarity, modest changes have occurred during the diversification of this miRNA family. The alignment of hairpin precursor sequences shows the relatedness of the members (Fig. 2 A). Focusing on the mature sequences, miR-200b and miR-200c are identical, as are miR-200a and miR-141 (Fig. 2 B). The 5′ end of the founder miRNA, miR-8, is most similar to miR-200b, miR-200c, and miR-429 (Fig. 2 B). This region is referred to as the seed sequence and plays an important role in target pairing (Lewis et al., 2003).
Figure 2.
Knockdown of miR-8 miRNAs by morpholino inhibition. (A) Phylogeny of zebrafish miR-8 family by alignment of miRNA precursor hairpin sequences. (B) Alignment of mature miRNA sequences from the miR-8 family in zebrafish. Identical nucleotides are shown in yellow, with those matching the founding member in Drosophila (dme-miR-8) indicated with asterisks. (C) Design of targeting antisense morpholino oligonucleotides against mature miR-200b (miR-200bMO1), mature miR-200a (miR-200aMO1), and the loop sequences from miR-200b (miR-200bMO2) and miR-200a (miR-200aMO2). (D) Expression of miR-200 family members at 36 hpf after injection of morpholinos into single-cell zebrafish embryos. RNA was isolated from embryos (injected as indicated), and Northern blots were probed with an oligonucleotide against miR-200b. Based on the hybridization conditions and because of the sequence similarity between the different family members, the resulting signals indicate the levels of all miR-200 family members, not just miR-200b. The numbers below the blot represent the ratio of miR-200b levels to U6 levels using densitometry.
Antisense technology has been widely used to interfere with miRNA function (Krutzfeldt et al., 2006). In zebrafish, antisense morpholino oligonucleotides have been used to inhibit miRNA function for up to 72 h postfertilization (hpf; Flynt et al., 2007; Kloosterman et al., 2007). To target the miR-8 family in zebrafish, we designed two sets of antisense morpholinos. The first set is complementary to the mature sequence of miR-200b (BMO1) and the mature sequence of miR-200a (AMO1; Fig. 2 C). A second control set was prepared that consists of morpholinos complementary to the seed and loop regions of both miR-200b (BMO2) and miR-200a (AMO2). Both sets are designed to effectively target members of the miR-8 family.
To determine the effectiveness of the morpholinos alone and in combination, we performed Northern blotting against miR-200 family members with RNA extracted from 36-hpf embryos that were injected at the single-cell stage with different combinations of morpholinos (Fig. 2 D). The greatest knockdown was achieved through injection of either the AMO1 + BMO1 (ABMO1) or AMO2 + BMO2 (ABMO2) combination of morpholinos. Injection of single morpholinos or a scrambled morpholino did not result in significant decreases in miRNA levels except for BMO2 (Fig. 2 D). Even though detectable levels of miR-8 family members were still observed when combinations of morpholinos were used, the resulting decreases were sufficient to generate phenotypic effects on ionocyte function (see next section).
miR-8 function and osmotic stress
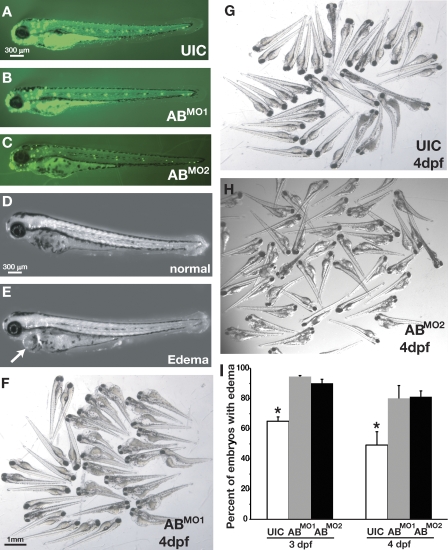
Next, we sought to determine the effects of knockdown of the miR-8 family on zebrafish development. Injection of the ABMO1 and ABMO2 combinations did not result in detectable defects in gross zebrafish embryo morphology at 36 hpf. Uninjected control (UIC) embryos and those injected with the ABMO1 or ABMO2 combination were virtually indistinguishable when examined under either light microscopy (not depicted) or after DASPEI staining (Fig. 3, A–C). Thus, at this time point and with this level of knockdown, there was no apparent defect in either overall development or in the specification of mitochondria-rich ionocytes. Because normal morphology and cell specification appeared intact, we next sought to test whether the miR-8 family functions to regulate the physiology of ionocytes.
Figure 3.
Loss of miR-8 miRNAs affects osmotic stress response. (A–C) DASPEI staining to show morphology of zebrafish embryos in UIC and ABMO1 - and ABMO2 -injected embryos. (D and E) 36 hpf, embryos subjected to osmotic stress were either unaffected (D) or exhibited edema (E). The arrow indicates swelling of the epicardium, which is typical of edema. (F–H) Severity of edema in UIC and ABMO1 and ABMO2 morphants at 4 dpf. (I) Percentage UIC or ABMO1 - or ABMO2 -injected embryos exhibiting edema at 3 and 4 dpf. Statistically significant differences were determined by ANOVA and are indicated by asterisks (α = 0.5); error bars show SEM.
To test whether the miR-8 family regulates ionocyte function, we subjected embryos injected with the ABMO1 or ABMO2 combination to osmotic stress. Injected morphants were allowed to develop in 1× Danieau buffer for the first 24 h of embryogenesis, after which they were transferred to high-salt buffer (10× Danieau buffer) for 24 h followed by a final transfer to distilled water. The transitions between dramatically different salt concentrations induced severe osmotic stress, and the morphological effects of such stress were documented 24 and 48 h after the final transfer to distilled water (Fig. 3, D–H). Consistent with the idea that the miR-8 family functions to regulate the physiology of ionocytes, zebrafish embryos injected with either the ABMO1 or the ABMO2 combination exhibited increased sensitivity to osmotic stress, displaying significantly increased edema compared with UIC embryos, both in severity and frequency (Fig. 3, C, D, and E). Interestingly, when UIC or ABMO1- or ABMO2-injected embryos were transferred to distilled water after equilibrating for 24 h in 1× buffer (i.e., no exposure to high salt), no observable defects were detected (unpublished data). Similarly, neither UIC embryos nor ABMO1 morphants raised continuously at high salt exhibited obvious developmental defects (unpublished data). This suggests that the reduction in the levels of miR-8 family members results in an inability to manage fluctuations that induce extreme osmotic stress and is consistent with a role for miR-8 family members in ionocyte physiology.
The miR-8 family participates in the regulation of Na+ accumulation in ionocytes
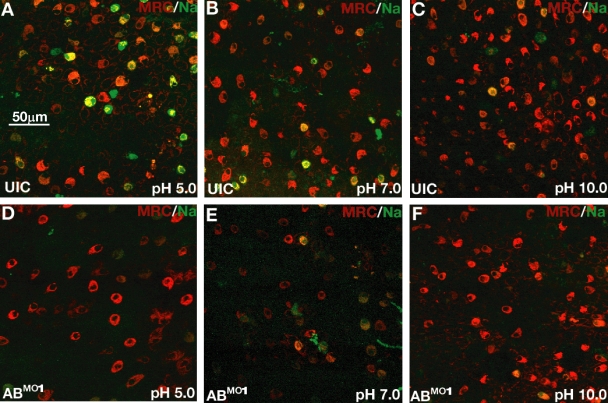
Next, we sought to determine whether changes in ion homeostasis could be observed in control and morphant embryos. To examine the accumulation of Na+ in HRC ionocytes, we used Sodium green, which emits fluorescence in correlation with increasing Na+ concentration (Esaki et al., 2007). As with DASPEI and MitoTracker red, Sodium green is cell permeable and can be used to stain live embryos. After a 60-min incubation of embryos in the presence of Sodium green, Na+ accumulation in ionocytes was readily observed using fluorescence microscopy (Fig. 4). We used a combination of Sodium green and MitoTracker red to visualize ionocytes in normal zebrafish embryos at three different pHs in 1× buffer (Fig. 4, A–C). The combination of dyes allowed verification that the Sodium green fluorescence was indeed derived from ionocytes. The accumulation of Na+ in zebrafish embryos depended on the pH of the culture water, with embryos raised at low pH exhibiting the greatest accumulation (Fig. 4 A). This is because Na+ accumulation in HRCs depends on the function of Na+/H+ exchangers (NHEs) and, therefore, is linked to H+ efflux (Esaki et al., 2007; Horng et al., 2007). These antiporters are important for ion movement and pH homeostasis in several different organisms (Claiborne et al., 2002). Interestingly, acidosis increases localization of NHEs at the apical membranes of mammalian renal cells, which, in turn, leads to enhanced rates of Na+/H+ exchange (Claiborne et al., 2002). A similar phenomenon is apparently occurring in zebrafish HRCs, in which the need for increased acid secretion is balanced by Na+ accumulation. This is evident from the increased number of Sodium green–positive cells at decreasing pH (Fig. 4, A–C).
Figure 4.
Loss of miR-8 miRNAs blocks Na+ accumulation in ionocytes. (A–C) Live wild-type zebrafish embryos were incubated with Sodium green (green) and MitoTracker red (red) at pH 5.0, pH 7.0, or pH 10.0. Na+ accumulation is indicated by green-stained cells. (D–F) Live embryos injected with ABMO1 were visualized by Sodium green and MitoTracker red at three pHs as in A–C.
Next, we sought to determine whether a change in Na+ accumulation could be observed in embryos injected with either the ABMO1 or ABMO2 combination. Decreased Na+ accumulation was observed under both conditions (Fig. 4, D–F and Fig. S1). The change in Na+ accumulation was most pronounced when comparing the ABMO1 or ABMO2 morphants with UICs at pH 5.0 (Fig. 4, A and D, and Fig. S1 and see Fig. 6 E). These results are consistent with a role for the miR-8 family in regulating ion homeostasis in ionocytes. For subsequent experiments, we focused on the observed differences at pH 5.0 before visualization of labeled embryos, and we only examined ABMO1 morphants because both sets of morpholinos gave similar or identical results.
Figure 6.
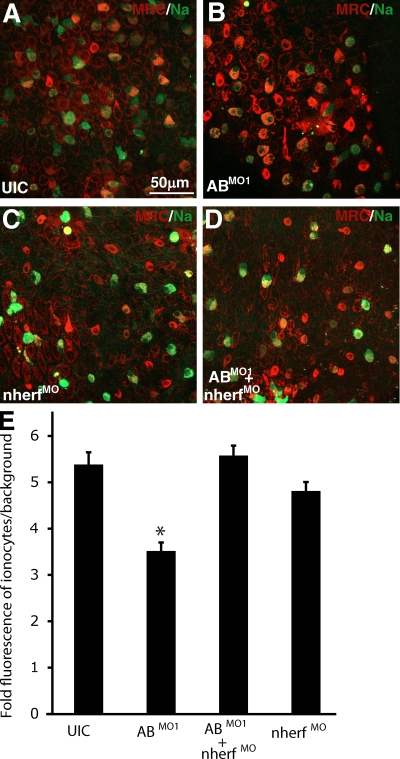
Rescue of Na+ accumulation defects in ABMO1 morphants by repression of nherf1. (A–D) Visualization of Sodium green (green) and MitoTracker red (red) in UIC, ABMO1-injected embryos, nherf1MO-injected embryos, and ABMO1- and nherf1MO-coinjected embryos was performed on live embryos incubated at pH 5. (E) Quantification of Sodium green fluorescence levels from embryos injected in A–D. Mean fluorescence was divided by local background. Statistical significance determined by ANOVA at α≤0.05 is indicated by the asterisk; n = 20 from five different embryos from three independent experiments. Error bars represent SEM.
nherf1 is a target of the miR-8 family
To better understand how the miR-8 family influences the physiology and function of ionocytes, we sought to identify miR-8 target genes that could be responsible for regulating Na+ accumulation. A variety of algorithms have been created to predict the targets of specific miRNAs based on sequence complementarity, sequence context, and conservation across species (Lewis et al., 2003; Chen et al., 2005a; Grimson et al., 2007). One of the predicted targets for both miR-200a and miR-200b is slc9a3r2 (located on chromosome 12 bp 30,682,734–30,726,868), which is also known as Nherf1 (Chen et al., 2005b). This gene encodes a phosphoprotein containing two N-terminal PDZ domains that interacts with a variety of membrane-associated partners, including NHEs and other ion transporters (Yun et al., 1997; Murthy et al., 1998; Lederer et al., 2003; Morales et al., 2007; Wheeler et al., 2007). The C-terminal domain of Nherf1 interacts with the cytoskeletal proteins merlin, ezrin, radixin, and moesin, enabling Nherf1 to serve as an adapter molecule linking membrane proteins to cytoskeletal actin filaments (Fig. 5 A; Weinman et al., 2000; Morales et al., 2007). There are multiple mammalian Na+/H+ exchange regulatory factor isoforms that are similar in domain structure but associate with different partners and exhibit tissue-specific expression patterns (Yun et al., 1997; Weinman et al., 2000). Like mammals, zebrafish possess multiple Na+/H+ exchange regulatory factor isoforms, most of which are uncharacterized. nherf1 is expressed in several regions of the brain, pronephros, and epidermis (Thisse et al., 2001). In addition to being an excellent candidate based on the regulation of Na+ accumulation by Nherf1, the MREs in the nherf1 3′ UTR are exceptionally strong, matching the current criteria described for efficient targeting by miRNAs (Fig. 5 B). These criteria include nearby adenine uracil–rich elements and targeting by tightly coexpressed miRNAs, which is consistent with the nherf1 3′ UTR structure and the polycistronic arrangement of miR-8 family members (Fig. S2; Grimson et al., 2007).
Figure 5.
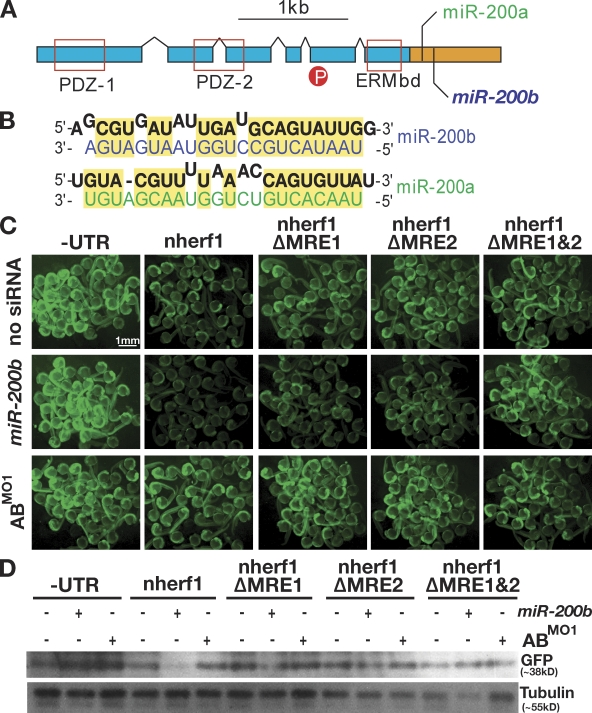
nherf1 is a target of the miR-8 family. (A) Diagram indicating the domain structure of Nherf1. The ORF is indicated by blue boxes, and the 3′UTR is shown in orange. Conserved protein domains, PDZ-1, PDZ-2, and the ERM-binding domain (ERMbd), are indicated by red boxes. The location of a phosphorylated residue is indicated by the red circle. Positions of the miR-8 family MREs are as indicated. (B) miR-8 family MREs predicted by the miRanda algorithm. The nherf1 mRNA is shown in black, and miR-200a and miR-200b sequences are shown in green and blue, respectively. (C) Single-cell embryos were injected with mRNAs derived from GFP reporters lacking a UTR (−UTR), fused to the full-length nherf1 UTR (nherf1), or mutant versions of the nherf1 UTR lacking individual MREs (nherf1 ΔMRE1 and nherf1 ΔMRE2) or both MREs (nherf1 ΔMRE1 and 2). Single-cell embryos were injected in the presence or absence of exogenous miR-200b or the ABMO1 morpholinos. Fluorescence levels were examined at 1 dpf, and clusters of embryos were photographed. (D) Embryo lysates were prepared from embryos treated as in C, and GFP protein levels were determined by Western blotting.
As a first test of whether nherf1 is targeted by the miR-8 family, we constructed a GFP reporter bearing the entire 3′ UTR of nherf1 as well as reporters containing deletions of one or both MREs (Fig. 5, C and D). Synthetic mRNAs prepared from these reporters were injected into single-cell embryos in the presence or absence of miR-200b or the ABMO1 morpholinos. By simple examination of the GFP levels in injected embryos at 1 d postfertilization (dpf), it is clear that embryos coinjected with the full-length GFP nherf1 3′ UTR mRNA and miR-200b resulted in down-regulation of GFP levels when compared with a specific GFP construct alone (Fig. 5 C). Only modest silencing could be observed in embryos coinjected with miR-200b and reporters that had only a single MRE (nherf1 ΔMRE1 and nherf1 ΔMRE2). No silencing was observed when both MREs were deleted (nherf1 ΔMRE1 and 2). The increase in fluorescence upon injection of the entire nherf1 3′ UTR and the ABMO1 morpholinos shows the effect of knockdown of endogenous levels of the miR-8 family and also serves as a specificity control. For all injections, detection of GFP protein levels via Western blotting of embryo lysates confirmed the trends of GFP fluorescence (Fig. 5). Together, the results are consistent with targeting of nherf1 by miR-8 family members.
Epistatic interaction between nherf1 and miR-8 family members
If nherf1 is indeed a target of miR-8 family members, the defect in Na accumulation in the ABMO1 morphants should be rescued by direct repression of nherf1. Nherf1 has been shown to be a negative regulator of NHE activity by promoting phosphorylation and subsequent internalization of NHEs (Yun et al., 1997; Murthy et al., 1998). To repress nherf1, we designed a morpholino complementary to the translation start site of nherf1 (nherf1MO). These morphants exhibited mild edema (similar to Fig. 3, E and G), suggesting compromised osmoregulation (not depicted). Thus, we monitored Na+ accumulation in ionocytes with Sodium green and MitoTracker red in embryos injected with the ABMO1 combination, the nherf1MO, or all three morpholinos (Fig. 6, A–D). Consistent with nherf1 expression being up-regulated in ABMO1 morphants because of a lack of repression by the miR-8 family, repression of nherf1 by the nherf1MO allowed restoration of Na+ accumulation in ABMO1 morphants (Fig. 6, B and D). To verify this result, we quantified Sodium green fluorescence (Fig. 6 E). The mean pixel intensity at 488 nm was determined for individual ionocytes and divided by the local background to determine the fold increase in Na+ accumulation. This analysis showed no significant differences in Na+ accumulation between UIC and nherf1MO- and ABMO1+ nherf1MO– injected embryos. In contrast, Na+ accumulation in ABMO-injected embryos was significantly decreased. These results are consistent with targeting of nherf1 by miR-8 miRNAs in HRC ionocytes.
Regulation of membrane trafficking by nherf1 and miR-8 family members
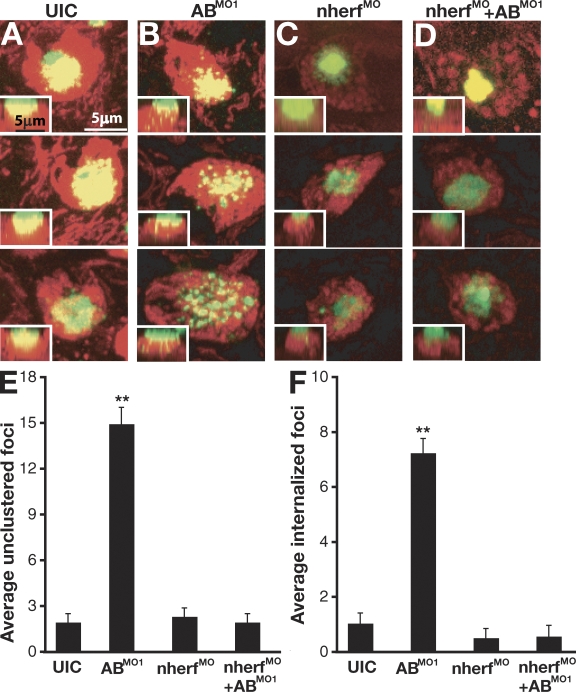
Besides PKA regulation of Na accumulation, Nherf1 has also been shown to regulate the trafficking and membrane localization of a variety of proteins, including ion channels, G protein–coupled receptors, and other glycosylated transmembrane proteins (Voltz et al., 2001; Morales et al., 2007; Theisen et al., 2007). To test whether defects in membrane localization occur in ABMO1 morphants, we examined ionocytes after staining with FITC-conjugated Con A (FITC–Con A). Embryos were incubated briefly with FITC–Con A, and apical membranes of HRCs were examined using fluorescent microscopy. Immediate visualization of ionocyte membranes after Con A staining showed little difference between UIC embryos and ABMO1 morphants (unpublished data). However, after 1 h, considerable differences in Con A distribution were observed (Fig. 7, A and B). In control embryos, Con A distribution was mostly localized to apical membranes of HRC ionocytes in dense, clustered structures. In contrast, a radical redistribution of Con A–labeled glycoproteins was observed in ABMO1 morphants. In addition to a more punctate appearance, the apical character of these ionocytes was disrupted, and increased levels of internalized FITC–Con A signals could be observed along the z axis. This is consistent with a role for Nherf1 in controlling membrane trafficking and internalization of specific receptors (Yun et al., 1997). To ensure that the defect was specific, we again used the nherf1MO to determine whether repression of elevated nherf1 expression in the ABMO1 morphants could rescue the change in localization of Con A–labeled glycoproteins (Fig. 7, C and D). As shown in the previous section, repression of nherf1 expression rescued the alteration of Con A localization seen when miR-8 function was blocked. To quantify the differences in distribution of Con A localization, we counted the number of FITC–Con A–labeled foci using two criteria. The first was whether the staining resulted in foci that were either clustered or ungrouped, and the second was whether there was an increase in the number of internalized Con A–labeled foci along the z axis toward the basolateral surface. As shown, the ABMO1 morphants showed statistically significant increases in both measurements (Fig. 7, E and F). This is consistent with a role for the miR-8 family in regulating membrane dynamics and trafficking of transmembrane proteins through regulation of Nherf1.
Figure 7.
Loss of miR-8 miRNAs alters apical membrane trafficking. (A–D) FITC–Con A (green) and MitoTracker red (red) staining was performed to examine the apical membranes of H+ pump–rich ionocytes. Con A staining in UIC, ABMO1-injected embryos, nherf1MO-injected embryos, and ABMO1- and nherf1MO-coinjected embryos. Live embryos were imaged with each panel showing a view of cells on the surface of the yolk sac and the inset showing a side view of the z stack with apical basal localization of Con A staining. (E and F) Mean number of ungrouped (E) or internalized (F) foci of Con A staining from embryos in A–D. Error bars represent SEM. Statistical significance is indicated by the asterisks and was determined by ANOVA at α ≤ 0.05 (n = 14 for UIC, n = 11 for ABMO1, n = 13 for nherf1MO, and n = 10 for nherf1MO+ ABMO1). Data were gathered from three independent experiments.
Discussion
miR-8 family miRNAs regulate nherf1 in zebrafish ionocytes
In this study, we demonstrate a role for the miR-8 family of miRNAs in zebrafish osmoregulation. These miRNAs modulate the expression of nherf1, which plays a role in regulating Na+/H+ exchange activity. Nherf1 negatively regulates NHE3 in a cAMP-dependent manner by recruiting activated PKA to phosphorylate NHE3 (Weinman et al., 2000). Phosphorylation results in the internalization of NHE3, thereby down-regulating ion exchange across the membrane. Interestingly, cAMP production is coupled to a variety of stress responses. Among these are hypertonicity, hypotonicity, and acidosis, all of which increase cAMP levels several fold (Disthabanchong et al., 2002; Orlic et al., 2002; Sheikh-Hamad and Gustin, 2004). Increased cAMP levels are thought to play an important role in the response to osmotic stress by abrogating the negative effects of stress-responsive genes whose activation can induce apoptosis (Pascual-Ahuir et al., 2001; Saran et al., 2002; d'Anglemont de Tassigny et al., 2004). If cAMP levels are elevated in ionocytes experiencing osmotic stress, this should (through a Nherf1-dependent mechanism) result in the inhibition of Na+/H+ exchange activity. This would be a deleterious outcome because NHE activity is required to balance Na+ accumulation and H+ efflux as well as for the retention of Na+ in hypotonic solution. The miR-8 family may function to ameliorate cAMP-mediated inhibition of NHEs during stress. This would allow Na+/H+ exchange to occur independently of protective cAMP elevation.
We have also shown that regulation of nherf1 by the miR-8 family is responsible for maintaining the apical character of ionocytes. The apical domains of ionocytes were revealed using FITC–Con A staining. Although the exact identity of the specific zebrafish glycoproteins that are recognized by Con A remains to be determined, the overall resemblance of the ionocytes studied in these experiments to mammalian renal brush border cells is striking (Tyska et al., 2005). In brush border cells, Nherf1 has been shown to be recruited to apical membranes by overexpression of podocalyxin, which is an obligate apical glycoprotein (Nielsen et al., 2007). Because of the large number of apical glycoproteins on the membranes of HRC ionocytes, Nherf1 may be constitutively recruited to the membranes of these cells. This would necessitate attenuation of nherf1 expression to permit NHE activity in these cells. In zebrafish, neuromasts and the nasal epithelium are also strongly labeled by Con A (unpublished data). Down-regulation of nherf1 may be essential for the appropriate presentation of specific glycoproteins on the apical membranes of these cell types.
nherf1 is predicted to be a target of miR-200b in mammals
The miRanda algorithm predicts that miR-200b should target both zebrafish and mammalian nherf1 (John et al., 2004). In mammals, miR-200b is expressed in the colon, kidney, prostate, pancreas, and thymus, all of which contain polarized secretory cells (Beauchamp et al., 2007). In the colon and kidney, Nherf1 is known to be an active participant in the regulation of many ion transporters in addition to Na+/H+ exchange (Stemmer-Rachamimov et al., 2001). Both of these organs contain brush border membranes that are reactive to Con A staining (Tyska et al., 2005; Nielsen et al., 2007). If miR-200b regulation of nherf1 in the colon and kidney has effects similar to our observations in zebrafish ionocytes, it will be critical to determine whether expression of miR-200b is restricted to specific cell types within these organs. Nherf1 expression in the colon is restricted, suggesting precise regulation of expression between cell types, potentially through the activity of miR-200b in these tissues (Stemmer-Rachamimov et al., 2001). Additionally, the cells of both the prostate and pancreas, which express miR-200b, are highly secretory and similarly reactive to Con A, requiring apical localization of multiple membrane proteins (Gheri et al., 1997; Arenas et al., 1999). It is also noteworthy that Nherf1 is up-regulated in proliferative endometrium compared with secretory endometrium (Stemmer-Rachamimov et al., 2001). Down-regulation of nherf1 by miR-200b may be essential for secretory epithelial cells to adjust their physiology toward a permanently differentiated state. Indeed, increased expression of Nherf1 has been observed in breast and liver cancer cells (Stemmer-Rachamimov et al., 2001).
Recently, miR-8 family members were shown to play a role in terminal olfactory differentiation in zebrafish (Choi et al., 2008). In this study, we did not observe defects in ionocyte differentiation in the absence of miR-8 family members, with the caveat that our knockdowns were not complete. Nevertheless, we observed a striking effect on ionocyte physiology, suggesting these miRNAs may have cell type–specific functions and that miR-8 family members may play key roles both during development and after terminal differentiation. It will be interesting to determine whether Nherf1 is expressed during olfactory differentiation and whether targeting by miR-8 family members affects membrane trafficking of olfactory receptors.
Other studies have shown a role for the miR-8 family in promoting epithelial fate in mammalian cells (Bracken et al., 2008; Burk et al., 2008; Gregory et al., 2008; Korpal et al., 2008). These miRNAs operate in a genetic bistable loop configuration with ZEB1 and ZEB2 transcription factors. We did not see a loss of ionocytes when inhibiting the miR-8 family. If ionocytes were losing their epithelial character, one might expect them to be extruded from the epidermis. It will be interesting to investigate whether these miRNAs take on such a function during later development.
miRNAs and stress
The function of the miR-8 family may be required for mounting appropriate stress responses in mammalian cells, as we have shown in zebrafish. During our efforts to describe the role of the miR-8 family in zebrafish, we attempted to determine whether the expression of miR-200b changes in response to salt concentration or pH. The results of these experiments demonstrated little alteration in the level of miR-8 family expression in whole embryo RNA extracts, at least at the time points tested. However, this may be caused by a lack of sensitivity when comparing whole embryos with ionocytes, especially given the high expression levels observed in nasal epithelium.
Originally, miRNAs were found to regulate developmental timing in worms, and a role for miRNAs in development is a continuous theme, translating into other phyla (Bartel and Chen, 2004). However, miRNAs have been found to have diverse functions beyond regulating development. Experiments in Drosophila uncovered a role for miR-14 in fat metabolism and stress (Xu et al., 2003), and miRNAs have been shown to play a role in triggering cardiac hypertrophy in response to stress (van Rooij et al., 2006). Additionally, the activity of CAT-1 (cationic amino acid transporter 1) is controlled by miR-122 in response to nutrient starvation (Bhattacharyya et al., 2006). The expression of a subset of miRNAs also appears to be up-regulated by p53 in response to oncogenic stress (He et al., 2007; Raver-Shapira et al., 2007). When coupled to our findings related to osmotic stress, a clear theme emerges in which a major function of miRNAs is to regulate the response to a variety of cellular stresses (Leung and Sharp, 2007).
Subcellular localization of Argonaute proteins, the effectors of RNA-induced silencing complexes and binding partners of miRNAs, shows localization with cytoplasmic foci called processing bodies (Zamore et al., 2000; Carmell et al., 2002; Hutvagner and Zamore, 2002; Liu et al., 2005). Argonaute proteins also accumulate in a stress-dependent manner in separate cytoplasmic foci called stress granules (SGs; Leung et al., 2006). Interestingly, miRNAs localize to SGs and have been shown to dynamically accumulate and dissociate from SGs in a stress-dependent manner. The unique mechanism of miRNA-mediated gene regulation may be used as a method of effecting rapid changes in gene expression, particularly during stress. Regulation of nherf1 by the miR-8 family serves as a particularly crucial stress response in that it links extracellular events to membrane trafficking, enabling sensitive and precise control of gene expression caused by changes in environmental cues and stresses.
Materials and methods
Live imaging of zebrafish embryos
Embryos were raised in egg water (0.03% Instant Ocean marine salt mix) for the initial 24 h of development (Esaki et al., 2007). After 24 h, embryos were transferred to 1× Danieau buffer (58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca[NO3]2, and 10 mM Hepes; Solnica-Krezel et al., 1994). The pH of the solution was controlled by buffering Hepes before the addition of component salts. For fluorescent staining, embryos were incubated in 1× Danieau buffer containing 0.25 nM DASPEI (Molecular Imaging; Harris et al., 2003), 0.5 µM MitoTracker deep red 633, or 10 µM Sodium green (Invitrogen; Esaki et al., 2007). Embryos were rinsed briefly three times and mounted in 1× Danieau buffer. Fluorescent staining was visualized by using a 40× objective on a laser-scanning microscope (LSM 510 Meta; Carl Zeiss, Inc.). Images were processed, and Z projections were made with LSM 510 software (Carl Zeiss, Inc.) before import into Photoshop (Adobe) for orientation and cropping. Fig. S1 was achieved using the same conditions on a confocal microscope (FV-1000; Olympus) with FV10-ASW 1.6 software (Olympus). All image acquisition was performed at room temperature. Quantification of Na+ accumulation in ionocytes was accomplished using ImageJ software (National Institutes of Health). Z projections of images of Sodium green fluorescence were imported into ImageJ. The mean intensity of fluorescent ionocytes was calculated and divided by local background to yield fold fluorescence of ionocyte/background (Figs. 1, 4, and 5) or with a dissecting microscope (MZFIII; Leica; Figs. 1 A and 3).
In situ hybridization and Northern blots
Detection of mature miR-200b was accomplished by in situ hybridization using a digoxigenin-labeled LNA oligonucleotide, 5′-TCATCATTACCAGGCAGTATTA-3′ (Wienholds et al., 2005; Kloosterman et al., 2006b). Visualization of miR-200b expression was performed after nitro blue tetrazolium (NBT)/5-bromo-4-chloro-3-indolyl phosphate (BCIP) color development. Embryos were mounted in 50% glycerol and photographed in color with a 20× objective in Fig. 1 (B′, C′, and D′). miR-200b localization was detected by in situ hybridization using a digoxigenin-labeled LNA probe and Cy3-labeled antidigoxigenin secondary antibodies (Jackson ImmunoResearch Laboratories). Images of embryos stained by NBT/BCIP were acquired by using a compound microscope (Axiophot; Carl Zeiss, Inc.) and a digital camera (Axiocam; Carl Zeiss, Inc.). Images were acquired with Axiovision software (Carl Zeiss, Inc.) and imported into Photoshop for orientation. Northern blotting was performed as described previously (Sempere et al., 2003; Flynt et al., 2007).
Microinjection
Fertilized single-cell zebrafish embryos were injected with 1-nl volumes. Morpholino oligonucleotides were injected as follows per embryo: 2 ng AMO1 5′-ACATCGTTACCAGACAGTGTTA-3′, 2 ng BMO1 5′-TCATCATTACCAGGCAGTATTA-3′, 2 ng AMO2 5′-GACAAAAGATTGTGACAGACCATTG-3′, 2 ng BMO2 5′-TGAAAAAGATTATGACGGACCATTA-3′, and 1 ng nherf1MO1 5′-CCTGAGGTCGCTGGACATTTT-3′. 40 pg in vitro–synthesized, capped mRNA encoding either GFP without a UTR (−UTR) or GFP fused to the nherf1 UTR (GFPnherfUTR) were injected alone or with 200 pg synthetic miR-200b into embryos.
Induction of osmotic stress
Embryos were raised in 1× egg water for the initial 24 h of development before transfer into 10× Danieau buffer. After 24 h, embryos were transferred to distilled water by multiple brief washes. The percentage of embryos exhibiting edema after transfer to distilled water was calculated after 24 and 48 h. Paired Student's t tests were performed to determine statistical differences between embryos exhibiting edema. Images acquired with the MZFIII dissecting microscope using an Axiocam digital camera were captured with Axiovision software.
GFP reporter analyses
Reporter analyses and Western blotting were performed as described previously (Flynt et al., 2007). To generate the nherf1 GFP reporter, the GFP ORF was fused to the 3′ UTR sequence of zebrafish nherf1. The nherf1 UTR was cloned from zebrafish embryo RNA extracts using oligo (dT) primed reverse transcription followed by PCR amplification with gene-specific primers (5′-GCCTCCTGCGTGC-3′ and 5′-GACTTTTCATAATATTTAATAACAAAAATCAT-3′). Internal deletions of the nherf1 3′ UTR were created using the QuikChange Site-Directed Mutagenesis kit (Agilent Technologies) with the following primers: Nherf1-MRE1 forward, 5′-GATTAGAAAACCCTTTACGTTCTGCTTGAGATTTTCC-3′; Nherf1-MRE1 reverse, 5′-GGAAAATCTCAAGCAGAACGTAAAGGGTTTTCTAATC-3′; Nherf1-MRE2 forward, 5′-GTATATTTTCTTGCTTCGCTTTGACCCTTCAAGAGCGAG-3′; and Nherf1-MRE2 reverse, 5′-CTCGCTCTTGAAGGGTCAAAGCGAAGCAAGAAAATATAC-3′. Images were acquired with an MZFIII dissecting microscope equipped with a fluorescent laser using a cooled mono 12-bit camera (Retiga 1300; QImaging) with QCapture 3.1.1 software (QImaging) and were imported into Photoshop for orientation and cropping.
Con A labeling
Embryos were incubated for 30 min in 1× Danieau buffer containing MitoTracker red. 50 µg/ml FITC-conjugated Con A was then added for an additional 10 min (Esaki et al., 2007). Excess Con A was removed by several brief washes in 1× Danieau buffer. After 1 h, embryos were mounted in 1× Danieau buffer, and FITC–Con A–labeled cells were visualized by fluorescent confocal microscopy using a 100× objective on an LSM 510 laser-scanning confocal microscope. The mean number of unclustered and internalized Con A foci was determined by examining Z stacks. In both assays, statistical differences between UIC and embryos injected with ABMO, nherf1MO, and nherf1MO + ABMO were determined by analysis of variance (ANOVA) at α ≤ 0.05.
Online supplemental material
Fig. S1 shows that loss of miR-8 miRNAs blocks Na+ accumulation in ionocytes. Fig. S2 shows the genomic organization of zebrafish miR-8 miRNAs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200807026/DC1.
Acknowledgments
We would like to thank Rachel Ostroff for assistance with experiments.
This work was supported by National Institutes of Health grant GM 075790 to J.G.Patton. A.S. Flynt was supported in part by National Institutes of Health grant T32 GM 008554.
Footnotes
Abbreviations used in this paper: ANOVA, analysis of variance; BCIP, 5-bromo-4-chloro-3-indolyl phosphate; dpf, day postfertilization; hpf, hour postfertilization; HRC, H+ pump–rich cell; LNA, locked nucleic acid; miRNA, microRNA; MRC, mitochondria-rich cell; MRE, miRNA recognition element; NBT, nitro blue tetrazolium; NHE, Na+/H+ exchanger; NRC, Na+-K+ pump–rich cell; SG, stress granule; UIC, uninjected control; UTR, untranslated region.
References
- Ambros V. 2003. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing.Cell. 113:673–676 [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T. 2003. The small RNA profile during Drosophila melanogaster development.Dev. Cell. 5:337–350 [DOI] [PubMed] [Google Scholar]
- Arenas M.I., Romo E., de Gaspar I., de Bethencourt F.R., Sanchez-Chapado M., Fraile B., Paniagua R. 1999. A lectin histochemistry comparative study in human normal prostate, benign prostatic hyperplasia, and prostatic carcinoma.Glycoconj. J. 16:375–382 [DOI] [PubMed] [Google Scholar]
- Bartel D.P., Chen C.Z. 2004. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs.Nat. Rev. Genet. 5:396–400 [DOI] [PubMed] [Google Scholar]
- Beauchamp L., Conrad R., Labourier E., Powers P. 2007. Ambion Poster: New Tools for miRNA and siRNA Analysis. Applied Biosystems. http://www.ambion.com/techlib/posters/miRNA_0309.html (accessed October 1, 2007)
- Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference.Nature. 409:363–366 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S.N., Habermacher R., Martine U., Closs E.I., Filipowicz W. 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress.Cell. 125:1111–1124 [DOI] [PubMed] [Google Scholar]
- Bracken C.P., Gregory P.A., Kolesnikoff N., Bert A.G., Wang J., Shannon M.F., Goodall G.J. 2008. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition.Cancer Res. 68:7846–7854 [DOI] [PubMed] [Google Scholar]
- Brennecke J., Stark A., Russell R.B., Cohen S.M. 2005. Principles of microRNA target recognition.PLoS Biol. 3:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk U., Schubert J., Wellner U., Schmalhofer O., Vincan E., Spaderna S., Brabletz T. 2008. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells.EMBO Rep. 9:582–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell M.A., Xuan Z., Zhang M.Q., Hannon G.J. 2002. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis.Genes Dev. 16:2733–2742 [DOI] [PubMed] [Google Scholar]
- Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., et al. 2005a. Real-time quantification of microRNAs by stem-loop RT-PCR.Nucleic Acids Res. 33:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.Y., Manninga H., Slanchev K., Chien M., Russo J.J., Ju J., Sheridan R., John B., Marks D.S., Gaidatzis D., et al. 2005b. The developmental miRNA profiles of zebrafish as determined by small RNA cloning.Genes Dev. 19:1288–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi P.S., Zakhary L., Choi W.Y., Caron S., Alvarez-Saavedra E., Miska E.A., McManus M., Harfe B., Giraldez A.J., Horvitz H.R., et al. 2008. Members of the miRNA-200 family regulate olfactory neurogenesis.Neuron. 57:41–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claiborne J.B., Edwards S.L., Morrison-Shetlar A.I. 2002. Acid-base regulation in fishes: cellular and molecular mechanisms.J. Exp. Zool. 293:302–319 [DOI] [PubMed] [Google Scholar]
- d'Anglemont de Tassigny A., Ghaleh B., Souktani R., Henry P., Berdeaux A. 2004. Hypo-osmotic stress inhibits doxorubicin-induced apoptosis via a protein kinase A-dependent mechanism in cardiomyocytes.Clin. Exp. Pharmacol. Physiol. 31:438–443 [DOI] [PubMed] [Google Scholar]
- Disthabanchong S., Martin K.J., McConkey C.L., Gonzalez E.A. 2002. Metabolic acidosis up-regulates PTH/PTHrP receptors in UMR 106-01 osteoblast-like cells.Kidney Int. 62:1171–1177 [DOI] [PubMed] [Google Scholar]
- Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. 2003. MicroRNA targets in Drosophila.Genome Biol. 5:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaki M., Hoshijima K., Kobayashi S., Fukuda H., Kawakami K., Hirose S. 2007. Visualization in zebrafish larvae of Na(+) uptake in mitochondria-rich cells whose differentiation is dependent on foxi3a.Am. J. Physiol. Regul. Integr. Comp. Physiol. 292:R470–R480 [DOI] [PubMed] [Google Scholar]
- Flynt A.S., Li N., Thatcher E.J., Solnica-Krezel L., Patton J.G. 2007. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate.Nat. Genet. 39:259–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheri G., Gheri Bryk S., Sgambati E. 1997. Glycoconjugate saccharidic moieties of the exocrine and endocrine pancreas in the chick embryo, newborn and adult.Biotech. Histochem. 72:158–167 [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Cinalli R.M., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F. 2005. MicroRNAs regulate brain morphogenesis in zebrafish.Science. 308:833–838 [DOI] [PubMed] [Google Scholar]
- Gregory P.A., Bert A.G., Paterson E.L., Barry S.C., Tsykin A., Farshid G., Vadas M.A., Khew-Goodall Y., Goodall G.J. 2008. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1.Nat. Cell Biol. 10:593–601 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. 2004. The microRNA Registry.Nucleic Acids Res. 32:D109–D111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. 2006. miRBase: microRNA sequences, targets and gene nomenclature.Nucleic Acids Res. 34:D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing.Mol. Cell. 27:91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J.A., Cheng A.G., Cunningham L.L., MacDonald G., Raible D.W., Rubel E.W. 2003. Neomycin-induced hair cell death and rapid regeneration in the lateral line of zebrafish (Danio rerio).J. Assoc. Res. Otolaryngol. 4:219–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., He X., Lim L.P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D., et al. 2007. A microRNA component of the p53 tumour suppressor network.Nature. 447:1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horng J.L., Lin L.Y., Huang C.J., Katoh F., Kaneko T., Hwang P.P. 2007. Knockdown of V-ATPase subunit A (atp6v1a) impairs acid secretion and ion balance in zebrafish (Danio rerio).Am. J. Physiol. Regul. Integr. Comp. Physiol. 292:R2068–R2076 [DOI] [PubMed] [Google Scholar]
- Hsiao C.D., You M.S., Guh Y.J., Ma M., Jiang Y.J., Hwang P.P. 2007. A positive regulatory loop between foxi3a and foxi3b is essential for specification and differentiation of zebrafish epidermal ionocytes.PLoS ONE. 2:e302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G., Zamore P.D. 2002. A microRNA in a multiple-turnover RNAi enzyme complex.Science. 297:2056–2060 [DOI] [PubMed] [Google Scholar]
- Ishizuka A., Siomi M.C., Siomi H. 2002. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins.Genes Dev. 16:2497–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicke M., Carney T.J., Hammerschmidt M. 2007. Foxi3 transcription factors and Notch signaling control the formation of skin ionocytes from epidermal precursors of the zebrafish embryo.Dev. Biol. 307:258–271 [DOI] [PubMed] [Google Scholar]
- John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. 2004. Human microRNA targets.PLoS Biol. 2:e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonz M.G., Nurse C.A. 2006. Epithelial mitochondria-rich cells and associated innervation in adult and developing zebrafish.J. Comp. Neurol. 497:817–832 [DOI] [PubMed] [Google Scholar]
- Ketting R.F., Fischer S.E., Bernstein E., Sijen T., Hannon G.J., Plasterk R.H. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans.Genes Dev. 15:2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman W.P., Plasterk R.H. 2006. The diverse functions of microRNAs in animal development and disease.Dev. Cell. 11:441–450 [DOI] [PubMed] [Google Scholar]
- Kloosterman W.P., Steiner F.A., Berezikov E., de Bruijn E., van de Belt J., Verheul M., Cuppen E., Plasterk R.H. 2006a. Cloning and expression of new microRNAs from zebrafish.Nucleic Acids Res. 34:2558–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman W.P., Wienholds E., de Bruijn E., Kauppinen S., Plasterk R.H. 2006b. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes.Nat. Methods. 3:27–29 [DOI] [PubMed] [Google Scholar]
- Kloosterman W.P., Lagendijk A.K., Ketting R.F., Moulton J.D., Plasterk R.H. 2007. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development.PLoS Biol. 5:e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpal M., Lee E.S., Hu G., Kang Y. 2008. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2.J. Biol. Chem. 283:14910–14914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt J., Poy M.N., Stoffel M. 2006. Strategies to determine the biological function of microRNAs.Nat. Genet. 38:S14–S19 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. 2001. Identification of novel genes coding for small expressed RNAs.Science. 294:853–858 [DOI] [PubMed] [Google Scholar]
- Lai E.C. 2002. Micro RNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation.Nat. Genet. 30:363–364 [DOI] [PubMed] [Google Scholar]
- Lederer E.D., Khundmiri S.J., Weinman E.J. 2003. Role of NHERF-1 in regulation of the activity of Na-K ATPase and sodium-phosphate co-transport in epithelial cells.J. Am. Soc. Nephrol. 14:1711–1719 [DOI] [PubMed] [Google Scholar]
- Lee Y., Jeon K., Lee J.T., Kim S., Kim V.N. 2002. MicroRNA maturation: stepwise processing and subcellular localization.EMBO J. 21:4663–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., Kim V.N. 2003. The nuclear RNase III Drosha initiates microRNA processing.Nature. 425:415–419 [DOI] [PubMed] [Google Scholar]
- Leung A.K., Sharp P.A. 2007. microRNAs: a safeguard against turmoil? Cell. 130:581–585 [DOI] [PubMed] [Google Scholar]
- Leung A.K., Calabrese J.M., Sharp P.A. 2006. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules.Proc. Natl. Acad. Sci. USA. 103:18125–18130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. 2003. Prediction of mammalian microRNA targets.Cell. 115:787–798 [DOI] [PubMed] [Google Scholar]
- Lin L.Y., Horng J.L., Kunkel J.G., Hwang P.P. 2006. Proton pump-rich cell secretes acid in skin of zebrafish larvae.Am. J. Physiol. Cell Physiol. 290:C371–C378 [DOI] [PubMed] [Google Scholar]
- Liu J., Valencia-Sanchez M.A., Hannon G.J., Parker R. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies.Nat. Cell Biol. 7:719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska E.A., Alvarez-Saavedra E., Townsend M., Yoshii A., Sestan N., Rakic P., Constantine-Paton M., Horvitz H.R. 2004. Microarray analysis of microRNA expression in the developing mammalian brain.Genome Biol. 5:R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales F.C., Takahashi Y., Momin S., Adams H., Chen X., Georgescu M.M. 2007. NHERF1/EBP50 head-to-tail intramolecular interaction masks association with PDZ domain ligands.Mol. Cell. Biol. 27:2527–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy A., Gonzalez-Agosti C., Cordero E., Pinney D., Candia C., Solomon F., Gusella J., Ramesh V. 1998. NHE-RF, a regulatory cofactor for Na(+)-H+ exchange, is a common interactor for merlin and ERM (MERM) proteins.J. Biol. Chem. 273:1273–1276 [DOI] [PubMed] [Google Scholar]
- Nielsen C.B., Shomron N., Sandberg R., Hornstein E., Kitzman J., Burge C.B. 2007. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs.RNA. 13:1894–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Ishizuka A., Siomi H., Siomi M.C. 2004. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways.Genes Dev. 18:1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlic T., Loomis W.H., Shreve A., Namiki S., Junger W.G. 2002. Hypertonicity increases cAMP in PMN and blocks oxidative burst by PKA-dependent and -independent mechanisms.Am. J. Physiol. Cell Physiol. 282:C1261–C1269 [DOI] [PubMed] [Google Scholar]
- Pascual-Ahuir A., Posas F., Serrano R., Proft M. 2001. Multiple levels of control regulate the yeast cAMP-response element-binding protein repressor Sko1p in response to stress.J. Biol. Chem. 276:37373–37378 [DOI] [PubMed] [Google Scholar]
- Raver-Shapira N., Marciano E., Meiri E., Spector Y., Rosenfeld N., Moskovits N., Bentwich Z., Oren M. 2007. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis.Mol. Cell. 26:731–743 [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans.Nature. 403:901–906 [DOI] [PubMed] [Google Scholar]
- Saran S., Meima M.E., Alvarez-Curto E., Weening K.E., Rozen D.E., Schaap P. 2002. cAMP signaling in Dictyostelium. Complexity of cAMP synthesis, degradation and detection.J. Muscle Res. Cell Motil. 23:793–802 [DOI] [PubMed] [Google Scholar]
- Sempere L.F., Sokol N.S., Dubrovsky E.B., Berger E.M., Ambros V. 2003. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity.Dev. Biol. 259:9–18 [DOI] [PubMed] [Google Scholar]
- Sempere L.F., Freemantle S., Pitha-Rowe I., Moss E., Dmitrovsky E., Ambros V. 2004. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation.Genome Biol. 5:R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh-Hamad D., Gustin M.C. 2004. MAP kinases and the adaptive response to hypertonicity: functional preservation from yeast to mammals.Am. J. Physiol. Renal Physiol. 287:F1102–F1110 [DOI] [PubMed] [Google Scholar]
- Solnica-Krezel L., Schier A.F., Driever W. 1994. Efficient recovery of ENU-induced mutations from the zebrafish germline.Genetics. 136:1401–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer-Rachamimov A.O., Wiederhold T., Nielsen G.P., James M., Pinney-Michalowski D., Roy J.E., Cohen W.A., Ramesh V., Louis D.N. 2001. NHE-RF, a merlin-interacting protein, is primarily expressed in luminal epithelia, proliferative endometrium, and estrogen receptor-positive breast carcinomas.Am. J. Pathol. 158:57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher E.J., Flynt A.S., Li N., Patton J.R., Patton J.G. 2007. MiRNA expression analysis during normal zebrafish development and following inhibition of the Hedgehog and Notch signaling pathways.Dev. Dyn. 236:2172–2180 [DOI] [PubMed] [Google Scholar]
- Theisen C.S., Wahl J.K., III, Johnson K.R., Wheelock M.J. 2007. NHERF links the N-cadherin/catenin complex to the platelet-derived growth factor receptor to modulate the actin cytoskeleton and regulate cell motility.Mol. Biol. Cell. 18:1220–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B., Pflumio S., Fürthauer M., Lopin B., Heyer V., Degrave A., Woehl R., Lux A., Steffan T., Charbonnier X.Q., Thisse C. 2001. Expression of the zebrafish genome during embryogenesis. ZFIN Direct Data Submission (http://zfin.org)
- Tuschl T., Zamore P.D., Lehmann R., Bartel D.P., Sharp P.A. 1999. Targeted mRNA degradation by double-stranded RNA in vitro.Genes Dev. 13:3191–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyska M.J., Mackey A.T., Huang J.D., Copeland N.G., Jenkins N.A., Mooseker M.S. 2005. Myosin-1a is critical for normal brush border structure and composition.Mol. Biol. Cell. 16:2443–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E., Sutherland L.B., Liu N., Williams A.H., McAnally J., Gerard R.D., Richardson J.A., Olson E.N. 2006. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure.Proc. Natl. Acad. Sci. USA. 103:18255–18260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voltz J.W., Weinman E.J., Shenolikar S. 2001. Expanding the role of NHERF, a PDZ-domain containing protein adapter, to growth regulation.Oncogene. 20:6309–6314 [DOI] [PubMed] [Google Scholar]
- Weinman E.J., Steplock D., Donowitz M., Shenolikar S. 2000. NHERF associations with sodium-hydrogen exchanger isoform 3 (NHE3) and ezrin are essential for cAMP-mediated phosphorylation and inhibition of NHE3.Biochemistry. 39:6123–6129 [DOI] [PubMed] [Google Scholar]
- Wheeler D., Sneddon W.B., Wang B., Friedman P.A., Romero G. 2007. NHERF-1 and the cytoskeleton regulate the traffic and membrane dynamics of G protein-coupled receptors.J. Biol. Chem. 282:25076–25087 [DOI] [PubMed] [Google Scholar]
- Wienholds E., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H.A. 2005. MicroRNA expression in zebrafish embryonic development.Science. 309:310–311 [DOI] [PubMed] [Google Scholar]
- Xu P., Vernooy S.Y., Guo M., Hay B.A. 2003. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism.Curr. Biol. 13:790–795 [DOI] [PubMed] [Google Scholar]
- Yun C.H., Oh S., Zizak M., Steplock D., Tsao S., Tse C.M., Weinman E.J., Donowitz M. 1997. cAMP-mediated inhibition of the epithelial brush border Na+/H+ exchanger, NHE3, requires an associated regulatory protein.Proc. Natl. Acad. Sci. USA. 94:3010–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P.D., Tuschl T., Sharp P.A., Bartel D.P. 2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals.Cell. 101:25–33 [DOI] [PubMed] [Google Scholar]