Abstract
Metazoan cell movement has been studied extensively in vitro, but cell migration in living animals is much less well understood. In this report, we have studied the Caenorhabditis elegans Q neuroblast lineage during larval development, developing live animal imaging methods for following neuroblast migration with single cell resolution. We find that each of the Q descendants migrates at different speeds and for distinct distances. By quantitative green fluorescent protein imaging, we find that Q descendants that migrate faster and longer than their sisters up-regulate protein levels of MIG-2, a Rho family guanosine triphosphatase, and/or down-regulate INA-1, an integrin α subunit, during migration. We also show that Q neuroblasts bearing mutations in either MIG-2 or INA-1 migrate at reduced speeds. The migration defect of the mig-2 mutants, but not ina-1, appears to result from a lack of persistent polarization in the direction of cell migration. Thus, MIG-2 and INA-1 function distinctly to control Q neuroblast migration in living C. elegans.
Introduction
Cell migration is an essential feature of metazoan development and also plays important roles in the physiology of adult organisms (Lehmann, 2001; Locascio and Nieto, 2001). Impaired cell migration results in various developmental disorders such as congenital brain defects, and excessive migration also contributes to pathologies such as tumor metastasis (http://www.cellmigration.org; Yamaguchi et al., 2005; Kurosaka and Kashina, 2008).
Considerable progress has been made in identifying the protein machinery that drives migration and the extracellular signals that guide migration (Webb et al., 2002; Pollard and Borisy, 2003; Ridley et al., 2003; Jaffe and Hall, 2005; Simpson et al., 2008). However, molecular mechanisms that control the speed and distance of cell migration in vivo are largely unknown. Why do some cells migrate away from their birth place while others remain? For those migrating cells, why do some cells migrate faster and further than others? Knowledge obtained from in vitro cultured cells offers a good starting point for understanding cell migration in live animals. However, cellular behavior in a 3D tissue shows several distinct properties from a 2D culturing condition (Yamada and Cukierman, 2007). Thus, investigations of cell migration in live metazoans are necessary to understand how molecular pathways discovered through in vitro model systems operate in living organisms.
To best study cell migration in live animals, one needs single cell resolution so that one can follow a cell's trajectory and changes in speed. Ideally, one would like to compare different cells that have different migration patterns so that one might be able to identify molecular differences that might underlie these distinct migratory capacities. Caenorhabditis elegans Q neuroblasts have the potential of being such an attractive model system. Pioneering work of C. elegans post-embryonic development reported that descendants of Q neuroblasts migrate different distances during the L1 larva stage (Sulston and Horvitz, 1977). Previous observations of Q neuroblast development mainly relied on Nomarski optics (Sulston and Horvitz, 1977). In this study, we developed GFP-based live cell time-lapse imaging methodologies to document Q neuroblast development with spinning-disk confocal microscopy. We found that descendants in the Q neuroblast lineage have distinct migratory speeds and distances, making it an appealing model to discover the molecular differences among descendants of Q neuroblasts. We provide evidence that MIG-2, a Rho family GTPase mutant (Zipkin et al., 1997), and INA-1, an integrin α subunit mutant (Baum and Garriga, 1997), play important but distinct roles in defining the distinct migratory behavior of Q descendants.
Results and discussion
Descendants of Q neuroblasts have distinct migratory capacities
A pair of bilateral Q neuroblasts on the left side (QL) and right side (QR) of C. elegans undergoes three rounds of asymmetrical cell divisions during C. elegans L1 larva development (Fig. 1, A–C; Sulston and Horvitz, 1977). Previous work using Nomarski optics identified the starting and ending positions of the descendants of Q neuroblasts (Fig. 1, D and E; Sulston and Horvitz, 1977) and estimated the migration speeds of a subset of the cells. However, detailed documentation of their migration patterns has not been performed by time-lapse recordings. In this study, we developed GFP-based fluorescence microscopy techniques to image Q neuroblast development. Using an integrant C. elegans strain that stably expresses a GFP-tagged Rho family GTPase, MIG-2, under its endogenous promoter, we are able to visualize the plasma membrane of Q cells (Zipkin et al., 1997). In some experiments, we also used a strain that expresses a diffuse, cytosolic GFP for imaging both the cell perimeter and outline of the nucleus. For our long-term time-lapse imaging experiments, we optimized conditions to immobilize animals and used spinning-disk confocal microscopy (see Materials and methods).
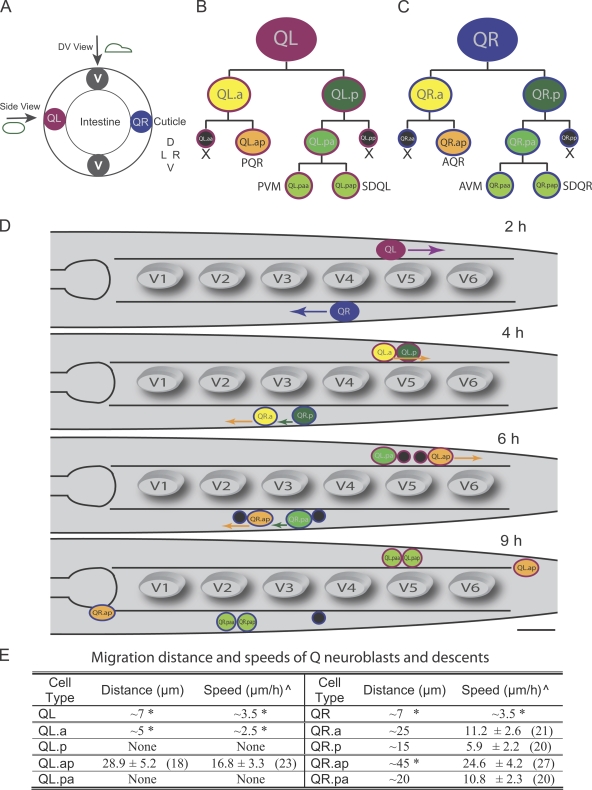
Figure 1.
Q neuroblasts lineage and the migration properties of the descendants. (A) The position of Q neuroblasts in a cross section of the nematode C. elegans. D, dorsal; V, ventral; L, left; R, right. (B and C) QL and QR neuroblasts lineage patterns, which produce three different neurons and two apoptotic cells (in black and marked by X) are shown. The products of these divisions give rise to ciliated sensory neurons (AQR and PQR), touch sensory neurons (AVM and PVM), and interneurons (SDQL and SDQR). (D) The migration of Q neuroblasts and their descendants in the L1 larva stage. Arrows indicate the migration direction. The time given is the hours after hatching. V1–V6, epithelial seam V1–V6 cells. Bar, 10 µm. (E) Summary of the migration distance and speeds of Q neuroblasts and their descendants. Asterisks denote values derived from the figures of previously published data (see Materials and methods). Other measurements are derived from our time-lapse observations. Carets indicate mean speed ± SEM (number of animals).
Our results show distinct migration patterns of four pairs of Q descendants from the first two rounds of cell division (Fig. 1, A–D; and Videos 1–6). The first division on the left side generates two cells: QL.a and QL.p. Previous work reported that QL.a and QL.p move past each other in opposite directions (Sulston and Horvitz, 1977). However, we found that QL.p remains stationary while QL.a migrates past QL.p (Fig. 1 D, 4 h; and Fig. S1). After the second division on the left side, QL.ap migrates at a rate of 16.8 µm/h for ∼30 µm toward the posterior, whereas QL.pa does not migrate (Fig. 1 D, 6 h). On the right side of the animal, QR.a migrates at a rate of 11.2 µm/h for ∼25 µm, and QR.p moves at 5.9 µm/h for ∼15 µm toward the anterior (Fig. 1 D, 4 h). After the second division, the migration rates of QR.ap and QR.pa increase but to different degrees. QR.ap migrates at 24.6 µm/h for ∼45 µm, whereas QR.p moves at 10.8 µm/h for ∼20 µm to the head (Fig. 1 D, 6 h). Thus, the Q neuroblasts and their descendants display a dramatic range of cell migration speeds and travel distances, migrating at speeds of 2.5–25 µm/h and for distances of 7–45 µm. The faster migrating cell in a pair of descendents always moves for the longer distance from their birth place (Fig. 1 E). Because one cell of the pair migrates faster and over a longer distance than the other, we refer to this cell as having a “higher migratory capacity.” Thus, QL/R.a versus QL/R.p and QL/R.ap versus QL/R.pa provide excellent model systems for studying molecular differences that control the distinct cell migratory capacities in a pair of daughter cells.
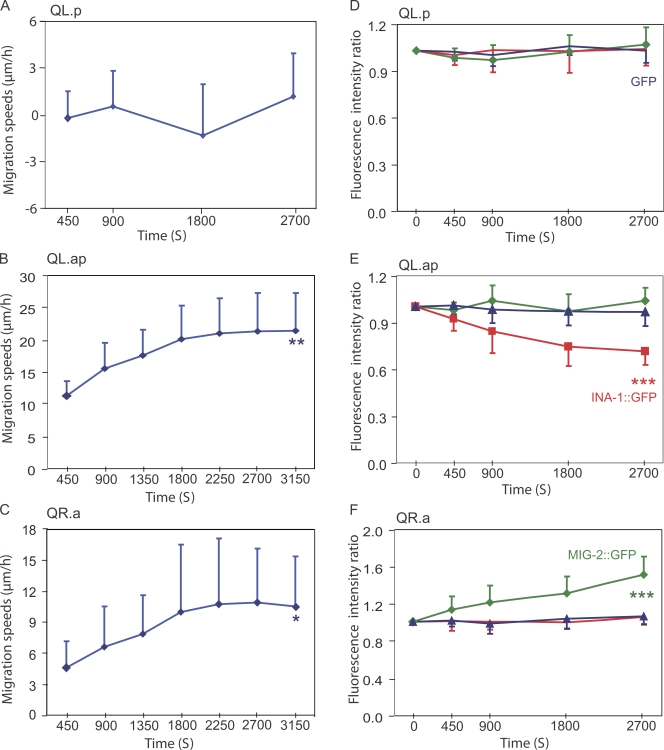
Figure 4.
Changes in MIG-2 and INA-1 protein levels during the migration of QL.p, QL.ap, and QR.a cells. (A–C) The migration speeds during different time periods measured for QL.p (n = 11), QL.ap (n = 9), and QR.a cells (n = 8). The QL.p is effectively stationary. (D–F) The fluorescence intensity ratio measured during different time periods of migration (compared with intensity at time 0) of MIG-2∷GFP (green; n = 8–11), INA-1∷GFP (red; n = 8–12), and soluble GFP (blue; n = 10–11). Error bars indicate mean ± SD. Statistical significances of the data (comparison between the start and end points of speed and fluorescence) are determined by Student's t test. *, P < 0.025; **, P < 0.005; ***, P < 0.001.
MIG-2 and INA-1 control Q descendants' migratory capacities
We next sought to uncover the molecular differences among the aforementioned four pairs of cells that might contribute to their different migratory capacities. As a starting point, we examined two C. elegans mutants that were already known to be defective in Q neuroblast development: mig-2 and ina-1 (Baum and Garriga, 1997; Zipkin et al., 1997). MIG-2 is classified as an Mtl Rac in the Rho GTPase family, whose members have been shown to stimulate actin cytoskeleton polymerization for cell migration, neuritogenesis, gastrulation, and cell corpse phagocytosis (Jaffe and Hall, 2005; Lundquist, 2006; Heasman and Ridley, 2008). In the constitutively active mig-2(rh17) mutant, QR cell migration was shown to be defective (Zipkin et al., 1997). Integrins are a family of heterodimeric transmembrane receptors consisting of α and β subunits that link the actin cytoskeleton to the ECM or the neighboring cell surface (Hynes, 2002; Avraamides et al., 2008). The ina-1 gene encodes an integrin α subunit that is associated with a β subunit, PAT-3, to form a functional integrin pair essential for cell migration, neuritogenesis, and tissue morphogenesis in C. elegans development (Baum and Garriga, 1997; Meighan and Schwarzbauer, 2007), and the point mutation in ina-1(gm144) reduces QR cell migration distance (Baum and Garriga, 1997).
In this study, we performed time-lapse observation of the QL.ap cell in mig-2(rh17) and ina-1(gm144) mutant animals, allowing us to observe both the rate and distance of QL.ap migration (Fig. 2, A and B). Compared with the migration distance of QL.ap in wild type (WT; 28.9 µm), QL.ap only migrates about half the distance in an ina-1 (13.3 µm) or mig-2 mutant (13.0 µm; Fig. 2 C). We also found that QL.ap migrates significantly slower in ina-1 (5.6 µm/h) and mig-2 (7.8 µm/h) mutant animals compared with QL.ap in WT (16.8 µm/h; Fig. 1 E and Fig. 2 D). Thus, our results showed that both INA-1 and MIG-2 regulate both the migration distance and speed of Q neuroblasts.
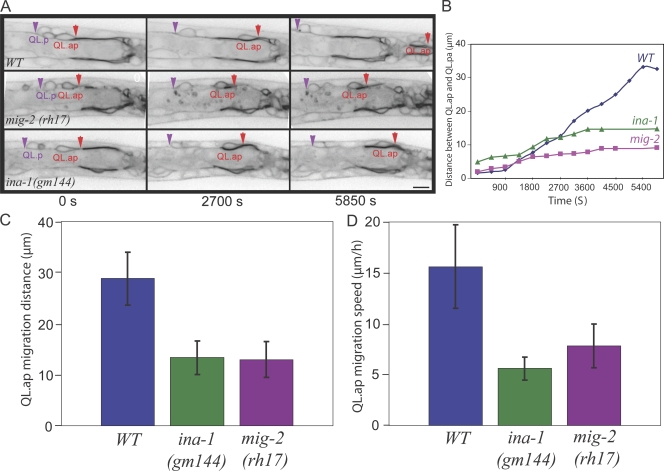
Figure 2.
MIG-2 and INA-1 control the cell migratory capacity of QL.ap. (A) QL.ap migrates toward the tail in a WT animal but fails to complete its posterior migration in mig-2(rh17) or ina-1(gm144) mutant animals. The plasma membrane of QL.ap is visualized by the expression of MIG-2∷GFP (image is inverted so that higher GFP intensity is black). The leading edge of QL.ap is marked by red arrows, and the rear of the stationary QL.p is marked by purple arrowheads. Bar, 5 µm. (B) The distance between QL.ap and QL.pa during migration in WT, ina-1(gm144), and mig-2(rh17). (C) The final migration distances of QL.ap in ina-1(gm144) (13.3 ± 3.3 µm, n = 22) and mig-2(rh17) (13.0 ± 3.5 µm, n = 11) mutants are shorter than that in WT (28.9 ± 5.2 µm, n = 18). Error bars indicate SEM. (D) The migration speeds of QL.ap in ina-1(gm144) (5.6 ± 1.2 µm/h, n = 12) and mig-2(rh17) (7.8 ± 2.1 µm/h, n = 11) mutants are slower than in WT (16.8 ± 3.3 µm/h, n = 23).
Dynamics of MIG-2 and INA-1 protein levels during Q cell migration
We next examined whether the levels of MIG-2 and INA-1 proteins change during Q cell migration. To assess this, we performed time-lapse imaging of MIG-2∷GFP and INA-1∷GFP proteins expressed from their endogenous promoters on genes that were stably integrated into the C. elegans genome (Fig. 3). Surprisingly, we found that MIG-2 and INA-1 protein levels change in most migrating Q neuroblasts, as described in detail in Fig. 3 and Fig. S3. In contrast, the fluorescence intensity of a soluble GFP protein (see Materials and methods) remained constant during Q cell migration (Fig. 3, E and F; Figs. S1 and S2; Videos 3 and 6).
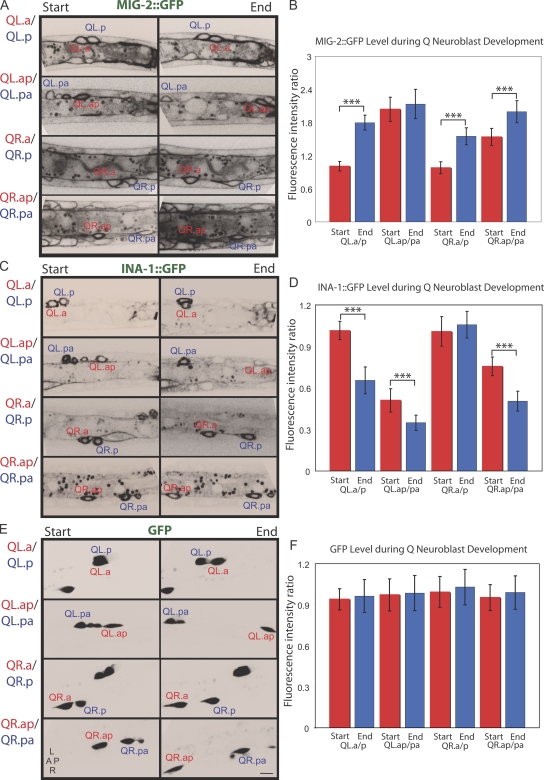
Figure 3.
Changes in MIG-2 and INA-1 protein levels during Q cell migration. (A–F) MIG-2∷GFP (A and B), INA-1∷GFP (C and D), and soluble GFP (E and F) were imaged at the start and end stage of migration of the indicated four pairs of Q neuroblast descendants. (A, C, and E) Images of Q cells at the start and the end of migration are shown. (B, D, and F) Fluorescence intensities of MIG-2∷GFP (n = 17–29), INA-1∷GFP (n = 17–33), and GFP (n = 12) expressed as a ratio between four Q descendant pairs are shown. ***, statistical significance of difference in the fluorescence intensity ratio between the start and the end (P < 0.001, Student's t test). Error bars indicate SEM. The absolute protein levels for each cell are shown in Fig. S3 (A–C). L, left; R, right; A, anterior; P, posterior. Bar, 5 µm.
Initially, the two recently divided Q daughter cells of the left (QL.a and QL.p) and right (QR.a and QR.p) sides received similar amounts of MIG-2∷GFP and INA-1∷GFP (GFP quantitatively measured in Fig. 3, B and D; and Fig. S3, A and B). However, QL.a, migrating past QL.p (Videos 1–3), increases its MIG-2∷GFP fluorescence by 1.8-fold (Fig. 3, A and B; and Fig. S1) while decreasing its INA-1∷GFP fluorescence by 0.7-fold compared with QL.p at the end of migration (Fig. 3, C and D; and Fig. S1). On the right side, MIG-2∷GFP increases by 1.5-fold in the fast migrating QR.a compared with QR.p at the end of migration; however, the relative protein level of INA-1∷GFP did not change in this cell pair (Fig. 3, A–D; and Fig. S3, A and B). Similar general trends were noted in the next Q descendants. Specifically, QL.ap, which migrates toward the posterior (Videos 4–6), decreases its INA-1∷GFP levels between the start and end of migration relative to the stationary QL.pa (MIG-2∷GFP does not change; Fig. 3, A–D; Fig. S2; Fig. S3, A and B). QR.ap increases its MIG-2∷GFP levels and decreases its INA-1∷GFP compared with the slower migrating QR.pa (Fig. 3, A–D). We also show that up-regulation of MIG-2∷GFP occurs normally in the ina-1(gm144) mutant, and down-regulation of INA-1∷GFP occurs normally in a mig-2(rh17) mutant (Fig. S3, D–G). In summary, Q descendants with higher migratory capacities either up-regulate MIG-2 levels during migration (QR.a/QR.p), down-regulate INA-1 levels (QL.ap/QL.pa), or do both (QL.a/QL.p and QR.ap/QR.pa).
Q neuroblast speed correlates with up-regulation of MIG-2 or down-regulation of INA-1
To obtain further evidence of a relationship between cell migratory capacities and the expression of MIG-2 and INA-1, we measured migration speed and MIG-2 or INA-1 protein levels over time for the QL.p, QL.ap, and QR.a cells. In the QL.p cell, which does not migrate, MIG-2, INA-1, and soluble GFP protein levels did not change over time (Fig. 4, A and D). In contrast, in the QL.ap cell, INA-1∷GFP levels progressively decrease over time as the cell increases its migration speed (Fig. 4, B and E). In the QR.a cell, which also increases its migration speed, the MIG-2∷GFP levels steadily increased over time (Fig. 4, C and F). As a control, we found that soluble GFP levels remain constant in these cells during migration (Fig. 4, E and F). These data are consistent with the view that up-regulation of MIG-2 and down-regulation of INA-1 may contribute to an increase in the speed of cell migration. However, protein levels of MIG-2 and INA-1 may not be tightly coupled to cell speed because GFP fluorescence appears ∼30 min after the protein is produced, as a result of maturation of fluorophore. Also, MIG-2∷GFP and INA-1∷GFP are expressed from multiple copies of integrated plasmids and yet move at similar speeds to nonexpressing cells or cells expressing soluble GFP.
MIG-2, but not INA-1, controls the proper polarization of migrating Q neuroblasts
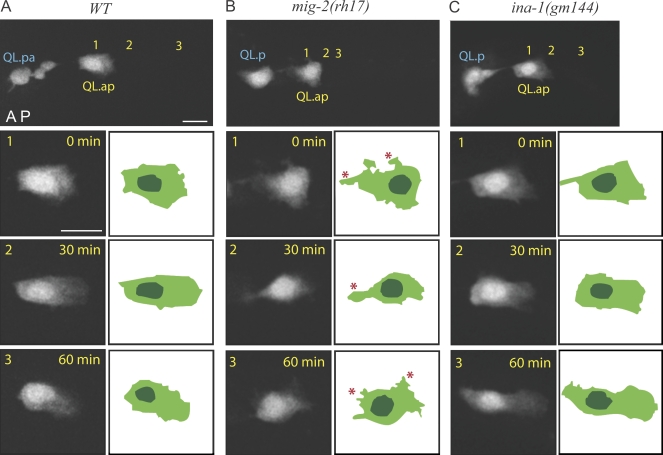
We next addressed how MIG-2 and INA-1 control migration by examining the morphology of migrating WT and mutant Q cells. For this study, we imaged soluble GFP in the QL.ap cell, which illuminates the cytoplasm and thus defines the boundaries of the cell perimeter and the nucleus (Fig. 1 A and Fig. 5). In WT animals, QL.ap elongates in its direction of migration, extending a lamellae toward the animal tail and positioning the nucleus toward the posterior (100% of cells elongated toward the tail, n = 15; Fig. 5 A and Video 7). The QL.ap maintained this morphology during the 60 min of migration that we examined. Strikingly, in the mig-2(rh17), the QL.ap showed a distinct morphology from WT animals. Notably, QL.ap randomly extended processes to the anterior or dorsal/ventral side of the animal over time and did not maintain a persistent elongated polarization toward the tail (only 1/10 animals showed what might be scored as persistent posterior polarization; Fig. 5 B and Video 8). In contrast, QL.ap in ina-1(gm144) mutants showed normal posterior polarization and nuclear positioning, even though it had a similar reduced migration speed to mig-2(rh17) animals (92% of animals showed normal QL.ap polarization to the tail, n = 12; Fig. 5 C and Video 9). These observations suggest that MIG-2 and INA-1 proteins control Q cell migration speed and distance by different mechanisms. MIG-2 is required for the polarization of migrating QL.ap, and the lack of persistent polarization (extending processes in the wrong direction) may likely explain the slower speed of migration in the mig-2(rh17) mutant animals. In contrast, INA-1 appears to regulate QL.ap migration speed by a mechanism independent of polarity.
Figure 5.
MIG-2, but not INA-1, polarizes migrating Q neuroblasts. (A–C) The morphology of migrating QL.ap cell visualizing from the left/right lateral side of C. elegans L1 larva expressing soluble GFP in WT (A), mig-2(rh17) (B), and ina-1(gm144) mutants (C). This GFP marker stains the Q cell periphery and the nucleus. The top panels show the first frames of QL.ap migration from Videos 7–9. Numbers 1, 2, and 3 indicate the QL.ap positions at 0 (1; the first frame), 30 (2), or 60 min (3) during migration from Videos 7–9. QL.ap in mig-2(rh17) (B) or ina-1(gm144) (C) migrates slower and shorter than its migration in WT (A). The bottom three panels show the magnified views of cell morphology of migrating QL.ap paired with schematic diagrams from the top panels or frames in Videos 7–9. Migrating QL.ap properly polarizes the lamellae toward the posterior in WT (A; 100%, n = 15) and in ina-1(gm144) (C; 92%, n = 12). However, in mig-2(rh17), QL.ap forms protrusions in random directions, marked by red asterisks. The anterior (A)–posterior (P) axis is the left to right. Light green, cytoplasm; dark green, nuclei. Bars, 5 µm.
Conclusion
In summary, our study shows that C. elegans Q neuroblasts provide an attractive model for visualizing cell migration in vivo and that tools, such as genetic mutations and expression of GFP-tagged genes, enable a molecular dissection of this process. In this study, we show that pairs of Q descendants migrate at different speeds and distances and that a higher cell migratory capacity correlates with the up-regulation of MIG-2 and down-regulation of INA-1. By examining cell morphology directly during migration in living animals, we also show that MIG-2 is essential for the polarization of migration Q cells, but INA-1 is involved in speed control but not polarity.
We find that cells with higher cell migratory capacity (QL/R.a vs. QL/R.p or QL/R.ap vs. QL/R.pa) have significantly more MIG-2 protein on the plasma membrane and that up-regulation of MIG-2 correlates with QR.a acceleration. To our knowledge, this is the first demonstration in metazoan development that a Rac GTPase dynamically changes its protein level during cell migration. However, in cell culture, overexpression of Rac1 was shown to promote the motility of T47D mammary epithelial cells (Keely et al., 1997), which is consistent with our findings.
Our study also shows that MIG-2 is involved in controlling cell polarity during migration in living animals. The mig-2(rh17) mutant animal bears a point mutant that constitutively activates MIG-2 by locking the protein in a GTP state (Zipkin et al., 1997). An inability to selectively activate MIG-2 in one part of the cell may explain why the QL.ap cell extends processes in many directions and has trouble in maintaining a persistent lamellae. The lack of persistent polarization likely explains the slower speed of migration of mig-2(rh17) rather than a defect in the actin machinery or interaction with the ECM (Fig. 5 B). Previous work using a fibroblast cell line has shown that increasing the amount of active Rac1 causes random migration, whereas the reduction of active Rac supports directionality (Pankov et al., 2005). Our results suggest that a similar paradigm may apply in living animals. However, MIG-2 and its Drosophila melanogaster homologues, the Mtl Racs, are distinct from vertebrate Rac1 (CED-10 and RAC-2 are the closest C. elegans homologues of vertebrate Rac1). RhoG in vertebrates was suggested to be the functional equivalent of Mtl Rac, but its function is largely unknown (Lundquist, 2006; de Curtis, 2008). Drosophila Mtl Rac is required for axon branching, guidance, and growth (Ng et al., 2002). It will be interesting to examine whether Mtl or RhoG is also involved in establishing polarity of migrating cells as we find for the C. elegans homologue, MIG-2.
We also find that cells with higher migratory capacity have significantly lower INA-1 protein and that down-regulation of INA-1 is associated with QL.ap acceleration. During tissue morphogenesis and remodeling, integrins have been reported to undergo temporal and spatial changes to control cell rearrangement by transcriptional or posttranscriptional mechanisms. For example, INA-1 protein levels were shown to decrease in the distal tip cell of the C. elegans gonad near the completion of migration, and blocking INA-1 down-regulation in vab-3(mw105) mutant produces an over migration phenotype (Meighan and Schwarzbauer, 2007). Similarly, we find that INA-1 levels decrease in several Q neuroblasts, although in our case we observe this decrease at the start of migration.
Integrins are critical for building focal adhesions that connect the ECM with the actin cytoskeleton. We find that INA-1 is required for cell migration in a polarity-independent mechanism (Fig. 5). We favor the idea that a reduction in integrin protein levels reduces the strength of focal adhesions, allowing for faster migration. In support of the notion, cultured cell lines migrate faster at intermediate than at high ECM concentrations (Palecek et al., 1997), which may be a result of an optimal integrin engagement of the ECM and retrogradely moving actin (Gupton and Waterman-Storer, 2006).
Although this study focuses on two molecular markers of cell migration, MIG-2 and INA-1, levels of many other proteins are likely to be regulated during cell migration. Systematic and quantitative screening of the expression of many GFP-tagged proteins might be possible and would provide a more complete molecular signature of the dynamics of cell migration. In addition, the clarity of imaging in this study suggests that readouts of protein activity (e.g., using fluorescence resonance energy transfer readout from fluorescent protein sensors) might be possible to perform. Time-lapse imaging of the dynamics of subcellular structures (e.g., centrosomes, chromosomes, and cytoskeleton) could also be performed. Finally, the microscopy methodologies developed for imaging Q cell migration can be applied to investigate other developmental processes in C. elegans larva, such as asymmetrical cell division and programmed cell death (Figs. S1 and S2).
Materials and methods
Strains
C. elegans strains were grown on NGM plates seeded with the Escherichia coli strain OP50 at 20°C using standard methods. Some strains were provided by the Caenorhabditis Genetics Center (funded by the National Institutes of Health National Center for Research Resources). The previously generated fluorescence markers used in this study, MIG-2∷GFP, INA-1∷GFP, and the soluble GFP, were integrated into C. elegans chromosomes so that the variation of fluorescence intensity among individual animals was minimized. MIG-2∷GFP and INA-1∷GFP were expressed under the control of their endogenous promoters, and the transgenes were used to rescue the corresponding mutants, indicating that they are functional (Baum and Garriga, 1997; Zipkin et al., 1997). The soluble GFP used in this study of an integration of the GFP under the control of egl-17 promoter, a fibroblast growth factor–like gene that is expressed in Q neuroblasts but not essential for Q cell migration (Burdine et al., 1997; Branda and Stern, 2000). This integration made by Branda and Stern (2000) fused the first 11 amino acids of egl-17 gene and seven amino acids of a nuclear localization signal (PKKKRKV) before the GFP protein. Fluorescence markers were introduced into mutants by genetic crosses, and Table S1 summarizes genotypes of strains used in this study. Genetic nomenclature for C. elegans was followed in this study (Horvitz et al., 1979).
Fluorescence microscopy
Images were collected by a microscope (Axiovert 200M; Carl Zeiss, Inc.) equipped with a 100× 1.45 NA oil objective and a cooled charge-coupled device camera (Orca II ERG; Hamamatsu Photonics) or an EM charge-coupled device camera (C9100-13; Hamamatsu Photonics) and the 488-nm line of an argon laser attached to a spinning-disk confocal scan head (CSU10; Yokogawa; obtained from Solamere, Inc.). Time-lapse images acquired at 10–20 sites were imaged simultaneously every 90 s using µManager software (www.micro-manager.org).
Imaging Q neuroblasts
Q neuroblast development in C. elegans L1 larva was recorded using a modification of a protocol for studying neuronal transport in adult worms (Ou et al., 2005). The fluorescent transgenic nematodes were anesthetized with 0.1–1 mM levamisole in M9 buffer, mounted on 2% agar pads, and maintained at 22°C. Q neuroblast development was imaged from either dorsal to ventral direction (Figs. 2, 3, S1, and S2 and Videos 1–6) or from a left to right angle (Fig. 5 and Videos 7–9) of C. elegans in L1 larva stage (Fig. 1 A). A pair of bilateral Q neuroblasts localize between intestine and cuticle on both QL and QR of the animal. Observing the dorsal to ventral direction, the side morphology of QL and QR can be visualized on the same focal plane below or above the plane of seam cell. From the left or right side of nematodes, the bottom of either the QL or QR cell, but not both, are visible on one focal plane.
Fluorescence quantitation
Images of Q neuroblasts were processed using ImageJ software (National Institutes of Health). Figs. 2 and 3 and Fig. S3 (D–G) are contrast-inverted images by Photoshop (CS3; Adobe), and the original frames and videos are available in the online supplemental material. We calculated fluorescence intensities of MIG-2, INA-1, and EGL-17 as the fluorescence divided by area. ImageJ software was used to circumscribe the fluorescence field and measure the interior GFP fluorescence and the size of area. Fluorescence intensity was measured from a single focus plane. This was reasonable because of the small sizes and z projection of Q neuroblasts (e.g., the depth of QL.a is 3.4 ± 0.4 µm, n = 10; the depth of QL.ap is 3.0 ± 0.4 µm, n = 12). Single section acquisition also minimized photobleaching and photodamage of Q neuroblast development over long imaging periods (2–3 h). Previous measurements from a single plane were shown to be valid in determining the sizes of Q neuroblasts before and after asymmetrical cell division (Cordes et al., 2006). To calculate the fluorescence intensity ratio, background fluorescence outside of Q neuroblasts in the same frame was subtracted.
Speed measurement
Migration speeds were measured from the first frame in which the Q cell started to move until the frame in which the Q cell stopped migrating. Our microscopy setup with a 100× objective and a camera with 1× binning provided an image field of 45.1 × 45.1 µm. Once a Q neuroblast moved out of the imaging area, we manually recentered the Q cells. Different stationary fiduciary markers were used for speed measurements. The myoblast cell does not move during L1 larva stage and is close to the QL.p or QL.pa cell, and we used the myoblast cell to show that QL.p and QL.pa are immotile. The QL.pa cell, but not the myoblast cell, is in the same imaging field as the QL.ap cell, and thus, we chose the QL.pa cell as an internal stationary marker for QL.ap speed measurements. Proper fiduciary maker for QR migration was not available, so we measured the migration speeds of QR cells within the field of view. Although this can contribute error as a result of animal or stage movement, in practice, we found negligible movement of the stationary myoblast and QL.pa cells with a frame over time, thus validating our measurement of QR movement. We estimated migration distances of QL, QL.a, and all QR cells by tracking the center of each cell in Fig. 13 of Sulston and Horvitz (1977) using the reported scale bar. Migration speeds of QL, QL.a, and QR were estimated from the same figure..
Online supplemental material
Figs. S1 and S2 show dynamics of MIG-2, INA-1, and soluble GFP as QL.a migrates past QL.p (Fig. S1) or QL.ap migrates toward the tail (Fig. S2). Fig. S3 shows MIG-2, INA-1, and GFP protein levels during Q cell migration. Videos 1–6 show migration of QL.a (Videos 1–3) and QL.ap (Videos 4–6) in L1 larva expressing MIG-2∷GFP (Videos 1 and 4), INA-1∷GFP (Videos 2 and 5), and soluble GFP (Videos 3 and 6). Videos 7–9 show the lateral views of QL.ap migrating to the tail in WT (Video 7), mig-2(rh17) (Video 8), or ina-1(gm144) (Video 9). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200812077/DC1.
Acknowledgments
We thank C. Kenyon and G. Garriage and members in their laboratories for encouragement, advice, and strains. We are grateful to N. Stuurman for microscopy assistance and Vale laboratory members for stimulating discussion. C. Adler, J.-W. Tsai, A. Carter, and T. Huckaba provided critical comments on the manuscript.
G. Ou received a postdoctoral fellowship from the Damon Runyon Cancer Research Foundation, and R.D. Vale received support from the National Institutes of Health, Cell Migration Consortium (GM064346), and the Howard Hughes Medical Institute.
Footnotes
Abbreviation used in this paper: WT, wild type.
References
- Avraamides C.J., Garmy-Susini B., Varner J.A. 2008. Integrins in angiogenesis and lymphangiogenesis.Nat. Rev. Cancer. 8:604–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum P.D., Garriga G. 1997. Neuronal migrations and axon fasciculation are disrupted in ina-1 integrin mutants.Neuron. 19:51–62 [DOI] [PubMed] [Google Scholar]
- Branda C.S., Stern M.J. 2000. Mechanisms controlling sex myoblast migration in Caenorhabditis elegans hermaphrodites.Dev. Biol. 226:137–151 [DOI] [PubMed] [Google Scholar]
- Burdine R.D., Chen E.B., Kwok S.F., Stern M.J. 1997. egl-17 encodes an invertebrate fibroblast growth factor family member required specifically for sex myoblast migration in Caenorhabditis elegans.Proc. Natl. Acad. Sci. USA. 94:2433–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes S., Frank C.A., Garriga G. 2006. The C. elegans MELK ortholog PIG-1 regulates cell size asymmetry and daughter cell fate in asymmetric neuroblast divisions.Development. 133:2747–2756 [DOI] [PubMed] [Google Scholar]
- de Curtis I. 2008. Functions of Rac GTPases during neuronal development.Dev. Neurosci. 30:47–58 [DOI] [PubMed] [Google Scholar]
- Gupton S.L., Waterman-Storer C.M. 2006. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration.Cell. 125:1361–1374 [DOI] [PubMed] [Google Scholar]
- Heasman S.J., Ridley A.J. 2008. Mammalian Rho GTPases: new insights into their functions from in vivo studies.Nat. Rev. Mol. Cell Biol. 9:690–701 [DOI] [PubMed] [Google Scholar]
- Horvitz H.R., Brenner S., Hodgkin J., Herman R.K. 1979. A uniform genetic nomenclature for the nematode Caenorhabditis elegans.Mol. Gen. Genet. 175:129–133 [DOI] [PubMed] [Google Scholar]
- Hynes R.O. 2002. Integrins: bidirectional, allosteric signaling machines.Cell. 110:673–687 [DOI] [PubMed] [Google Scholar]
- Jaffe A.B., Hall A. 2005. Rho GTPases: biochemistry and biology.Annu. Rev. Cell Dev. Biol. 21:247–269 [DOI] [PubMed] [Google Scholar]
- Keely P.J., Westwick J.K., Whitehead I.P., Der C.J., Parise L.V. 1997. Cdc42 and Rac1 induce integrin-mediated cell motility and invasiveness through PI(3)K.Nature. 390:632–636 [DOI] [PubMed] [Google Scholar]
- Kurosaka S., Kashina A. 2008. Cell biology of embryonic migration.Birth Defects Res. C Embryo Today. 84:102–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R. 2001. Cell migration in invertebrates: clues from border and distal tip cells.Curr. Opin. Genet. Dev. 11:457–463 [DOI] [PubMed] [Google Scholar]
- Locascio A., Nieto M.A. 2001. Cell movements during vertebrate development: integrated tissue behaviour versus individual cell migration.Curr. Opin. Genet. Dev. 11:464–469 [DOI] [PubMed] [Google Scholar]
- Lundquist E.A. 2006. Small GTPases.WormBook. 17:1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meighan C.M., Schwarzbauer J.E. 2007. Control of C. elegans hermaphrodite gonad size and shape by vab-3/Pax6-mediated regulation of integrin receptors.Genes Dev. 21:1615–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J., Nardine T., Harms M., Tzu J., Goldstein A., Sun Y., Dietzl G., Dickson B.J., Luo L. 2002. Rac GTPases control axon growth, guidance and branching.Nature. 416:442–447 [DOI] [PubMed] [Google Scholar]
- Ou G., Blacque O.E., Snow J.J., Leroux M.R., Scholey J.M. 2005. Functional coordination of intraflagellar transport motors.Nature. 436:583–587 [DOI] [PubMed] [Google Scholar]
- Palecek S.P., Loftus J.C., Ginsberg M.H., Lauffenburger D.A., Horwitz A.F. 1997. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness.Nature. 385:537–540 [DOI] [PubMed] [Google Scholar]
- Pankov R., Endo Y., Even-Ram S., Araki M., Clark K., Cukierman E., Matsumoto K., Yamada K.M. 2005. A Rac switch regulates random versus directionally persistent cell migration.J. Cell Biol. 170:793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D., Borisy G.G. 2003. Cellular motility driven by assembly and disassembly of actin filaments.Cell. 112:453–465 [DOI] [PubMed] [Google Scholar]
- Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., Horwitz A.R. 2003. Cell migration: integrating signals from front to back.Science. 302:1704–1709 [DOI] [PubMed] [Google Scholar]
- Simpson K.J., Selfors L.M., Bui J., Reynolds A., Leake D., Khvorova A., Brugge J.S. 2008. Identification of genes that regulate epithelial cell migration using an siRNA screening approach.Nat. Cell Biol. 10:1027–1038 [DOI] [PubMed] [Google Scholar]
- Sulston J.E., Horvitz H.R. 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans.Dev. Biol. 56:110–156 [DOI] [PubMed] [Google Scholar]
- Webb D.J., Parsons J.T., Horwitz A.F. 2002. Adhesion assembly, disassembly and turnover in migrating cells–over and over and over again.Nat. Cell Biol. 4:E97–E100 [DOI] [PubMed] [Google Scholar]
- Yamada K.M., Cukierman E. 2007. Modeling tissue morphogenesis and cancer in 3D.Cell. 130:601–610 [DOI] [PubMed] [Google Scholar]
- Yamaguchi H., Wyckoff J., Condeelis J. 2005. Cell migration in tumors.Curr. Opin. Cell Biol. 17:559–564 [DOI] [PubMed] [Google Scholar]
- Zipkin I.D., Kindt R.M., Kenyon C.J. 1997. Role of a new Rho family member in cell migration and axon guidance in C. elegans.Cell. 90:883–894 [DOI] [PubMed] [Google Scholar]