Abstract
The Salt Overly Sensitive (SOS) pathway plays an important role in the regulation of Na+/K+ ion homeostasis and salt tolerance in Arabidopsis thaliana. Previously, we reported that the calcium binding proteins SOS3 and SOS3-LIKE CALCIUM BINDING PROTEIN8 (SCaBP8) nonredundantly activate the protein kinase SOS2. Here, we show that SOS2 phosphorylates SCaBP8 at its C terminus but does not phosphorylate SOS3. In vitro, SOS2 phosphorylation of SCaBP8 was enhanced by the bimolecular interaction of SOS2 and SCaBP8 and did not require calcium ions. In vivo, this phosphorylation was induced by salt stress, occurred at the membrane, stabilized the SCaBP8-SOS2 interaction, and enhanced plasma membrane Na+/H+ exchange activity. When a Ser at position 237 in the SCaBP8 protein (the SOS2 phosphorylation target) was mutated to Ala, SCaBP8 was no longer phosphorylated by SOS2 and the mutant protein could not fully rescue the salt-sensitive phenotype of the scabp8 mutant. By contrast, when Ser-237 was mutated to Asp to mimic the charge of a phosphorylated Ser residue, the mutant protein rescued the scabp8 salt sensitivity. These data demonstrate that calcium sensor phosphorylation is a critical component of SOS pathway regulation of salt tolerance in Arabidopsis.
INTRODUCTION
Maintenance of Na+/K+ ion homeostasis is essential for plant growth and development in saline conditions where accumulation of salts leads to significant reductions in plant growth. The Salt Overly Sensitive (SOS) pathway in Arabidopsis thaliana plays an important role in regulating Na+/ K+ ion homeostasis during plant growth in high salt conditions. SOS3 (Liu and Zhu, 1998), a calcium binding protein, perceives an increase in cellular calcium levels induced by salt stress. Upon sensing the calcium signal, SOS3 interacts with, activates, and recruits SOS2, a protein kinase, to the plasma membrane (Halfter et al., 2000; Quintero et al., 2002). This complex further activates SOS1 (Shi et al., 2000), a Na+/H+ exchanger (antiporter), to regulate Na+/K+ ion homeostasis and prevent the accumulation of toxic levels of Na+ (Zhu et al., 1998; Zhu, 2002).
The Arabidopsis genome encodes nine additional SOS3-LIKE CALCIUM BINDING PROTEINS (SCaBPs), also known as CALCINEURIN B-LIKE (CBL) calcium binding proteins (Luan et al., 2002; Gong et al., 2004; Kolukisaoglu et al., 2004). SOS3 is also known as CBL4 (Kim et al., 2000; see Supplemental Figure 1 online for a protein alignment of the SCaBP/CBL family members). Four SCaBP/CBL proteins have been shown to regulate ion homeostasis. SCaBP1/CBL2 interacts with and activates SOS2-LIKE PROTEIN KINASE5 (PKS5), and this complex negatively regulates the activity of AHA2, a plasma membrane H+-ATPase (Fuglsang et al., 2007). CBL1/SCaBP5 and CBL9/SCaBP7 interact with and activate CBL-INTERACTING PROTEIN KINASE23 (CIPK23); this complex in turn activates the activity of AKT1, a plasma membrane K+ channel, to regulate K+ homeostasis (Xu et al., 2006; Cheong et al., 2007). Recently, we identified SCaBP8/CBL10 as another regulator of SOS2 that, in contrast with SOS3, which mainly functions in root tissue, preferentially protects shoot tissue from salt stress. Like SOS3, SCaBP8 interacts with, activates, and recruits SOS2 to the plasma membrane to activate SOS1 in the plant plasma membrane or SOS1 expressed in yeast (Quan et al., 2007).
A number of SCaBP/CBL proteins contain the four helix-loop-helix structural domains (EF-hands) found in many calcium binding proteins. These SCaBP proteins share significant sequence similarity with the B subunit of calcineurin in yeast and a neuronal calcium sensor in animals, with sequences only differing at their N and C termini (Gong et al., 2004). Both the B subunit of calcineurin and neuronal calcium sensor proteins are myristoylated in vivo; however, it is not known if this modification is required for their function (Zhu et al., 1995). Similar protein domains have been shown to regulate the localization or activities of several SCaBP proteins. Three out of 10 of the SCaBP/CBL family proteins have the MGXXXS/T (K) myristoylation signature sequence (Gong et al., 2004; Kolukisaoglu et al., 2004; Batistic et al., 2008). This N-terminal myristoylation and acylation were shown to be required for plasma membrane localization of CBL1/SCaBP5 (Batistic et al., 2008), while a myristoylation domain at the SOS3 N terminus was shown to be important for SOS3 function in salt tolerance (Ishitani et al., 2000). While SCaBP8 lacks an N-terminal myristoylation motif, an N-terminal 25–amino acid hydrophobic domain is required for SCaBP8 plasma membrane localization (Quan et al., 2007). Both SOS3 and SCaBP8 activate SOS2 and SOS1; however, they cannot replace each other in reciprocal mutant complementation assays, suggesting that each has a unique function(s) in the response of Arabidopsis to growth in salt.
In this study, we report that SOS2 phosphorylates SCaBP8, but not SOS3, and that this modification is important for SCaBP8 function in salt tolerance by stabilizing the membrane localization of the SCaBP8-SOS2 complex.
RESULTS
SOS2 Phosphorylates SCaBP8 at Its C Terminus
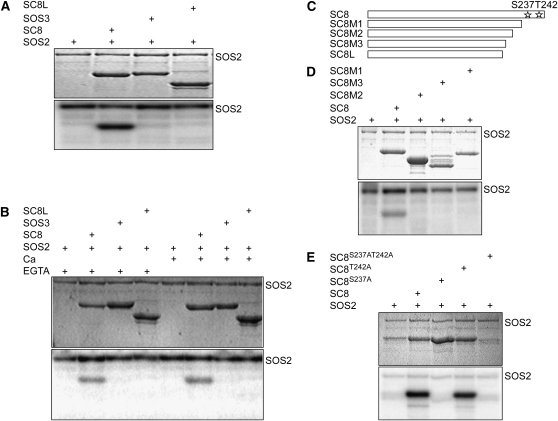
The SOS3 and SCaBP8 calcium sensors have been shown to specifically function in the salt stress response of Arabidopsis (Liu and Zhu, 1998; Quan et al., 2007) but perform unique functions in the regulation of SOS2. One possible mechanism underlying this difference in regulation is that the SCaBP8 protein may require an additional modification, such as phosphorylation by SOS2. To test this, we monitored SOS2 phosphorylation of SOS3 or SCaBP8 in vitro and, surprisingly, found that SOS2 phosphorylated SCaBP8 but not SOS3 (Figure 1A). SCaBP8L, a mis-spliced cDNA of SCaBP8 that retains the seventh intron and encodes a 211–amino acid protein truncated at its C terminus (Quan et al., 2007), was not phosphorylated by SOS2 (Figure 1A), indicating that the phosphorylation site in SCaBP8 is located in its C terminus. Because SCaBP8 is a calcium binding protein that is thought to sense calcium signals triggered by salt stress to activate SOS2 (Harper et al., 2004; Quan et al., 2007), we determined if calcium is required for SOS2 phosphorylation of SCaBP8. To do this, 0.5 mM calcium or 5 mM EGTA were included in the SOS2 kinase assays. The results indicate that phosphorylation is not enhanced by the presence of calcium (Figure 1B).
Figure 1.
SOS2 Phosphorylates SCaBP8.
(A) SOS2 phosphorylates SCaBP8 but not SOS3. Kinase assay was initiated by adding radiolabeled ATP to the mixture of SOS2 kinase and SOS3/SCaBP8. SDS-PAGE gel with Coomassie blue–stained SOS2 and SCaBP8 (SC8) proteins (top panel); autoradiograph showing SCaBP8 phosphorylation by SOS2 (bottom panel, bottom band) and SOS2 autophosphorylation (bottom panel, top bands).
(B) Calcium does not affect SCaBP8 phosphorylation. SDS-PAGE gel with Coomassie blue–stained SOS2, SOS3, and SCaBP8 proteins (top panel); autoradiograph showing SCaBP8 phosphorylation (bottom panel, bottom bands) and SOS2 autophosphorylation (bottom panel, top bands).
(C) Model of SCaBP8 C-terminal deletions.
(D) SOS2 phosphorylates the SCaBP8 C terminus. SDS-PAGE with Coomassie blue–stained SOS2 and SCaBP8 proteins (top panel); autoradiograph showing SCaBP8 phosphorylation (bottom panel, bottom band) and SOS2 autophosphorylation (bottom panel, top bands).
(E) SOS2 phosphorylates SCaBP8 at Ser-237. SDS-PAGE with Coomassie blue–stained SOS2 and SCaBP8 proteins (top panel); autoradiograph showing SCaBP8 phosphorylation (bottom panel).
SC8, SCaBP8; SC8L, SCaBP81-211aa; SC8M1, SCaBP81-234aa; SC8M2, SCaBP81-213aa; SC8M3, SCaBP81-201aa; SC8S237A, SCaBP8 with S237 mutated to A; SC8T242A, SCaBP8 with T242 mutated to A; SC3S237AT242A, SCaBP8 with S237 and T242 mutated to A.
SOS2 Phosphorylates SCaBP8 at Ser-237
To determine the SOS2 phosphorylation site in SCaBP8, we generated three additional SCaBP8 truncated proteins with C-terminal deletions: glutathione S-transferase (GST)-SCaBP81-201, GST-SCaBP81-213, and GST-SCaBP81-234 (Figure 1C). None of these proteins were phosphorylated by SOS2 (Figure 1D). These results indicated that the phosphorylation site must be in the last 12 amino acids of the SCaBP8 C terminus as the SCaBP81-234 construct had only these 12 amino acids removed. To map the phosphorylation site, we mutated all of the Ser and Thr residues in this region to Ala. When Ser-237 was changed to Ala (SCaBP8S237A), the mutant protein was no longer phosphorylated by SOS2 (Figure 1E). As a control, Thr-242 was also mutated to Ala. While SCaBP8T242A was still phosphorylated by SOS2, the double mutant SCaBP8S237A/T242A (containing both the T242A and S237A mutations) was not (Figure 1E). Our results demonstrate that Ser-237 is the only amino acid phosphorylated by SOS2 in vitro. When we compared the C-terminal sequences of the SCaBP/CBL family members, Ser-237 was found to be conserved in eight of the SCaBP proteins, including SOS3 (see Supplemental Figure 1 online).
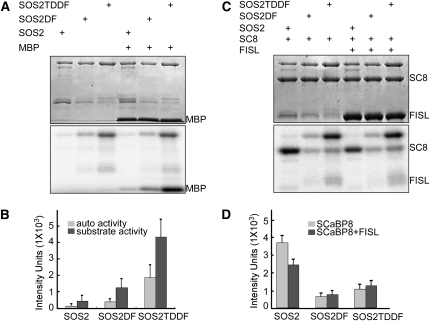
SOS2 Phosphorylation of SCaBP8 Is Enhanced by SCaBP8-SOS2 Interaction
SCaBP8 has been shown to interact with SOS2 via a FISL motif in the SOS2 kinase (Quan et al., 2007). Deletion of this motif or a change in a Thr in the SOS2 kinase loop to Asp (T/D) has been shown to increase SOS2 activity (Guo et al., 2001). To test if the interaction between SCaBP8 and SOS2 is required for phosphorylation, recombinant SOS2 protein, SOS2 protein with the FISL motif deleted (SOS2DF), and SOS2 protein with the combined FISL deletion and T/D change (SOS2T/DDF) were used in our in vitro kinase assays. SOS2T/DDF had the highest activity both in terms of autophosphorylation and phosphorylation of the substrate Myelin Basic Protein (MBP), followed by SOS2DF and SOS2 (Figures 2A and 2B). These results are consistent with data described by Guo et al. (2001). However, when SCaBP8 was used as a substrate, the SCaBP8 phosphorylation level was significantly higher in the assay with wild-type SOS2 protein compared with the assays with SOS2DF or SOS2T/DDF protein (Figures 2C and 2D). These data suggest that, while SOS2-SCaBP8 interaction is not essential for SOS2 phosphorylation of SCaBP8, this interaction dramatically enhances the phosphorylation. Additional support for this conclusion came from experiments in which we added FISL peptide to our SOS2-SCaBP8 kinase assays. The results showed that blocking the SOS2–SCaBP8 interaction with added FISL peptide significantly reduced SOS2 phosphorylation of SCaBP8 (Figures 2C and 2D).
Figure 2.
SOS2–SCaBP8 Interaction Enhances SCaBP8 Phosphorylation.
(A) Highest levels of autophosphorylation and MBP phosphorylation are seen with SOS2T/DDF. SDS-PAGE with Coomassie blue–stained MBP and SOS2 proteins (top panel); autoradiograph showing SOS2 autophosphorylation (bottom panel, top bands) and MBP (bottom panel, bottom bands) phosphorylation.
(B) Quantification of data shown in (A). The signals were quantified by ImageQuant 5.0 software.
(C) Addition of exogenous FISL motif peptide reduces SOS2 phosphorylation of SCaBP8. SDS-PAGE with Coomassie blue–stained SCaBP8, FISL-GST, and SOS2 proteins (top panel); autoradiograph showing SOS2 autophosphorylation (bottom panel, top bands) and SCaBP8 phosphorylation (bottom panel, bottom bands).
(D) Quantification of data shown in (C). The signals were quantified by ImageQuant 5.0 software.
All data represent means ± se of at least three replicate experiments. Auto, SOS2 autophosphorylation; substrate activity, MBP phosphorylation; SOS2DF, SOS2 protein with the FISL motif deleted; SOS2T/DDF, SOS2 protein with the FISL motif deleted and a Thr-to-Asp mutation in the kinase domain.
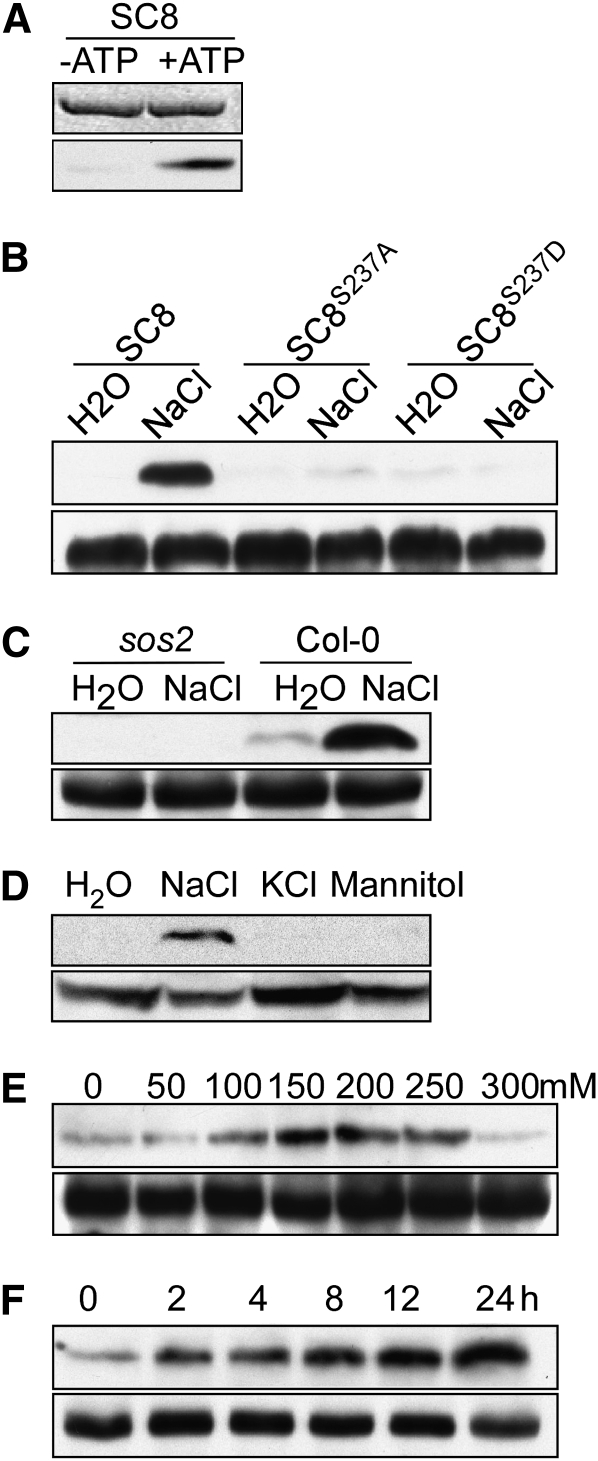
SOS2 Phosphorylation of SCaBP8 Is Induced by NaCl in Planta
To determine if SCaBP8 is phosphorylated by SOS2 in planta, phosphorylation site-specific antibodies were generated by immunizing rabbits with the chemically synthesized phosphorylated peptide C-TAFPpSFIFNTE-NH2 (phosphorylated form). To select the phospho-specific antibodies, the serum was first incubated with the phosphopeptides that had been coupled to SulfoLink resin. After washing the column, the phospho-specific antibodies were eluted at pH 2.7 and immediately neutralized. The antibodies were then run over a column containing the unphosphorylated polypeptide C-TAFPSFIFNTE-NH2 to remove antibodies not specific for the phosphoserine. The flow-through was collected and characterized (Miermont et al., 2000). To evaluate the specificity of the antibodies for the Ser-237 phosphorylation site, SCaBP8 proteins were incubated with SOS2 protein in kinase buffer in the presence or absence of ATP. The proteins were then separated using SDS-PAGE and detected by immunoblotting. A strong cross-reaction was detected only when SCaBP8 was incubated with both SOS2 and ATP (Figure 3A). In the absence of ATP, a very weak signal appeared only after a significantly longer exposure. These results suggest that the antibodies specifically recognize phosphorylated Ser-237 in SCaBP8.
Figure 3.
SOS2 Phosphorylates SCaBP8 in Vivo.
(A) Anti-phosphoserine237 antibodies detect SCaBP8Ser237 phosphorylation in vitro. SDS-PAGE with Coomassie blue–stained SCaBP8 proteins (top panel); autoradiograph showing SCaBP8 phosphorylation detected by anti-phosphoserine237 antibodies (bottom panel).
(B) Anti-phosphoserine237 antibodies detect SCaBP8 but not SCaBP8S237A or SCaBP8S237D in transgenic wild-type plants under salt stress. Proteins extracted from the transgenic plants expressing 6myc-SCaBP8, 6myc-SCaBP8S237A, and 6myc-SCaBP8S237D were analyzed with anti-phosphoserine237 (top panel) or anti-myc (bottom panel) antibodies. Seedlings were treated with NaCl or water for 12 h.
(C) No SCaBP8Ser237 phosphorylation is detected in sos2 mutant plants. Proteins extracted from the wild-type (Col-0) or sos2 plants expressing 6myc-SCaBP8 were analyzed with anti-phosphoserine237 (top panel) or anti-myc (bottom panel) antibodies.
(D) Only NaCl induces SCaBP8Ser237 phosphorylation in vivo. Wild-type (Col-0) seedlings expressing 6myc-SCaBP8 were treated with 100 mM NaCl, 100 mM KCl, 100 mM mannitol, or water for 3 h. Proteins were extracted and analyzed using anti-phosphoserine237 (top panel) or anti-myc (bottom panel) antibodies.
(E) NaCl concentration affects SCaBP8 phosphorylation. Wild-type (Col-0) seedlings expressing 6myc-SCaBP8 were treated with a range of NaCl concentrations. Proteins were extracted and analyzed with anti-phosphoserine237 (top panel) or anti-myc (bottom panel) antibodies.
(F) SCaBP8 phosphorylation increased with longer NaCl treatment. Wild-type (Col-0) seedlings expressing 6myc-SCaBP8 were treated with 100 mM NaCl for the indicated times. Proteins were extracted and analyzed with anti-phosphoserine237 (top panel) or anti-myc (bottom panel) antibodies.
SC8, SCaBP8; SC8S237A, SCaBP8 in which S237 was changed to A; SC8S237D, SCaBP8 in which S237 was changed to D.
To examine SCaBP8 phosphorylation in vivo, we generated 35S:6myc-SCaBP8, 35S:6myc-SCaBP8S237A, and 35S:6myc-SCaBP8S237D transgenic lines in wild-type Arabidopsis (Columbia-0 [Col-0] background) and 35S:6myc-SCaBP8 transgenic lines in the sos2 mutant background. Total proteins were extracted from 10-d-old seedlings of these four transgenic lines and subjected to immunoblots using either anti-myc or anti-phosphoserine237 antibodies. Anti-cmyc (anti-myc) antibody readily detected myc-labeled SCaBP8 in all transgenic plants, but no signal was observed when anti-phosphoserine237 antibodies were used. Because the SOS pathway specifically functions in the response of Arabidopsis to growth in salt, we reasoned that SOS2 phosphorylation of SCaBP8 might be induced by salt stress. To test this, transgenic plants were treated with 100 mM NaCl or water for 12 h, and proteins were extracted and assayed by immunoblotting. The phosphoserine237 signal was detected only in protein extracts from wild-type plants expressing myc-SCaBP8 treated with NaCl (Figures 3B and 3C). No phosphoserine237 signal was seen in wild-type plants expressing either myc-SCaBP8S237D or myc-SCaBP8S237A (Figure 3B) or in sos2 mutant plants expressing myc-SCaBP8 (Figure 3C). Nearly equal amounts of myc-SCaBP8, myc-SCaBP8S237A, or myc-SCaBP8S237D protein were present in the assays as demonstrated by immunoblots with anti-myc antibodies (Figures 3B and 3C). Our data demonstrate that SOS2 specifically phosphorylates SCaBP8S237 and that this phosphorylation is induced by salt stress.
To further determine whether the phosphorylation is specific to NaCl stress, the 35S:6myc-SCaBP8 transgenic plants in the wild-type background were treated with NaCl, KCl, or mannitol. Immunoblots showed that phosphorylation was detected only after NaCl treatment even when similar amounts of myc-SCaBP8 protein were present in each treatment (Figure 3D). To determine if calcium affects this phosphorylation, transgenic plants were treated with 5 or 50 mM CaCl2, and proteins were analyzed by immunoblot assays. Calcium treatment did not significantly enhance phosphorylation (see Supplemental Figure 2 online). SOS2 phosphorylation of SCaBP8 reached a maximum level at NaCl concentrations between 150 and 200 mM and declined at higher NaCl concentrations (Figure 3E). The SCaBP8 phosphorylation level increased gradually with longer NaCl treatments (Figure 3F).
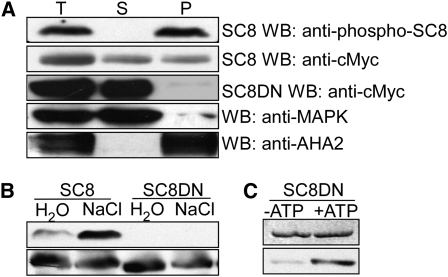
SOS2 Phosphorylates Membrane-Bound SCaBP8
SCaBP8 recruits SOS2 to the plasma membrane in yeast and Arabidopsis (Quan et al., 2007). To determine if SOS2 phosphorylates membrane-bound SCaBP8, the 35S:6myc-SCaBP8 transgenic plants were treated with NaCl, and membrane and soluble proteins were isolated and used in immunoblot assays with anti-phosphoserine237 antibody. The phosphorylation of SCaBP8S237 was detected in total protein extract and in the membrane protein fraction, but not in the soluble protein fraction (Figure 4A). Arabidopsis AHA2 and MAPK3 antibodies were used as markers to label the membrane and soluble fractions, respectively. Using anti-myc antibodies, myc-SCaBP8 protein was detected in all three fractions. These results suggest that the phosphorylated SCaBP8 protein is membrane bound.
Figure 4.
SCaBP8 Phosphorylation Takes Place at the Membrane.
(A) SCaBP8Ser237 phosphorylation was detected only in membrane fractions. Seedlings of wild-type (Col-0) plants expressing 6myc-SCaBP8 or 6myc-SCaBP8DN, in which a 25–amino acid hydrophobic domain in the N terminus of SCaBP8 that is required for SCaBP8 plasma membrane localization was removed, were treated with 100 mM NaCl for 3 h. Membrane and soluble fractions were separated by centrifugation at 4°C for 60 min at 140,000g. Equal aliquots of total (T), supernatant (S), and pellet (P) proteins were separated by SDS-PAGE followed by analysis with anti-myc (loading control), anti-phosphoserine237, anti-MAPK3 (loading control for a soluble-protein fraction), or anti-AHA2 antibodies (loading control for a cell membrane fraction).
(B) SCaBP8Ser237 phosphorylation was not detected in 6myc-SCaBP8DN transgenic plants. Seedlings of wild-type (Col-0) plants expressing 6myc-SCaBP8 or 6myc-SCaBP8DN were treated with 100 mM NaCl or water for 3 h. Proteins were extracted and analyzed with anti-phosphoserine237 (top panel) or anti-myc (bottom panel) antibodies.
(C) Anti-phosphoserine237 antibodies detect SCaBP8Ser237 phosphorylation of SCaBP8DN in vitro. SDS-PAGE with Coomassie blue–stained SCaBP8 proteins (top panel); autoradiograph showing SCaBP8DN phosphorylation detected by anti-phosphoserine237 antibodies (bottom panel).
SC8DN, SCaBP8DN in which a 25–amino acid hydrophobic domain in the N terminus has been deleted; AHA2, isoform of the plasma membrane H+-ATPAse; MAPK3, mitogen-activated protein kinase3.
Previously, we demonstrated that a 25–amino acid hydrophobic domain in the N terminus of SCaBP8 is required for plasma membrane localization in yeast and onion cells (Quan et al., 2007). To determine the importance of plasma membrane localization for SCaBP8 function in salt tolerance, we removed this peptide from SCaBP8 (SCaBP8DN) using PCR-based mutagenesis. The truncated protein was fused to a myc-tag, expressed under the control of the 35S promoter, and transformed into wild type and the scabp8 mutant. T2 seed from >50 independent T1 transgenic lines in the scabp8 background were tested for salt tolerance; none could rescue the mutant phenotype, suggesting that this peptide is essential for SCaBP8 function in salt tolerance (see Supplemental Figure 3 online). To further demonstrate that this peptide is essential for SCaBP8 localization to the membrane in Arabidopsis, we separated membrane and soluble protein fractions from the transgenic plants expressing SCaBP8DN. The myc-SCaBP8DN protein could be detected in the total protein extract and soluble protein fraction but was below detection levels in the membrane fraction (Figure 4A). When we examined the phosphorylation status of SCaBP8DN in transgenic scabp8 plants, the phosphorylated protein was also below the level of detection (Figure 4B). As a control, we purified SCaBP8DN and SOS2 proteins from Escherichia coli, incubated them in kinase buffer with or without ATP, and analyzed the phosphorylation using our anti-phosphoserine237 antibodies. As shown in Figure 4C, SOS2 phosphorylated SCaBP8DN in vitro. Together, these results indicate that the phosphorylation event occurs after SCaBP8 reaches the membrane.
Additional support for SCaBP8 phosphorylation after it reaches the membrane came from studies in which we demonstrated that phosphorylation of SCaBP8 is not required for targeting to the plasma membrane. These studies were based on analysis of the subcellular localization of SCaBP8S237A and SCaBP8S237D mutant proteins in yeast using the Sos Recruitment System (Quintero et al., 2002). This system can monitor the interaction between a plasma membrane–located protein and a cytoplasmic protein fused to the human hSos protein, a functional homolog of the yeast Ras GEF CDC25. When two proteins interact at the plasma membrane, the human hSos protein restores Ras activation and rescues the thermosensitive yeast mutant cdc25-2 (Aronheim et al., 1997). We have shown previously that SCaBP8 recruits SOS2 to the plasma membrane of yeast (Quan et al., 2007). Mutant proteins SCaBP8S237A and SCaBP8S237D were coexpressed with the reporter fusion protein SOS2:hSos to test whether recruitment of the protein kinase to the plasma membrane was affected by mutation of Ser-237 in SCaBP8. As shown in Supplemental Figure 4 online, the S237A and S237D mutations did not significantly affect the targeting of SCaBP8 to the plasma membrane or the recruitment of SOS2. Moreover, when the targeting of SCaBP8 and SCaBP8S237A to the plasma membrane was monitored directly in the presence of the wild-type SOS2 protein or an inactive SOS2 kinase (with a mutation in an essential Lys residue at position 40; SOS2K40N), no difference in subcellular localization was found.
SCaBP8 Phosphorylation Is Required for Salt Tolerance
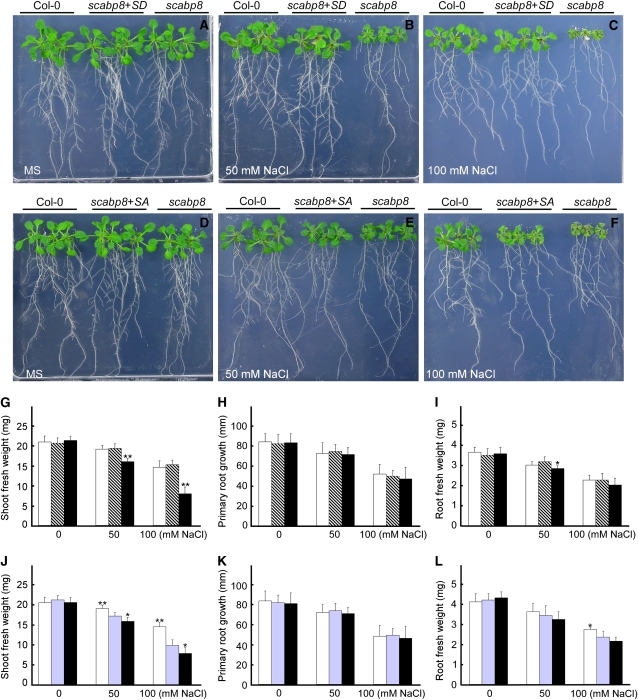
To determine if phosphorylation of SCaBP8 at Ser-237 is required for salt tolerance in Arabidopsis, we transformed 35S:6myc-SCaBP8S237A and 35S:6myc-SCaBP8S237D into the scabp8 mutant. T2 seed from >100 independent T1 transgenic lines from each transformation were tested for their growth in salt. In 93 lines out of a total of 112 independent transgenic lines harboring SCaBP8S237D, the salt-sensitive phenotype was completely suppressed. Partial rescue was seen in 14 lines, while scapb8 mutant levels of sensitivity were seen in only five lines. These numbers are comparable to the complementation rates reported previously for the 35SP:SCaBP8 construct (Quan et al., 2007). These results suggest that mimicking the phosphorylation status of Ser-237 made SCaBP8 fully functional in Arabidopsis. By contrast, no transgenic line was found among 121 T2 lines harboring SCaBP8S237A in which the salt-sensitive phenotype of scabp8 was fully suppressed. Complementation was partial in 78 lines and negligible in the remaining 43 lines. These results demonstrate that phosphorylation of SCaBP8 is essential for salt tolerance (Figure 5). RT-PCR analysis showed that expression of SCaBP8 was similar in the 35S:6myc-SCaBP8S237A, 35S:6myc-SCaBP8S237D, and 35S:6myc-SCaBP8DN transgenic lines (see Supplemental Figure 5 online), demonstrating that differences in complementation were not due to differences in levels of transgene expression.
Figure 5.
SCaBP8S237 Phosphorylation Is Required for SCaBP8 Function in Salt Tolerance.
Growth of scabp8 expressing SCaBP8S237D and SCaBP8S237A. Five-day-old wild-type, transgenic scabp8 plants containing SCaBP8S237D or SCaBP8S237A and scabp8 mutant seedlings were transferred onto Murashige and Skoog (MS) medium ([A] and [D]), MS medium containing 50 mM NaCl ([B] and [E]), or 100 mM NaCl ([C] and [F]). Shoot fresh weight ([G] and [J]), primary root growth ([H] and [K]), and root fresh weight ([I] and [L]) were measured 10 d after transfer. Open bars, wild type; gray bars, SCaBP8S237D transgenic plants; light-blue bars, SCaBP8S237A transgenic plants; black bars, scabp8. All data represent means ± se of at least three replicate experiments. Asterisks indicate significant difference between the transgenic line and the wild type or between the transgenic line and scabp8 (Student's t test, *P < 0.05 and **P < 0.01).
SCaBP8 Phosphorylation Enhances Its Interaction with SOS2
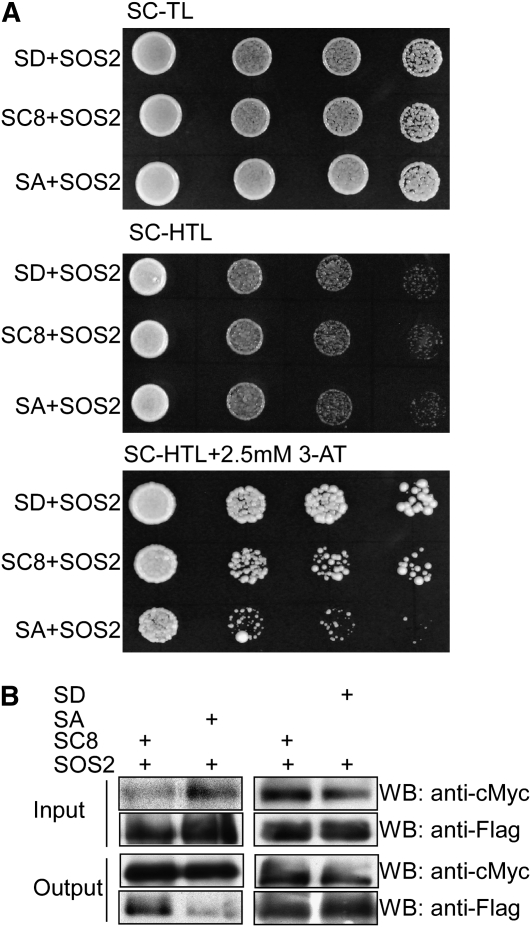
To uncover the biological function of SOS2 phosphorylation of SCaBP8, we determined whether phosphorylation affected the ability of SCaBP8 to bind calcium and to interact with, activate, or recruit SOS2 to the plasma membrane. In vitro, we did not detect a significant difference between SCaBP8 and SCaBP8S237A in calcium binding assays or the activation of SOS2. In yeast, both SCaBP8 and SCaBP8S237A recruited SOS2 to the plasma membrane (see Supplemental Figure 4 online). However, in yeast two-hybrid assays, mutation of Ser to Ala in SCaBP8S237A significantly decreased the interaction between the mutant protein and SOS2 when compared with wild-type SCaBP8 (Figure 6A). By contrast, the SCaBP8S237D mutation, which mimics a phosphorylated Ser residue, enhanced the interaction with SOS2 (Figure 6A). These interaction differences were not due to differences in levels of SCaBP8, SCaBP8S237A, or SCaBP8S237D expression (see Supplemental Figure 6 online). To examine this interaction in planta, we generated 35S:6myc-SOS2 transgenic plants and transformed 35S:3flag-SCaBP8, 35S:3flag-SCaBP8S237A, or 35S:3flag-SCaBP8S237D into protoplasts of a T3 homozygous line expressing 35S:6myc-SOS2 (Sheen, 2001). Protein extracts were sonicated, myc-SOS2 was immunoprecipitated, and flag-SCaBP8 was detected via immunoblot analysis. In comparison to SCaBP8 and SCaBP8S237D, the amount of SCaBP8S237A protein pulled down by SOS2 was substantially reduced (Figure 6B).
Figure 6.
SCaBP8 Phosphorylation Affects Its Interaction with SOS2.
(A) The SCaBP8S237A mutation reduces SCaBP8 interaction with SOS2 in yeast. Yeast strain AH109 harboring SOS2 in the pGADT7 vector was transformed with SCaBP8, SCaBP8S237D, or SCaBP8S237A in the pGBKT7 vector. Growth of the transformed yeast was assayed on media minus Trp and Leu (top panel), minus Trp, Leu, and His (middle panel), or minus Trp, Leu, and His with 2.5 mM 3-amino-1,2,4-triazole (bottom panel).
(B) The SCaBP8S237A mutation reduces SCaBP8 interaction with SOS2 in Arabidopsis. Flag-SCaBP8, flag-SCaBP8S237D, and flag-SCaBP8S237A were expressed in protoplasts from myc-SOS2 transgenic plants. Total protein was analyzed using anti-myc and anti-flag antibodies (Input). After immunoprecipitation with anti-myc conjugated agarose, the components in the pellet were detected via immunoblot analysis (Output). SC8, SCaBP8; SD, SCaBP8S237D; SA, SCaBP8S237A.
SCaBP8S237 Phosphorylation Is Required for Activation of SOS1, the Plasma Membrane Na+/H+ Antiporter
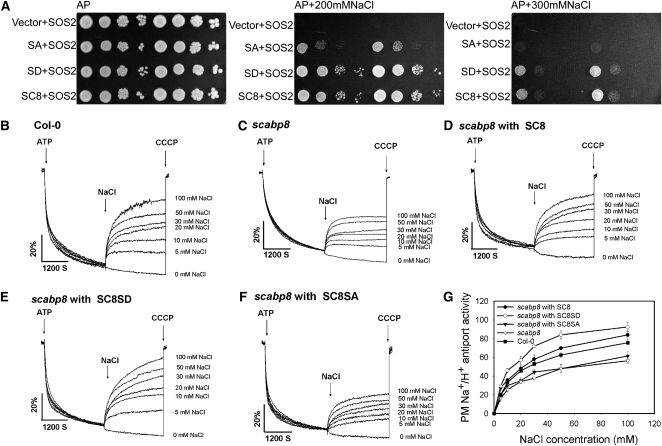
SCaBP8 recruits SOS2 to the plasma membrane, and this complex activates SOS1 in yeast (Quan et al., 2007). To determine if phosphorylation of SCaBP8 affects the capacity of the SCaBP8-SOS2 complex to activate SOS1, yeast cells expressing SOS1 and SOS2 were transformed with empty vector or the SCaBP8, SCaBP8S237A, or SCaBP8S237D construct. The transformed yeast strains were incubated on AP media containing 200 or 300 mM NaCl for 3 d (Figure 7A). SCaBP8S237D in combination with SOS2 activated SOS1 to levels similar to what has been seen for the wild-type SCaBP8-SOS2 complex. However, the activity of SOS1, measured as yeast growth in NaCl, was significantly reduced when SCaBP8S237A mutant protein was used (Figure 7A). These results suggest that full activation of SOS1 by SCaBP8-SOS2 requires the phosphorylation of SCaBP8.
Figure 7.
The Phosphorylation of SCaBP8 Affects Plasma Membrane Na+/H+ Antiport Activity.
(A) SCaBP8S237A cannot activate the SOS pathway in yeast. Yeast AXT3K cells lack the Na+ efflux proteins ENA1-4 and NHA1 and the vacuolar Na+/H+ antiporter NHX1. Expression of SOS1 activated by SOS2-SOS3/SCaBP8 complex can fully restore the NaCl tolerance. Yeast AXT3K cells expressing Arabidopsis SOS1 and transformed with the indicated combinations of Arabidopsis genes were grown overnight in liquid medium with 1 mM KCl. Five microliters of serial decimal dilutions were spotted onto plates of the same medium or media supplemented with 200 or 300 mM NaCl. Each plate has two replicates. Plates were incubated at 28°C for 4 d. AP, Arg phosphate.
(B) to (G) Plasma membrane–enriched vesicles were isolated from NaCl-treated wild-type (Col-0) and scabp8 plants. The Na+ transport assay was initiated by the addition of ATP, and formation of ΔpH was measured at excitation and emission wavelengths of 430 and 500 nm, respectively. When the pH gradient (ΔpH) reached steady state, NaCl was added over a range of final NaCl concentrations from 0 to 100 mM, and initial rates of Na+-induced dissipation of ΔpH were measured.
(B) and (C) Plasma membrane Na+/H+ antiport activity is reduced in the scabp8 mutant. Wild-type (Col-0) (B) and scabp8 (C) plasma membrane Na+/H+ antiport activity as a function of NaCl concentration.
(D) to (F) Recombinant SCaBP8 (D) and SCaBP8S237D (E) proteins stimulate plasma membrane Na+/H+ antiport activity in the scabp8 mutant, while SCaBP8S237A (F) does not.
(G) Na+/H+ antiport activity is reduced in the scabp8 mutant compared with the wild type over a range of NaCl concentrations. The units of Na+/H+ antiport activity are Δ%F/min/mg protein.
For (B) to (F), one representative experiment of three replicates is shown. For (G), data represent means ± se of at least three replicate experiments. Each replicate experiment was performed using independent membrane preparations. SC8, SCaBP8 protein; SC8SD, SCaBP8S237D protein; SC8SA, SCaBP8S237A protein; CCCP, m-chlorophenylhydrazone; PM, plasma membrane.
To determine whether SCaBP8 affects the activity of SOS1 in Arabidopsis, Na+-induced dissipation of a pH gradient (ΔpH), which is monitored by a Na+-induced increase in quinacrine fluorescence that is positively related to Na+/H+ exchange activity, was examined. Plasma membrane vesicles were isolated from 5-week-old plants of the wild type and the scabp8 mutant treated with 250 mM NaCl for 3 d. As shown in Figures 7B and 7C, plasma membrane Na+/H+ antiport activity was reduced in scabp8 plants compared with the wild type. The initial rate of dissipation of ΔpH by 100 mM NaCl was 75 units/min/mg protein in wild-type plants compared with 55 units/min/mg protein in scabp8. Over a range of NaCl concentrations between 0 and 100 mM, plasma membrane Na+/H+ antiport activity was consistently lower in scabp8 than in the wild type. Together with our yeast data, reduced Na+/H+ transport activity in scabp8 plants demonstrates that SCaBP8 is involved in the regulation of SOS1 activity.
Next, we analyzed whether the phosphorylation of SCaBP8 also affects SOS1 activity. Plasma membrane vesicles were isolated from the scabp8 mutant, and purified SCaBP8, SCaBP8S237D, or SCaBP8S237A protein was added to the Na+/H+ antiport assays. In the presence of 250 ng/mL SCaBP8, Na+/H+ antiport activity in scabp8 plants was slightly higher than activity in wild-type plants (Figure 7). Stimulation of Na+/H+ antiport activity in scabp8 mutant vesicles by SCaBP8S237D was greater than of SCaBP8; however, little effect of SCaBP8S237A was observed. Boiled recombinant SCaBP8, SCaBP8S237D, or SCaBP8S237A proteins did not have any effect on antiport activity. These data demonstrate that the phosphorylation of SCaBP8 is critical for the regulation of plasma membrane Na+/H+ antiport activity.
DISCUSSION
SCaBP calcium sensor activation of its interacting SOS2-like protein kinase is critical for its function in regulating ion transport in Arabidopsis (Qiu et al., 2002; Shi et al., 2002; Xu et al., 2006, Fuglsang et al., 2007, Quan et al., 2007). While SCaBP proteins do not have predicted transmembrane domains, some family members locate at and recruit PKS kinases to cellular membranes to perform their function. Lipid modifications of the SCaBP proteins have been shown to be essential for their function or membrane localization in planta (Ishitani et al., 2000; Batistic et al., 2008). N-terminal myristoylation of SOS3 is important for its function in salt tolerance, and, in yeast, it is required to recruit SOS2 to the plasma membrane (J.M. Pardo, unpublished results). Whether myristoylation is required for SOS3 plasma membrane localization in Arabidopsis remains to be determined. Like SOS3, SCaBP8 activates SOS2 kinase activity and recruits SOS2 to the plasma membrane, but it lacks a myristoylation signature sequence (MGXXXS/K). SCaBP8 protein associates with the plasma membrane due to an N-terminal hydrophobic domain (Quan et al., 2007). This association is very stable because high concentrations of KCl or Triton X-100 do not remove it from the plasma membrane (Quan et al., 2007). In this study, we determined that, in Arabidopsis, SCaBP8 is phosphorylated by SOS2 and that this modification stabilizes the SOS2-SCaBP8 complex at the membrane by enhancing the interaction of these two proteins. This finding identifies a posttranslational modification and an additional level of regulation within the SCaBP family.
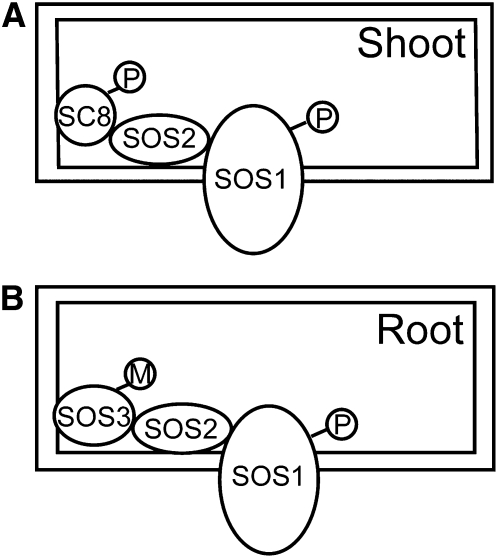
It is believed that once the calcium signal is induced by stress stimuli, calcium binds to and activates SCaBP proteins (Harper et al., 2004). Some of the SCaBP proteins in turn interact with, activate, and recruit PKS protein kinases to cellular membranes and regulate their target activities. Both SOS3 and SCaBP8 function in Arabidopsis salt tolerance by activating SOS2 in the plasma membrane (Liu et al., 2000; Quan et al., 2007). However, different modifications are required for their full function in vivo. It is likely that myristoylation of SOS3 targets the SOS3-SOS2 complex to the plasma membrane in roots, phosphorylation of SCaBP8 by SOS2 stabilizes SOS2 at the membrane in shoots, and SOS3/SCaBP8-SOS2 phosphorylates SOS1 and enhances the activity of this plasma membrane Na+/H+ antiporter (Figure 8).
Figure 8.
A Working Model of Activation of SOS2 by SCaBP8 in Shoots and by SOS3 in Roots.
(A) In shoot tissue, SCaBP8 recruits SOS2 to the plasma membrane and then the salt-induced phosphorylation strengthens the SCaBP8-SOS2 complex at the plasma membrane. SOS2 then phosphorylates and activates SOS1.
(B) In root tissue, the N-terminal myristoylation of SOS3 recruits SOS2 to the plasma membrane. SOS2 phosphorylates and activates SOS1.
SC8, SCaBP8; M, myristoylation; P, phosphorylation.
The SOS2 FISL motif is essential for the interaction of SOS2 with either SOS3 or SCaBP8 (Guo et al., 2001; Quan et al., 2007). The FISL motif inserts into a crevice of SOS3 that is surrounded by four EF-hands (Sanchez-Barrena et al., 2007). SOS3 and SCaBP8 share high sequence similarity in their EF-hand domains and but have distinct N- and C-terminal domains. In studies of SOS3 crystals, the C-terminal peptide was disordered and could not be detected in the structure (Sanchez-Barrena et al., 2005). Although the three-dimensional structure of SCaBP8 is not yet available, its C terminus shares higher sequence similarity with SCaBP1/CBL2 than SOS3. In SCaBP1, the C-terminal peptide spans and covers the crevice generated by the EF-hand domains (Nagae et al., 2003; Akaboshi et al., 2008). By analogy, a similar mechanism in SCaBP8 might obstruct the interaction between SOS2 and SCaBP8. Upon phosphorylation by SOS2 under salt stress, the C terminus of SCaBP8 might be displaced to allow a better fit for the FISL motif of SOS2 into the crevice. This is consistent with our data indicating that the phosphorylation affects, above all, the strength of the interaction between SOS2 and SCaBP8.
The finding that SCaBP8 protein is present in both soluble and membrane fractions (Figure 4; Quan et al., 2007) while the phosphorylated form of SCaBP8 is found exclusively in the membrane fraction is intriguing. Because the truncated, cytosolic mutant SCaBP8DN protein was phosphorylated in vitro by SOS2 but not in vivo (Figure 4B) and mutation of Ser-237 did not compromise the targeting of SCaBP8 to the plasma membrane (see Supplemental Figure 4 online), we conclude that phosphorylation of SCaBP8 is not a prerequisite for its targeting to the plasma membrane. Rather, phosphorylation appears to take place only after SCaBP8 reaches the membrane. This would, in turn, suggest that the phosphorylation event is triggered by additional input(s) beyond the interaction between SCaBP8 and SOS2 which could, theoretically, also take place in the cytoplasm. We speculate that it is the interaction of the SCaBP8-SOS2 complex with its downstream target that brings about the phosphorylation of SCaBP8 and stabilization of the ternary complex. This model is consistent with the fact that SCaBP8S237A, which cannot be phosphorylated, was still targeted to the plasma membrane but was incompetent to mediate full activation of SOS1 in yeast cells and plasma membrane–enriched vesicles from Arabidopsis (Figure 7) or to confer salt tolerance in planta (Figure 5). In mammalian cells, calcineurin B homologous proteins regulate Na+/H+ exchange activity by interacting with the cytoplasmic domain of the transporter (Pang et al., 2001). We cannot exclude the possibility that SCaBP8 directly associates with the SOS1 and that the phosphorylation of SCaBP8 by SOS2 strengthens this interaction. It is worth noting that SOS2 has been shown to activate CAX1, the vacuolar Ca2+/H+ exchanger, through protein–protein interaction that is independent of protein phosphorylation (Cheng et al., 2004). It is tempting to speculate that phosphorylation of the SCaBP subunit is an alternative way to regulate the activity of the target protein rather than by phosphorylation of the target. If this is indeed the case, it might offer a reasonable explanation to the puzzling observation that SCaBP8 and SOS3 cannot replace each other in reciprocal mutant complementation assays (Quan et al., 2007). While the SOS3-SOS2 complex regulates its target protein by phosphorylation of the target (Quintero et al., 2002; Fujii and Zhu, 2009), the SCaBP8-SOS2 complex may do so by stabilization of the ternary complex after phosphorylation of SCaBP8.
Kim et al. (2007) reported that the CBL10 (SCaBP8)-CIPK24 (SOS2) complex is also associated with the vacuolar membrane (tonoplast) and regulates vacuolar sequestration of Na+. In addition, SOS2 has been shown indirectly to regulate Na+/H+ antiport activity in tonoplast-enriched vesicles from Arabidopsis (Qiu et al., 2004). SOS2 is known to physically interact with two ion transporters at the tonoplast, the vacuolar H+-ATPase regulatory subunits B1 and B2 and CAX1 (Cheng et al., 2004; Batelli et al., 2007). H+ transport activity in tonoplast vesicles isolated from sos2-2 mutant cells was reduced relative to vesicles from wild-type cells. However, whether SCaBP8 is required for these transport processes occurring at the tonoplast was not determined. Our unpublished results (J.M. Pardo) indicate that Na+/H+ exchange in tonoplast vesicles is not altered in the scabp8 mutant. By contrast, we provide compelling evidence that Na+/H+ exchange at the plasma membrane of Arabidopsis is severely reduced in the scabp8 mutant (Figure 7). SOS1 is the only known protein responsible for Na+/H+ exchange at the plasma membrane (Qiu et al., 2002), in keeping with our data showing that SOS1 is a primary target of the SCaBP8-SOS2 kinase complex (Figure 7; Quan et al., 2007).
METHODS
Plasmid Construction
To produce SCaBP8L, SCaBP81-201, SCaBP81-213, SCaBP81-234, SCaBP8S237A, SCaBP8T242A, and SCaBP8S237AT242A GST fusion proteins, the DNA fragments were amplified with the primers listed in Supplemental Table 1 online. The SOS2 FISL motif was amplified from the pGEX-2TK-SOS2 plasmid using the primer pairs 5′-CGGGATCCATGATGAATGCCTTTGAGATG-3′ and 5′-AGCGTCGACTCAGTCAAATAGTGCAGATAAA-3′. These PCR products were cloned into the pGEX-6P-1 vector at the BamHI and SalI sites. All primers and plasmid constructs used in these studies are listed in Supplemental Table 1 online.
Protein Purification and Kinase Assays
All constructs were transformed into Escherichia coli strain BL21 (DE3).
The recombinant proteins (including SCaBP8, SOS3, and SOS2) were purified with glutathione sepharose (Amersham Pharmacia) according to the manufacturer's protocol.
Kinase buffer included 20 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 10 μM ATP, and 1 mM DTT. Kinase assays (total volume of 20 μL) were started by adding 1 μg protein and 0.5 μL of [γ-32P]ATP (5 μCi), and the mixtures were incubated at 30°C for 30 min (Quan et al., 2007). Reactions were terminated by the addition of 6× SDS loading buffer and were incubated at 95°C for 5 min. The proteins were separated using 12% (w/v) SDS-PAGE and stained with Coomassie Brilliant Blue R 250 followed by exposure to a phosphor screen (Amersham Biosciences). Signals were captured by a Typhoon 9410 phosphor imager (Amersham Biosciences), and signals were quantified by ImageQuant 5.0 software.
Preparation of Anti-Phosphoserine237 SCaBP8 Polyclonal Antibodies
Anti-phosphoserine237 polyclonal antibodies were made by AbMart (www.ab-mart.com.cn). Two 9–amino acid peptides (corresponding to amino acids 232 to 243 of SCaBP8) with N-terminal Cys residues, C-TAFPpSFIFNTE-NH2 (phosphorylated form) and C-TAFPSFIFNTE-NH2 (nonphosphorylated form), were also synthesized by AbMart. The Ser-237 phosphospecific peptide (C-TAFPpSFIFNTE-NH2) was used to produce polyclonal phosphospecific antibodies, and the nonphosphorylated peptide (C-TAFPSFIFNTE-NH2) was used for screening and purification.
SCaBP8Ser237 Phosphorylation in Planta
35S:6myc-SCaBP8, 35S:6myc-SCaBP8S237A, 35S:6myc-SCaBP8S237D, and 35S:6myc-SCaBP8DN plasmids were constructed by excising the DNA fragments with BamHI and SalI from GST-SCaBP8, GST-SCaBP8S237A, GST-SCaBP8S237D, and GST-SCaBP8DN plasmids and cloning them into the pCAMBIA1307-6myc binary vector downstream of the myc tag. The resulting constructs were introduced into Agrobacterium tumefaciens GV3101 and transformed into Arabidopsis thaliana Col-0, scabp8, or the sos2 mutant. T3 homozygous lines were treated with different concentrations of NaCl for the times indicated or with 100 mM KCl or 100 mM mannitol for 12 h. Plant protein was extracted using 2× cold extraction buffer containing 10 mM Tris-Cl, 150 mM NaCl, 2 mM EDTA, 0.5% (v/v) Nonidet P-40, 2× protease inhibitor (Roche), and 2× phosphatase inhibitor (Roche). The resulting samples were then analyzed by SDS-PAGE and blotted onto a polyvinylidene difluoride membrane (Millipore). The blots were probed with primary anti-SCaBP8-phosphoserine237 or anti-cmyc (Sigma-Aldrich; M4439) antibodies, and chemiluminescence signals were detected by film.
Isolation of Soluble and Membrane Fractions
Transgenic plants were homogenized with extraction buffer. Total crude homogenate was centrifuged at 4°C for 10 min at 10,000g, and the resulting supernatant was centrifuged at 4°C for 60 min at 140,000g to generate microsomal membrane (a mixture of small membrane vesicles and fragments arising from the plasma membrane, tonoplast, endoplasmic reticulum, and Golgi apparatus) and soluble protein fractions.
Salt Sensitivity Assays
Seeds of wild-type Arabidopsis, transgenic plants, and the scabp8 mutant were sterilized in a solution containing 20% (v/v) sodium hypochlorite and 0.1% (v/v) Triton X-100 for 10 min, washed five times with sterile water, and sown on MS medium with 0.6% (w/v) Phytagel agar (Sigma-Aldrich). The plates were placed at 4°C for 2 d, and then the seeds were germinated vertically at 23°C under continual illumination. Four-day-old seedlings were transferred onto MS medium with salts added as described.
Yeast Two-Hybrid Assays
SOS2 coding sequence was amplified with the following primers: 5′-TGGAATTCATGACAAAGAAAATGAGAAG-3′ and 5′-CGGGATCCTCAAAACGTGATTGTTCTGAG-3′. The PCR product was digested with EcoRI and BamHI and then cloned into pGADT7. SCaBP8, SCaBP8S237A, and SCaBP8S237D were amplified from the GST-SCaBP8, GST-SCaBP8S237A, and GST-SCaBP8S237D plasmids with the primer pairs 5′-TGGAATTCATGGAACAAGTTTCCTCTAG-3′ and 5′-AGCGTCGACTCAGTCTTCAACCTCAGTGTT-3′ and cloned into pGBKT7 with EcoRI and SalI. Yeast strain AH109 expressing pGADT7-SOS2 (prey) was transformed with pGBKT7-SCaBP8, pGBKT7-SCaBP8S237A, or pGBKT7-SCaBP8S237D (baits). Transformed yeast cells were selected on synthetic complete medium lacking Trp and Leu. Empty prey or bait vector was transformed with SOS2 or different SCaBP8 proteins as negative controls. Interaction was determined on synthetic complete medium lacking Trp, Leu, and His and supplemented with 2.5 mM 3-amino-1,2,4-triazole (Sigma-Aldrich). All bait proteins were tested for self-activation; none were found to activate the two reporter genes His3 or LacZ.
Coimmunoprecipitation Assays
The coding sequences of SOS2, SCaBP8, SCaBP8S237A, and SCaBP8S237D were translationally fused downstream of the c-myc or flag tags and cloned into the pCAMBIA1205 vector. The plasmids were purified by CsCl gradient centrifugation and transformed into Arabidopsis mesophyll protoplasts. After overnight incubation, the protoplasts were lysed, sonicated, and centrifuged. Ten microliters of anti-myc agarose conjugate (Sigma-Aldrich) was incubated with the extract supernatant overnight at 4°C. After washing five times in 1 mL of extraction buffer, the coimmunoprecipitation products were detected via immunoblot analysis. Both anti-myc (Sigma-Aldrich) and anti-flag (Sigma-Aldrich) antibodies were used at 1:5000 dilutions, and chemiluminescence signals were detected by film.
Plasma Membrane Isolation
Plasma membrane–enriched vesicles were isolated from 5-week-old plants using aqueous two-phase partitioning as described by Qiu et al. (2002). All steps were performed at 4°C or on ice. Plants were homogenized in isolation buffer containing 0.33 M sucrose, 10% (w/v) glycerol, 0.2% (w/v) BSA, 5 mM EDTA, 5 mM DTT, 5 mM ascorbate, 0.2% (w/v) casein, 0.6% (w/v) polyvinylpyrrolidone, 1 mM PMSF, 1× protease inhibitor, and 50 mM HEPES-KOH, pH 7.5. Two to four milliliters of homogenization buffer were used per gram of tissue. The homogenate was filtered through one layer of miracloth and centrifuged at 13,000g for 10 min. The supernatant then was centrifuged for 50 min at 80,000g to obtain a microsomal pellet that was resuspended in a buffer containing 0.33 M sucrose, 3 mM KCl, 0.1 mM EDTA, 1 mM DTT, 1 mM PMSF, 1× protease inhibitor, and 5 mM potassium phosphate, pH 7.8. The suspension was added to a phase mixture to obtain a phase system consisting of 6.2% (w/w) Dextran T-500 and 6.2% (w/w) polyethylene glycol 3350 in 5 mM potassium phosphate, pH 7.8, buffer containing 0.33 M sucrose and 3 mM KCl. The final upper phases were collected, diluted with resuspension buffer containing 0.33 M sucrose, 10% (w/v) glycerol, 0.1% (w/v) BSA, 0.1 mM EDTA, 2 mM DTT, 1× protease inhibitor, and 20 mM HEPES-KOH, pH 7.5, and centrifuged for 50 min at 100,000g. The resulting pellet was collected and resuspended with the above-described resuspension buffer containing 1 mM EDTA.
Plasma Membrane Na+/H+ Antiport Assays
Na+/H+ antiport activity was measured as a Na+-induced dissipation of the pH gradient (ΔpH; i.e., a Na+-induced increase in quinacrine fluorescence) as described by Qiu et al. (2002). Recombinant SCaBP8, SCaBP8S237D, or SCaBP8S237A protein (250 ng/mL) was preincubated for 10 min at room temperature with plasma membrane vesicles isolated from scabp8 mutant plants. An inside-acid ΔpH was formed in the vesicles by the activity of the H+-ATPase and measured as a decrease (quench) in the fluorescence of quinacrine (a pH-sensitive fluorescent probe). Assays (2 mL) contained 5 μM quinacrine, 3 mM MgSO4, 100 mM KCl, 25 mM 1,3-bis[Tris(hydroxylmethyl)methylamino]propane-HEPES, pH 6.5, 250 mM mannitol, and 50 μg/mL of plasma membrane protein. Reactions were mixed by inversion several times and then placed in a dark chamber in a fluorescence spectrophotometer (Hitachi F-4500). Reactions were equilibrated in the dark with stirring for 5 min before beginning fluorescence readings. The assay was initiated by the addition of ATP to a final concentration of 3 mM, and formation of ΔpH was measured at excitation and emission wavelengths of 430 and 500 nm, respectively. When the maximum ΔpH was formed (reached steady state), NaCl was added to initiate Na+ transport. At the end of each reaction, 10 μM (final concentration) of the protonophore m-chlorophenylhydrazone (CCCP) was added to dissipate any remaining ΔpH. Specific activity was calculated by dividing the initial rate by the mass of plasma membrane protein in the reaction (Δ%F/min per mg of protein). Unless indicated, all data represent means ± se of at least three replicate experiments.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession numbers At5g24270 (SOS3) and At4g33000 (SCaBP8).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Comparison of the C-Terminal Sequence of the SCaBP/CBL Family Members.
Supplemental Figure 2. Calcium Does Not Enhance the Phosphorylation of SCaBP8.
Supplemental Figure 3. A 25–Amino Acid Hydrophobic Peptide in the N Terminus of SCaBP8 Is Required for Salt Tolerance.
Supplemental Figure 4. Ser-237 Is Not Required for Targeting SCaBP8 to the Plasma Membrane or for Recruitment of SOS2.
Supplemental Figure 5. Expression of SCaBP8 in Wild-Type and scabp8 Mutant Plants.
Supplemental Figure 6. Expression of SCaBP8 in Yeast.
Supplemental Table 1. Primers Used for Plasmid Construction.
Supplemental Data Set 1. Text File Corresponding to Alignment in Supplemental Figure 1.
Supplementary Material
Acknowledgments
We thank Xing Wang Deng and Jianmin Zhou for critical reading of the manuscript and stimulating discussions, Quansheng Qiu for help with the measurement of Na+/H+ antiport activity, Anja T. Fuglsang and Michael G. Palmgren for providing anti-AHA2 antibodies, and Cheng Zhan and Jun Zhang for excellent technical assistance. This work was supported by National Basic Research Program of China Grant 2006CB100100 and National High Technology Research and Development Program of China 863 Grant 2003AA210100 to Y.G., by Spanish Ministry of Science and Innovation Grant BFU2006-06968 to J.M.P., and by U.S. Department of Energy/Energy Biosciences Grant DE-FG02-04ER15616 to K.S.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Yan Guo (guoyan@nibs.ac.cn).
Online version contains Web-only data.
References
- Akaboshi, M., Hashimoto, H., Ishida, H., Saijo, S., Koizumi, N., Sato, M., and Shimizu, T. (2008). The crystal structure of plant-specific calcium-binding protein AtCBL2 in complex with the regulatory domain of AtCIPK14. J. Mol. Biol. 377 246–257. [DOI] [PubMed] [Google Scholar]
- Aronheim, A., Zandi, E., Hennemann, H., Elledge, S.J., and Karin, M. (1997). Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol. Cell. Biol. 17 3094–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batelli, G., Verslues, P.E., Agius, F., Qiu, Q., Fujii, H., Pan, S., Schumaker, K.S., Grillo, S., and Zhu, J.-K. (2007). SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Mol. Cell. Biol. 27 7781–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batistic, O., Sorek, N., Schultke, S., Yalovsky, S., and Kudla, J. (2008). Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell 20 1346–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, N.H., Pittman, J.K., Zhu, J.-K., and Hirschi, K.D. (2004). The protein kinase SOS2 activates the Arabidopsis H+/Ca2+ antiporter CAX1 to integrate calcium transport and salt tolerance. J. Biol. Chem. 279 2922–2926. [DOI] [PubMed] [Google Scholar]
- Cheong, Y.H., Pandey, G.K., Grant, J.J., Batistic, O., Li, L., Kim, B.-G., Lee, S.-C., Kudla, J., and Luan, S. (2007). Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 52 223–239. [DOI] [PubMed] [Google Scholar]
- Fuglsang, A.T., Guo, Y., Cuin, T.A., Qiu, Q., Song, C.P., Kristiansen, K.A., Bych, K., Schulz, A., Shabala, S., Schumaker, K.S., Palmgren, M.G., and Zhu, J.-K. (2007). Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19 1617–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, H., and Zhu, J.-K. (2009). An autophosphorylation site of the protein kinase SOS2 is important for salt tolerance in Arabidopsis. Mol. Plant 2 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, D., Guo, Y., Schumaker, K.S., and Zhu, J.-K. (2004). The SOS3 family of calcium sensors and SOS2 family of protein kinases in Arabidopsis. Plant Physiol. 134 919–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y., Halfter, U., Ishitani, M., and Zhu, J.-K. (2001). Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13 1383–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter, U., Ishitani, M., and Zhu, J.-K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 97 3735–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J.F., Breton, G., and Harmon, A. (2004). Decoding Ca2+ signals through plant protein kinases. Annu. Rev. Plant Biol. 55 263–288. [DOI] [PubMed] [Google Scholar]
- Ishitani, M., Liu, J., Halfter, U., Kim, C.S., Shi, W., and Zhu, J.-K. (2000). SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, B.-G., Waadt, R., Cheong, Y.H., Pandey, G.K., Dominguez-Solis, J.R., Schultke, S., Lee, S.C., Kudla, J., and Luan, S. (2007). The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J. 52 473–484. [DOI] [PubMed] [Google Scholar]
- Kim, K.N., Cheong, Y.H., Gupta, R., and Luan, S. (2000). Interaction specificity of Arabidopsis calcineurin B-like calcium sensors and their target kinases. Plant Physiol. 124 1844–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolukisaoglu, U., Weinl, S., Blazevic, D., Batistic, O., and Kudla, J. (2004). Calcium sensors and their interacting protein kinases: Genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 134 43–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., and Zhu, J.-K. (1998). A calcium sensor homolog required for plant salt tolerance. Science 280 1943–1945. [DOI] [PubMed] [Google Scholar]
- Liu, J., Ishitani, M., Halfter, U., Kim, C.-S., and Zhu, J.-K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. USA 97 3730–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan, S., Kudla, J., Rodriguez-Concepcion, M., Yalovsky, S., and Gruissem, W. (2002). Calmodulins and calcineurin B-like proteins: Calcium sensors for specific signal response coupling in plants. Plant Cell 14 S389–S400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miermont, A.M., Mohamed, A.S., and Swope, S.L. (2000). Generation of phosphorylation state-specific SRC-class kinase antibodies for analysis of kinase activation. J. Immunol. Methods 246 203–215. [DOI] [PubMed] [Google Scholar]
- Nagae, M., Nozawa, A., Koizumi, N., Sano, H., Hashimoto, H., Sato, M., and Shimizu, T. (2003). The crystal structure of the novel calcium-binding protein AtCBL2 from Arabidopsis thaliana. J. Biol. Chem. 278 42240–42246. [DOI] [PubMed] [Google Scholar]
- Pang, T., Su, X., Wakabayashi, S., and Shigekawa, M. (2001). Calcineurin homologous protein as an essential cofactor for Na+/H+ exchangers. J. Biol. Chem. 276 17367–17372. [DOI] [PubMed] [Google Scholar]
- Qiu, Q.S., Guo, Y., Dietrich, M.A., Schumaker, K.S., and Zhu, J.-K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 99 8436–8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, Q.S., Guo, Y., Quintero, F.J., Pardo, J.M., Schumaker, K.S., and Zhu, J.-K. (2004). Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J. Biol. Chem. 279 207–215. [DOI] [PubMed] [Google Scholar]
- Quan, R., Lin, H., Mendoza, I., Zhang, Y., Cao, W., Yang, Y., Shang, M., Chen, S., Pardo, J.M., and Guo, Y. (2007). SCaBP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19 1415–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero, F.J., Ohta, M., Shi, H., Zhu, J.-K., and Pardo, J.M. (2002). Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. USA 99 9061–9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Barrena, M.J., Fujii, H., Angulo, I., Martinez-Ripoll, M., Zhu, J.-K., and Albert, A. (2007). The structure of the c-terminal domain of the protein kinase AtSOS2 bound to the calcium sensor AtSOS3. Mol. Cell 26 427–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Barrena, M.J., Martinez-Ripoll, M., Zhu, J.-K., and Albert, A. (2005). The structure of the Arabidopsis thaliana SOS3: Molecular mechanism of sensing calcium for salt stress response. J. Mol. Biol. 345 1253–1264. [DOI] [PubMed] [Google Scholar]
- Sheen, J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127 1466–1475. [PMC free article] [PubMed] [Google Scholar]
- Shi, H., Ishitani, M., Kim, C., and Zhu, J.-K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 97 6896–6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H., Quintero, F.J., Pardo, J.M., and Zhu, J.-K. (2002). The putative plasma membrane Na(+)/H(+) antiporter SOS1 controls long-distance Na(+) transport in plants. Plant Cell 14 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., Li, H.D., Chen, L.Q., Wang, Y., Liu, L.L., He, L., and Wu, W.H. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125 1347–1360. [DOI] [PubMed] [Google Scholar]
- Zhu, D., Cardenas, M.E., and Heitman, J. (1995). Myristoylation of calcineurin B is not required for function or interaction with immunophilin–immunosuppressant complexes in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 270 24831–24838. [DOI] [PubMed] [Google Scholar]
- Zhu, J.-K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.-K., Liu, J., and Xiong, L. (1998). Genetic analysis of salt tolerance in Arabidopsis: Evidence for a critical role of potassium nutrition. Plant Cell 10 1181–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.