Abstract
We previously demonstrated that the primary cilium coordinates platelet-derived growth factor (PDGF) receptor (PDGFR) α–mediated migration in growth-arrested fibroblasts. In this study, we investigate the functional relationship between ciliary PDGFR-α and the Na+/H+ exchanger NHE1 in directional cell migration. NHE1 messenger RNA and protein levels are up-regulated in NIH3T3 cells and mouse embryonic fibroblasts (MEFs) during growth arrest, which is concomitant with cilium formation. NHE1 up-regulation is unaffected in Tg737orpk MEFs, which have no or very short primary cilia. In growth-arrested NIH3T3 cells, NHE1 is activated by the specific PDGFR-α ligand PDGF-AA. In wound-healing assays on growth-arrested NIH3T3 cells and wild-type MEFs, NHE1 inhibition by 5′-(N-ethyl-N-isopropyl) amiloride potently reduces PDGF-AA–mediated directional migration. These effects are strongly attenuated in interphase NIH3T3 cells, which are devoid of primary cilia, and in Tg737orpk MEFs. PDGF-AA failed to stimulate migration in NHE1-null fibroblasts. In conclusion, stimulation of directional migration in response to ciliary PDGFR-α signals is specifically dependent on NHE1 activity, indicating that NHE1 activation is a critical event in the physiological response to PDGFR-α stimulation.
Introduction
The process of cell migration is pivotal to many fundamental physiological processes, including embryonic and fetal development, immune responses, wound healing, and tissue homeostasis. Consequently, altered function of components of the migratory machinery is associated with severe pathophysiological conditions, including developmental defects, chronic inflammatory diseases, and cancer metastasis (Ridley et al., 2003; Vicente-Manzanares et al., 2005; Schwab et al., 2007).
Cell migration is a complex multistep process involving extensive cytoskeletal rearrangement and the concerted action of multiple ion transport proteins and membrane receptors. These processes result in the formation of lamellipodia and other protrusive structures and in local changes of cell–matrix interactions and cell volume, which collectively enable the cell to move forward (Ridley et al., 2003; Vicente-Manzanares et al., 2005; Schwab et al., 2007). Directional migration relies on the ability of the cell to sense and react to chemosensory stimuli (Wu, 2005) such as PDGF (Heldin and Westermark, 1999; Jechlinger et al., 2006). Notably, exposure to PDGF activates the ubiquitous plasma membrane Na+/H+ exchanger NHE1 (Cassel et al., 1983; Ma et al., 1994; Yan et al., 2001), which is a central player in the regulation of proliferation, survival, and migration (Putney et al., 2002; Pedersen, 2006; Stock and Schwab, 2006). We and others have demonstrated an essential role for NHE1 in cell migration and invasion in many cell types, including a variety of cancer cell types (Lagana et al., 2000; Denker and Barber, 2002; Stock et al., 2005, 2007; Cardone et al., 2005b; Stuwe et al., 2007; Hayashi et al., 2008). In migrating cells such as fibroblasts or epithelial cells, NHE1 localizes predominantly to the leading edge (Denker et al., 2000; Lagana et al., 2000; Cardone et al., 2005a; Stock et al., 2007), and findings in fibroblasts indicate that NHE1 activity and attachment to the ERM (ezrin/radixin/moesin) family of plasma membrane F-actin linker proteins are required for cell polarity and migration (Denker and Barber, 2002). This is of substantial interest in the context of human cancers, in which metastatic capacity is linked to increased expression of NHE1 (Cardone et al., 2005b). However, the possible link between PDGF signaling and NHE1 in migration/chemotaxis has not previously been addressed.
PDGF exists as homo- or heterodimers of PDGF-A, -B, -C, and -D chains. Although PDGF-BB activates both homo- and heterodimers of PDGF receptor (PDGFR) α and PDGFR-β, PDGF-AA is specific for the PDGFR-α homodimer (Heldin and Westermark, 1999). Both PDGF-AA and PDGF-BB have been shown to stimulate migration in various cell types (Shure et al., 1992; Hayashi et al., 1995; Yu et al., 2001). Recently, we showed that PDGFR-α signaling is coordinated by the primary cilium in mammalian fibroblasts (Schneider et al., 2005). PDGFR-α, which is encoded by a growth arrest–specific (gas) gene (Lih et al., 1996), is targeted to the primary cilium, where ligand-dependent activation of the receptor and of the ERK1/2 and protein kinase B/Akt pathways is initiated (Schneider et al., 2005; unpublished data). The primary cilium is an essential sensory organelle in most growth-arrested mammalian cells and coordinates a series of critical signal transduction pathways in development and tissue homeostasis, which, in addition to PDGFR-α signaling, include the Hedgehog and Wnt pathways (Christensen et al., 2007). In NIH3T3 fibroblasts, we previously showed that primary cilia are virtually absent in nonconfluent cycling interphase cells but grow on almost all confluent quiescent growth-arrested cells within 24 h of serum starvation (Schneider et al., 2005). Similarly, serum-starved wild-type (WT) mouse embryonic fibroblasts (MEFs) grow normal primary cilia that coordinate PDGFR-α–mediated signal transduction and directional cell migration, whereas defects in assembly of the primary cilium in Tg737orpk MEFs block these events (Schneider et al., 2005; unpublished data). These results indicate that the primary cilium is part of the positioning machinery that coordinates cell polarity, which is essential for wound healing and developmental processes (Schneider and Haugh, 2006).
In this study, we investigated the interactions between the primary cilium and NHE1 in PDGFR-α–dependent directional cell migration. We demonstrate that NHE1 mRNA and protein levels are increased during growth arrest independent of the formation of the primary cilium. In contrast, NHE1 activity is strongly attenuated by serum deprivation of NIH3T3 fibroblasts, which is coincident with the time course of primary cilia appearance (Schneider et al., 2005). NHE1 activity is partly restored by stimulation with PDGF-AA, which also stimulates directional cell migration (unpublished data). Inhibition of NHE1 activity by 5′-(N-ethyl-N-isopropyl) amiloride (EIPA) potently inhibits PDGF-AA–mediated directional cell migration in growth-arrested WT MEFs in which primary cilia are present, whereas migration in growth-arrested Tg737orpk mutant MEFs is unaffected by either PDGF-AA or EIPA. Furthermore, PDGF-AA fails to stimulate migration in NHE1-deficient fibroblasts. We conclude that in fibroblasts, PDGFR-α signaling initiated in the primary cilium controls directional migration responses via up-regulation of NHE1 activity, indicating that NHE1 activation is a critical event in the physiological response to activation of PDGFR-α.
Results
Expression and localization of NHE1 in NIH3T3 fibroblasts and in WT and Tg737orpk MEFs
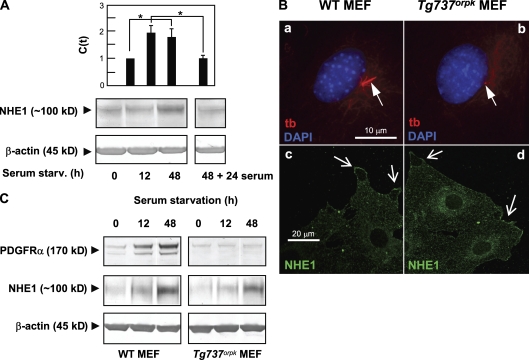
We first asked whether NHE1 is up-regulated during growth arrest in NIH3T3 fibroblasts (i.e., concomitant with formation of the primary cilium). NIH3T3 cells were serum starved for 0, 12, and 48 h, and mRNA and protein levels were measured by quantitative PCR using specific NHE1 TaqMan probes and Western blotting, respectively. As shown in Fig. 1 A (top), the NHE1 mRNA level was up-regulated about twofold after both 12 and 48 h of serum starvation. The protein level increased more slowly and was nearly tripled after 48 h of serum starvation (Fig. 1 A, bottom). NHE1 up-regulation was reversible upon reentry into the cell cycle (Fig. 1 A).
Figure 1.
Expression and localization of NHE1. (A) Quantitative data from real-time PCR analysis (top) and Western blotting (bottom) show up-regulation of both mRNA and protein levels of NHE1 during growth arrest in NIH3T3 cells exposed to serum starvation for 0 (interphase cells), 12, and 48 h. The up-regulation was reversible, as levels of both mRNA and protein were reduced to interphase levels after the readdition of serum for 24 h. The data shown represent 3–11 independent experiments for each condition. Error bars represent the SEM value of the relative C(t) value from each condition (n = 3–5). Data were analyzed using parametric or nonparametric ANOVA, and the level of significance is shown (*, P < 0.05). C(t), threshold cycle. (B, a and b) Epifluorescence microscopy analysis of primary cilia (red; anti–acetylated α-tubulin [tb]; arrows) in growth-arrested WT and Tg737orpk MEFs (serum starved for 48 h). (c and d) Confocal imaging of NHE1 localization (green) in growth-arrested WT (c) and Tg737orpk (d) MEFs. Arrows indicate lamellipodial regions of cells. Note that intracellular NHE1 localization is also seen. (C) Western blot analysis (n = 3) of PDGFR-α and NHE1 protein levels in WT and Tg737orpk MEFs after 0, 12, and 48 h of serum starvation. β-Actin was used as a loading control. As seen, growth arrest leads to up-regulation of both PDGFR-α and NHE1 protein levels in both WT and Tg737orpk MEFs.
To test whether NHE1 up-regulation during growth arrest depends on the formation of the primary cilium, we performed similar experiments in WT MEFs, which, as previously reported, grow cilia at quiescence, and in Tg737orpk mutant MEFs, which form no or only very short primary cilia (Fig. 1 B, a and b; Schneider et al., 2005). Similar to the findings in NIH3T3 cells, the NHE1 protein level was nearly tripled at time 48 h after serum starvation in both WT and Tg737orpk MEFs (Fig. 1 C). Collectively, these data indicate that NHE1 is a gas protein but that, in contrast to the up-regulation of PDGFR-α during growth arrest (Schneider et al., 2005), up-regulation of NHE1 is independent of assembly of the primary cilium. In growth-arrested WT and Tg737orpk MEFs, NHE1 localized to the plasma membrane, in particular to lamellipodia-like structures at the edges of the cells and intracellularly, in perinuclear membranes and in a cytoplasmic vesicular-like pattern (Fig. 1 B, c and d). A similar pattern of localization was observed in NIH3T3 cells (unpublished data). The NHE1 localization pattern was not detectably different in WT or Tg737orpk MEFs after 1 h of exposure to 50 ng/ml PDGF-AA (n = 3; unpublished data). Collectively, these results demonstrate that NHE1 expression is up-regulated concomitant with formation of the primary cilium and that neither expression nor localization of NHE1 are compromised in Tg737orpk MEFs.
NHE1 activity in NIH3T3 cells
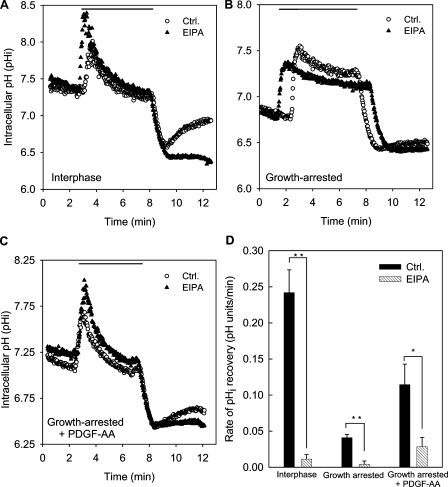
Because NHE1 activity is very tightly regulated (Pedersen, 2006), an up-regulation of NHE1 expression does not mean that NHE1 activity is also up-regulated. To address this point, NHE1 activity was determined in NIH3T3 fibroblasts using an NH4Cl prepulse, which is a classical protocol for evaluating intracellular pH (pHi) recovery after an acid load (Boron, 2004). In this protocol, the cells initially rapidly alkalinize as NH3 diffuses across the plasma membrane and combines with cytosolic H+. This is followed by a second phase of slow acidification as NH4+ enters the cells (e.g., through K+ channels) and, finally, a rapid acidification when NH3 rushes out as extracellular NH4Cl is removed (Fig. 2). In interphase cells under control conditions, the rate of pHi recovery after acidification was 0.24 ± 0.03 pH units/min (n = 3), and recovery was nearly abolished in the presence of 5 µM of the Na+/H+ exchanger inhibitor EIPA (Fig. 2, A and D). Because we previously established that NHE2, a related Na+/H+ exchanger isoform that is also partially EIPA sensitive, is not detectable in NIH3T3 fibroblasts (Rentsch et al., 2007), this indicates that NHE1 is the major effector in pHi regulation after an acid load in these cells.
Figure 2.
Roles of NHE1 and PDGFR-α in pHi regulation in NIH3T3 cells during interphase and growth arrest. (A–C) Original tracings of pHi recovery measurements in the absence and presence of 5 µM EIPA. 10 mM NH4Cl was present in the Ringer’s solution as indicated by the horizontal line. Cells grown in the presence of serum (A), cells which had been serum starved for 48 h (B), and serum-starved cells treated with 50 ng/ml PDGF-AA for 1 h before the experiment (C) are shown. It may be noted that the rate of slow acidification in the presence of NH4Cl, representing NH4+ influx via either K+ channels or other transporters with substrate affinity for NH4+ (Boron, 2004), is reduced in the growth-arrested cells in the absence but not in the presence of PDGF-AA. This indicates that these transporters are regulated during the cell cycle and also means that the total acidification after NH4Cl removal is less in the growth-arrested cells. However, this does not affect the comparison between the recovery rates in the absence and presence of EIPA. (D) Summary of the pHi recovery rates under the conditions shown. The rate of pHi recovery (in pH units/minute) was calculated from the slope of the initial linear part of the curve after NH4Cl removal. Data shown are representative traces (A–C) or mean ± SEM of three to five experiments per condition. Data were analyzed using parametric or nonparametric ANOVA, and the level of significance is shown (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Ctrl., control.
In growth-arrested NIH3T3 fibroblasts, recovery after acid loading was strongly reduced to ∼15% of that in the presence of serum, and under these conditions, inhibition of NHE1 abolished the remaining pHi recovery (Fig. 2, B and D). These results demonstrate that in spite of the up-regulation at the mRNA and protein levels, NHE1 activity is strongly reduced in growth-arrested NIH3T3 cells. Consistent with this conclusion, steady-state pHi was 7.24 ± 0.09 (n = 3) in interphase cells and 6.82 ± 0.06 (n = 4) in growth-arrested cells.
Incubation of growth-arrested NIH3T3 cells with 50 ng/ml PDGF-AA for 1 h increased steady-state pHi to 7.10 ± 0.09 (n = 8) and increased the rate of pHi recovery to ∼50% of that seen in interphase cells (Fig. 2, C and D). This effect was abolished by 5 µM EIPA, demonstrating that stimulation of ciliary PDGFR-α partially restored NHE1 activity in growth-arrested fibroblasts (Fig. 2, C and D).
Migration of fibroblasts with primary cilia: growth-arrested NIH3T3 cells and WT MEFs
Having established that NHE1 is up-regulated concomitant with formation of the primary cilium and stimulated by ciliary PDGFR-α signaling, we next addressed the possible functional relationship between ciliary PDGFR-α signaling and NHE1 activity in control of directional fibroblast migration. To this end, we performed wound-healing assays and determined two parameters: (1) the translocation of the cell during the course of the experiment, i.e., the distance covered by the cell over 5 h, and (2) the distance covered by the cell specifically in the direction perpendicular to the wound, which is a measure of the directionality of movement and allows the calculation of the velocity of movement into the wound. A summary of the translocation data for NIH3T3 cells and MEFs can be found in Table I, and details and original traces are given in Figs. 3 and 5 for growth-arrested and interphase NIH3T3 cells and in Figs. 4 and 6 for WT and Tg737orpk MEFs, respectively.
Table I.
Summary of translocation data
| Cell type | Primary cilium | PDGF-AA | EIPA | Translocation |
| µm | ||||
| NIH3T3 | No (interphase) | − | − | 60 ± 8.7 (17) |
| NIH3T3 | No (interphase) | − | + | 30 ± 3.0 (25) |
| NIH3T3 | No (interphase) | + | − | 60 ± 5.4 (26) |
| NIH3T3 | No (interphase) | + | + | 33 ± 2.7 (25) |
| NIH3T3 | Yes | − | − | 21 ± 3.6 (23) |
| NIH3T3 | Yes | − | + | 11± 1.8 (23) |
| NIH3T3 | Yes | + | − | 45 ± 5.7 (25) |
| NIH3T3 | Yes | + | + | 13 ± 1.5 (26) |
| WT MEF | Yes | − | − | 45 ± 4.2 (53) |
| WT MEF | Yes | − | + | 27 ± 4.4 (22) |
| WT MEF | Yes | + | − | 111 ± 5.4 (23) |
| WT MEF | Yes | + | + | 15 ± 1.8 (20) |
| Tg737orpk MEF | No or very short | − | − | 75 ± 5.7 (21) |
| Tg737orpk MEF | No or very short | − | + | 63 ± 5.7 (19) |
| Tg737orpk MEF | No or very short | + | − | 87 ± 7.8 (34) |
| Tg737orpk MEF | No or very short | + | + | 57 ± 5.7 (30) |
The table summarizes the translocation data obtained for NIH3T3 cells, WT MEFs, and Tg737orpk MEFs in 5-h wound-healing assays as a function of (a) the presence or absence of a primary cilium, (b) the presence or absence of 50 ng/ml PDGF-AA, and (c) the presence or absence of 10 µM of the NHE1 inhibitor EIPA. The absence or presence of primary cilia was assessed by visual inspection and has also been previously described (Schneider et al., 2005). Translocation data are shown as mean ± SEM , with the number of cells shown in parenthesis.
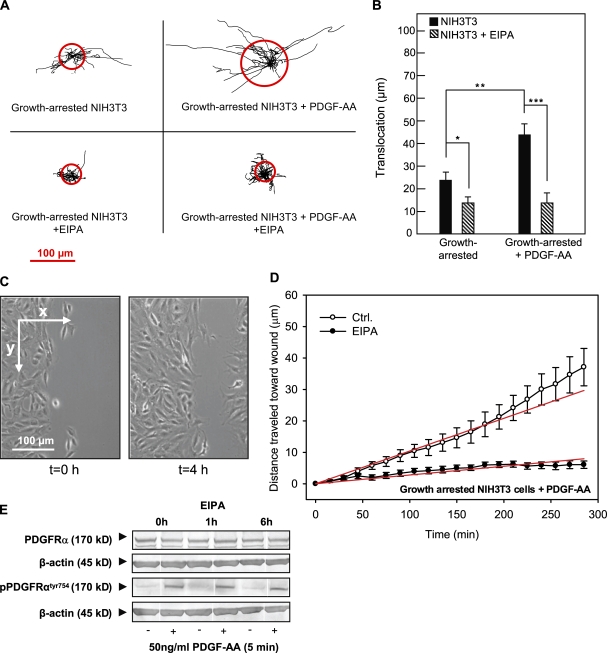
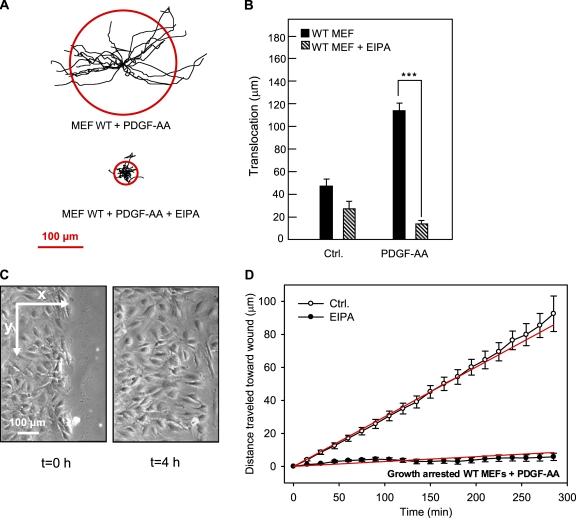
Figure 3.
Effects of PDGF-AA and EIPA in wound-healing assays on growth-arrested NIH3T3 fibroblasts. (A) Trajectories (normalized to common starting points) of growth-arrested NIH3T3 cells with and without 50 ng/ml PDGF-AA or 10 µM EIPA, as indicated. The radii of the red circles illustrate the mean translocation of the cells within a 5-h time period (n = 17–25). (B) Translocation (in micrometers and calculated as the distance between the position of the cell center at the beginning and at the end of the experiment) in the absence or presence of PDGF-AA or EIPA as indicated. (A and B) Data are individual trajectories (A) or means ± SEM (B) of 23–26 cells in three independent experiments. Data were analyzed using parametric or nonparametric ANOVA, and the level of significance is shown (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (C) Images of the left side of the wound in a confluent monolayer of growth-arrested NIH3T3 cells (serum starved for 24 h) at time 0 and 4 h after the wound was made. The arrows indicate the x and y directions of movement, which were used for estimating directional migration. (D) Velocity of migration perpendicular to the wound (x direction) in PDGF-AA–stimulated, growth-arrested NIH3T3 cells in the absence (open circles) or presence (closed circles) of EIPA. Directionality of movement was estimated by plotting the mean of the distance covered by each cell in the x direction as a function of time. The slope of this line is the velocity in x, in micrometers/minute, which is taken as a measure of directionality. In PDGF-AA–stimulated, growth-arrested cells, the velocity of movement into the wound is 0.104 ± 0.004 µm/min, and in the presence of 10 µM EIPA, this value is reduced to 0.028 ± 0.001 µm/min. Data shown are means ± SEM of 23–26 cells in three independent experiments. The velocity in the presence of EIPA is significantly different from that in the absence of EIPA (P < 0.0001). Ctrl., control. (E) NIH3T3 cells were serum starved for 24 h to induce growth arrest, exposed to 5 µM EIPA for 0, 1, or 6 h as indicated, and treated with 50 ng/ml PDGF-AA for 5 min as indicated (+). Cells were lysed, and Western blots of total and Tyr754-phosphorylated PDGFR-α were performed as described in Materials and methods. β-Actin was used as a loading control. Data shown are representative of three independent experiments.
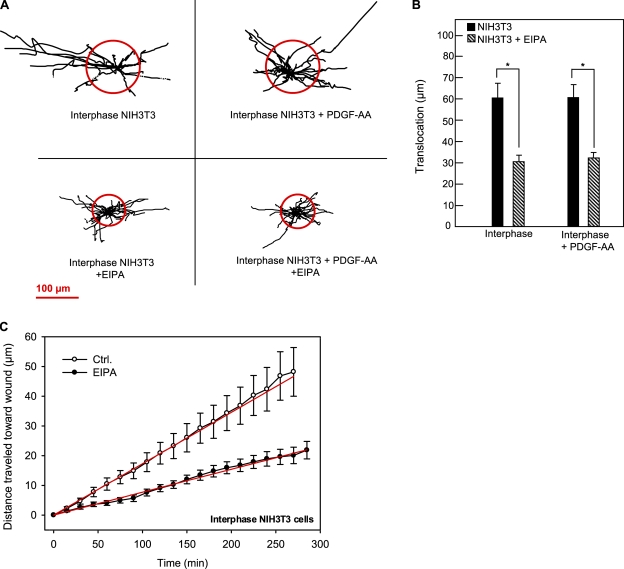
Figure 5.
Effect of EIPA in wound-healing assays on interphase NIH3T3 fibroblasts. (A) Trajectories (normalized to common starting points) of interphase NIH3T3 cells in the absence or presence of 10 µM EIPA and 50 ng/ml PDGF-AA, as indicated. The radii of the red circles illustrate the mean translocation of the cells within a 5-h time period. (B) Translocation (shown in micrometers and calculated as the distance between the position of the cell center at the beginning and at the end of the experiment) of interphase and growth-arrested NIH3T3 cells with or without EIPA, as indicated. (A and B) Data are individual trajectories (A) or means ± SEM (B) of 17–26 cells in three to four independent experiments. Data were analyzed using parametric or nonparametric ANOVA, and the level of significance is shown (*, P < 0.05). (C) Velocity of migration perpendicular to the wound in interphase NIH3T3 cells in the absence (open circles) or presence (closed circles) of 10 µM EIPA obtained as described in the legend for Fig. 3. Data shown are means ± SEM of 17–26 cells in three to four independent experiments. The velocity in the presence of EIPA is significantly different from that in the absence of EIPA (P < 0.0001). Ctrl., control.
Figure 4.
Effects of PDGF-AA and EIPA in wound-healing assays on growth-arrested WT MEFs. (A) Trajectories (normalized to common starting points) of growth-arrested WT MEFs in the presence of 50 ng/ml PDGF-AA and in the absence or presence of 10 µM EIPA, as indicated. The radii of the red circles illustrate the mean translocation of the cells within a 5-h time period. (B) Translocation (in micrometers) of growth-arrested WT MEFs in the absence or presence of 50 ng/ml PDGF-AA and 10 µM EIPA, as indicated, calculated as described in the legend for Fig. 3. (A and B) Data shown are individual trajectories (A) or means ± SEM (B) of 20–53 cells in three to seven independent experiments. Data were analyzed using parametric or nonparametric ANOVA, and the level of significance is shown (***, P < 0.001). (C) Images of the left side of the wound in a confluent monolayer of growth-arrested WT MEFs at time 0 and 4 h after the wound was made. The arrows indicate the x and y directions of movement, which were used for estimating directional migration. (D) Velocity of migration perpendicular to the wound (x direction) in PDGF-AA–stimulated, growth-arrested WT MEFs in the absence (open circles) or presence (closed circles) of EIPA. Directionality of movement was estimated by plotting the mean of the distance covered by each cell in the x direction as a function of time. The slope of this line yields the velocity in the x direction (in micrometers/minute), which is a measure of directionality. The velocity in growth-arrested, PDGF-AA–treated WT MEFs was 0.301 ± 0.003 µm/min in the absence and 0.030 ± 0.004 µm/min in the presence of 10 µM EIPA. Data shown are means ± SEM of 20–53 cells in three to seven independent experiments. The velocity in the presence of EIPA is significantly different from that in the absence of EIPA (P < 0.0001). Ctrl., control.
Figure 6.
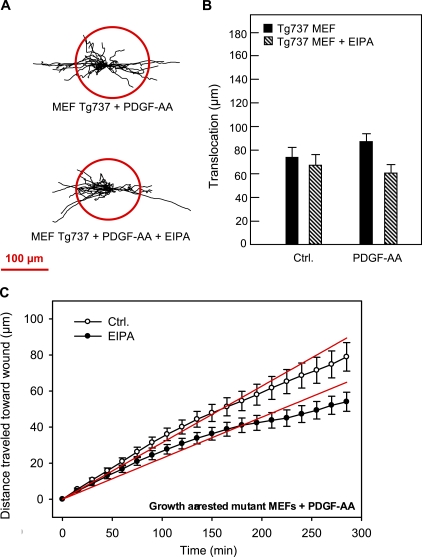
Effects of PDGF-AA and EIPA on translocation of growth-arrested Tg737orpk MEFs in wound-healing assays. (A) Trajectories (normalized to common starting points) of growth-arrested Tg737orpk MEFs in the presence of 50 ng/ml PDGF-AA and in the absence or presence of 10 µM EIPA, as indicated. The radii of the red circles illustrate the mean translocation of the cells within a 5-h time period. (B) Translocation (in micrometers) of growth-arrested Tg737orpk MEFs in the absence or presence of 50 ng/ml PDGF-AA and 10 µM EIPA, as indicated, calculated as described in the legend for Fig. 3. (A and B) Data shown are individual trajectories (A) or means ± SEM (B) of 19–34 cells in two to four independent experiments. (C) Velocity of migration perpendicular to the wound of Tg737orpk MEFs in the absence (open circles) or presence (closed circles) of EIPA. Data shown are means ± SEM of 19–34 cells in two to four independent experiments. The velocity in the presence of EIPA is significantly different from that in the absence of EIPA (P < 0.0001). Ctrl., control.
We first determined the effect of EIPA on migration of two types of fibroblasts with primary cilia: growth-arrested NIH3T3 cells and growth-arrested WT MEFs. Fig. 3 A shows trajectories of growth-arrested NIH3T3 cells in the absence and presence of 50 ng/ml PDGF-AA normalized to common starting points. The radii of the red circles represent the mean distances covered within the experimental periods of 5 h. Fig. 3 B provides a statistical summary of these experiments. In growth-arrested NIH3T3 cells, translocation was 21 ± 3.6 µm and was reduced by ∼50% by EIPA to 11 ± 1.8 µm. In the presence of PDGF-AA, translocation was increased to 45 ± 5.7 µm, and under these conditions, translocation was reduced by more than two thirds by EIPA to 13 ± 1.5 µm (Fig. 3 B).
Next, we assessed the role of NHE1 specifically in directional migration of the growth-arrested NIH3T3 cells in the presence of PDGF-AA. The directionality of movement was estimated by plotting the mean distance covered by the cells in the direction perpendicular to the wound edge (Fig. 3 C, the x direction) as a function of time. The slope of this curve, obtained by linear regression, corresponds to the velocity of directional movement into the wound. The slope of the corresponding plots for the movement parallel to the wound edge (Fig. 3 C, the y direction) was always close to zero (not depicted), indicating that movement in this direction was random and essentially nonprogressing so that directional movement occurred only perpendicular to the wound edge. The directional migration data for growth-arrested, PDGF-AA–treated NIH3T3 cells are summarized in Fig. 3 D. As seen (Fig. 3 D, legend), the cells moved into the wound at a velocity of 0.104 ± 0.004 µm/min, whereas in the presence of 10 µM EIPA, this velocity was reduced by ∼75% to 0.028 ± 0.001 µm/min.
To ascertain that the effect of EIPA on PDGF-AA–stimulated migration was not caused by an inhibition of PDGFR-α activation, we monitored Tyr754 phosphorylation of PDGFR-α, corresponding to receptor activation, after a 5-min exposure to 50 ng/ml PDGF-AA in the absence of EIPA and after 1 and 6 h of exposure to 5 µM EIPA (Fig. 3 E). As seen, the level of PDGFR-α expression and the ability of PDGF-AA to induce Tyr754 phosphorylation of PDGFR-α were unaffected by the presence of EIPA. Furthermore, neither EIPA (Fig. S1 A) nor siRNA-mediated knockdown of NHE1 (Fig. S1 B) significantly affected the ability of fibroblasts to form primary cilia. These results show that the inhibitory effect of EIPA on PDGF-AA–induced cell migration is not caused by an effect of cilia formation and that EIPA exerts its effect on NHE1 downstream from activation of PDGFR-α. Finally, confirming that the effect of EIPA on cell migration reflected inhibition of NHE1, siRNA-mediated knockdown of NHE1 inhibited cell migration (Fig. S1 C).
Next, we performed similar experiments on WT MEFs. Fig. 4 A shows the trajectories of growth-arrested WT MEFs in the presence of PDGF-AA and in the absence and presence of EIPA, and Fig. 4 B provides statistical summaries (see also Videos 1–3). Exposure to PDGF-AA nearly tripled the translocation of WT MEFs from 45 ± 4.2 µm to 111 ± 5.4 µm. Similar to what was seen in the growth-arrested NIH3T3 cells, the effect of EIPA was substantially greater in the presence of PDGF-AA, but in WT MEFs, this effect was even more pronounced than in NIH3T3 cells. Inhibition of NHE1 by 10 µM EIPA had no statistically significant effect on translocation in WT MEFs in the absence of PDGF-AA but nearly abolished translocation in the PDGF-AA–treated WT MEFs (Fig. 4 B).
Fig. 4 C shows WT MEFs migrating into the wound and illustrates the x and y axes used to estimate directionality, and Fig. 4 D summarizes the directionality data. The growth-arrested, PDGF-AA–treated WT MEFs traveled at a rate of 0.301 ± 0.003 µm/min in the direction perpendicular to the wound. Upon inhibition of NHE1, this directional migration of WT MEFs was reduced by 90% to 0.030 ± 0.004 µm/min (Fig. 4 D). Collectively, these data indicate that NHE1 activity plays a pivotal role in mediating directional migration in response to PDGFR-α signaling.
Migration of fibroblasts deficient in primary cilia: interphase NIH3T3 cells and Tg737orpk mutant MEFs
To evaluate whether the role of NHE1 in cell migration depends on signaling via the primary cilium, we next performed wound-healing assays on two types of fibroblasts deficient in primary cilia: interphase NIH3T3 cells, which lack primary cilia, and Tg737orpk mutant MEFs, which have no or only very short primary cilia and exhibit impaired PDGFR-α expression (Schneider et al., 2005) and decreased directional migration in response to chemotactic stimuli (unpublished data). As seen in Fig. 5 (A and B), the translocation of interphase NIH3T3 cells was 60.0 ± 8.7 µm (17 cells in three experiments) under control conditions. Inhibition of NHE1 by 10 µM EIPA reduced translocation by ∼50% to 30.0 ± 3.0 µm (25 cells in three experiments). Importantly, in the interphase NIH3T3 cells, stimulation with PDGF-AA did not increase translocation, and NHE1 inhibition similarly reduced translocation after PDGF-AA stimulation by ∼50% (Table I). This indicated that the role of NHE1 in translocation was reduced in interphase NIH3T3 cells compared with growth-arrested, PDGF-AA–treated cells, in which EIPA inhibited translocation by ∼70% (compare with Fig. 3 B and Table I). Therefore, we next addressed whether this was also the case for directional migration. As seen in Fig. 5 C, in interphase NIH3T3 cells, migration into the wound proceeded at a velocity of 0.173 ± 0.002 µm/min under control conditions and was reduced by ∼55% to 0.077 ± 0.001 µm/min in the presence of EIPA compared with 75% in the growth-arrested cells (compare with Fig. 3 D). In other words, inhibition of NHE1 was less efficient in reducing both translocation and directional migration in interphase NIH3T3 cells than in growth-arrested, PDGF-AA–treated NIH3T3 cells.
To test the hypothesis that this difference reflected the presence (in growth-arrested NIH3T3 cells) versus absence (in interphase NIH3T3 cells) of a primary cilium, we next performed wound-healing assays on the primary cilia–deficient Tg737orpk mutant MEFs (Fig. 6). Tg737orpk MEFs exhibited a higher translocation in the absence of PDGF-AA than did WT MEFs (compare Fig. 6 B with Fig. 4 B). This is in accordance with our previous findings and indicates that the primary cilium may restrain excessive cell migration (unpublished data).
In another study (unpublished data), we have shown that translocation of Tg737orpk MEFs was unaffected by PDGF-AA (Fig. 6 B). Importantly, translocation of Tg737orpk MEFs was not significantly affected by NHE1 inhibition by 10 µM EIPA, neither in the presence nor in the absence of PDGF-AA (Fig. 6, A and B). With respect to directional migration, the velocity of migration of Tg737orpk MEFs into the wound was 0.31 ± 0.01 µm/min in the absence of EIPA and reduced by only 28% to 0.23 ± 0.01 µm/min in the presence of EIPA (Fig. 6 C). Compared with the 90% reduction in the WT MEFs (Fig. 4 D), this demonstrates that the inhibitory effect of EIPA is strongly blunted in the cilia-deficient Tg737orpk MEFs. Collectively, these data indicate that NHE1 plays a major role in directional migration, in a manner highly dependent on the presence of the primary cilium and specifically on ciliary PDGFR-α signaling. NHE1 also plays a role, albeit a lesser role, in the migration of interphase cells lacking a primary cilium, suggesting that several pathways regulate NHE1 during cell migration.
Migration of NHE1-null and hNHE1-expressing PS120 fibroblasts
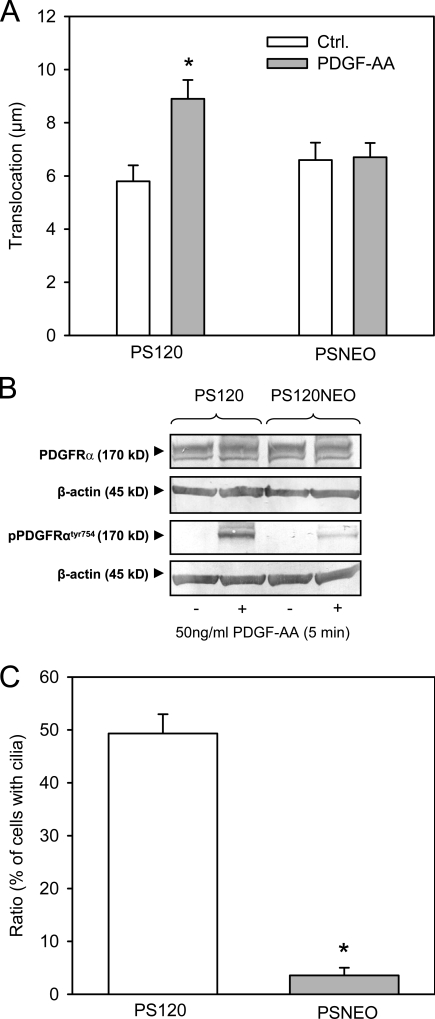
Although EIPA is a widely used and relatively specific inhibitor of NHE1 activity, we wished to further test the involvement of NHE1 using a more specific approach. To this end, we performed 5-h wound-healing assays similar to those performed on NIH3T3 cells and MEFs on growth-arrested NHE1-null PS120 fibroblasts (PSNEO) and on growth-arrested PS120 fibroblasts stably expressing NHE1 (PS120-NHE1; Denker et al., 2000). In the growth-arrested state, these cells migrated relatively slowly in wound-healing assays. As seen in Fig. 7 A, the mean translocation of the NHE1-expressing cells was 5.8 ± 0.60 µm (n = 52 cells in six independent experiments) and 8.9 ± 0.71 µm (n = 63 cells in six independent experiments) in the absence and presence of PDGF-AA stimulation, respectively (i.e., exposure to PDGF-AA–stimulated translocation; P < 0.01). In the NHE1-null cells, the mean translocation in the 5-h wound-healing assay was 6.6 ± 0.65 µm (n = 45 cells in four independent experiments) and 6.7 ± 0.54 µm (n = 64 cells in four independent experiments) in the absence and presence of PDGF-AA stimulation, respectively (i.e., in these cells, there was no effect of 50 ng/ml PDGF-AA on translocation). Thus, despite the fact that growth-arrested PS120 fibroblasts migrate slower than NIH3T3 cells and WT MEFs, these results confirm our conclusion that cells with primary cilia only respond to PDGF-AA stimulation when NHE1 can be activated. However, we noted that, compared with NHE1-expressing cells, NHE1-null cells exhibited reduced, although not abolished, PDGFR-α activation in response to PDGF-AA (Fig. 7 B), reflecting a significantly reduced number of primary cilia in the NHE1-null cells (Fig. 7 C). Therefore, the extent to which these factors contribute to the lack of PDGF-AA–mediated stimulation of migration in NHE1-null cells requires further investigation. Importantly, this is not a concern for the EIPA-treated cells because EIPA affected neither the ability of cells to form primary cilia (Fig. S1 A) nor the activation of PDGFR-α by PDGF-AA (Fig. 3 E).
Figure 7.
Migration, PDGFR-α activation, and cilia formation in NHE1-null and NHE1-expressing cells. (A) PS120 (expressing NHE1) and PSNEO (deficient for NHE1) cells were serum starved for 48 h to induce growth arrest, and migration was analyzed in 4-h wound-healing assays in the absence and presence of 50 ng/ml PDGF-AA, as indicated. The graph shows the mean translocation ± SEM of 45–64 cells in four to six independent experiments. Ctrl., control. (B) PS120 and PSNEO (PS120NEO) cells were serum starved for 48 h, exposed to 50 ng/ml PDGF-AA, and lysed, and Western blots of total and Tyr754-phosphorylated PDGFR-α were performed as described in Materials and methods. β-Actin was used as a loading control. As seen, PDGFR-α expression was similar in the two cell types, whereas PDGF-AA–induced phosphorylation of PDGFR-α was reduced, although not abolished, in the PSNEO cells. Data shown are representative of three independent experiments for each cell line. (C) PS120 and PSNEO cells were serum starved for 48 h to induce growth arrest, paraformaldehyde fixed, and labeled for acetylated α-tubulin to visualize primary cilia. Approximately 300 randomly chosen cells from each experiment were analyzed for the presence or absence of primary cilia, and the results shown are the mean ± SEM of the percentage of cells with cilia in six independent sets of paired experiments. (A and C) Data were analyzed using parametric or nonparametric ANOVA, and the level of significance is shown (*, P < 0.05).
Discussion
We have previously shown that the primary cilium plays a major role in PDGFR-α–mediated regulation of cell growth control (Schneider et al., 2005), cell migration, and chemosensory responses (unpublished data). In this study, we demonstrate that the expression but not the activity of the ubiquitous Na+/H+ exchanger NHE1 is up-regulated upon quiescence in serum-free medium, identifying NHE1 as a gas protein. Moreover, we show that NHE1 activity is required for control of directional cell migration by ciliary PDGFR-α signaling, indicating that NHE1 activation is a critical event in the physiological response to activation of PDGFR-α.
The marked, reversible up-regulation of NHE1 protein levels upon growth arrest observed in fibroblasts appears to occur at the transcriptional level because NHE1 mRNA levels were increased to a similar extent. A similar level of up-regulation of NHE1 expression upon serum deprivation was previously reported in kidney epithelial cells (Carraro-Lacroix et al., 2006). Up-regulation of NHE1 expression during growth arrest in Tg737orpk MEFs occurs at a level comparable with that in WT MEFs, indicating that up-regulation proceeds through mechanisms that are independent of ciliary assembly. This is in sharp contrast to the up-regulation of PDGFR-α during growth arrest, which is blocked in Tg737orpk cells (Schneider et al., 2005). In a previous study in NIH3T3 cells stably expressing a fragment of the NHE1 promoter, it was reported that the addition of serum (from a serum level of 0.5 to 10%) increased NHE1 promoter activity (Besson et al., 1998). This could indicate that the growth arrest–induced increase in mRNA and protein levels observed in this study reflects increased mRNA stability. However, inhibition of extracellular signal-regulated kinase activity, which is a well-known consequence of serum deprivation in NIH3T3 cells (Schneider et al., 2005), was reported to stimulate NHE1 promoter activity (Besson et al., 1998), which appears to be in accordance with the present finding that NHE1 expression is increased during growth arrest. The promoter region of NHE1 contains binding sites for several transcription factors, which are known or likely known to be regulated by serum deprivation, including AP-1 and cAMP response element–binding protein (Miller et al., 1991); however, the potential roles of these proteins in the up-regulation of NHE1 observed in this study remain to be tested.
In contrast to the up-regulation of NHE1 expression in growth-arrested cells, NHE1 activity in NIH3T3 cells was strongly reduced during growth arrest, a finding which is in agreement with studies on other cell types (Reshkin et al., 2000; Lehoux et al., 2001). Thus, in spite of increased expression, NHE1 activity was lower in quiescent cells, presumably reflecting a very tight control of NHE1 activity under these conditions. Recent work has demonstrated a permissive role for an NHE1-mediated increase in pHi in cell cycle progression, apparently through regulation of cyclin D expression and Cdc2 kinase activity and, thus, G2/M entry and transition (Putney and Barber, 2003). It is likely that up-regulation of NHE1 expression, but tight repression of its activity, serves to prepare the cell such that it can elicit a rapid intracellular alkalinization at the appropriate time for reentry into the cell cycle upon delivery of the relevant signal. Thus, the dual control of NHE1 at the expression and posttranscriptional levels may serve as a form of coincidence detection, ensuring that these processes are tightly regulated both by cell cycle stage and by the availability of the relevant ligand. A similar mechanism likely pertains to the control of other NHE1-dependent processes (a case in point being the directional migration process addressed in this study). Finally, although our findings clearly show that the increase in NHE1 expression was not dependent on the formation of the primary cilium, the close temporal correlation between these two important consequences of growth arrest warrants future investigations.
Exposure of growth-arrested NIH3T3 cells to the specific PDGFR-α ligand PDGF-AA partially restored NHE1 activity, which is measured as the global, cytoplasmic NHE1-dependent pHi recovery after acidification. Although activation of NHE1 by nonisoform-discriminating PDGFR ligands has been demonstrated in several cell types (Cassel et al., 1983; Ma et al., 1994; Yan et al., 2001), this is, to our knowledge, the first study to show that a specific PDGFR-α ligand activates NHE1.
Consistent with the marked decrease in NHE1 activity in quiescent cells, migration was strongly reduced in growth-arrested NIH3T3 cells. In addition to stimulating NHE1 activity, PDGF-AA markedly stimulated directional cell migration in growth-arrested NIH3T3 cells in a manner fully dependent on NHE1 activity. In growth-arrested WT MEFs, which, similar to growth-arrested NIH3T3 cells, have a primary cilium, both the net translocation and the directionality of PDGF-AA–mediated migration were similarly dependent on NHE1 activity. In congruence with the involvement of NHE1, PDGF-AA had no effect on translocation in NHE1-null fibroblasts. In the NHE1-null fibroblasts, reduced PDGFR-α activation may also have contributed to this effect because these cells have problems forming primary cilia as compared with their NHE1-expressing counterparts. As neither EIPA nor acute siRNA-mediated NHE1 knockdown affected cilia formation in NIH3T3 cells, this clearly does not represent a role for NHE1 in cilia formation but appears to be an anomaly reflecting long-term genetic alterations in the PSNEO cells, which was not further addressed in this study (Putney and Barber, 2004).
In marked contrast, in Tg737orpk MEFs, which are deficient in primary cilia and, thus, in PDGFR-α expression (Schneider et al., 2005), migration was unaffected by PDGF-AA and only very modestly affected by EIPA, further supporting the notion of a PDGFR-α–NHE1 axis being required for directional migration. Because PDGF-AA activates only the homodimeric PDGFR-α receptor, which is virtually only localized in the primary cilium, whereas NHE1 is activated at the leading edge of the cell, this poses the question of how activation of NHE1 may act at a distance. NHE1 in both WT and Tg737orpk MEFs localized to the cell surface in the leading edge, which is in agreement with previous studies (Lagana et al., 2000; Denker and Barber, 2002; Stock et al., 2005, 2007; Frantz et al., 2007; Stuwe et al., 2007), but also to intracellular vesicle-like structures. It remains a possibility that when PDGF-AA is added to the medium, NHE1 may be transported to the lamellipodial plasma membrane at a higher level in quiescent WT cells, presumably by vesicular exocytosis. However, our initial observations have not convincingly shown an increase in overall plasma membrane localization of NHE1 after PDGF-AA treatment.
Several mechanisms may underlie the linkage between ciliary PDGFR-α signaling and NHE1 activation. One possible pathway is through ERK1/2 and its effector p90RSK, which activates NHE1 by direct phosphorylation on Ser703 of its C-terminal tail, after several types of stimuli (Takahashi et al., 1999; Cuello et al., 2007). Consistent with such a mechanism, we have previously shown that ERK1/2 is activated in NIH3T3 cells and WT MEFs but not in Tg737orpk MEFs in response to PDGFR-α activation in the primary cilium (Schneider et al., 2005). The PI3K–Akt pathway is also activated in MEFs downstream of PDGFR-α activation (unpublished data) and could also contribute to NHE1 stimulation, e.g., by direct phosphorylation of NHE1 (Ser648 of human NHE1 being a high-probability consensus site for phosphorylation by Akt1) and/or by inducing actin and microtubule cytoskeletal rearrangements.
In contrast to the almost complete ablation of PDGF-AA–mediated migration by EIPA in cells with primary cilia (i.e., growth-arrested NIH3T3 cells and WT MEFs), migration of cells lacking cilia (i.e., interphase NIH3T3 cells and Tg737orpk MEFs) had a substantial EIPA-insensitive component. This was also the case for unstimulated, growth-arrested cells and points to redundancy and/or compensatory mechanisms in basal cell migration. In accordance with this view, translocation in the absence of PDGF-AA was similar in growth-arrested NHE1-null and NHE1-expressing PS120 fibroblasts. In contrast, during interphase growth, the rate of cell migration was previously found to be ∼40% lower in NHE1-null compared with NHE1-expressing PS120 fibroblasts (Denker and Barber, 2002), which is comparable with the difference between growth-arrested and interphase NIH3T3 cells seen in this study. The difference between interphase and growth-arrested cells presumably reflects the fact that although NHE1 expression is up-regulated in growth-arrested cells, NHE1 activity is essentially absent (shown in Fig. 2 for NIH3T3 cells). Collectively, these data indicate that the relative role of NHE1 in directional cell migration is cell cycle state and cell type dependent and that other ion transport proteins can fulfill roles similar to that of NHE1 under some conditions. A major candidate is Na+-HCO3− cotransport, for which a role in cell migration was previously demonstrated (Schwab et al., 2005).
A recent study showed that Na+-dependent HCO3− transport appeared more important than NHE1 in pHi regulation in both WT and Tg737orpk choroid plexus epithelia in the mouse (Banizs et al., 2007). This only partial role for NHE1 in pHi regulation contrasts with the fact that during growth arrest when primary cilia were present, both NIH3T3 cells and WT MEFs were fully dependent on NHE1 activity for PDGF-AA–mediated migration. Notably, NHE1 played a greater role in global pHi recovery after acidification in interphase NIH3T3 cells than in growth-arrested, PDGF-AA–treated NIH3T3 cells (Fig. 2), but the relative contribution of NHE1 to both translocation and directional migration was greater in the latter. In other words, there is no strong correlation between the effect of NHE1 on global pHi and its role in directional migration. These data reinforce the possibility that the role of NHE1 in directional migration in response to ciliary PDGFR-α signaling involves a local signaling circuit at the leading edge of the migrating cells and does not reflect the contribution of the transporter to global pHi recovery. We can envision a sequences as follows: when PDGF-AA is added, (a) important ciliary PDGFR-α signaling components are activated, (b) the activation of downstream signaling components (e.g., ERK1/2 or Akt) activates NHE1 in the cell membrane for ion transport or alternatively promotes increased lamellipodial localization of NHE1, and (c) at the leading edge, NHE1, in turn, is pivotal for the F-actin organization required for cell migration. The effect of NHE1 on F-actin organization may be mediated via effects on Cdc42 activity (Frantz et al., 2007), on protein kinase activity (Pedersen et al., 2007a), or as a consequence of direct pH sensitivity of F-actin regulatory proteins such as cofilin or talin (Srivastava et al., 2007). Another possible pathway involves the interaction of NHE1 with ERM proteins, which has been found to play a role in cell migration (Denker and Barber, 2002) as well as in Akt activation after some (Wu et al., 2004) albeit not all (Rasmussen et al., 2008) stimuli.
In a few experiments, we observed by immunofluorescence microscopy analysis a weak localization of NHE1 to primary cilia of NIH3T3 cells (unpublished data), although, in most cases, we could not detect increased NHE1 labeling in the cilium. NHE1 contains two unique ciliary targeting peptide motifs, including an N-terminal sequence motif (R14IFP), which is similar to that required for ciliary targeting of the transient receptor potential ion channel, and polycystin-2 (R6VXP; Geng et al., 2006). Although speculative at this point, it is thus possible that NHE1 has dual functions, i.e., in the primary cilium, where PDGFR-α–mediated signal transduction is initiated, and in the plasma membrane, where NHE1 is activated at the leading edge of migrating cells.
Thus, we have shown in this study that NHE1 activity is specifically required for PDGF-AA–mediated stimulation of fibroblast migration in a manner downstream from activation of PDGFR-α and not reflecting a role for NHE1 in formation of the primary cilium. Although a role for leading-edge NHE1 activity in cell polarity and directional migration has been demonstrated in a variety of cell types (Lagana et al., 2000; Denker and Barber, 2002; Patel and Barber, 2005; Stock et al., 2005, 2007; Frantz et al., 2007; Stuwe et al., 2007), this study is, to our knowledge, the first to demonstrate a pivotal role of NHE1 in the response to chemical stimuli detected and transmitted through the primary cilium. These findings support the notion that the primary cilium functions as a cellular positioning detector in directional cell migration. Therefore, the interplay between the primary cilium and NHE1 may be critical to the physiological response to PDGFR-α signaling in development processes and tissue homeostasis.
Materials and methods
Cell culture and reagents
Experiments were performed on NIH3T3 fibroblasts, WT and Tg737orpk mutant MEFs, NHE1-null PS120 fibroblasts (denoted PSNEO), and PS120 fibroblasts stably expressing NHE1 (provided by D. Barber, University of California, San Francisco, San Francisco, CA; Denker et al., 2000). Tg737 encodes the protein polariz/IFT88, which is part of the intraflagellar transport protein complex responsible for assembly and maintenance of the primary cilium (Rosenbaum and Witman, 2002), and Tg737orpk MEFs form no or only very short primary cilia (Schneider et al., 2005). PS120 fibroblasts are Chinese hamster lung fibroblasts deficient in NHE1 and were transfected either with rat NHE1 or control vector (Denker et al., 2000). NIH3T3 cells were grown in DME (Invitrogen) with 10% fetal calf serum (Invitrogen) and 1% penicillin-streptomycin (Invitrogen). MEFs were grown in 50% DME and 50% F12 Ham (Invitrogen) with 10% fetal calf serum and 1% penicillin-streptomycin. PS120 cells were grown in DME with 5% fetal calf serum, 1% penicillin-streptomycin, and 400 µg/ml G418. All cells were maintained at 37°C, 5% CO2, and 95% humidity. Cells were examined at either 70% confluency in the presence of serum (interphase cells) or at 90% confluency followed by serum starvation for 12 or 48 h to induce growth arrest, or serum was readded to the 48-h serum-starved cells for 24 h to stimulate cell cycle entrance. PDGF-AA was purchased from R&D systems, and stock solutions were prepared at 50 µg/µl in 4 mM HCl/0.1% BSA. EIPA was purchased from Invitrogen and was prepared as a 5-mM stock in double-distilled H2O, and BCECF-AM (2′,7′-bis-[2-carboxyethyl]-5,6-carboxyfluorescein, tetraacetoxymethylester), which was also purchased from Invitrogen, was prepared as a 1.6-mM stock in desiccated DMSO.
Real-time PCR
Cells were grown in Petri dishes and washed with ice-cold PBS, and RNA was purified using the RNeasy Mini kit (QIAGEN). RNA was converted to cDNA by reverse transcription using Superscript RT II (Invitrogen) and 250 µg/µl random primers (Invitrogen) according to the manufacturer’s instructions. Real-time PCR set up was performed as triplets of each individual sample using 1 µg cDNA and 19 µl master mix (2 µM forward primer 5′-CGAACTGCTACACATCCTTGTCTT-3′, 2 µM reverse primer 5′-GTCATAGCTGGCAAATTCCTC-3′, 2 µM NHE1 TaqMan probe 5′-TGCCGTCACTGTGGTCCTCTATCACCT-3′, 30 nM reference dye [Agilent Technologies Inc.], and Brilliant QPCR master mix [Agilent Technologies Inc.]). 18S was used as a reference gene (18S master mix: 2 µM forward primer 5′-TTTAATATACGCTATTGGAGCTGGAA-3′, 2 µM reverse primer 5′-GGATCCATTGGAGGGCAAGT-3′, 2 µM NHE1 TaqMan probe 5′-TACCGCGGCTGCTGGCACC-3′, 30 nM reference dye, and Brilliant QPCR master mix). Quantitative PCR was performed using a real-time thermal cycler (Agilent Technologies, Inc.; 95°C for 10 min, 40 cycles of 95°C for 30 s, and 58°C for 1 min), and data were analyzed with Mx4000 software (Agilent Technologies, Inc.).
SDS-PAGE and Western blot analysis
Cells grown in Petri dishes were washed in ice-cold PBS, lysed in 100 µl boiling lysis buffer with 0.1% SDS, scraped off, and transferred 10–15 times through a 27-gauge needle. Lysates were centrifuged at 16,000 g to precipitate debris, and protein concentrations were estimated using a BCA kit (Thermo Fisher Scientific). Proteins were analyzed by SDS-PAGE and Western blot analysis using the XCell and XCell II blot module systems (Novex) as previously described (Christensen et al., 2001). In brief, proteins were separated by gel electrophoresis under denaturing and reducing conditions on 10% NuPAGE Bis-Tris gels using NuPAGE MOPS SDS running buffer (NP0002) and Fermentas protein standards. Separated proteins were electrophoretically transferred to nitrocellulose membranes, which were stained in 1% Ponceau S red solution, and incubated for 2 h at room temperature in blocking buffer (5% nonfat dry milk in TBST [0.01 M Tris-HCl, 0.15 M NaCl, 0.1% Tween 20, pH 7.4]) before incubation with primary antibodies in blocking buffer overnight at 4°C. The antibodies used were rabbit polyclonal anti–PDGFR-α (1:600; Santa Cruz Biotechnology, Inc.), rabbit polyclonal anti–phospho-Tyr754–PDGFR-α (1:200; Santa Cruz Biotechnology, Inc.), anti-NHE1 (1:500; Xb-17 rabbit polyclonal was a gift of M. Musch, University of Chicago, Chicago, IL), and mouse monoclonal anti–β-actin (1:10,000; Sigma-Aldrich). Labeling was detected by incubation with alkaline phosphatase-coupled secondary antibodies (1:200) in blocking buffer for 1 h followed by development using 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (Kirkegaard and Perry Laboratories, Gaithersburg, MD).
Immunofluorescence analysis
Cells were grown on glass coverslips in 6-well test plates (Cellstar) and fixed in 4% paraformaldehyde for 15 min at room temperature followed by permeabilization in 0.2% Triton X-100. Cells were quenched in PBS with 2% BSA for 30 min and incubated with the following primary antibodies at room temperature for 2 h: anti–acetylated α-tubulin (1:5,000 mouse antibody; Sigma-Aldrich), anti–PDGFR-α (Santa Cruz Biotechnology, Inc.), and anti-NHE1 (Xb-17; both rabbit antibodies were used at 1:500). Cells were washed in PBS and incubated with DAPI (Invitrogen) and Alexa Fluor 568–conjugated goat anti–mouse IgG and Alexa Fluor 488–conjugated goat anti–rabbit IgG (both at 1:1,000; Invitrogen) for 1 h. Preparations were mounted in N-propyl-gallate at 2% wt/vol in PBS/glycerin and were visualized at room temperature. For standard epifluorescence imaging, cells were visualized on a microscope (Eclipse E600; Nikon) with EPI-FL3 filters, a 100× 1.25 NA objective, and a cooled charge-coupled device camera (MagnaFire; Optronics), For confocal imaging, cells were visualized using a 40× 1.25 NA Plan-Apochromat objective and the 488-nm Ar/Kr laser line of a microscope (DMIRB/E; Leica) with a confocal laser-scanning unit (TSC NT; Leica). Optical slice thickness was 1 µm, and pinhole size was 1 airy disc. Images shown are frame averaged and presented in RGB pseudocolor. No or negligible labeling was detectable in the absence of primary antibody. Digital images were processed (overlays and brightness/contrast adjustment only) using Photoshop CS2 version 9.0.2 (Adobe).
Measurements of pHi
pHi was monitored at 37°C essentially as previously described (Pedersen et al., 2007b). In brief, NIH3T3 cells or WT or Tg737orpk MEFs were seeded on glass coverslips 24–48 h before experiments. Confluency at the time of experiments was 80–95%. Cells were loaded with 1.2 µM BCECF-AM in Ringer’s solution (150 mM NaCl, 1 mM Na2HPO4, 1 mM CaCl2, 10 mM Hepes, and 5 mM glucose, pH 7.4), and mounted in the cuvette of a spectrophotometer (Ratiomaster; PTI). Emission was detected at 525 nm after excitation at 445 and 495 nm (NIH3T3 fibroblasts) or 445 and 500 nm (MEFs). The 445:495 nm ratio was calculated after background subtraction, and calibration to pHi was performed using the 7-point nigericin/high K+ technique, essentially as previously described (Boyarsky et al., 1988). The high K+ Ringer solution was similar to the standard Ringer solution, except that 140 mM NaCl was replaced with KCl. The ability of the cells to recover after an acid load was evaluated by the NH4Cl prepulse technique: cells were exposed to 10 mM NH4Cl in Ringer’s solution for 5 min followed by NH4Cl removal to induce intracellular acidification, the recovery from which is a measure of the capacity for pHi regulation after an acid load. To estimate the contribution of NHE1, experiments were performed in the absence and presence of 5 µM EIPA. The effect of PDGF-AA was estimated by preincubating the cells with 50 ng/ml PDGF-AA for 1 h before the NH4Cl prepulse. Because initial pHi was comparable in all experiments for a given cell type, pHi recovery was compared by calculating recovery rates (pH units/minute) from the slope of the initial linear part of the curve after NH4Cl removal.
Wound-healing assays
Migration of individual NIH3T3 fibroblasts, MEFs, and PS120 fibroblasts was assessed with wound-healing assays. Fibroblasts (interphase or quiescent as indicated) were grown to 80–90% confluency in 12.5-cm2 culture flasks, and a wound was scratched in the cell layer using a fine sterile pipette tip. The culture medium was changed to fresh, serum-free medium, and the cells were allowed to recover for 1 h at 37°C/5% CO2 before recording. The cells were placed in a heated chamber (37°C) on the stage of an inverted microscope (Axiovert 25 or Axiovert 40C; Carl Zeiss, Inc.) equipped with 10× or 20× objectives. Migration into the wound was monitored for ∼5 h with a video camera (XT-ST70CE and XC-77CE; Hamamatsu Photonics) controlled by HiPic software (Hamamatsu Photonics). In some experiments, cells were stimulated with 50 ng/ml PDGF-AA in the absence or presence of 10 µM EIPA. Images were taken at 15-min intervals and stored as stacks of tiff files. For data analysis, the circumferences of the cells were labeled at each time step throughout the entire image stacks using Amira software (Mercury Computer Systems; http://www.tgs.com/) as described previously (Dreval et al., 2005; Dieterich et al., 2008). Migration was quantified as the movement of the cell center. The x and y coordinates of the cell center (in micrometers) were determined as geometric means of equally weighted pixel positions within the cell outlines. We used two parameters to quantify migration. The translocation (in micrometers) was calculated as the mean distance between the position of the cell center at the beginning and at the end of the experiment for the observed group of cells. Translocation summarizes stochastic movements of the cells (Dreval et al., 2005; Dieterich et al., 2008) and contributions from directed migration induced by the wound-healing condition and is a measure of sustained migration (Schwab et al., 2006). The directionality of movement was estimated by plotting the mean of the numerical distance of the cells traveled in the direction perpendicular to the wound (the x direction) as a function of time. The mean positions of cells were fitted to the functions x(t) = vx × t and y(t) = vy × t. The velocities vx and vy and their uncertainties were estimated with the least-square fit routine of gnuplot (http://www.gnuplot.info/). The slope of the x distance versus time plot is the velocity in the x direction (in micrometers/minute) and is a measure of directionality in our experimental setup. The corresponding plots for directionality in the y direction (parallel to the wound) were always close to zero, indicating that in this experimental setup, directional movement is measured only perpendicular to the wound.
siRNA-mediated knockdown of NHE1
NHE1 knockdown in NIH3T3 cells was performed using SMARTpool siRNA (Thermo Fisher Scientific) against mouse NHE1 (NHE1) or scrambled oligonucleotides of comparable GC content (mock), both at 50 nM, and FECT transfection agent (Thermo Fisher Scientific) according to the manufacturer’s instructions. The day after siRNA treatment, the cells were serum starved as described in Cell culture and reagents and were used for experiments 48 or 72 h after siRNA treatment as indicated. Cilia counting experiments were always accompanied by parallel Western blots against NHE1 to verify knockdown.
Statistics
All experiments were repeated 3–11 times, and data are presented as representative individual experiments or as mean values ± SEM. The data were tested for significance using analysis of variance (ANOVA) or Kruskal Wallis test (nonparametric ANOVA). The level of significance was set at P < 0.05 except where otherwise stated.
Online supplemental material
Fig. S1 shows the effects of EIPA and NHE1 knockdown in NIH3T3 fibroblasts. Video 1 shows the migration of growth-arrested WT MEFs in a wound-healing assay in the absence of EIPA and PDGF-AA. Videos 2 and 3 show the migration of PDGF-AA–stimulated, growth-arrested WT MEFs in a wound-healing assay in the absence or presence of EIPA, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200806019/DC1.
Acknowledgments
PS120 and PSNEO fibroblasts were a gift from Diane Barber. We are grateful to Sabine Mally and Iben Rønn Veland for expert experimental assistance.
This work was supported by the Danish National Research Council (grants 21-04-0535 and 272-07-0530), the Lundbeck Foundation (grant R9-A969), the Danish Cancer Society (grant DP05072 to S.F. Pedersen and S.T. Christensen), Deutsche Forschungsgemeinschaft (Schw 407/10-1,/9-3), Interdisziplinäres Zentrum für Klinische Forschung Münster (Schw 2/030/08 to A. Schwab), the National Institute of Diabetes and Digestive and Kidney Diseases (grants DK41918 and DK41296 to P. Satir), and funds from the University of Copenhagen (to L. Schneider).
Footnotes
Abbreviations used in this paper: ANOVA, analysis of variance; EIPA, 5′-(N-ethyl-N-isopropyl) amiloride; gas, growth arrest specific; MEF, mouse embryonic fibroblast; PDGFR, PDGF receptor; pHi, intracellular pH; WT, wild type.
References
- Banizs B., Komlosi P., Bevensee M.O., Schwiebert E.M., Bell P.D., Yoder B.K. 2007. Altered pHi regulation and Na+/HCO3- transporter activity in choroid plexus of cilia-defective Tg737(orpk) mutant mouse. Am. J. Physiol. Cell Physiol. 292:C1409–C1416 [DOI] [PubMed] [Google Scholar]
- Besson P., Fernandez-Rachubinski F., Yang W., Fliegel L. 1998. Regulation of Na+/H+ exchanger gene expression: mitogenic stimulation increases NHE1 promoter activity. Am. J. Physiol. 274:C831–C839 [DOI] [PubMed] [Google Scholar]
- Boron W.F. 2004. Regulation of intracellular pH. Adv. Physiol. Educ. 28:160–179 [DOI] [PubMed] [Google Scholar]
- Boyarsky G., Ganz M.B., Sterzel R.B., Boron W.F. 1988. pH regulation in single glomerular mesangial cells. I. Acid extrusion in absence and presence of HCO3-. Am. J. Physiol. 255:C844–C856 [DOI] [PubMed] [Google Scholar]
- Cardone R.A., Bagorda A., Bellizzi A., Busco G., Guerra L., Paradiso A., Casavola V., Zaccolo M., Reshkin S.J. 2005a. Protein kinase A gating of a pseudopodial-located RhoA/ROCK/p38/NHE1 signal module regulates invasion in breast cancer cell lines. Mol. Biol. Cell. 16:3117–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone R.A., Casavola V., Reshkin S.J. 2005b. The role of disturbed pH dynamics and the Na+/H+ exchanger in metastasis. Nat. Rev. Cancer. 5:786–795 [DOI] [PubMed] [Google Scholar]
- Carraro-Lacroix L.R., Ramirez M.A., Zorn T.M., Reboucas N.A., Malnic G. 2006. Increased NHE1 expression is associated with serum deprivation-induced differentiation in immortalized rat proximal tubule cells. Am. J. Physiol. Renal Physiol. 291:F129–F139 [DOI] [PubMed] [Google Scholar]
- Cassel D., Rothenberg P., Zhuang Y.X., Deuel T.F., Glaser L. 1983. Platelet-derived growth factor stimulates Na+/H+ exchange and induces cytoplasmic alkalinization in NR6 cells. Proc. Natl. Acad. Sci. USA. 80:6224–6228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S.T., Guerra C., Wada Y., Valentin T., Angeletti R.H., Satir P., Hamasaki T. 2001. A regulatory light chain of ciliary outer arm dynein in Tetrahymena thermophila. J. Biol. Chem. 276:20048–20054 [DOI] [PubMed] [Google Scholar]
- Christensen S.T., Pedersen L.B., Schneider L., Satir P. 2007. Sensory cilia and integration of signal transduction in human health and disease. Traffic. 8:97–109 [DOI] [PubMed] [Google Scholar]
- Cuello F., Snabaitis A.K., Cohen M.S., Taunton J., Avkiran M. 2007. Evidence for direct regulation of myocardial Na+/H+ exchanger isoform 1 phosphorylation and activity by 90-kDa ribosomal S6 kinase (RSK): effects of the novel and specific RSK inhibitor fmk on responses to alpha1-adrenergic stimulation. Mol. Pharmacol. 71:799–806 [DOI] [PubMed] [Google Scholar]
- Denker S.P., Barber D.L. 2002. Cell migration requires both ion translocation and cytoskeletal anchoring by the Na-H exchanger NHE1. J. Cell Biol. 159:1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denker S.P., Huang D.C., Orlowski J., Furthmayr H., Barber D.L. 2000. Direct binding of the Na–H exchanger NHE1 to ERM proteins regulates the cortical cytoskeleton and cell shape independently of H+ translocation. Mol. Cell. 6:1425–1436 [DOI] [PubMed] [Google Scholar]
- Dieterich P., Klages R., Preuss R., Schwab A. 2008. Anomalous dynamics of cell migration. Proc. Natl. Acad. Sci. USA. 105:459–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreval V., Dieterich P., Stock C., Schwab A. 2005. The role of Ca2+ transport across the plasma membrane for cell migration. Cell. Physiol. Biochem. 16:119–126 [DOI] [PubMed] [Google Scholar]
- Frantz C., Karydis A., Nalbant P., Hahn K.M., Barber D.L. 2007. Positive feedback between Cdc42 activity and H+ efflux by the Na-H exchanger NHE1 for polarity of migrating cells. J. Cell Biol. 179:403–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng L., Okuhara D., Yu Z., Tian X., Cai Y., Shibazaki S., Somlo S. 2006. Polycystin-2 traffics to cilia independently of polycystin-1 by using an N-terminal RVxP motif. J. Cell Sci. 119:1383–1395 [DOI] [PubMed] [Google Scholar]
- Hayashi H., Aharonovitz O., Alexander R.T., Touret N., Furuya W., Orlowski J., Grinstein S. 2008. Na+/H+ exchange and pH regulation in the control of neutrophil chemokinesis and chemotaxis. Am. J. Physiol. Cell Physiol. 294:C526–C534 [DOI] [PubMed] [Google Scholar]
- Hayashi N., Takehara K., Soma Y. 1995. Differential chemotactic responses mediated by platelet-derived growth factor alpha- and beta-receptors. Arch. Biochem. Biophys. 322:423–428 [DOI] [PubMed] [Google Scholar]
- Heldin C.H., Westermark B. 1999. Mechanism of action and in vivo role of platelet-derived growth factor. Physiol. Rev. 79:1283–1316 [DOI] [PubMed] [Google Scholar]
- Jechlinger M., Sommer A., Moriggl R., Seither P., Kraut N., Capodiecci P., Donovan M., Cordon-Cardo C., Beug H., Grunert S. 2006. Autocrine PDGFR signaling promotes mammary cancer metastasis. J. Clin. Invest. 116:1561–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagana A., Vadnais J., Le P.U., Nguyen T.N., Laprade R., Nabi I.R., Noel J. 2000. Regulation of the formation of tumor cell pseudopodia by the Na+/H+ exchanger NHE1. J. Cell Sci. 113:3649–3662 [DOI] [PubMed] [Google Scholar]
- Lehoux S., Abe J., Florian J.A., Berk B.C. 2001. 14-3-3 Binding to Na+/H+ exchanger isoform-1 is associated with serum-dependent activation of Na+/H+ exchange. J. Biol. Chem. 276:15794–15800 [DOI] [PubMed] [Google Scholar]
- Lih C.J., Cohen S.N., Wang C., Lin-Chao S. 1996. The platelet-derived growth factor alpha-receptor is encoded by a growth-arrest-specific (gas) gene. Proc. Natl. Acad. Sci. USA. 93:4617–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.H., Reusch H.P., Wilson E., Escobedo J.A., Fantl W.J., Williams L.T., Ives H.E. 1994. Activation of Na+/H+ exchange by platelet-derived growth factor involves phosphatidylinositol 3′-kinase and phospholipase C gamma. J. Biol. Chem. 269:30734–30739 [PubMed] [Google Scholar]
- Miller R.T., Counillon L., Pages G., Lifton R.P., Sardet C., Pouyssegur J. 1991. Structure of the 5′-flanking regulatory region and gene for the human growth factor-activatable Na/H exchanger NHE-1. J. Biol. Chem. 266:10813–10819 [PubMed] [Google Scholar]
- Patel H., Barber D.L. 2005. A developmentally regulated Na-H exchanger in Dictyostelium discoideum is necessary for cell polarity during chemotaxis. J. Cell Biol. 169:321–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen S.F. 2006. The Na+/H+ exchanger NHE1 in stress-induced signal transduction: implications for cell proliferation and cell death. Pflugers Arch. 452:249–259 [DOI] [PubMed] [Google Scholar]
- Pedersen S.F., Darborg B.V., Rasmussen M., Nylandsted J., Hoffmann E.K. 2007a. The Na+/H+ exchanger, NHE1, differentially regulates mitogen-activated protein kinase subfamilies after osmotic shrinkage in Ehrlich Lettre Ascites cells. Cell. Physiol. Biochem. 20:735–750 [DOI] [PubMed] [Google Scholar]
- Pedersen S.F., King S.A., Nygaard E.B., Rigor R.R., Cala P.M. 2007b. NHE1 inhibition by amiloride- and benzoylguanidine-type compounds. Inhibitor binding loci deduced from chimeras of NHE1 homologues with endogenous differences in inhibitor sensitivity. J. Biol. Chem. 282:19716–19727 [DOI] [PubMed] [Google Scholar]
- Putney L.K., Barber D.L. 2003. Na-H exchange-dependent increase in intracellular pH times G2/M entry and transition. J. Biol. Chem. 278:44645–44649 [DOI] [PubMed] [Google Scholar]
- Putney L.K., Barber D.L. 2004. Expression profile of genes regulated by activity of the Na-H exchanger NHE1. BMC Genomics. 5:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putney L.K., Denker S.P., Barber D.L. 2002. The changing face of the Na+/H+ exchanger, NHE1: structure, regulation, and cellular actions. Annu. Rev. Pharmacol. Toxicol. 42:527–552 [DOI] [PubMed] [Google Scholar]
- Rasmussen M., Alexander R.T., Darborg B.V., Mobjerg N., Hoffmann E.K., Kapus A., Pedersen S.F. 2008. Osmotic cell shrinkage activates ezrin/radixin/moesin (ERM) proteins: activation mechanisms and physiological implications. Am. J. Physiol. Cell Physiol. 294:C197–C212 [DOI] [PubMed] [Google Scholar]
- Rentsch M.L., Ossum C.G., Hoffmann E.K., Pedersen S.F. 2007. Roles of Na+/H+ exchange in regulation of p38 mitogen-activated protein kinase activity and cell death after chemical anoxia in NIH3T3 fibroblasts. Pflugers Arch. 454:649–662 [DOI] [PubMed] [Google Scholar]
- Reshkin S.J., Bellizzi A., Albarani V., Guerra L., Tommasino M., Paradiso A., Casavola V. 2000. Phosphoinositide 3-kinase is involved in the tumor-specific activation of human breast cancer cell Na+/H+ exchange, motility, and invasion induced by serum deprivation. J. Biol. Chem. 275:5361–5369 [DOI] [PubMed] [Google Scholar]
- Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., Horwitz A.R. 2003. Cell migration: integrating signals from front to back. Science. 302:1704–1709 [DOI] [PubMed] [Google Scholar]
- Rosenbaum J.L., Witman G.B. 2002. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3:813–825 [DOI] [PubMed] [Google Scholar]
- Schneider I.C., Haugh J.M. 2006. Mechanisms of gradient sensing and chemotaxis: conserved pathways, diverse regulation. Cell Cycle. 5:1130–1134 [DOI] [PubMed] [Google Scholar]
- Schneider L., Clement C.A., Teilmann S.C., Pazour G.J., Hoffmann E.K., Satir P., Christensen S.T. 2005. PDGFRalphaalpha signaling is regulated through the primary cilium in fibroblasts. Curr. Biol. 15:1861–1866 [DOI] [PubMed] [Google Scholar]
- Schwab A., Rossmann H., Klein M., Dieterich P., Gassner B., Neff C., Stock C., Seidler U. 2005. Functional role of Na+-HCO3- cotransport in migration of transformed renal epithelial cells. J. Physiol. 568:445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab A., Wulf A., Schulz C., Kessler W., Nechyporuk-Zloy V., Romer M., Reinhardt J., Weinhold D., Dieterich P., Stock C., Hebert S.C. 2006. Subcellular distribution of calcium-sensitive potassium channels (IK1) in migrating cells. J. Cell. Physiol. 206:86–94 [DOI] [PubMed] [Google Scholar]
- Schwab A., Nechyporuk-Zloy V., Fabian A., Stock C. 2007. Cells move when ions and water flow. Pflugers Arch. 453:421–432 [DOI] [PubMed] [Google Scholar]
- Shure D., Senior R.M., Griffin G.L., Deuel T.F. 1992. PDGF AA homodimers are potent chemoattractants for fibroblasts and neutrophils, and for monocytes activated by lymphocytes or cytokines. Biochem. Biophys. Res. Commun. 186:1510–1514 [DOI] [PubMed] [Google Scholar]
- Srivastava J., Barber D.L., Jacobson M.P. 2007. Intracellular pH sensors: design principles and functional significance. Physiology (Bethesda). 22:30–39 [DOI] [PubMed] [Google Scholar]
- Stock C., Schwab A. 2006. Role of the Na+/H+ exchanger NHE1 in cell migration. Acta Physiol. (Oxf.). 187:149–157 [DOI] [PubMed] [Google Scholar]
- Stock C., Gassner B., Hauck C.R., Arnold H., Mally S., Eble J.A., Dieterich P., Schwab A. 2005. Migration of human melanoma cells depends on extracellular pH and Na+/H+ exchange. J. Physiol. 567:225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock C., Mueller M., Kraehling H., Mally S., Noel J., Eder C., Schwab A. 2007. pH nanoenvironment at the surface of single melanoma cells. Cell. Physiol. Biochem. 20:679–686 [DOI] [PubMed] [Google Scholar]
- Stuwe L., Muller M., Fabian A., Waning J., Mally S., Noel J., Schwab A., Stock C. 2007. pH dependence of melanoma cell migration: protons extruded by NHE1 dominate protons of the bulk solution. J. Physiol. 585:351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi E., Abe J., Gallis B., Aebersold R., Spring D.J., Krebs E.G., Berk B.C. 1999. p90(RSK) is a serum-stimulated Na+/H+ exchanger isoform-1 kinase. Regulatory phosphorylation of serine 703 of Na+/H+ exchanger isoform-1. J. Biol. Chem. 274:20206–20214 [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Webb D.J., Horwitz A.R. 2005. Cell migration at a glance. J. Cell Sci. 118:4917–4919 [DOI] [PubMed] [Google Scholar]
- Wu D. 2005. Signaling mechanisms for regulation of chemotaxis. Cell Res. 15:52–56 [DOI] [PubMed] [Google Scholar]
- Wu K.L., Khan S., Lakhe-Reddy S., Jarad G., Mukherjee A., Obejero-Paz C.A., Konieczkowski M., Sedor J.R., Schelling J.R. 2004. The NHE1 Na+/H+ exchanger recruits ezrin/radixin/moesin proteins to regulate Akt-dependent cell survival. J. Biol. Chem. 279:26280–26286 [DOI] [PubMed] [Google Scholar]
- Yan W., Nehrke K., Choi J., Barber D.L. 2001. The Nck-interacting kinase (NIK) phosphorylates the Na+-H+ exchanger NHE1 and regulates NHE1 activation by platelet-derived growth factor. J. Biol. Chem. 276:31349–31356 [DOI] [PubMed] [Google Scholar]
- Yu J., Moon A., Kim H.R. 2001. Both platelet-derived growth factor receptor (PDGFR)-alpha and PDGFR-beta promote murine fibroblast cell migration. Biochem. Biophys. Res. Commun. 282:697–700 [DOI] [PubMed] [Google Scholar]