Abstract
Live-cell imaging studies aided by mathematical modeling have provided unprecedented insight into assembly mechanisms of multiprotein complexes that control genome function. Such studies have unveiled emerging properties of chromatin-associated systems involved in DNA repair and transcription.
Introduction
Essential functions of the genome, such as transcription, replication, and DNA repair, are controlled by a variety of molecular mechanisms that each involve a dynamic interplay between multiple protein factors and specific genomic locations in a structurally ordered and time-dependent fashion. Recent developments in live-cell imaging and progress in quantitative fluorescence microscopy have provided novel insight into the dynamic interplay of multiprotein complexes with chromatin. In vivo studies have revealed that many proteins that control genome function rapidly diffuse inside the mammalian nucleus in the absence of molecular interactions, with apparent diffusion rates ranging between ∼0.1 and 15 µm2/s, depending on the shape and the size of the molecule. Binding to static structures such as chromatin usually lowers the mobility of a protein substantially (Houtsmuller et al., 1999; Phair and Misteli, 2000). Many nuclear proteins rapidly exchange between the freely mobile and the chromatin-bound immobile state on the time scale of seconds to minutes (Houtsmuller and Vermeulen, 2001; Gorski et al., 2006). Although the binding kinetics of many individual proteins have been measured, little is known about how proteins assemble into the functional multiprotein complexes that are involved in genome function.
In this mini-review, we focus on general mechanisms and kinetics of the in vivo assembly of chromatin-associated multiprotein complexes. We discuss how proteins find their target site on the genome and give an overview of the binding kinetics of proteins involved in transcription and DNA repair. Finally, we discuss how several live-cell studies, aided by kinetic modeling, have unveiled novel properties of assembly of multiprotein complexes on the chromatin fiber. We anticipate that future approaches aimed at combining live-cell kinetics and mathematical modeling will continue to provide detailed insight into the temporal organization and molecular mechanism of multiprotein complexes that control genome function.
How do site-specific proteins find target sites on the DNA?
Essentially all processes that control genome function are performed by complexes containing multiple proteins that assemble on specific sites on the DNA. Formation of such multiprotein complexes is often initiated by recognition proteins with affinity for a specific sequence or structure of the DNA, such as a promoter or a DNA lesion. The affinity of such proteins for DNA is determined by the ratio of its binding rate to (on-rate, kon) and its dissociating rate from these sites (off-rate, koff). Proteins often bind to specific and nonspecific sites with similar on-rates, whereas affinity for specific sites is mostly determined by a lower dissociation rate, resulting in a longer retention time on the specific site (Hopfield, 1974; Qian, 2008). How do site-specific proteins find their correct target sites in a high excess of nonspecific binding sites? Several studies support a model in which a protein that binds to a specific DNA sequence freely diffuses through the nucleus and transiently interacts with chromatin. Because nonspecific (i.e., low affinity) sites are usually present in large excess over specific sites, most binding events will be at nonspecific sites (Misteli, 2008). Typically, if a protein interacts with nonspecific, low affinity sites on chromatin, it will rapidly dissociate and rebind until it encounters a high affinity (i.e., specific) site from which it dissociates more slowly (Gorski et al., 2006).
Interestingly, some proteins associate with their target sites in vitro several orders of magnitude faster (rates up to 1010 M−1s−1) than expected from diffusion-limited binding (∼108 M−1s−1; Berg et al., 1981; Gorman and Greene, 2008). Some models explain this rapid rate of association by movement of the protein from an initial nonspecific site to its target site by 1D diffusion along the DNA by a sliding mechanism, which involves electrostatic DNA–protein interactions (Berg et al., 1981; Halford and Marko, 2004; Elf et al., 2007). Several DNA-binding proteins are able to move along DNA without dissociating from it, including restriction enzymes, transcription factors, and DNA repair proteins (Elf et al., 2007; Gorman and Greene, 2008). Structural studies support a model in which target binding is coupled to a conformational change in the protein and/or substrate. Such a scenario would reconcile fast 1D diffusion with strong specific interaction of proteins with target sites (Erie et al., 1994; Kalodimos et al., 2004; Gorman and Greene, 2008). Because sliding is mainly caused by electrostatic interactions, the sliding properties of a protein are determined by the distribution of (mainly positively) charged residues on the protein surface that interact with DNA. A protein that displays 1D diffusion is thought to track the major groove of DNA, thus spiraling around the helix as it diffuses along the DNA (Gorman and Greene, 2008). Another possibility is that proteins diffuse freely on the DNA surface (termed 2D diffusion). Experiments revealed that such a mechanism is used by some DNA-binding proteins and could allow a protein to bypass obstacles such as nucleosomes (Kampmann, 2004). Therefore, 2D diffusion of proteins might be more relevant in a chromatin context. At physiological ionic strength, proteins only diffuse along the DNA over distances of ∼50 bp, as the ionic strength reduces electrostatic DNA–protein interactions and thus 1D diffusion (Gowers et al., 2005; Gorman and Greene, 2008). This suggests that 1D and 2D diffusion are not the main mode of translocation of DNA-binding proteins.
Although 3D diffusion alone is not sufficient to explain the high rates (>108 M−1s−1) at which some proteins appear to associate with specific target sites on the genome, many other proteins appear to have much lower association rate constants (Gabdoulline and Wade, 2002), which are consistent with target finding by 3D diffusion. It should be noted that the physiological relevance of 1D or 2D diffusion in vivo is currently unclear. Most single-molecule studies that address this issue have analyzed protein binding to naked DNA in vitro (for review see Gorman and Greene, 2008). In contrast, a recent study in living bacteria supports 1D diffusion by the lactose repressor in vivo (Elf et al., 2007). Moreover, a p53 mutant deficient in 1D diffusion in vitro was unable to bind promoters in vivo (McKinney et al., 2004), which suggests that 1D diffusion might be relevant in mammalian cells. In summary, the contribution of 3D, 2D, and 1D diffusion to finding a target site in vivo will depend on the biophysical properties of the protein, the number and nature of the binding sites, and the concentration of the binding protein.
Assembly of multi-protein complexes that control genome function
Finding a target site by a site-specific recognition protein is only the starting point. After recognition of a DNA lesion or a promoter, a multiprotein complex is assembled that, for instance, carries out transcription or DNA repair. Remarkably little is known of how these multiprotein complexes are formed and how they function inside the cell. Recent pioneering studies have used an interdisciplinary systems biology approach to unveil kinetic properties of such complex systems in vivo. In this mini-review, we give an overview of the initial attempts to understand the kinetic properties of multiprotein complexes that carry out DNA repair and transcription.
Assembly of DNA repair complexes
To protect the integrity of the genome, multiple DNA repair mechanisms have evolved to deal with specific DNA injuries (Hoeijmakers, 2001; Essers et al., 2006). For example, nucleotide excision repair (NER) removes helix-distorting injuries that affect one of the DNA strands, whereas homologous recombination (HR) and nonhomologous end joining repair double strand breaks (DSBs). NER involves the assembly of repair complexes containing up to 10 protein factors, which cooperate in space and time. In the absence of damage, core NER proteins xeroderma pigmentosum group A protein (XPA), replication protein A, XPG, and ERCC1/XPF display rapid diffusion rates, which are mainly dominated by free diffusion of the individual repair components (Houtsmuller et al., 1999; Essers et al., 2006; Hoogstraten et al., 2008). Although transient interactions between NER factors may occur in the absence of damage, their different mobilities exclude stable interactions between NER proteins. Concurringly, these proteins do not display high affinity for DNA lesions or to each other; rather, they only bind to repair intermediates (Volker et al., 2001). In contrast to other NER factors, the damage recognition protein XPC moves much slower inside the nucleus as it continuously binds nonspecifically to chromatin with a residence time of ∼0.3 s (Hoogstraten et al., 2008). At any moment, about half of the XPC molecules are freely mobile and half are bound. If damaged DNA sites are present, XPC occasionally encounters a helix-distorting lesion to which it binds more stably (t1/2 = 25 s; Hoogstraten et al., 2008). Binding of XPC to a DNA lesion triggers assembly of the NER complex from freely diffusing proteins. NER proteins XPG, transcription factor II H (TFIIH), and ERCC1/XPF rapidly bind to and dissociate from (t1/2 ≈ 1 min) repair complexes, whereas XPA exchanges somewhat slower (t1/2 ≈ 2 min; Houtsmuller et al., 1999; Essers et al., 2006; Luijsterburg et al., 2007).
Rapid association–dissociation kinetics make the formation of a functional multiprotein DNA repair complex that contains the correct set of proteins to carry out a specific chromatin-associated process a low-probability event. As a result, a large fraction of the protein complexes will contain an incomplete set of repair factors. Consequently, only a small fraction of the complexes will be enzymatically active, containing the necessary components to trigger a specific chromatin-associated event, such as unwinding or incising the DNA. In this scenario, progression of the NER process would then be achieved by the sequential formation of different repair intermediates, each serving as a substrate for the assembly of subsequent repair factors necessary to carry out the next chromatin-associated event. Effectively, this means that the NER process is split up in several subprocesses (recognition, unwinding, incision, and resynthesis) that are performed sequentially. Kinetic modeling of the NER system suggests that most of the repair time by far is spent on the formation of functional complexes, which is an inherent property of large multiprotein complexes (unpublished data). Consequently, modulating the efficiency of complex assembly may provide a logical mechanism to regulate the rate of chromatin-associated processes (Gorski et al., 2008). In addition, several cycles of building up and tearing down protein complexes before an actual chromatin-associated event is catalyzed might provide a form of quality control by a mechanism known as kinetic proofreading (see “Kinetic proofreading”; Qian, 2008).
Repair of DSBs by HR involves assembly of a protein complex that is initiated by binding of the Mre11–Rad50–Nbs1 complex to the damaged site, and subsequent formation of a Rad51 nucleoprotein filament aided by binding of additional repair proteins such as Rad54, Rad52, and replication protein A (San Filippo et al., 2008). Live-cell imaging revealed that Rad51 filaments are highly stable and that the residence time of Rad51 proteins is in the order of hours. The residence times of Rad52 (∼1 min) and of Rad54 (∼10 s) on chromatin are much shorter (Essers et al., 2002). Thus, Rad51 seems to be a strongly bound component during HR that possibly serves as a binding platform for several other repair proteins that exchange rapidly. DSBs can also be repaired by nonhomologous end joining in the absence of a sister chromatid (e.g., in G1), which involves the ring-shaped Ku70/80 dimer and the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs). The Ku complex recruits LigIV via XRCC4 to broken DNA ends, which in turn joins the broken ends (Mari et al., 2006). Binding of the Ku complex to broken DNA ends is reversible, and the exchange between bound and soluble pools occurs on scale of seconds (∼40 s; Mari et al., 2006). Similarly, DNA-PKcs exchanges between soluble and DNA bound pools within 1 min. When DNA-PKcs cannot be phosphorylated or perform its kinase activity, a much larger fraction of the DNA-PKcs pool is bound for longer times (Mari et al., 2006; Uematsu et al., 2007). These studies highlight the importance of posttranslational modification of repair proteins, and show that, in case of DNA-PKcs, phosphorylation decreases the residence time of this repair protein at the repair site.
Collectively, these live-cell imaging studies are in agreement with a model in which most repair factors assemble at sites of DNA damage from freely diffusing components and form short-lived complexes on damaged chromatin (Houtsmuller et al., 1999; Hoogstraten et al., 2002; Essers et al., 2006; Mari et al., 2006). Affinity differences between specific and nonspecific sites (both for protein–protein and for protein–DNA interactions) are often relatively small because specific and nonspecific sites are often structurally similar. Therefore, it is likely that if the affinity of a protein for its substrate would increase, this would also result in higher affinity for the structural analogue, which is not a true substrate and is often present in large excess. In this light, it is likely that the affinity of repair proteins is tuned such that transient binding is sufficient to assemble complexes at specific (in this case damaged) sites with an acceptable rate, whereas at the same time, the low affinity ensures that complex assembly at nonspecific sites is limited. Indeed, a recent study showed that tethering of DSB repair proteins Mre11, Rad50, or Nbs1 to chromatin, thus artificially increasing their affinity for DNA, elicits a DNA damage response at undamaged sites that includes activation of Chk1/Chk2 and cell cycle arrest (Soutoglou and Misteli, 2008). This indicates that binding of a single repair protein with high affinity to nonspecific sites (i.e., undamaged DNA) is sufficient to trigger a cellular DNA damage response.
Assembly of transcription initiation and elongation complexes
Transcription involves assembly of a multiprotein transcription initiation complex on the chromatin fiber. Various live-cell imaging studies in combination with kinetic modeling have unveiled that, like DNA repair proteins, many transcription factors, coactivators, and RNA polymerases (RNA pol) bind rapidly and reversibly to target sites (Dundr et al., 2002; Hager et al., 2006; Darzacq et al., 2007; Gorski et al., 2008). Occasionally, these factors assemble in a way that leads to transcription initiation and the production of RNA (Darzacq et al., 2007). Several transcription factors and coactivators (e.g., GR, GRIP-1, p53, TFIIB, and TFIIH) diffuse rapidly inside the nucleus. At any given time, 15–25% of these proteins are bound for 3–5 s to chromatin (Hoogstraten et al., 2002; Gorski et al., 2006). Although short residence times (with a time scale of a few seconds) on chromatin are common for transcription factors, some have residence times in the order of 1 min (e.g., androgen receptor and TATA box-binding protein; Chen et al., 2002; Farla et al., 2004), and others appear to be very stable bound to promoters (Nalley et al., 2006; Yao et al., 2006). Measurements on the dynamics of RNA polymerase II molecules at sites of transcription have shown that, out of 100 RNA pol II molecules that interact with a gene, 84 will do so very transiently, with a residence time of a few seconds, whereas 15 molecules are bound a little longer (about a minute), and only 1 molecule will engage in elongation producing an mRNA molecule (Darzacq et al., 2007). These live-cell studies combined with kinetic modeling revealed that the majority of RNA pol II–promoter interactions are not productive (Darzacq et al., 2007). This onset of transcription, which is inefficient at first sight, indicates that assembly of an active transcription initiation complex at a promoter is slow, similar to complex assembly during DNA repair. It is likely that promoter DNA exists in several functional states (e.g., closed, unwound, containing specific posttranslational modifications of, for instance, histones), which are produced as transcription initiation progresses (Hager et al., 2006). In analogy to DNA repair, each of these states may serve as the substrate for a specific set of transcription factors and coregulators. The stepwise and sequential transitions through these different of states may help to drive the process to completion (Fig. 1). More in vivo studies are necessary to decide whether the kinetic properties of transcription complexes can be generalized.
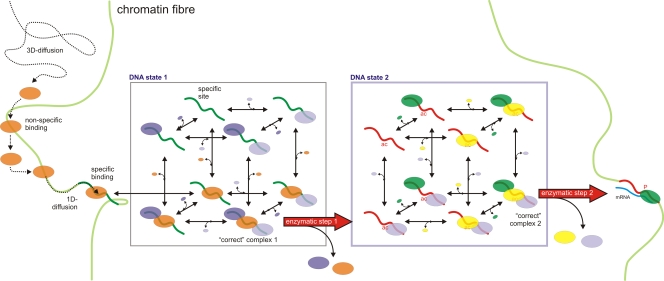
Figure 1.
Model for binding of a site-specific protein to a target site, and subsequent assembly of a multiprotein complex on that site. A site-specific protein (orange oval) diffuses rapidly inside the nucleus and binds nonspecifically to chromatin (represented by the light green line), dissociates and subsequently rebinds. Alternatively, it moves along chromatin by 1D diffusion and encounters a specific site (dark green) to which the protein binds more stably. The orange protein mediates the assembly of a complex consisting of two additional proteins (light purple and dark purple). Binding of all these proteins is stochastic, and eight different assembly states can be formed on the specific site consisting of one or a combination of the three proteins, or the site can be devoid of any protein. Once the “correct” complex 1, containing all three proteins, is formed, the specific site is modified (e.g., acetylated shown in red), resulting in dissociation of the orange and dark purple protein, while the light purple protein remains bound (because it has affinity for the altered state, whereas the other proteins do not). The red arrow reflects an enzymatic step (in this case acetylation). This altered state is the substrate for a new set of proteins (the green and yellow protein and the light purple protein from the last box) to bind to. Complex assembly is again stochastic, and eight different assembly states can be formed. Assembly of the “correct” complex 2, containing all three proteins in the second box, results in an enzymatic step that produces mRNA and subsequent dissociation of the yellow and light purple proteins. The probability of the overall reaction (i.e., binding of five different proteins to the same site) is increased by splitting the reactions in assembly of complex 1 and complex 2 separated by an enzymatic reaction. The enzymatic step drives the reaction forward. Completion of processes involving more proteins can be kinetically driven by multiple enzymatic reactions.
Transcription of ribosomal RNA (rRNA) genes by the RNA pol I system is also a highly dynamic process (Dundr et al., 2002). The majority of preinitiation factors (UBF1 and -2) and transcription factors (TAFI48) rapidly exchange within 5 s at rRNA genes, whereas TFIIH exchange in nucleoli is considerably slower (∼25 s; Hoogstraten et al., 2002). Although polymerases have often been described as preformed complexes (Seither et al., 1998), results from live-cell imaging experiments strongly suggest that pol I is assembled from its individual components at the site of its activity, where the subunits are rapidly exchanged (∼5 s; Dundr et al., 2002; Gorski et al., 2008). Similar to RNA pol II transcription, only 1–3% of the RNA pol I binding events result in elongation, which is inefficient in terms of association/dissociation steps needed to initiate transcription. However, with several dozens of transcription factors binding events per second, such an “inefficient” mechanism still results in ∼5,000 ribosomal transcripts per minute, sustaining ribosomal production rates that ensure cell viability (Dundr et al., 2002). Interestingly, RNA pol I subunits exchange on rRNA promoters approximately four times slower in S phase, during which the rRNA transcriptional output is much higher than in G1. This suggests that longer retention times of individual RNA pol I subunits to promoters are directly related to a more efficient formation of transcriptionally active RNA pol I complexes (Gorski et al., 2008). A dominant-negative mutant of one of the initiation factors lowered the retention time of pol I subunits, leading to a decreased transcriptional output. Thus, modulation of RNA pol I assembly/disassembly kinetics may be an elegant mechanism to control the transcriptional output of rRNA genes (Gorski et al., 2008). In conclusion, these studies indicate that the formation of active transcription initiation complexes is the slowest step and involves many binding and dissociation events of the individual proteins, similar to the formation of repair complexes. Additionally, the apparently inefficient complex assembly during transcription initiation may serve as a regulatory mechanism that reduces the incorporation of wrong (i.e., nonspecifically binding) proteins in the complex, a process named kinetic proofreading (see “Kinetic proofreading”).
Understanding assembly and functioning of genome-controlling complexes
Live-cell studies of GFP-tagged proteins involved in chromatin-associated processes generate large and complex sets of data that generally are difficult to interpret and integrate without the aid of kinetic modeling. Mathematical modeling of quantitative in vivo datasets is a powerful tool in obtaining mechanistic insight into genome-associated processes. It allows estimation of biophysical parameters of proteins and their interactions, such as diffusion coefficients and association/dissociation rate constants that cannot be determined directly in vivo. Moreover, modeling of the kinetic properties of chromatin-associated systems as a whole rather than their individual components provides detailed insight into the properties of such systems (unpublished data; Dundr et al., 2002; Politi et al., 2005; Darzacq et al., 2007). Advanced mathematical tools are available to describe the kinetics of protein diffusion, binding, and reaction processes, and to determine the model parameters from experimental data (Phair and Misteli, 2001). Reaction–diffusion models have been developed that allow the determination of diffusion coefficients as well as protein binding and dissociation rate constants from FRAP, fluorescence loss in photobleaching, and FCS data based on differential equations or Monte Carlo simulations (Zotter et al., 2006; Wachsmuth et al., 2008; van Royen et al., 2009). Additionally, ordinary differential equation models for protein complex formation on chromatin are being developed that quantitatively account for multiprotein complex formation processes, while considering diffusion to be very rapid on the time-scale at which association dissociation reactions take place (Dundr et al., 2002; Politi et al., 2005; Darzacq et al., 2007; Gorski et al., 2008). Pioneering live-cell imaging studies combined with kinetic modeling revealed that proteins only occasionally form an active multiprotein protein complex on chromatin (unpublished data; Dundr et al., 2002; Darzacq et al., 2007; Gorski et al., 2008). These initial attempts to describe multiprotein complex assembly quantitatively suggest that a low probability to assemble an enzymatically active protein complex may be a shared characteristic of genome-associated processes.
These findings suggest a scenario in which proteins do not bind in a fixed order to assemble a multiprotein complex. Rather, an ensemble of complexes with different protein composition is formed. A complex containing the correct set of proteins, required to catalyze a specific chromatin-associated event (e.g., unwinding, incision, DNA synthesis, or a histone modification), is formed with a low probability. This “probabilistic” view on complex assembly (depicted in Fig. 1) is different from the often-held view that complex assembly on chromatin occurs through an ordered mechanism in which each protein is incorporated in a stepwise fashion into a chromatin-bound complex. Future studies should be directed at examining whether the mechanisms outlined here for DNA repair and transcription can be extended to other biological systems in the nucleus.
Kinetic proofreading
The interplay of reversible protein complex assembly and often ATP-driven irreversible steps catalyzed by multiprotein complexes can increase the specificity of genome-controlling processes beyond the binding specificity of its individual components by a mechanism known as kinetic proofreading (Hopfield, 1974). In such a mechanism, a protein–DNA complex is taken through a series of high-energy intermediate states, selecting for components with a relatively low dissociation rate, thus leading to a more faithful discrimination between specific binding to true substrates and nonspecific binding to nonsubstrates. For instance, the NER protein XPC has a remarkably small difference in affinity between undamaged sites (i.e., the wrong substrate) and DNA lesions (the correct substrate). Nonetheless, the NER system as a whole discriminates with high specificity between lesions and undamaged DNA. Kinetic modeling shows that kinetic proofreading can dramatically increase the specificity of a system for the correct substrate beyond the ability of the recognition protein to discriminate correct substrates from false ones (Hopfield, 1974; Qian, 2008). Kinetic proofreading of damage recognition by the NER system may involve ATP hydrolysis by the helicase TFIIH and several cycles of association/dissociation of XPC and TFIIH (Giglia-Mari et al., 2006). Importantly, many genome-associated processes involve enzymatic reactions including ATP hydrolysis, unwinding, incision, ligation, and posttranslational modification of proteins (e.g., histones), which can drive progression of genome-associated processes (see Fig. 1).
In conclusion, live-cell imaging combined with kinetic modeling seems an essential tool for studying the choreography of proteins that make up multiprotein systems on the chromatin fiber. This systems biology approach, pioneered by the studies outlined in this mini-review, will provide detailed and comprehensive insight into the orchestration of genome functions in vivo.
Acknowledgments
This work was in part supported by two grants from the Dutch research council: a ZonMW grant (912-03-012) and a Netherlands Organisation for Scientific Research Rubicon grant (2007/09198/ALW/825.07.042 to M.S. Luijsterburg).
Footnotes
Abbreviations used in this paper: DSB, double strand break; HR, homologous recombination; NER, nucleotide excision repair; rRNA, ribosomal RNA; TFII, transcription factor II; XP, xeroderma pigmentosum protein.
References
- Berg O.G., Winter R.B., von Hippel P.H. 1981. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory.Biochemistry. 20:6929–6948 [DOI] [PubMed] [Google Scholar]
- Chen D., Hinkley C.S., Henry R.W., Huang S. 2002. TBP dynamics in living human cells: constitutive association of TBP with mitotic chromosomes.Mol. Biol. Cell. 13:276–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X., Shav-Tal Y., de Turris V., Brody Y., Shenoy S.M., Phair R.D., Singer R.H. 2007. In vivo dynamics of RNA polymerase II transcription.Nat. Struct. Mol. Biol. 14:796–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Hoffmann-Rohrer U., Hu Q., Grummt I., Rothblum L.I., Phair R.D., Misteli T. 2002. A kinetic framework for a mammalian RNA polymerase in vivo.Science. 298:1623–1626 [DOI] [PubMed] [Google Scholar]
- Elf J., Li G.W., Xie X.S. 2007. Probing transcription factor dynamics at the single-molecule level in a living cell.Science. 316:1191–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erie D.A., Yang G., Schultz H.C., Bustamante C. 1994. DNA bending by Cro protein in specific and nonspecific complexes: implications for protein site recognition and specificity.Science. 266:1562–1566 [DOI] [PubMed] [Google Scholar]
- Essers J., Houtsmuller A.B., van Veelen L., Paulusma C., Nigg A.L., Pastink A., Vermeulen W., Hoeijmakers J.H., Kanaar R. 2002. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage.EMBO J. 21:2030–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J., Vermeulen W., Houtsmuller A.B. 2006. DNA damage repair: anytime, anywhere? Curr. Opin. Cell Biol. 18:240–246 [DOI] [PubMed] [Google Scholar]
- Farla P., Hersmus R., Geverts B., Mari P.O., Nigg A.L., Dubbink H.J., Trapman J., Houtsmuller A.B. 2004. The androgen receptor ligand-binding domain stabilizes DNA binding in living cells.J. Struct. Biol. 147:50–61 [DOI] [PubMed] [Google Scholar]
- Gabdoulline R.R., Wade R.C. 2002. Biomolecular diffusional association.Curr. Opin. Struct. Biol. 12:204–213 [DOI] [PubMed] [Google Scholar]
- Giglia-Mari G., Miquel C., Theil A.F., Mari P.O., Hoogstraten D., Ng J.M., Dinant C., Hoeijmakers J.H., Vermeulen W. 2006. Dynamic interaction of TTDA with TFIIH is stabilized by nucleotide excision repair in living cells.PLoS Biol. 4:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J., Greene E.C. 2008. Visualizing one-dimensional diffusion of proteins along DNA.Nat. Struct. Mol. Biol. 15:768–774 [DOI] [PubMed] [Google Scholar]
- Gorski S.A., Dundr M., Misteli T. 2006. The road much traveled: trafficking in the cell nucleus.Curr. Opin. Cell Biol. 18:284–290 [DOI] [PubMed] [Google Scholar]
- Gorski S.A., Snyder S.K., John S., Grummt I., Misteli T. 2008. Modulation of RNA polymerase assembly dynamics in transcriptional regulation.Mol. Cell. 30:486–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowers D.M., Wilson G.G., Halford S.E. 2005. Measurement of the contributions of 1D and 3D pathways to the translocation of a protein along DNA.Proc. Natl. Acad. Sci. USA. 102:15883–15888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager G.L., Elbi C., Johnson T.A., Voss T., Nagaich A.K., Schiltz R.L., Qiu Y., John S. 2006. Chromatin dynamics and the evolution of alternate promoter states.Chromosome Res. 14:107–116 [DOI] [PubMed] [Google Scholar]
- Halford S.E., Marko J.F. 2004. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 32:3040–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers J.H. 2001. Genome maintenance mechanisms for preventing cancer.Nature. 411:366–374 [DOI] [PubMed] [Google Scholar]
- Hoogstraten D., Nigg A.L., Heath H., Mullenders L.H., van Driel R., Hoeijmakers J.H., Vermeulen W., Houtsmuller A.B. 2002. Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo.Mol. Cell. 10:1163–1174 [DOI] [PubMed] [Google Scholar]
- Hoogstraten D., Bergink S., Verbiest V.H., Luijsterburg M.S., Geverts B., Raams A., Dinant C., Hoeijmakers J.H., Vermeulen W., Houtsmuller A.B. 2008. Versatile DNA damage detection by the global genome nucleotide excision repair protein XPC.J. Cell Sci. 121:2850–2859 [DOI] [PubMed] [Google Scholar]
- Hopfield J.J. 1974. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity.Proc. Natl. Acad. Sci. USA. 71:4135–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtsmuller A.B., Vermeulen W. 2001. Macromolecular dynamics in living cell nuclei revealed by fluorescence redistribution after photobleaching.Histochem. Cell Biol. 115:13–21 [DOI] [PubMed] [Google Scholar]
- Houtsmuller A.B., Rademakers S., Nigg A.L., Hoogstraten D., Hoeijmakers J.H., Vermeulen W. 1999. Action of DNA repair endonuclease ERCC1/XPF in living cells.Science. 284:958–961 [DOI] [PubMed] [Google Scholar]
- Kalodimos C.G., Biris N., Bonvin A.M., Levandoski M.M., Guennuegues M., Boelens R., Kaptein R. 2004. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes.Science. 305:386–389 [DOI] [PubMed] [Google Scholar]
- Kampmann M. 2004. Obstacle bypass in protein motion along DNA by two-dimensional rather than one-dimensional sliding.J. Biol. Chem. 279:38715–38720 [DOI] [PubMed] [Google Scholar]
- Luijsterburg M.S., Goedhart J., Moser J., Kool H., Geverts B., Houtsmuller A.B., Mullenders L.H., Vermeulen W., van Driel R. 2007. Dynamic in vivo interaction of DDB2 E3 ubiquitin ligase with UV-damaged DNA is independent of damage-recognition protein XPC.J. Cell Sci. 120:2706–2716 [DOI] [PubMed] [Google Scholar]
- Mari P.O., Florea B.I., Persengiev S.P., Verkaik N.S., Bruggenwirth H.T., Modesti M., Giglia-Mari G., Bezstarosti K., Demmers J.A., Luider T.M., et al. 2006. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4.Proc. Natl. Acad. Sci. USA. 103:18597–18602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney K., Mattia M., Gottifredi V., Prives C. 2004. p53 linear diffusion along DNA requires its C terminus.Mol. Cell. 16:413–424 [DOI] [PubMed] [Google Scholar]
- Misteli T. 2008. Physiological importance of RNA and protein mobility in the cell nucleus.Histochem. Cell Biol. 129:5–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalley K., Johnston S.A., Kodadek T. 2006. Proteolytic turnover of the Gal4 transcription factor is not required for function in vivo.Nature. 442:1054–1057 [DOI] [PubMed] [Google Scholar]
- Phair R.D., Misteli T. 2000. High mobility of proteins in the mammalian cell nucleus.Nature. 404:604–609 [DOI] [PubMed] [Google Scholar]
- Phair R.D., Misteli T. 2001. Kinetic modelling approaches to in vivo imaging.Nat. Rev. Mol. Cell Biol. 2:898–907 [DOI] [PubMed] [Google Scholar]
- Politi A., Moné M.J., Houtsmuller A.B., Hoogstraten D., Vermeulen W., Heinrich R., van Driel R. 2005. Mathematical modeling of nucleotide excision repair reveals efficiency of sequential assembly strategies.Mol. Cell. 19:679–690 [DOI] [PubMed] [Google Scholar]
- Qian H. 2008. Cooperativity and specificity in enzyme kinetics: a single-molecule time-based perspective.Biophys. J. 95:10–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J., Sung P., Klein H. 2008. Mechanism of eukaryotic homologous recombination.Annu. Rev. Biochem. 77:229–257 [DOI] [PubMed] [Google Scholar]
- Seither P., Iben S., Grummt I. 1998. Mammalian RNA polymerase I exists as a holoenzyme with associated basal transcription factors.J. Mol. Biol. 275:43–53 [DOI] [PubMed] [Google Scholar]
- Soutoglou E., Misteli T. 2008. Activation of the cellular DNA damage response in the absence of DNA lesions.Science. 320:1507–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu N., Weterings E., Yano K., Morotomi-Yano K., Jakob B., Taucher-Scholz G., Mari P.O., van Gent D.C., Chen B.P., Chen D.J. 2007. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks.J. Cell Biol. 177:219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Royen M.E., Farla P., Mattern K.A., Geverts B., Trapman J., Houtsmuller A.B. 2009. Fluorescence recovery after photobleaching (FRAP) to study nuclear protein dynamics in living cells.Methods Mol. Biol. 464:363–385 [DOI] [PubMed] [Google Scholar]
- Volker M., Moné M.J., Karmakar P., van Hoffen A., Schul W., Vermeulen W., Hoeijmakers J.H., van Driel R., van Zeeland A.A., Mullenders L.H. 2001. Sequential assembly of the nucleotide excision repair factors in vivo.Mol. Cell. 8:213–224 [DOI] [PubMed] [Google Scholar]
- Wachsmuth M., Caudron-Herger M., Rippe K. 2008. Genome organization: balancing stability and plasticity.Biochim. Biophys. Acta. 1783:2061–2079 [DOI] [PubMed] [Google Scholar]
- Yao J., Munson K.M., Webb W.W., Lis J.T. 2006. Dynamics of heat shock factor association with native gene loci in living cells.Nature. 442:1050–1053 [DOI] [PubMed] [Google Scholar]
- Zotter A., Luijsterburg M.S., Warmerdam D.O., Ibrahim S., Nigg A., van Cappellen W.A., Hoeijmakers J.H., van Driel R., Vermeulen W., Houtsmuller A.B. 2006. Recruitment of the nucleotide excision repair endonuclease XPG to sites of UV-induced DNA damage depends on functional TFIIH.Mol. Cell. Biol. 26:8868–8879 [DOI] [PMC free article] [PubMed] [Google Scholar]