Abstract
The binding of fluorescently tagged proteins to tandem DNA arrays has been instrumental in understanding nuclear organization and function. Through the use of more natural tandem DNA arrays, Hu et al. (Hu, Y., I. Kireev, M. Plutz, N. Ashourian, and A.S. Belmont. 2009. J. Cell Biol. 185:87–100) gain new insights into chromatin organization and dynamics, and into the association of splicing factors with active genes.
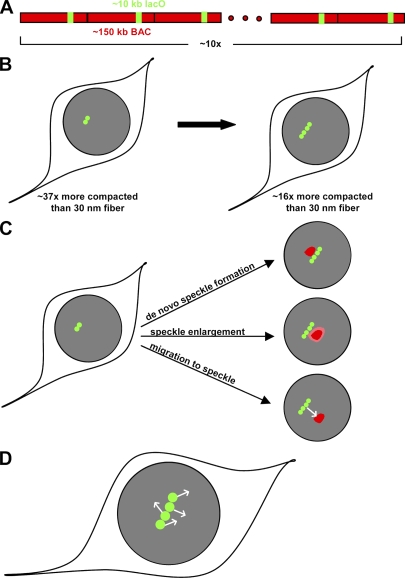
Tandem DNA arrays, which are composed of repeated DNA segments, have been successfully used to visualize genes in living cells. This is accomplished by expressing a fluorescently tagged DNA-binding protein in a cell containing a tandem array that incorporates the protein's binding site within each repeated segment (Rafalska-Metcalf and Janicki, 2007). This yields a locally high concentration of fluorescence, enabling the locus to be detected in the light microscope. Although cells contain some endogenous gene arrays that have been exploited by a few groups (Dundr et al., 2002; Karpova et al., 2008), most studies have used synthetic arrays because these can be optimally engineered. Synthetic arrays are typically constructed from hundreds of short repeated segments of plasmid DNA, yielding inserts up to many megabases in length. These arrays not only oversimplify the complexity of the mammalian genome, they are also subject to heterochromatin formation, a more repressive state of chromatin in which genes can be transcriptionally silenced. In this issue, Hu et al. (see p. 87 of this issue) construct arrays (Fig. 1 A) comprised of long stretches of almost exclusively mammalian DNA (∼150 kb) incorporated at a low copy number (∼10 copies) into chromosomes, thereby overcoming many of the limitations of previous synthetic arrays. Each repeat in the new arrays contains a small region of lac operator sites that will bind to a GFP-tagged lac repressor, which permits visualization of these sequences. Analysis of these new arrays provides strong evidence that transcription may occur in relatively condensed DNA and further suggests that active genes may seek out factors required for mRNA splicing.
Figure 1.
Visualizing chromatin organization and dynamics during transcription. (A) The arrays are composed of a single human or mouse BAC with lac operator sites incorporated for visualization. (Red dots indicate continuation of the repeat for 10 copies). (B) Decondensation occurs upon transcriptional induction of one or more of the genes in the BAC, yielding a linear trajectory of spots. Even after decondensation, compaction levels are still far above those of a 30-nm fiber. (C) Three different types of association with speckles were observed. (D) Independent, random motion of individual GFP spots was detected (for visibility, the arrows depicting this motion are larger than the actual movements).
A major focus of the current study was to use more natural arrays to investigate chromatin decondensation upon transcription induction. It is widely believed that chromatin is extensively compacted within nuclei, but that transcriptionally active regions decondense to the level of DNA wrapped around nucleosomes, namely a 10-nm fiber. To investigate chromatin organization in a transcriptionally active region, the authors constructed their arrays from bacterial artificial chromosomes (BACs) that contained known inducible mammalian genes. Consistent with several previous studies (Tumbar et al., 1999; Müller et al., 2001, 2004; Janicki et al., 2004), induction of transcription in these arrays caused them to decondense into a series of colinear spots (Fig. 1 B). By measuring the distance between these spots, the authors discovered that the degree of decondensation is not large: on average, the array decondenses to a level that is ∼96-fold more compacted than a 10-nm fiber.
A distinguishing feature of the current study is that several important controls have been performed to rule out other explanations for the limited decondensation observed. The authors find that their induced genes are expressed at close to endogenous levels. Further, they show that the induced genes are transcribed in most of the repeats, and that limited decondensation is observed regardless of whether the lac marker is positioned within or outside of the induced genes. Using electron microscopy, the authors are unable to detect any further decondensation of DNA not visible in the light microscope. Thus, this work provides the strongest evidence to date that transcription can occur at high levels of chromatin compaction.
It is of course difficult to prove with complete certainty that further decondensation of chromatin does not occur at the array. Conceivably, highly decondensed chromatin might be fragile and destroyed by the preparation for electron microscopy, or the apparently thicker fibers detected by light microscopy might actually reflect a serpentine path of thinner, underlying fibers. In this regard, it will be very interesting to examine this array with other forms of high-resolution microscopy, such as electron spectroscopic imaging. This technique reproducibly detects 10-nm fibers. It also detects the next level of DNA folding, the 30-nm fiber. Interestingly, this approach has not found evidence for any further compaction of chromatin beyond the 30-nm fiber (Dehghani et al., 2005), an apparent contradiction to the new arrays which appear to be at least 16-fold more compacted than a 30-nm fiber.
In the course of their studies, the authors discovered that one of their arrays (which contained the Hsp70 gene) was consistently associated with a speckle, a nuclear body enriched for mRNA splicing factors (Hu et al., 2009). Active genes are found at the edge of speckles, but it has not been clear how the association between speckles and active genes occurs. Previous studies had suggested that splicing factors could be recruited from the speckles to an active gene (Dirks et al., 1997; Misteli et al., 1997). To directly investigate how this association occurred in their system, the authors performed double-label live cell imaging to visualize both speckles and the activated gene array (Hu et al., 2009). They observed several types of association (Fig. 1 C): de novo speckle formation at the array, relatively uniform enlargement of preexisting speckles to contact the array, and migration of the array to the speckle. This last case is especially important for two reasons. It provides another example of longer range migration of gene loci within the nucleus (Chuang et al., 2006; Dundr et al., 2007), and it bolsters the view that speckles are not simply storage depots but rather functional sites that can recruit selected genes (Shopland et al., 2003; Brown et al., 2008).
Time-lapse analysis of the arrays also revealed another interesting property. The individual GFP spots comprising each array “jiggled” independently of each other (Fig. 1 D). This jiggling motion is consistent with previous studies indicating that chromatin undergoes constrained diffusion (Marshall et al., 1997). What is new in the current study is that adjacent regions of the same transcriptionally active chromosomal segment moved independently of each other. This argues against attachment of the transcriptional machinery to a stable nuclear scaffold (Jackson et al., 1984), at least one that extends over a large area of the nucleus. The current observations, however, could be explained by either a more dynamic or a more flexible scaffold.
Certainly, these observations will be followed by further insights as the full potential of this new array system is exploited. We can also look forward to seeing other more natural engineered arrays in the future, and ultimately, relatively noninvasive tagging of a single, endogenous gene.
References
- Brown J.M., Green J., das Neves R.P., Wallace H.A., Smith A.J., Hughes J., Gray N., Taylor S., Wood W.G., Higgs D.R., Iborra F.J., Buckle V.J. 2008. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment.J. Cell Biol. 182:1083–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C.H., Carpenter A.E., Fuchsova B., Johnson T., de Lanerolle P., Belmont A.S. 2006. Long-range directional movement of an interphase chromosome site.Curr. Biol. 16:825–831 [DOI] [PubMed] [Google Scholar]
- Dehghani H., Dellaire G., Bazett-Jones D.P. 2005. Organization of chromatin in the interphase mammalian cell.Micron. 36:95–108 [DOI] [PubMed] [Google Scholar]
- Dirks R.W., de Pauw E.S., Raap A.K. 1997. Splicing factors associate with nuclear HCMV-IE transcripts after transcriptional activation of the gene, but dissociate upon transcription inhibition: evidence for a dynamic organization of splicing factors.J. Cell Sci. 110:515–522 [DOI] [PubMed] [Google Scholar]
- Dundr M., Hoffmann-Rohrer U., Hu Q., Grummt I., Rothblum L.I., Phair R.D., Misteli T. 2002. A kinetic framework for a mammalian RNA polymerase in vivo.Science. 298:1623–1626 [DOI] [PubMed] [Google Scholar]
- Dundr M., Ospina J.K., Sung M.H., John S., Upender M., Ried T., Hager G.L., Matera A.G. 2007. Actin-dependent intranuclear repositioning of an active gene locus in vivo.J. Cell Biol. 179:1095–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Kireev I., Plutz M., Ashourian N., Belmont A.S. 2009. Large-scale chromatin structure of inducible genes: transcription on a condensed, linear template.J. Cell Biol. 185:87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D.A., McCready S.J., Cook P.R. 1984. Replication and transcription depend on attachment of DNA to the nuclear cage.J. Cell Sci. Suppl. 1:59–79 [DOI] [PubMed] [Google Scholar]
- Janicki S.M., Tsukamoto T., Salghetti S.E., Tansey W.P., Sachidanandam R., Prasanth K.V., Ried T., Shav-Tal Y., Bertrand E., Singer R.H., Spector D.L. 2004. From silencing to gene expression: real-time analysis in single cells.Cell. 116:683–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova T.S., Kim M.J., Spriet C., Nalley K., Stasevich T.J., Kherrouche Z., Heliot L., McNally J.G. 2008. Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter.Science. 319:466–469 [DOI] [PubMed] [Google Scholar]
- Marshall W.F., Straight A., Marko J.F., Swedlow J., Dernburg A., Belmont A., Murray A.W., Agard D.A., Sedat J.W. 1997. Interphase chromosomes undergo constrained diffusional motion in living cells.Curr. Biol. 7:930–939 [DOI] [PubMed] [Google Scholar]
- Misteli T., Caceres J.F., Spector D.L. 1997. The dynamics of a pre-mRNA splicing factor in living cells.Nature. 387:523–527 [DOI] [PubMed] [Google Scholar]
- Müller W.G., Walker D., Hager G.L., McNally J.G. 2001. Large-scale chromatin decondensation and recondensation regulated by transcription from a natural promoter.J. Cell Biol. 154:33–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W.G., Rieder D., Kreth G., Cremer C., Trajanoski Z., McNally J.G. 2004. Generic features of tertiary chromatin structure as detected in natural chromosomes.Mol. Cell. Biol. 24:9359–9370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalska-Metcalf I.U., Janicki S.M. 2007. Show and tell: visualizing gene expression in living cells.J. Cell Sci. 120:2301–2307 [DOI] [PubMed] [Google Scholar]
- Shopland L.S., Johnson C.V., Byron M., McNeil J., Lawrence J.B. 2003. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods.J. Cell Biol. 162:981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T., Sudlow G., Belmont A.S. 1999. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain.J. Cell Biol. 145:1341–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]