Abstract
Stress-induced phosphorylation of eIF2α inhibits global protein synthesis to conserve energy for repair of stress-induced damage. Stress-induced translational arrest is observed in cells expressing a nonphosphorylatable eIF2α mutant (S51A), which indicates the existence of an alternative pathway of translational control. In this paper, we show that arsenite, heat shock, or ultraviolet irradiation promotes transfer RNA (tRNA) cleavage and accumulation of tRNA-derived, stress-induced small RNAs (tiRNAs). We show that angiogenin, a secreted ribonuclease, is required for stress-induced production of tiRNAs. Knockdown of angiogenin, but not related ribonucleases, inhibits arsenite-induced tiRNA production and translational arrest. In contrast, knockdown of the angiogenin inhibitor RNH1 enhances tiRNA production and promotes arsenite-induced translational arrest. Moreover, recombinant angiogenin, but not RNase 4 or RNase A, induces tiRNA production and inhibits protein synthesis in the absence of exogenous stress. Finally, transfection of angiogenin-induced tiRNAs promotes phospho-eIF2α–independent translational arrest. Our results introduce angiogenin and tiRNAs as components of a phospho-eIF2α–independent stress response program.
Introduction
The survival of mammalian cells exposed to adverse environmental conditions requires a radical reprogramming of protein translation (Yamasaki and Anderson, 2008). Stress-induced translational arrest of mRNAs encoding “housekeeping” proteins is triggered by a family of eIF2α kinases that reduce the availability of eIF2α–GTP–tRNAiMet ternary complexes required for translation initiation (Anderson and Kedersha, 2008). Under these conditions, translation of a subset of mRNAs encoding upstream open reading frames (uORF; e.g., ATF4) is selectively enhanced, a consequence of uORF “read-through” (Lu et al., 2004). The reprogramming of protein translation is part of an integrated stress response that promotes the survival of cells subjected to adverse environmental conditions (Ron and Walter, 2007).
The finding that low-dose oxidative stress inhibits protein translation in cells expressing nonphosphorylatable eIF2α suggested the existence of a phospho-eIF2α–independent translation control pathway (McEwen et al., 2005). We hypothesized that stress-induced cleavage of tRNA, a phenomenon first described as a starvation response in Tetrahymena thermophila (Lee and Collins, 2005), and later observed in bacteria (Haiser et al., 2008), fungi (Jochl et al., 2008), and mammalian cells (Thompson et al., 2008), may contribute to stress-induced translational arrest. Sequence analysis reveals that tRNAs are cleaved in or near the anticodon loop. In both T. thermophila (Lee and Collins, 2005) and Saccharomyces cerevisiae (Thompson et al., 2008), many 3′ fragments possess “CCA” additions characteristic of mature tRNA. Moreover, these fragments lack tRNA introns, which indicates that they are not derived from pre-tRNAs. In most cases, the appearance of tRNA fragments is not accompanied by a significant depletion of mature tRNA, indicating that a small subset of the total tRNA population is cleaved during stress. In mammalian cells, analogous tRNA-derived fragments comprise a small subset of PIWI-associated RNAs (piRNAs), which suggests that tRNA anticodon cleavage may lead to the assembly of specific RNP complexes (Brennecke et al., 2007). It is possible that tRNA fragment-containing RNPs contribute to a phospho-eIF2α–independent translation control pathway.
Angiogenin is a secreted ribonuclease that was first identified as an angiogenic factor found in tumor cell–conditioned medium (Fett et al., 1985). The secretion of angiogenin is enhanced by hypoxia, which indicates that it may be a component of a stress response program (Hartmann et al., 1999; Nakamura et al., 2006). Angiogenin binds to receptors on the surface of endothelial cells that facilitate its internalization and transport to the nucleolus (Hu et al., 1997; Hatzi and Badet, 1999; Wiedlocha, 1999). Remarkably, promotion of new blood vessel growth is dependent on its ribonuclease activity (Shapiro and Vallee, 1987). Although the RNA targets required for angiogenesis are unknown, in vitro studies have shown that tRNAs are preferred targets (Saxena et al., 1992). Angiogenin also promotes ribosomal RNA transcription and cellular proliferation (Tsuji et al., 2005), which suggests that it has multiple functions. We have discovered that angiogenin is a stress-activated ribonuclease that cleaves tRNA and inhibits protein translation. Our results introduce angiogenin and tRNA-derived stress-induced RNAs (tiRNAs) as previously unappreciated components of the mammalian stress response.
Results and discussion
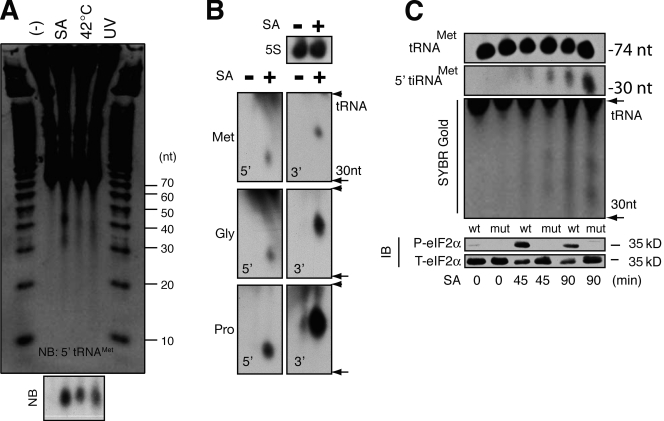
Extracts prepared from human U2OS cells exposed to arsenite-induced oxidative stress, heat shock, or UV irradiation were separated on a denaturing gel and developed with SYBR gold to visualize stress-induced small RNAs (Fig. 1 A). In stressed cells, two discrete bands corresponding to RNAs centered around 30 and 40 nucleotides were observed. Northern blotting using cDNA probes complementary to the 5′ end of tRNAMet (Fig. 1 A, bottom, NB) and the 5′ and 3′ ends of tRNAMet, tRNAGly, and tRNAPro (Fig. 1 B; 5S RNA is included as a loading control) confirms that the stress-induced RNAs are produced by tRNA cleavage. The size of these fragments requires that cleavage occur, as in T. thermophila (Lee and Collins, 2005), in or near the anticodon loop. These tiRNAs appear rapidly (within 20 min) in cells subjected to arsenite-induced oxidative stress, and persist for at least 11 h in cells allowed to recover from stress (Fig. S1 A). The phosphorylation and dephosphorylation of eIF2α over this time course provides a marker of stress and recovery from stress (Fig. S1 A). Arsenite-induced tiRNAs are observed in several different primate cell lines (Fig. S1 B). Importantly, tiRNAs are not observed in cells undergoing etoposide- or caffeine-induced apoptosis (Fig. S1 C), which indicates that tiRNA production is not a nonspecific consequence of cell death.
Figure 1.
Stress-induced production of tiRNA. (A) U2OS cells treated with sodium arsenite (SA; 500 µM, 2 h), heat (42°C, 2 h), or UV irradiation (200 J/m2, 12 h) were extracted with Trizol, and total RNA (10 µg) was separated on a 15% TBE-urea gel before processing with SYBR gold. The gel was also transferred to membrane and hybridized to a biotin probe complementary to the 5′ end of tRNAMet (NB). (B) Northern blotting analysis of RNA extracted from U2OS cells cultured in the absence (−) or presence (+) of sodium arsenite (500 µM, 2 h). Blots were hybridized to cDNAs complementary to the 5′ or 3′ fragments of the indicated tRNAs (bottom) or 5S RNA as a loading control (top). (C) MEFs derived from wild type (wt) or eIF2α (S51A) mutant (mut) mice were cultured in the absence (−) or presence (SA) of sodium arsenite (500 µM) for the indicated times before Trizol extraction. tRNA and 5′ tRNA fragments were quantified by Northern blotting (top) and SYBR gold staining (middle). Phospho- and total eIF2α was quantified by immunoblotting (IB; bottom).
To determine the potential for tiRNAs to mediate phospho-eIF2α–independent translational arrest, we compared their induction in mouse embryo fibroblasts (MEFs) derived from wild-type or eIF2α (S51A) mutant mice (Scheuner et al., 2001). The expression of mature tRNAMet is similar in wild-type and mutant (mut) cells in the absence or presence of arsenite (Fig. 1 C, SA). In both wild-type and mutant MEFs, tiRNAs are much less abundant than tRNAs. Although densitometric analysis may be misleading because of the relative overexpression of tRNAMet, the calculated ratios of tiRNAMet/tRNAMet are always <0.1, which indicates that depletion of tRNA is unlikely to contribute to the functional effects of tRNA cleavage. Interestingly, the induction of tiRNAMet is significantly greater in mutant cells compared with wild-type cells, indicating that phospho-eIF2α is not required for, and may inhibit, tiRNA production. We also quantified the production of tiRNAs in U2OS cells treated with control or heme-regulated initiation factor 2-α kinase (HRI)-specific siRNAs (Fig. S2). Knockdown of HRI, the eIF2α kinase activated by arsenite, increases the arsenite-induced production of tiRNAs. Collectively, these results indicate that the induction of tiRNAs does not require phospho-eIF2α. Moreover, phospho-eIF2α may suppress the induction of tiRNA.
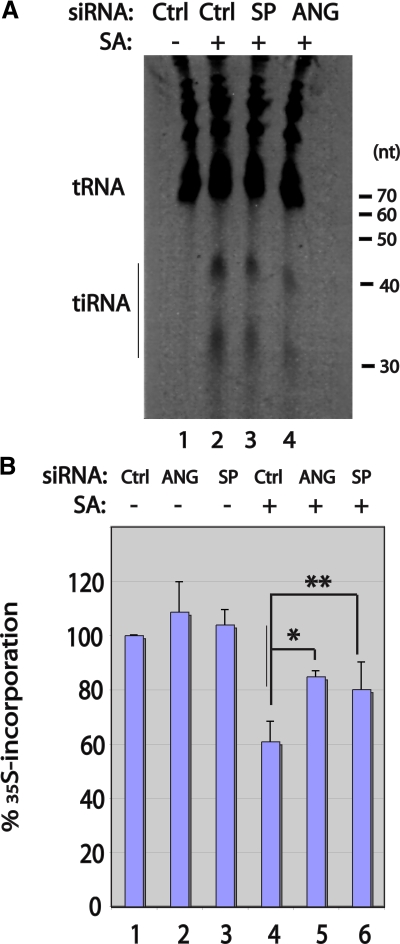
Ribonucleases that target the anticodon loop of tRNA are found in both prokaryotes and eukaryotes (Ardelt et al., 1991; Kaufmann, 2000; Suhasini and Sirdeshmukh, 2006). In Xenopus oocytes, a tRNA anticodon nuclease designated onconase has been found to inhibit protein synthesis and promote the death of selected tumor cells (Ardelt et al., 1991; Suhasini and Sirdeshmukh, 2006). The toxic effects of onconase are observed at doses that do not markedly deplete cellular tRNAs, which suggests that activation of the ribotoxic stress response may contribute to its toxic effects (Iordanov et al., 2000). Angiogenin is an onconase-related ribonuclease that selectively cleaves tRNA in mammalian cells (Saxena et al., 1992). To determine whether angiogenin is required for the stress-induced production of tiRNA, we used siRNA to knock down angiogenin expression before quantifying arsenite-induced tiRNA production and stress-induced translational repression. Transfection of a Dharmacon SMART pool targeting angiogenin reduces the expression of angiogenin mRNA (quantified using quantitative RT-PCR) to 63 ± 8% (n = 5) of the control level. Under these conditions, the arsenite-induced production of tiRNA is reduced to 55 ± 6% (n = 3) of the control level (Fig. 2 A, lane 3; representative of three independent experiments). This result was confirmed using an angiogenin-specific siRNA that more efficiently reduces angiogenin expression (Tsuji et al., 2005). This reagent reduces the expression of angiogenin mRNA (quantified using quantitative RT-PCR) to 27% ± 12 (n = 3) of the control level, and production of tiRNAs to 36 ± 8% (n = 3) of the control level (Fig. 2 A, lane 4; representative of three independent experiments). To determine the effect of angiogenin on stress-induced translational repression, U2OS cells were treated with the indicated siRNAs, then pulsed with [35S]methionine-containing medium in the absence or presence of sodium arsenite (Fig. 2 B, SA; 100 µM for 1 h) for 30 min. In all cases, [35S]methionine incorporation was normalized to that of cells treated with control siRNA. This analysis reveals that reduced expression of angiogenin significantly inhibits arsenite-induced translational repression (Fig. 2 B, compare lane 4 to lanes 5 and 6). These results are consistent with a role for angiogenin-induced tiRNAs in stress-induced translational repression, although cleavage of mRNA and/or rRNA could also contribute to inhibition of protein synthesis.
Figure 2.
Effect of endogenous angiogenin on tiRNA production and protein synthesis. (A) U2OS cells were transfected with control (D0, lanes 1 and 2), angiogenin-specific SMART pool (SP; lane 3), or angiogenin-specific individual (ANG; lane 4) siRNAs, then cultured in the absence (−; lane 1) or presence (+; lanes 2–4) of sodium arsenite (SA; 250 µM) for 1 h before Trizol extraction, separation on a 15% TBE-urea gel, and CYBR gold staining. The positions of tRNAs and tiRNAs are indicated in this representative gel. Densitometric scanning was used to quantify the relative expression of tiRNAs in three independent experiments. (B) U2OS cells were transfected with the indicated siRNAs, cultured in the absence (−; lanes 1–3) or presence (+; lanes 4–6) of sodium arsenite (100 µM, 1 h), and pulsed with [35S]methionine-containing medium for 60 min before protein extraction. [35S]methionine incorporation (mean ± SD, n = 3–5) is normalized to that observed in cells treated with control siRNA (designated 100%). *, P = 0.02 (n = 3); **, P = 0.04 (n = 5).
In S. cerevisiae, oxidative stress-induced tRNA cleavage is mediated by the RNaseT2 orthologue RNY1 (see Thompson et al., on p. 43 of this issue). In U2OS cells, knockdown of RNase T2 does not prevent arsenite-induced tiRNA production (Fig. S3), which indicates that yeast and humans use different enzymes to cleave tRNA. Knockdown of either RNase L or RNase Z (ELAC2) does not prevent arsenite-induced tiRNA cleavage (Fig. S3), which indicates that stress-induced tRNA cleavage is not a general property of ribonucleases.
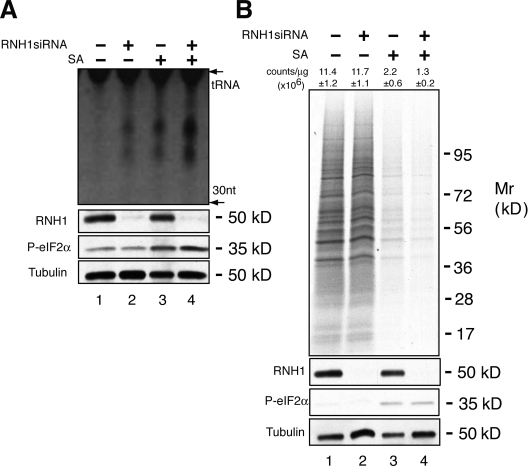
We found that the angiogenin inhibitor RNH1 (Naddeo et al., 2005) regulates the production of tiRNA in U2OS cells. Targeted knockdown of RNH1 induces the production of tiRNAs in the absence or presence of arsenite-induced oxidative stress (Fig. 3 A, lanes 2 and 4, respectively). This result suggests that angiogenin is constitutively expressed but held in an inactive state by RNH1. In cells subjected to arsenite-induced oxidative stress, knockdown of RNH1 enhances tiRNA production and promotes stress-induced translational silencing (Fig. 3 B, compare lanes 3 and 4). In unstressed cells, tiRNAs induced by RNH1 knockdown do not inhibit protein synthesis under the assayed conditions (Fig. 3 B, compare lanes 1 and 2). This may be due to a requirement for stress- or secreted angiogenin–induced cofactors. Further experiments will be needed to clarify this point.
Figure 3.
Effect of RNH1 on tiRNA production. (A) U2OS cells were treated with siRNA targeting RNH1 (lanes 2 and 4) 48 h before culturing cells in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of sodium arsenite (SA, 200 µM) for 2 h. Cells were extracted with Trizol, and RNA was separated by 15% PAGE and stained with SYBR gold. The location of tRNAs is indicated. tiRNAs migrate as small RNAs centered around 30 and 40 nucleotides. The expression of RNH1, phospho-eIF2α, and tubulin were quantified by Western blotting analysis (bottom). (B) U2OS cells were treated with siRNAs targeting RNH1 (lanes 2 and 4), then cultured in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of sodium arsenite (SA, 500 µM) for 1 h. Cells were then processed for gel electrophoresis and autoradiography (top) or TCA precipitation and scintillation counting (mean counts per microgram ± SD, n = 3, are shown over each lane). The expression of RNH1, phospho-eIF2α, and tubulin were quantified by Western blotting analysis (bottom).
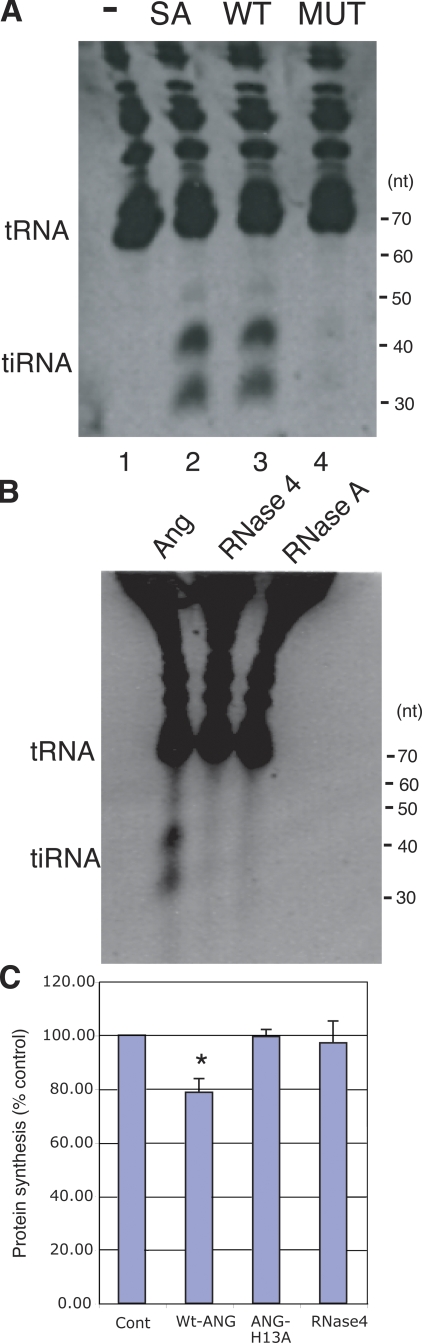
Because angiogenin is a stress-induced secreted protein that is taken up by adjacent cells, we tested the ability of purified recombinant angiogenin to induce the production of tiRNAs in U2OS cells. Wild-type angiogenin, but not an inactive mutant that has been implicated in the pathogenesis of amyotrophic lateral sclerosis (P112L; Fig 4 A, MUT; Wu et al., 2007), induces the production of tiRNA in U2OS cells (Fig. 4 A). The angiogenin-related ribonucleases RNase 4 and RNase A do not induce the production of tiRNA under these conditions (Fig. 4 B). Wild-type angiogenin, but not an inactive mutant (ANG-H13A; Shapiro et al., 1986), or RNase 4, significantly inhibits global protein synthesis in U2OS cells (Fig. 4 C). These results are consistent with a role for angiogenin in both stress-induced tRNA cleavage and stress-induced translational arrest.
Figure 4.
Effect of recombinant angiogenin on tiRNA production and protein synthesis. (A) U2OS cells were cultured in the absence (−; lane 1) or presence of sodium arsenite (SA; lane 2; 500 µM, 1 h), recombinant wild-type angiogenin (WT; 1 µg/ml, lane 3), or recombinant mutant angiogenin (MUT, 1 µg/ml, lane 4) for 1 h before Trizol extraction, separation by 15% TBE-urea gel, and CYBR gold staining. The positions of tRNAs and tiRNAs are indicated on the left. (B) U2OS cells were treated with the indicated ribonucleases (1 µg/ml) for 1 h before processing as described in A. (C) U2OS cells were treated with the indicated ribonucleases (1 µg/ml) for 30 min in the presence of [35S]methionine-containing medium before protein extraction. 35S counts in cells cultured in media alone (Cont) were normalized to 100%. Results are the means ± SD (n = 3). *, P = 0.02.
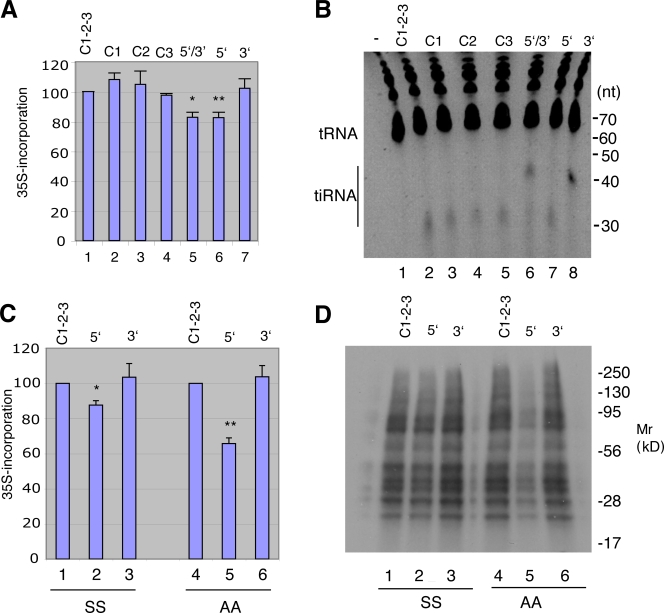
Endogenous tiRNAs corresponding to 5′ and 3′ tRNA fragments were purified from angiogenin-treated cells and transfected into U2OS cells. Pulse labeling with [35S]methionine reveals that 5′ tiRNAs, but not 3′ tiRNAs or synthetic control RNAs (sequences corresponding to PIWI-associated RNAs; see Materials and methods) significantly inhibit protein synthesis (Fig. 5 A). The combination of 5′ and 3′ tiRNAs also inhibits translation, indicating that 3′ tiRNA does not inhibit the activity of 5′ tiRNA. Importantly, the ratio of transfected tiRNA/tRNA (Fig. 5 B) is similar to the ratio of endogenous tiRNA/tRNA observed in arsenite-treated cells (Fig. 2 A). Similar results were obtained when 5′ and 3′ tiRNAs were transfected into wild-type (Fig. 5 C: SS) and S51A mutant MEFs (Fig. 5 C, AA), which indicates that inhibition of protein synthesis does not require phosphorylation of eIF2α, ruling out a primary role for PKR in this process. Autoradiographic analysis of 35S-labeled proteins reveals that 5′ tiRNAs inhibit global protein synthesis (Fig. 5 D).
Figure 5.
Effect of endogenous tiRNAs on protein synthesis. (A) Endogenous 5′ and 3′ (lane 5), 5′ (lane 6), or 3′ (lane 7) tiRNAs extracted from angiogenin-treated U2OS cells were transfected into U2OS cells using lipofectamine. After 6 h, cells were pulsed with [35S]methionine-containing medium for 30 min before protein extraction. Total counts per minute (cpm) per microgram of protein was normalized to cells treated with a combination of three PIWI-associated control RNAs (Control 1-2-3, lane 1; Control-1, lane 2; Control 3, lane 3) and expressed as a mean ± SD (n = 3). *, P = 0.01; **, P = 0.01. (B) U2OS cells were transfected with the indicated control RNAs (lanes 1–5) or endogenous 5′ (lane 7), 3′ (lane 8), or 5′ and 3′ (lane 6) tiRNAs. After 6 h, cells were washed, and Trizol extracts were separated on a 15% TBD-urea gel and stained with CYBR gold. (C) Wild-type (SS) and S51A mutant (AA) MEFs were transfected with control RNAs (lanes 1 and 4) or endogenous 5′ (lanes 2 and 5) or 3′ (lanes 3 and 6) tiRNAs, pulsed with [35S]methionine-containing medium, and extracted. Total cpm per microgram of protein was normalized to that of cells treated with control RNA and expressed as means ± SD (n = 3). *, P = 0.01; **, P = 0.003. (D) Samples from C were separated by 15% SDS-PAGE, transferred to nitrocellulose, and exposed for autoradiography. Migration of molecular size markers is shown at the right.
Angiogenin is a secreted protein that functions in the acute phase response induced by inflammatory stimuli (Olson et al., 1998). We have shown that: (1) recombinant angiogenin induces the production of tiRNAs in U2OS cells (while this manuscript was under review, similar results were reported by Fu et al. [2009]), (2) recombinant angiogenin inhibits protein synthesis in U2OS cells, (3) knockdown of angiogenin inhibits arsenite-induced tiRNA production and translational repression, (4) knockdown of RNH1 enhances tiRNA production and promotes arsenite-induced translational repression, and (5) transfection of purified, endogenous 5′ but not 3′ tiRNAs inhibits protein synthesis in U2OS cells as well as wild-type and S51A mutant MEFs. Collectively, these results strongly implicate angiogenin and tiRNAs in a process of stress-induced translational repression.
The finding that tRNA fragments possess posttranscriptionally added “CCA” residues and lack leaders, trailers, and introns suggests that tiRNAs are derived from mature tRNAs (Lee and Collins, 2005; Thompson et al., 2008; Fu et al., 2009). We have found that neomycin, a drug that prevents recombinant angiogenin from entering the nucleus, has no effect on angiogenin-induced tRNA cleavage, which suggests that angiogenin cleaves mature, cytoplasmic tRNAs (unpublished data). The finding that 5′ but not 3′ tiRNAs inhibit protein synthesis reveals a functional difference between these tRNA fragments. This may be a consequence of tRNA fragment size: smaller 5′ fragments (∼30 nucleotides) may bind to a protein cofactor more efficiently than larger 3′ fragments (∼40 nucleotides). Another important consideration is the nature of the 5′ and 3′ ends of the tRNA fragments. The 5′ ends of 5′ tiRNAs, like tRNA, are likely to be 5′ monophosphates. In contrast, the 5′ ends of 3′ tiRNAs are likely to be hydroxyl groups. As miRNAs and piRNAs have 5′ monophosphates, this modification may promote the stability and function of small RNAs. The 3′ ends of 5′ tiRNAs are likely to be 2′, 3′ cyclic phosphates, as angiogenin cleavage leaves this moiety at the 3′ ends of cleaved RNAs (Rybak and Vallee, 1988). This cyclic phosphate residue may be resolved to 2′ phosphate or 3′ phosphate groups. Whether this moiety is further modified to produce the protective 2′-O-methyl groups found at the termini of miRNAs and piRNAs (Horwich et al., 2007) is an important question for future research. In contrast, the 3′ ends of 3′ tiRNAs, like tRNAs, are likely to be hydroxyl groups. Thus, the nature of the 5′ and 3′ ends of these tRNA fragments may be an important determinant of their functional potential.
Because stress-induced reprogramming of protein translation can help cells survive adverse environmental conditions, secreted angiogenin may activate an “infectious” stress response program that allows stressed cells to warn their brethren of approaching noxious stimuli. This may occur at the organismal level by secretion of angiogenin from the liver (Olson et al., 1998), or at the tissue level by secretion of angiogenin from stressed cells within peripheral tissues. The local and systemic actions of angiogenin would be consistent with its expression from an upstream promoter that is expressed in all tissues and a downstream promoter that is expressed in liver cells (Dyer and Rosenberg, 2005). Just as interferons are virus-induced factors that protect adjacent or distant cells from virus infection, angiogenin may be a stress-induced factor that protects adjacent or distant cells from the deleterious effects of environmental stress.
Angiogenin has also been implicated in the pathogenesis of amyotrophic lateral sclerosis (ALS), a neurological disease caused by the death of motor neurons. Mutations in angiogenin are found in a small subset of patients with both familial and sporadic ALS (Greenway et al., 2006; Wu et al., 2007). ALS-associated angiogenin missense mutations have reduced ribonuclease and angiogenic activity (Greenway et al., 2006; Wu et al., 2007). It is therefore possible that the survival of motor neurons requires the angiogenin-induced stress response program.
Materials and methods
Cell culture and treatment
U2OS cells were cultured in DME (Invitrogen) supplemented with 10% fetal calf serum (Sigma-Aldrich), 100 U/ml antibiotics (penicillin), and 100 µg/ml streptomycin. Lipofectamine 2000 (Invitrogen) and Optimem medium (Invitrogen) were used for transfection of tiRNAs and siRNAs. The wild-type (SS) and S51A knock-in (AA) MEFs, a gift from D. Scheuner and R. Kaufman (University of Michigan Medical Center, Ann Arbor, MI), were cultured in DME with 10% fetal calf serum and antibiotics. For stress induction, the indicated doses of sodium arsenite (Sigma-Aldrich) were added in medium. The cells were washed with PBS twice before UV irradiation (UV cross-linker FB-UVXL-1000; Fisher Biotech). Heat shock was achieved incubating cells in a 42°C oven. U2OS cells were treated with recombinant wild-type or mutant angiogenin (Wu et al., 2007), RNase 4 (purified using the same methods used for angiogenin), or RNase A (0.5 µg/ml; QIAGEN) for 1 h before processing for quantification of tiRNAs.
RNA analysis
Total RNA was extracted by using Trizol (Invitrogen). RNA (10 µg per well) was analyzed using TBS-urea gels (Invitrogen) or 1.1% agarose/2% formaldehyde MOPS gels, transferred to Nytran Supercharge membranes (Schleicher and Schuell), and hybridized overnight at 50° with digoxigenin (DIG)-labeled DNA probes in DIG Easy Hyb solution (Roche). After washing at 60° with 2× SSC/0.1% SDS (10 min) and 0.5× SSC/0.1% SDS (twice for 20 min), the membranes were blocked in blocking reagent (Roche) for 30 min at room temperature, probed with alkaline phosphatase-labeled anti-digoxigenin antibody (Roche) for 30 min, and washed for 30 min with 130 mM TrisHCl, pH 7.5/100 mM NaCl/0.3% Tween 20. Signals were visualized with CDP-Star (Roche). DIG-labeled probes for 5S rRNA, tRNA, and tiRNAs were prepared using the DIG Oligonucleotide 3′-End Labeling kit (second generation; Roche) according to the manufacturer's instructions.
Metabolic labeling
In in vivo experiments, control RNA or tiRNAs were transfected into U2OS cells or MEFs in 24-well plates, and cultured for the indicated times. The cells were incubated with labeling medium (DME without l-glutamine, sodium pyruvate, l-methionine, or l-cystine [Invitrogen], supplemented with 5% dialyzed fetal bovine serum [Thermo Fisher Scientific]) for 30–60 min, replaced with fresh labeling medium containing ∼150–250 µCi of l-[35S]methionine per well (EasyTag EXPRESS 35S Protein Labeling Mix; PerkinElmer) and incubated for 30 min. Recombinant ribonucleases or arsenite was added together with the 35S labeling mix and incubated for 30 min or 60 min, respectively, before processing. After washing with PBS twice, cells were harvested in 400 µl of lysis buffer (2% SDS/20 mM Hepes, pH 7.4) and sonicated, and the protein was precipitated by the addition of 60% acetone. The proteins were resuspended in lysis buffer, and 10 µl of each sample in Ecoscint H (National Diagnostics) was counted using a liquid scintillation counter (Beckman Coulter). Protein concentration was determined by Protein Assay BCA Protein Assay kit (Pierce).
Antibodies and reagents
Antibodies against phospho-eIF2α or total eIF2α were obtained from Assay Designs or Santa Cruz Biotechnology, Inc., respectively. Polyclonal rabbit anti-RNH1 was obtained from Proteintech Group, Inc.
tiRNA isolation and transfection
U2OS cells (6.0 × 107) were treated with 0.5 µg/ml recombinant angiogenin or 500 µM sodium arsenite for 90 min before extraction with Trizol. 2 µg of total RNA was separated using four sets of 15% TBE-urea acrylamide gels. Gel fractions containing 5′ tiRNA and 3′ tiRNA visualized using SYBR gold nucleic acid gel stain (Invitrogen) were crushed and soaked in 20 ml of nuclease-free 1 M NaCl with rocking at 4°C overnight. After centrifugation, linear acrylamide (Applied Biosystems) and ethanol (to 60%) were added to each supernatant. The RNA mixtures were filtered through a MEGAclear filter cartridge (Applied Biosystems). After washing the filter with 4 ml of 80% ethanol, the MEGAclear filter cartridges were washed again with 500 µl of 80% ethanol. After removing excess ethanol, the bound tiRNAs were eluted by 80 µl of heated (95°C) elution solution.
U2OS cells or MEFs were transfected with 5′ or 3′ tiRNAs (1 µM) using Lipofectamine 2000. After 6 h, washed cells were metabolically labeled as described in “Metabolic labeling.”
tRNA sequences
The tRNA sequences were obtained from the RNA World Website (http://www.imb-jena.de/RNA.html) at the Leibniz Institute for Age Research, Fritz Lipmann Institute.
siRNA treatment
For angiogenin knockdown, HRI, RNH1, RNaseL, ELAC2, and RNase T2 siGENOME SMART pools for each molecule were purchased from Thermo Fisher Scientific. The individual angiogenin siRNA sequence was 5′-GACUUGCUUAUUCUUAGGUUU-3′. U2OS cells were transfected with 40 nM of siRNA using Lipofectamine 2000 12 h after plating cells. On the next day, the cells were replated, and a second siRNA transfection was performed, after which cells were cultured for 24 h. Total RNA was isolated using Trizol after a 90-min incubation with 500 µM sodium arsenite.
RNA oligos
ctrlRNA1 (piR58620, control RNA): 5′-UGUGAGUCACGUGAGGGCAGAAUCUGCUC-3′; ctrlRNA2 (piR006650, control RNA): 5′-UGAAGGGUUUUUUGUGUCUCUAUUUCCUUC-3′; ctrlRNA3 (piR016792, control RNA): 5′-CCUCCCAAAGUGCUGGGAUUACAGGCGUGAG-3′.
DNA oligos
SY173, probe for 5′ tiRNAMet: 5′-GGGCCCAGCACGCTTCCGCTGCGCCACTCTGC-3′; SY167, probe for 3′ tiRNAMet: 5′-TAGACAGAGGATGGTTTCGATCCATCGACC-3′; SY176, probe for 5′ tiRNAGly, piR-61648, 5′ half of tRNA128-GlyGCC: 5′-GGCAGGCGAGAATTCTACCACTGAACCACCAA-3′; SY186, probe for 3′ tiRNAGly, 3′ half of tRNA128-GlyGCC: 5′-GCCGGGAATCGAACCCGGGCCTCCCGCG-3′; SY180, probe for 5′ tiRNAPro, piR-36074, 5′ half of tRNA12-ProTGG: 5′-CCGAGAATCATACCCCTAGACCAACGAGCC-3′; SY181, probe for 3′ tiRNAPro, 3′ half of tRNA11-ProAGG: 5′-CTCGTCCGGGATTTGAACCCGGGACCTCTCGC-3′; SY161, probe for 5S ribosomal RNA: 5′-GGGTGGTATGGCCGTAGAC-3′.
Online supplemental material
Fig. S1 shows arsenite-induced production of tiRNAs. Fig. S2 shows that phospho-eIF2α is a negative regulator of tiRNA production. Fig. S3 shows the effect of endonucleases on tiRNA production. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200811106/DC1.
Acknowledgments
We thank the Anderson laboratory for helpful discussions and advice. We acknowledge Dr. Kaneyuki Tsuchimochi for helpful discussions and Dr. Randal Kaufman for providing S51A mutant MEFs.
This work was supported by National Institutes of Health grants AI065858, AI033600, and AR0514732.
Footnotes
Abbreviations used in this paper: MEF, mouse embryo fibroblast; piRNA, PIWI-associated RNA; tiRNA, transfer RNA-derived stress-induced RNA.
References
- Anderson P., Kedersha N. 2008. Stress granules: the Tao of RNA triage.Trends Biochem. Sci. 33:141–150 [DOI] [PubMed] [Google Scholar]
- Ardelt W., Mikulski S.M., Shogen K. 1991. Amino acid sequence of an anti-tumor protein from Rana pipiens oocytes and early embryos. Homology to pancreatic ribonucleases.J. Biol. Chem. 266:245–251 [PubMed] [Google Scholar]
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila.Cell. 128:1089–1103 [DOI] [PubMed] [Google Scholar]
- Dyer K.D., Rosenberg H.F. 2005. The mouse RNase 4 and RNase 5/ang 1 locus utilizes dual promoters for tissue-specific expression.Nucleic Acids Res. 33:1077–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fett J.W., Strydom D.J., Lobb R.R., Alderman E.M., Bethune J.L., Riordan J.F., Vallee B.L. 1985. Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells.Biochemistry. 24:5480–5486 [DOI] [PubMed] [Google Scholar]
- Fu H., Feng J., Liu Q., Sun F., Tie Y., Zhu J., Xing R., Sun Z., Zheng X. 2009. Stress induces tRNA cleavage by angiogenin in mammalian cells.FEBS Lett. 583:437–442 [DOI] [PubMed] [Google Scholar]
- Greenway M.J., Andersen P.M., Russ C., Ennis S., Cashman S., Donaghy C., Patterson V., Swingler R., Kieran D., Prehn J., et al. 2006. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis.Nat. Genet. 38:411–413 [DOI] [PubMed] [Google Scholar]
- Haiser H.J., Karginov F.V., Hannon G.J., Elliot M.A. 2008. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor.Nucleic Acids Res. 36:732–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann A., Kunz M., Kostlin S., Gillitzer R., Toksoy A., Brocker E.B., Klein C.E. 1999. Hypoxia-induced up-regulation of angiogenin in human malignant melanoma.Cancer Res. 59:1578–1583 [PubMed] [Google Scholar]
- Hatzi E., Badet J. 1999. Expression of receptors for human angiogenin in vascular smooth muscle cells.Eur. J. Biochem. 260:825–832 [DOI] [PubMed] [Google Scholar]
- Horwich M.D., Li C., Matranga C., Vagin V., Farley G., Wang P., Zamore P.D. 2007. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC.Curr. Biol. 17:1265–1272 [DOI] [PubMed] [Google Scholar]
- Hu G.F., Riordan J.F., Vallee B.L. 1997. A putative angiogenin receptor in angiogenin-responsive human endothelial cells.Proc. Natl. Acad. Sci. USA. 94:2204–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordanov M.S., Ryabinina O.P., Wong J., Dinh T.H., Newton D.L., Rybak S.M., Magun B.E. 2000. Molecular determinants of apoptosis induced by the cytotoxic ribonuclease onconase: evidence for cytotoxic mechanisms different from inhibition of protein synthesis.Cancer Res. 60:1983–1994 [PubMed] [Google Scholar]
- Jochl C., Rederstorff M., Hertel J., Stadler P.F., Hofacker I.L., Schrettl M., Haas H., Huttenhofer A. 2008. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis.Nucleic Acids Res. 36:2677–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann G. 2000. Anticodon nucleases.Trends Biochem. Sci. 25:70–74 [DOI] [PubMed] [Google Scholar]
- Lee S.R., Collins K. 2005. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila.J. Biol. Chem. 280:42744–42749 [DOI] [PubMed] [Google Scholar]
- Lu P.D., Harding H.P., Ron D. 2004. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response.J. Cell Biol. 167:27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen E., Kedersha N., Song B., Scheuner D., Gilks N., Han A., Chen J.J., Anderson P., Kaufman R.J. 2005. Heme-regulated inhibitor (HRI) kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 (eIF2) inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure.J. Biol. Chem. 280:16925–16933 [DOI] [PubMed] [Google Scholar]
- Naddeo M., Vitagliano L., Russo A., Gotte G., D'Alessio G., Sorrentino S. 2005. Interactions of the cytotoxic RNase A dimers with the cytosolic ribonuclease inhibitor.FEBS Lett. 579:2663–2668 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Yamabe H., Osawa H., Nakamura N., Shimada M., Kumasaka R., Murakami R., Fujita T., Osanai T., Okumura K. 2006. Hypoxic conditions stimulate the production of angiogenin and vascular endothelial growth factor by human renal proximal tubular epithelial cells in culture.Nephrol. Dial. Transplant. 21:1489–1495 [DOI] [PubMed] [Google Scholar]
- Olson K.A., Verselis S.J., Fett J.W. 1998. Angiogenin is regulated in vivo as an acute phase protein.Biochem. Biophys. Res. Commun. 242:480–483 [DOI] [PubMed] [Google Scholar]
- Ron D., Walter P. 2007. Signal integration in the endoplasmic reticulum unfolded protein response.Nat. Rev. Mol. Cell Biol. 8:519–529 [DOI] [PubMed] [Google Scholar]
- Rybak S.M., Vallee B.L. 1988. Base cleavage specificity of angiogenin with Saccharomyces cerevisiae and Escherichia coli 5S RNAs.Biochemistry. 27:2288–2294 [DOI] [PubMed] [Google Scholar]
- Saxena S.K., Rybak S.M., Davey R.T., Jr., Youle R.J., Ackerman E.J. 1992. Angiogenin is a cytotoxic, tRNA-specific ribonuclease in the RNase A superfamily.J. Biol. Chem. 267:21982–21986 [PubMed] [Google Scholar]
- Scheuner D., Song B., McEwen E., Liu C., Laybutt R., Gillespie P., Saunders T., Bonner-Weir S., Kaufman R.J. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis.Mol. Cell. 7:1165–1176 [DOI] [PubMed] [Google Scholar]
- Shapiro R., Vallee B.L. 1987. Human placental ribonuclease inhibitor abolishes both angiogenic and ribonucleolytic activities of angiogenin.Proc. Natl. Acad. Sci. USA. 84:2238–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro R., Riordan J.F., Vallee B.L. 1986. Characteristic ribonucleolytic activity of human angiogenin.Biochemistry. 25:3527–3532 [DOI] [PubMed] [Google Scholar]
- Suhasini A.N., Sirdeshmukh R. 2006. Transfer RNA cleavages by onconase reveal unusual cleavage sites.J. Biol. Chem. 281:12201–12209 [DOI] [PubMed] [Google Scholar]
- Thompson D.M., Parker R. 2009. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae.J. Cell Biol. 185:43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D.M., Lu C., Green P.J., Parker R. 2008. tRNA cleavage is a conserved response to oxidative stress in eukaryotes.RNA. 14:2095–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji T., Sun Y., Kishimoto K., Olson K.A., Liu S., Hirukawa S., Hu G.F. 2005. Angiogenin is translocated to the nucleus of HeLa cells and is involved in ribosomal RNA transcription and cell proliferation.Cancer Res. 65:1352–1360 [DOI] [PubMed] [Google Scholar]
- Wiedlocha A. 1999. Following angiogenin during angiogenesis: a journey from the cell surface to the nucleolus.Arch. Immunol. Ther. Exp. (Warsz.). 47:299–305 [PubMed] [Google Scholar]
- Wu D., Yu W., Kishikawa H., Folkerth R.D., Iafrate A.J., Shen Y., Xin W., Sims K., Hu G.F. 2007. Angiogenin loss-of-function mutations in amyotrophic lateral sclerosis.Ann. Neurol. 62:609–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki S., Anderson P. 2008. Reprogramming mRNA translation during stress.Curr. Opin. Cell Biol. 20:222–226 [DOI] [PMC free article] [PubMed] [Google Scholar]