Abstract
Plant roots show an impressive degree of plasticity in adapting their branching patterns to ever-changing growth conditions. An important mechanism underlying this adaptation ability is the interaction between hormonal and developmental signals. Here, we analyze the interaction of jasmonate with auxin to regulate lateral root (LR) formation through characterization of an Arabidopsis thaliana mutant, jasmonate-induced defective lateral root1 (jdl1/asa1-1). We demonstrate that, whereas exogenous jasmonate promotes LR formation in wild-type plants, it represses LR formation in jdl1/asa1-1. JDL1 encodes the auxin biosynthetic gene ANTHRANILATE SYNTHASE α1 (ASA1), which is required for jasmonate-induced auxin biosynthesis. Jasmonate elevates local auxin accumulation in the basal meristem of wild-type roots but reduces local auxin accumulation in the basal meristem of mutant roots, suggesting that, in addition to activating ASA1-dependent auxin biosynthesis, jasmonate also affects auxin transport. Indeed, jasmonate modifies the expression of auxin transport genes in an ASA1-dependent manner. We further provide evidence showing that the action mechanism of jasmonate to regulate LR formation through ASA1 differs from that of ethylene. Our results highlight the importance of ASA1 in jasmonate-induced auxin biosynthesis and reveal a role for jasmonate in the attenuation of auxin transport in the root and the fine-tuning of local auxin distribution in the root basal meristem.
INTRODUCTION
Root systems of higher plants mainly consist of an embryonic primary root and postembryonically developed lateral roots (LRs). Root branching or LR formation in higher plants is influenced by a wide range of environmental cues, such as nutrients and water availability in the soil (Malamy, 2005). The plasticity of LR formation is of critical importance for sessile plants, allowing them to compete for resources in the soil and adapt to constantly changing growth conditions. The mechanisms by which plants incorporate diverse regulatory signals into the developmental program of LRs are under active investigation.
Extensive studies in Arabidopsis thaliana have detailed the developmental processes associated with the initiation and later development of the lateral root primordium (LRP). A LRP is derived from the pericycle founder cell located in vascular tissues of the primary root (Malamy and Benfey, 1997; Casimiro et al., 2001; Dubrovsky et al., 2001). An early event at the onset of LRP initiation is that a small number of mature pericycle cells adjacent to the xylem poles divide asymmetrically to form daughter cells that are shorter than the flanking undivided pericycle cells. These daughter cells then undergo a series of precisely oriented cell divisions and expansions leading to the formation of a new organ. Eventually, an emerged LR contains a meristem that has essentially the same structure as that of the primary root meristem.
Despite structural and cytological changes occurring during LRP initiation and development, the underlying molecular mechanisms are not yet fully understood. The phytohormone auxin seems to play an important role during each stage of LR formation (Casimiro et al., 2003; De Smet et al., 2006; Fukaki et al., 2007; Nibau et al., 2008; Fukaki and Tasaka, 2009). Physiological studies have indicated that application of auxin stimulates LR formation (Celenza et al., 1995), whereas application of polar auxin transport (PAT) inhibitors prevents LR formation (Casimiro et al., 2001; Himanen et al., 2002). Consistently, Arabidopsis mutants with elevated endogenous auxin levels have an increased number of LRs; mutants with defective auxin transport, perception, or signaling consistently show reduced LR formation (Casimiro et al., 2003; De Smet et al., 2006; Nibau et al., 2008).
The essential role of auxin during LRP development has been confirmed and refined by recent in-depth investigations using the auxin-responsive reporter DR5:β-glucuronidase (GUS) (Sabatini et al., 1999; Benková et al., 2003; Dubrovsky et al., 2008). These studies elegantly demonstrated that auxin is the local instructive signal for specification of founder cells that give rise to LRPs. Furthermore, establishment of an auxin gradient with its maximum at the tip is important for proper LRP development. This gradient is dependent on PAT mediated by the PIN-FORMED (PIN) auxin efflux facilitators (De Smet et al., 2006; Tanaka et al., 2006; Michniewicz et al., 2007).
In contrast with auxin, less is known about the action of jasmonate on LR formation. As a stress hormone, jasmonate plays a well-established role in regulation of defense responses to herbivore attack and pathogen infection (Creelman and Mullet, 1997; Turner et al., 2002; Browse, 2005; Wasternack, 2007; Howe and Jander, 2008; Kazan and Manners, 2008). In addition, jasmonate also regulates many aspects of plant growth and development (Creelman and Mullet, 1997; Turner et al., 2002; Browse, 2005; Wasternack, 2007). For example, it has long been recognized that application of jasmonate inhibits root growth (Ueda and Kato, 1980). This action has been employed as the basis for the most prominent genetic assay to identify jasmonate-related mutants in Arabidopsis (Staswick et al., 1992; Turner et al., 2002; Wasternack, 2007), which have significantly advanced our understanding of the molecular mechanisms of the jasmonate signaling pathway (Creelman and Mullet, 1997; Turner et al., 2002; Browse, 2005; Wasternack, 2007; Howe and Jander, 2008; Kazan and Manners, 2008).
Here, we report the characterization of an Arabidopsis mutant, jasmonate-induced defective lateral root1 (jdl1/asa1-1), which essentially does not form LRs in the presence of 20 μM MeJA, the methyl ester of jasmonic acid. Using the mitotic promoter-reporter construct CYCB1;1:GUS as an LRP reporter, we show that while MeJA promotes LR formation in the wild type, it represses LRP initiation in jdl1/asa1-1. Gene cloning studies indicate that JDL1 encodes the auxin biosynthetic gene ANTHRANILATE SYNTHASE α1 (ASA1), leading to our hypothesis that jasmonate regulates LR formation through crosstalk with auxin and effects on auxin biosynthesis. Our results not only highlight the importance of ASA1 in mediating jasmonate-induced auxin biosynthesis, but also reveal a hitherto masked ability of jasmonate to downregulate PIN1 and PIN2 protein levels. We propose that jasmonate both controls ASA1-dependent auxin biosynthesis and attenuates PIN-dependent auxin transport, providing a fine-tuned regulation of local auxin accumulation in the root basal meristem that is essential for LR formation.
RESULTS
jdl1/asa1-1 Is a Jasmonate Response Mutant in LR Formation
The inhibitory effect of jasmonate on primary root growth has been well recognized and widely employed as a genetic assay to identify jasmonate-related mutants in Arabidopsis. We found that jasmonate promotes LR formation in wild-type Arabidopsis plants. Under our growth conditions, MeJA at 1 μM, a concentration that did not significantly affect primary root growth, increased the number of total LRs (including LRPs and emerged LRs) by 83% (Figure 1B). MeJA, at concentrations of 10 or 20 μM, significantly inhibited primary root growth (Figure 1C) and increased total LRs by ∼33% (Figure 1B). When MeJA concentration was raised to 50 μM, primary root growth was severely inhibited (Figure 1C), and the promotional effect on LR formation was not significant (Figure 1B). To explore the mechanisms of jasmonate action on LR formation, we performed a forward genetic screen for seedlings that did not show LR formation when grown on medium containing 20 μM MeJA (see Methods). Seedlings that appeared similar to the wild type without MeJA but had reduced LR formation in the presence of MeJA were identified as jdl mutants. Here, we describe the characterization of the jdl1 (also designated as asa1-1; see below) mutant.
Figure 1.
jdl1/asa1-1 Shows Defective LR Formation in Response to MeJA Treatment.
(A) LR phenotypes of 10-d-old Col-0 and jdl1/asa1-1 seedlings grown vertically on medium without MeJA (Control) or containing 20 μM MeJA (MeJA). Note that LR formation in the jdl1/asa1-1 mutant was blocked by exogenous MeJA.
(B) Quantification of total LRs (LRPs plus emerged LRs) per primary root. Seven-day-old seedlings grown on Murashige and Skoog medium plates containing indicated concentrations of MeJA were counted for LR formation. Data show average and sd of 15 seedlings and are representative of at least three independent experiments. Asterisks denote t test significance compared with untreated plants: *P < 0.05; **P < 0.01.
(C) Measurement of primary root length of 10-d-old Col-0 and jdl1/asa1-1 seedlings grown on medium containing indicated concentrations of MeJA. Data show average and sd of 15 seedlings and are representative of at least three independent experiments.
(D) to (G) proCYCB1;1:GUS staining assays showing that MeJA represses LRP initiation in jdl1/asa1-1. Photographs show typical roots of 6-d-old seedlings. Arrows show LRP or emerged LR, as indicated by proCYCB1;1:GUS staining.
(D) and (E) Expression pattern of proCYCB1;1:GUS at LRP initiation sites of representative Col-0 roots grown on MeJA-free medium (D) or on medium containing 20 μM MeJA (E).
(F) and (G) Expression pattern of proCYCB1;1:GUS at LRP initiation sites of representative jdl1/asa1-1 roots grown on MeJA-free medium (F) or on medium containing 20 μM MeJA (G).
In the absence of MeJA, LR formation in jdl1/asa1-1 was only slightly reduced compared with the wild type (Figures 1A and 1B). Detailed morphological comparison indicated that, when grown on MeJA-free medium, LR development in jdl1/asa1-1 was largely normal (see Supplemental Figure 1 online). To gain insight into the developmental basis for jasmonate action on LR formation, wild-type and jdl1/asa1-1 seedlings were grown on medium containing a range of concentrations of MeJA, and total LRs were counted. Interestingly, while MeJA application led to increased numbers of total LRs in the wild type, it exerted a repressive, rather than promotional, effect on LR formation in jdl1/asa1-1 (Figures 1A and 1B). Under our experimental conditions, 20 μM MeJA was sufficient to block the overall ability of jdl1/asa1-1 to form LR (Figures 1A and 1B; see Supplemental Figure 1 online). Therefore, jdl1/asa1-1 represents a jasmonate response mutant for LR formation.
To determine which stage of LR formation is blocked by MeJA in jdl1/asa1-1, the proCYCB1;1:GUS reporter was introduced into the jdl1/asa1-1 mutant background through genetic crossing. The proCYCB1;1:GUS reporter marks the first divisions in pericycle cells during LR initiation and serves as a useful tool to visualize the LRP initiation and later development processes (Casimiro et al., 2001; Himanen et al., 2002; Laplaze et al., 2007). Microscopic monitoring of proCYCB1;1:GUS expression indicated that MeJA perturbs the initial anticlinal division of pericycle cells leading to LRP initiation in the jdl1/asa1-1 mutant. For example, in the presence of 20 μM MeJA, GUS was expressed in the primary root tip and LRP initiation sites in the wild-type root (Figures 1D and 1E). In jdl1/asa1-1 root, however, GUS expression was only found in the root tip, but not in pericycle cells (Figures 1F and 1G). Therefore, jdl1/asa1-1 is impaired in LRP initiation in response to jasmonate.
Although jdl1/asa1-1 was defective in MeJA-induced LR formation, this mutant showed a normal response to MeJA in primary root growth (Figure 1C), suggesting that JDL1 represents a critical component required for MeJA-regulated LR formation, but not for MeJA-induced inhibition of primary root growth.
JDL1 Codes for ASA1, a Rate-Limiting Enzyme for the Biosynthesis of Trp, a Precursor to Indole-3-Acetic Acid
The deficiency of jdl1/asa1-1 in MeJA-induced LR formation provides a facile assay to determine the molecular basis of this defect using a map-based cloning approach. Using 1621 individuals showing the jdl1/asa1-1 phenotype from an F2 mapping population, the jdl1/asa1-1 mutation was mapped to a region between two molecular markers on the BAC clone MJJ13 of chromosome 5 (Figure 2A). DNA sequencing and comparison of the predicted genes in this interval revealed a single G-to-A nucleotide substitution in the annotated gene At5g05730 (Figure 2B), which was previously shown to encode the α-subunit of ASA1 (Niyogi and Fink, 1992). Anthranilate synthase converts chorismate to anthranilate, a rate-limiting step for the biosynthesis of Trp, a metabolic intermediate for indole-3-acetic acid (IAA) biosynthesis (Radwanski and Last, 1995; Woodward and Bartel, 2005).
Figure 2.
Map-Based Cloning of the JDL1 Gene.
(A) Fine genetic and physical mapping of JDL1. The target gene was initially mapped to a genetic interval between markers ciw18 and CRE482478 on Arabidopsis chromosome 5. Analysis of a mapping population consisting of 1631 plants delimited the target gene to a region defined by markers MJJ-1 and CER438063 on the BAC clone MJJ13. Numbers in parentheses indicate the number of recombination events identified between markers and the target gene.
(B) Exon (box)-intron (line) structure of the ASA1 gene (At5g05730). Sequence analysis revealed that the ASA1 gene derived from the jdl1/asa1-1 mutant contains a G-to-A mutation, which results in substitution of the predicted Trp-175 to a premature stop codon. Two other T-DNA insertion alleles of the ASA1 gene, including Salk_040353 (asa1-2) and wei2-2 (asa1-3), are indicated by open triangles.
(C) RT-PCR analysis of ASA1 expression in the jdl1/asa1-1 mutant. Twelve-day-old Col-0 and jdl1/asa1-1 seedlings were harvested for RNA extraction and RT-PCR analysis. The primers used for PCR were ASA1F1 and ASA1R1 for ASA1 and ACTIN7F1 and ACTIN7R1 for ACTIN7 (see Supplemental Table 1 online).
(D) jdl1/asa1-1 is allelic to the T-DNA insertion mutant Salk_040353, which contains a T-DNA insertion in the ASA1 gene. Col-0, jdl1/asa1-1, Salk_040353, and F1 progenies from crosses between Salk_040353 and jdl1/asa1-1 or Col-0 were grown vertically for 10 d on medium without MeJA (Control) or containing 20 μM MeJA (MeJA).
(E) LR phenotypes of the wei2-2 mutant, which is a T-DNA insertion allele of ASA1 (Stepanova et al., 2005). Col-0 and wei2-2 seedlings were grown vertically for 10 d on medium without MeJA (Control) or with 20 μM MeJA (MeJA).
The G-to-A mutation in jdl1/asa1-1–derived ASA1 occurs in the third exon and results in a substitution of the predicted Trp-175 to a premature translational stop codon (Figure 2B). RT-PCR analysis indicated that this mutation impairs the transcriptional expression of ASA1 (Figure 2C). We then analyzed two independent mutant alleles of ASA1. A homozygous T-DNA line, Salk_040353, which contains a T-DNA insertion in the ninth exon of ASA1 (Figure 2B), showed a similar MeJA-induced LR phenotype as jdl1/asa1-1 (Figure 2D). F1 plants between Salk_040353 and the wild type (Columbia [Col-0]) showed wild-type response to MeJA in LR formation, suggesting that homozygous Salk_040353 harbors a recessive mutation (Figure 2D). F1 plants from a cross between jdl1/asa1-1 and Salk_040353 showed defective LR formation in the presence of exogenous MeJA, indicating that jdl1/asa1-1 is allelic to Salk_040353 (Figure 2D).
In addition, the wei2-2 mutant, recently shown to be a null allele of the ASA1 gene (Stepanova et al., 2005), also exhibited similar jasmonate-induced LR formation defect (Figure 2E). Together, these results demonstrated that the MeJA-induced LR formation defect of jdl1/asa1-1 is caused by mutation of the ASA1 gene. We therefore refer to JDL1 as ASA1, as this corresponds with the known biochemical function of this gene. Accordingly, the original jdl1 mutant was redesignated as asa1-1, Salk_040353 was redesignated as asa1-2, and wei2-2 was redesignated as asa1-3.
Several independent studies have demonstrated that loss of function of ASA1 results in defective Trp biosynthesis and, as a consequence, leads to a reduction of endogenous IAA levels (Rutherford et al., 1998; Ljung et al., 2005; Stepanova et al., 2005). In this context, we reasoned that the LR formation phenotype of jdl1/asa1-1 might be caused by defective auxin biosynthesis. In our feeding experiments, 10 mM exogenous anthranilate, a direct product of the ASA1 protein, fully rescued the LR formation defect of jdl1/asa1-1 in the absence or presence of MeJA (Figure 3). Exogenous application of Trp, a precursor of auxin biosynthesis, also rescued the jdl1/asa1-1 phenotype (Figure 3). Finally, IAA at 10 nM, a concentration that does not show a significant effect on root growth of wild-type seedlings (Rahman et al., 2001; Stepanova et al., 2005), was also able to partially restore the LR formation defect of jdl1/asa1-1 (Figure 3). Together, our feeding experiments support the notion that the LR formation phenotype of jdl1/asa1-1 is, in fact, caused by a defect in auxin biosynthesis in the presence of MeJA.
Figure 3.
Rescue of the LR Formation Defect of jdl1/asa1-1 by Anthranilate, Trp, and IAA.
Col-0 and jdl1/asa1-1 seedlings were grown vertically for 7 d on MeJA-free medium or medium containing 20 μM MeJA, with or without supplementation of 10 mM anthranilate (ANT), 10 mM Trp, or 10 nM IAA, and total LRs (LRPs plus emerged LRs) per primary root were counted. Values represent average and sd of 15 plants and are indicative of at least three independent experiments. Asterisks denote t test significance compared with untreated plants: *P < 0.05; **P < 0.01.
ASA1 Is Important for MeJA-Induced IAA Biosynthesis
To study the role of the ASA1 gene in jasmonate response in LR formation, we examined the MeJA-induced expression of ASA1. For this experiment, 12-d-old wild-type plants were treated with MeJA, and ASA1 transcript levels in whole seedlings were measured with quantitative RT-PCR (qRT-PCR). As shown in Figure 4A, ASA1 transcript elevation was seen as early as 0.5 h after MeJA treatment, with a maximum level reached after 2 h. The MeJA-induced ASA1 expression tended to decrease in a time course between 2 and 9 h after MeJA treatment (Figure 4A).
Figure 4.
MeJA Treatment Induces IAA Biosynthesis through Transcriptional Activation of ASA1 Expression.
(A) qRT-PCR analysis of ASA1 expression in wild-type seedlings during the time course after MeJA treatment. Twelve-day-old Col-0 seedlings were treated with 50 μM MeJA, and whole seedlings were harvested at different time points for RNA extraction and qRT-PCR analyses. The transcript levels of ASA1 were normalized to the ACTIN7 expression. Error bars represent the sd of triplicate reactions.
(B) RNA gel blot analyses of MeJA-induced ASA1 expression in the wild type (Col-0) and the coi1-1 mutant. Twelve-day-old Col-0 and coi1-1 seedlings were treated with 50 μM MeJA, and whole seedlings were harvested at the indicated times for RNA extraction. The top panel shows a RNA gel blot probed with 32P-labeled cDNA for ASA1, and the bottom panel shows amounts of rRNA as loading controls.
(C) qRT-PCR analyses of MeJA-regulated expression of ASA1 in shoot and root tissues of Col-0 and coi1-1. Twelve-day-old Col-0 and coi1-1 seedlings were treated with 50 μM MeJA for 3 h, and shoot and root tissues were harvested separately for RNA extraction and qRT-PCR analyses. The transcript levels of ASA1 were normalized to the ACTIN7 expression, and error bars represent the sd of triplicate reactions..
(D) to (G) MeJA-induced proASA1:GUS expression. Three-day-old seedlings of proASA1:GUS transgenic line grown on control medium were transferred either to medium without MeJA (D) or to medium containing 10 μM MeJA (E) for 2 d.
(F) Magnification of the framed region in (D). Bar = 50 μm.
(G) Magnification of the framed region in (E). Bar = 50 μm.
(H) Free IAA measurement of Col-0 and jdl1/asa1-1 seedlings in response to MeJA treatment. Twelve-day-old Col-0 and jdl1/asa1-1 seedlings were left untreated (Control) or treated with 100 μM MeJA (MeJA) for 2 d and then free IAA levels were measured. At least 10 biological replicates were analyzed. An analysis of variance followed by Fisher's Least Significant Difference (LSD) mean separation test (SPSS) was performed on the data. Samples with the different letters are significantly different at P < 0.01. Error bars represent se.
To study whether the MeJA-induced expression of ASA1 requires the function of COI1, a central regulator of jasmonate-mediated processes in Arabidopsis (Xie et al., 1998), we compared the MeJA-induced ASA1 expression in the wild type and the coi1-1 mutant (Xie et al., 1998). RNA gel blot analysis using RNAs from whole seedlings showed that the MeJA-induced expression of ASA1 is significantly reduced in coi1-1 (Figure 4B). We further examined the MeJA-induced ASA1 expression in shoot and root tissues by qRT-PCR and found that the coi1-1 mutation abolished MeJA-mediated induction of ASA1 expression in both shoot and root tissues (Figure 4C). These data indicated that MeJA activates the expression of ASA1 in a COI1-dependent manner.
The MeJA-induced ASA1 expression pattern was also examined with the ASA1 promoter fused with the GUS reporter (proASA1:GUS). At the 5-d-old seedling stage, proASA1:GUS was mainly expressed in the root tip and the hypocotyl/root junction (Figures 4D and 4F). MeJA treatment not only significantly enhanced proASA1:GUS expression in the root tip, but also expanded its expression domains to vascular tissues along the whole primary root (Figures 4E and 4G). In addition to ASA1, MeJA also increased the transcription levels of several other auxin biosynthesis-related genes (see Supplemental Figure 2 online). Among them, ASB1 (Stepanova et al., 2005), NIT3 (Kutz et al., 2002), and YUCCA2 (Cheng et al., 2006) have been shown to be directly involved in IAA biosynthesis. Our promoter-GUS assays indicated that MeJA significantly enhanced the expression of ASB1 (see Supplemental Figure 2D online), NIT3 (see Supplemental Figure 2E online), and YUCCA2 (see Supplemental Figure 2F online) in the root. These data suggest that MeJA induces IAA biosynthesis through transcriptional activation of ASA1 and other auxin biosynthetic genes. To test this possibility, we measured free IAA levels in 12-d-old wild-type and jdl1/asa1-1 seedlings. Without MeJA treatment, free IAA levels in jdl1/asa1-1 seedlings were slightly lower than those in the wild type (Figure 4H). After MeJA treatment, however, free IAA levels in the wild type were almost doubled, whereas free IAA levels in the mutant were only increased by 50% (Figure 4H), indicating that ASA1 is required for MeJA-mediated induction of IAA biosynthesis.
Taken together, these results demonstrated that ASA1 is required for jasmonate-induced auxin biosynthesis and led us to hypothesize that jasmonate regulates LR formation through activation of the ASA1-dependent auxin biosynthesis and subsequent response. Further support for this hypothesis came from our finding that jasmonate failed to promote LR formation in the slr1 (Fukaki et al., 2002) and arf7-1 arf19-2 (Okushima et al., 2005) double mutants (see Supplemental Figure 3 online), in which LRP initiation is abolished as a result of disrupted auxin signaling.
MeJA-Regulated proIAA2:GUS Expression in Wild-Type and jdl1/asa1-1 Roots
To explore how jasmonate interacts with auxin to differentially regulate LR formation in the wild type and jdl1/asa1-1, we examined the spatial distribution of the auxin response in wild-type and jdl1/asa1-1 roots using the auxin-responsive reporter proIAA2:GUS, which provides a greater spatial resolution of the auxin response in the root (Swarup et al., 2001; Marchant et al., 2002). In this experiment, we paid special attention to the proIAA2:GUS expression in a region at the transition between the meristem and the elongation zone, referred to as the root basal meristem, the site of LRP initiation (Beemster et al., 2003; De Smet et al., 2007). In untreated wild-type roots, proIAA2:GUS was expressed in the stele and columella cells (Figure 5A). Upon MeJA treatment, proIAA2:GUS expression was strongly enhanced in the stele and ectopically induced in the lateral root cap (LRC) and epidermal cells (Figure 5B), suggesting that MeJA upregulates auxin response in wild-type roots. As expected, the MeJA-mediated upregulation of proIAA2:GUS expression was significantly reduced in the jasmonate-insensitive mutant coi1-1 (Figures 5C and 5D).
Figure 5.
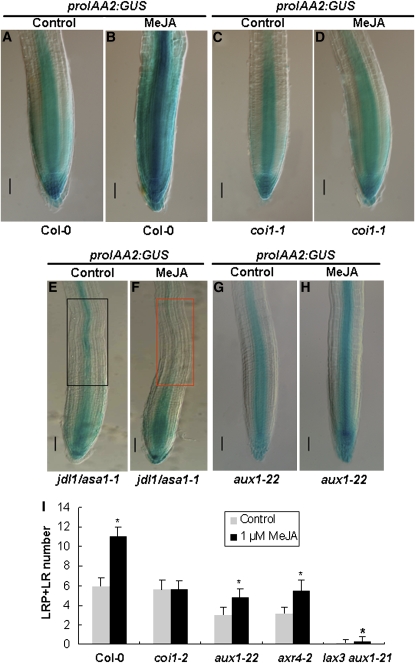
MeJA-Regulated proIAA2:GUS Expression in Roots of Col-0, coi1-1, jdl1/asa1-1, and aux1-22.
(A) and (B) Control (A) and MeJA-treated (B) Col-0 roots.
(C) and (D) Control (C) and MeJA-treated (D) coi1-1 roots.
(E) and (F) Control (E) and MeJA-treated (F) jdl1/asa1-1 roots. Framed regions indicate that MeJA reduces proIAA2:GUS expression in the basal meristem of jdl1/asa1-1 roots.
(G) and (H) Control (G) and MeJA treated (H) aux1-22 roots.
For (A) to (H), 4-d-old seedlings of the indicated genotypes grown on control medium were transferred to medium without MeJA (Control) or containing 10 μM MeJA (MeJA) for an additional 2 d, and the proIAA2:GUS expression was monitored. Bars = 50 μm.
(I) MeJA-mediated LR formation was affected in JA signaling and auxin transport mutants. Col-0, coi1-2, aux1-22, axr4-2, and lax3 aux1-21 seedlings were grown vertically on medium with or without 1 μM MeJA for 7 d, and total LRs (LRPs plus emerged LRs) per primary root were counted. Values show average and sd of 15 seedlings and are representative of at least three independent experiments. Asterisks denote t test significance compared with untreated plants: *P < 0.01.
Consistently, the MeJA-mediated induction of LR formation was largely abolished in coi1-2 (Xu et al., 2002), in which LR formation is largely normal in the absence of MeJA (Figure 5I). In jdl1/asa1-1 roots, the basal expression of proIAA2:GUS was somehow lower (Figure 5E), and, most surprisingly, MeJA treatment led to a significant reduction of proIAA2:GUS expression, particularly in the root basal meristem (Figure 5F). These results indicated that whereas MeJA increases auxin response in the basal meristem of wild-type roots, it attenuates auxin response in the basal meristem of jdl1/asa1-1 roots. The contrasting action of MeJA on the root auxin response between the wild type and jdl1/asa1-1 is consistent with the distinct effect of MeJA on LR formation between the two genotypes. As MeJA can actually increase free IAA levels by up to 50% in jdl1/asa1-1 whole seedlings (Figure 4H), a plausible explanation for the MeJA-induced reduction of proIAA2:GUS expression in jdl1/asa1-1 root is that MeJA might exert a negative effect on local auxin accumulation in the mutant root, especially in the root basal meristem.
Among the two physiologically distinct auxin transport pathways, both the phloem-based auxin transport from source leaves and the local PAT in the root contribute to auxin accumulation in the root basal meristem (Swarup et al., 2001, 2005; Blilou et al., 2005; Leyser, 2005). It has been proposed that the auxin influx carrier protein AUX1, which functions in both auxin transport routes, plays a role in connecting the phloem-based transport of IAA to the PAT system (Swarup et al., 2001; Marchant et al., 2002; De Smet et al., 2007). In this context, we first checked the jasmonate-induced expression of proIAA2:GUS expression in aux1-22 (Swarup et al., 2007), a null mutant disrupting the function of AUX1. The results indicated that the MeJA-triggered induction of proIAA2:GUS expression was largely abolished in aux1-22 (Figures 5G and 5H), suggesting that the jasmonate-mediated activation of auxin response in the root requires the function of AUX1. Next, we checked the LR promotional effect of jasmonate in several auxin transport mutants, including aux1-22 (Swarup et al., 2007), axr4-2 (Dharmasiri et al., 2006), and lax3 aux1-21 (Swarup et al., 2008). Due to defective auxin transport capacities, these mutants showed reduced LR formation in the absence of MeJA (Figure 5I). As expected, MeJA failed to restore LR formation of these mutants to wild-type levels (Figure 5I), indicating that MeJA-mediated induction of LR formation requires auxin transport.
The idea that MeJA affects auxin transport (both phloem-based transport and PAT) during LR formation was substantiated by comparative inspection of the auxin-responsive reporters DR5rev:green fluorescent protein (GFP) (Ottenschläger et al., 2003) and DR5:GUS (Ulmasov et al., 1997) in wild-type and jdl1/asa1-1 roots. In wild-type roots, MeJA upregulated the expression of these reporters in root tips (see Supplemental Figure 4 online) and at the LRP initiation sites along the primary root length (see Supplemental Figure 5 online). In jdl1/asa1-1 roots, however, MeJA downregulated the expression of these reporters in root tips (see Supplemental Figure 4 online) and at the LRP initiation sites along the primary root length (see Supplemental Figure 5 online).
Jasmonate Modulates Expression of Auxin Transport Components
To explore how jasmonate regulates auxin transport during LR formation, we investigated the effect of MeJA treatment on the expression of several important components of the auxin transport machinery. Using transgenic lines containing the proPIN1:GUS, proPIN2:GUS, and proAUX1:GUS reporters, which have been used previously to monitor the native expression of these genes (Vieten et al., 2005; Ruzicka et al., 2007), we observed a general induction effect of MeJA on the expression of these auxin efflux and infux genes at the transcriptional level. For example, when 5-d-old seedlings grown on control medium were transferred to medium containing 10 μM MeJA, upregulation of proPIN1:GUS (Figures 6A and 6B), proPIN2:GUS (Figures 6C and 6D), and proAUX1:GUS (Figures 6E and 6F) expression was detectable within 12 h after transfer.
Figure 6.
MeJA-Regulated Expression of Auxin Transport Components in the Roots of Wild-Type Plants.
(A) and (B) Control (A) and MeJA-treated (B) roots examined for proPIN1:GUS expression.
(C) and (D) Control (C) and MeJA-treated (D) roots examined for proPIN2:GUS expression.
(E) and (F) Control (E) and MeJA-treated (F) roots examined for proAUX1:GUS expression.
For (A) to (F), 5-d-old seedlings of transgenic Col-0 plants containing the indicated constructs were grown on control medium and then transferred to medium without MeJA (Control) or containing 10 μM MeJA (MeJA) for 12 h and then were collected for GUS staining. Bars = 50 μm.
(G) qRT-PCR analyses of MeJA-regulated expression of PIN1, PIN2, and AUX1 in Col-0 and jdl1/asa1-1 roots. Ten-day-old Col-0 and jdl1/asa1-1 seedlings were treated with 50 μM MeJA for 6 h, and roots were harvested for RNA extraction and qRT-PCR analyses. The transcript levels of PIN1, PIN2, and AUX1 were normalized to the ACTIN2 expression. Asterisks denote t test significance compared with untreated plants: *P < 0.05. Results of one of three independent experiments are shown. Error bars represent the sd of triplicate reactions.
(H) to (K) MeJA treatment downregulates the PIN1 and PIN2 protein levels in wild-type and jdl1/asa1-1 roots.
(H) PIN1 and PIN2 immunolocalization of untreated wild-type root.
(I) PIN1 and PIN2 immunolocalization of wild-type root treated with MeJA.
(J) PIN1 and PIN2 immunolocalization of untreated jdl1/asa1-1 root.
(K) PIN1 and PIN2 immunolocalization of jdl1/asa1-1 root treated with MeJA.
Four-day-old seedlings were transferred to medium without MeJA (Control) or with 20 μM MeJA (MeJA) for 48 h. Seedlings were fixed for PIN1 and PIN2 immunolocalization assays. Root tips were visualized and photographed with a laser scanning confocal microscope. Green, PIN1; red, PIN2. Bars = 50 μm. Images shown are representative of at least six independent experiments.
To quantitatively assess the effect of MeJA on the expression of these genes, we performed qRT-PCR analyses using RNAs extracted from MeJA-treated roots. To address whether the jdl1/asa1-1 mutation affects the action of MeJA on the regulation of these genes, we examined the expression of PIN1, PIN2, and AUX1 in roots of the wild type and jdl1/asa1-1. Similar to the promoter:GUS assays, MeJA treatment led to a general increase of PIN1, PIN2, and AUX1 mRNAs in wild-type roots (Figure 6G). Importantly, the MeJA-mediated induction of PIN1 and PIN2 expression was obviously reduced in jdl1/asa1-1 roots (Figure 6G). Notably, whereas MeJA upregulated AUX1 expression in wild-type roots, this hormone significantly downregulated AUX1 expression in jdl1/asa1-1 roots (Figure 6G). These results demonstrated that MeJA regulates the expression of the auxin transport-related genes at the transcriptional level and highlighted the importance of ASA1 in this regulation.
Finally, we asked whether jasmonate might regulate auxin transport-related genes at the protein level. For these experiments, 4-d-old wild-type and jdl1/asa1-1 seedlings grown on control medium were transferred to medium without or with 20 μM MeJA. Forty-eight hours after transfer, PIN1 and PIN2 protein levels were assessed by immunolocalization assays. As shown in Figures 6H and 6I, MeJA treatment led to substantial reduction of PIN1 and PIN2 protein levels in wild-type roots. Significantly, the MeJA-mediated reduction of PIN1 and PIN2 protein levels in jdl1/asa1-1 roots was much more severe than that observed in wild-type roots (Figures 6J and 6K), suggesting that the ASA1 gene function is important for jasmonate-mediated downregulation of PIN1 and PIN2 protein levels.
The Action of Jasmonate to Regulate LR Formation Is Independent of Ethylene
Recent reports have demonstrated that ethylene regulates LR formation through interaction with auxin (Negi et al., 2008; Ivanchenko et al., 2008). To test the possibility that jasmonate might act through the ethylene pathway to regulate LR formation, we examined MeJA-induced LR formation in the ethylene-insensitive mutants etr1-1 and ein2-5. ETR1 encodes one of the ethylene receptor in Arabidopsis (Chang et al., 1993). EIN2 encodes a membrane protein with an unknown biochemical function and plays a positive role in ethylene signaling (Alonso et al., 1999). Consistent with recent observations (Ivanchenko et al., 2008; Negi et al., 2008), etr1-1 and ein2-5 showed slightly increased LR formation in the absence of MeJA (Figure 7A). Given that etr1-1 and ein2-5 exhibited increased sensitivity to MeJA in primary root growth, we treated these plants with relatively low concentrations of MeJA and compared their LR formation with that of jdl1/asa1-1. As shown in Figure 7A, whereas 0.1 or 1 μM MeJA significantly reduced LR formation in jdl1/asa1-1, these concentrations of MeJA significantly increased LR formation in etr1-1, ein2-5, and the wild type. These results indicated that, in contrast with jdl1/asa1-1, which was defective in MeJA-induced LR formation, etr1-1 and eni2-5 showed near wild-type response to MeJA in LR formation, suggesting that the MeJA action on LR formation does not require ethylene signaling.
Figure 7.
The Relationship between Jasmonate and Ethylene in Controlling LR Formation through the ASA1 Gene.
(A) MeJA-induced LR formation of the ethylene-insensitive mutants etr1-1 and ein2-5. Seven-day-old seedlings of Col-0, jdl1/asa1-1, etr1-1, and ein2-5 grown on medium containing indicated concentrations of MeJA were counted for LR formation (LRPs plus emerged LRs). Data show average and sd of 15 seedlings and are representative of at least three independent experiments. Asterisks denote t test significance compared with untreated plants: *P < 0.05.
(B) ACC-induced LR formation in jdl1/asa1-1 and coi1-2. Seven-day-old seedlings of Col-0, jdl1/asa1-1, and coi1-2 grown on medium containing indicated concentrations of ethylene precursor ACC were counted for LR formation (LRPs plus emerged LRs). Data show average and sd of 15 seedlings and are representative of at least three independent experiments. Asterisks denote t test significance compared with untreated plants: *P < 0.05.
It has been shown that the ASA1 gene plays a role in ethylene-mediated inhibition of primary root growth (Stepanova et al., 2005; Ruzicka et al., 2007; Swarup et al., 2007). To test whether ASA1 is also important for the ethylene action in LR formation, jdl1/asa1-1 and the wild type were examined for their response to the ethylene precursor 1-amino-1-cyclopropane carboxylic acid (ACC) in LR formation. As shown in Figure 7B, a low concentration of ACC (0.02 μM) was able to promote LR formation in the wild type, and this promotional effect was impaired in jdl1/asa1-1. A higher concentration of ACC (0.2 μM) inhibited LR formation in the wild type, and there was a significant reduction in the ability of 0.2 μM ACC to inhibit LR formation in jdl1/asa1-1 (Figure 7B), consistent with a recent proposal that ASA1 plays a role in ethylene-mediated regulation of LR formation (Negi et al., 2008). In parallel, we examined the ethylene-mediated LR formation in the coi1-2 mutant, which is resistant to jasmonate but partially fertile and able to produce a small quantity of seeds (Xu et al., 2002). As shown in Figure 7B, the ethylene response of coi1-2 in LR formation was essentially similar to that of the wild type, suggesting that the ethylene action in regulating LR formation does not require the COI1-dependent jasmonate signaling.
Together, our results imply that the action of jasmonate to regulate LR formation is independent of ethylene. This hypothesis is also supported by the fact that loss-of-function of ASA1 led to an altered response to ethylene both in primary root growth (Stepanova et al., 2005; Ruzicka et al., 2007; Swarup et al., 2007) and in LR formation (Negi et al., 2008; Ivanchenko et al., 2008; Figure 7B). Here, we show that the jdl1/asa1-1 mutant only affects jasmonate-induced LR formation and does not affect MeJA-induced primary root growth inhibition.
DISCUSSION
JDL1/ASA1 Is an Interaction Node through Which Jasmonate Integrates Its Action with Auxin to Regulate LR Formation
It has been long recognized that the stress hormone jasmonate inhibits primary root growth (Wasternack, 2007). We report here that jasmonate also plays a role in regulating LR formation in Arabidopsis. Given the established role of auxin in LR formation (Casimiro et al., 2003; De Smet et al., 2006; Nibau et al., 2008, Fukaki and Tasaka, 2009) and the previous finding that jasmonate and auxin share common components in their signaling pathways (Tiryaki and Staswick, 2002), it is reasonable to speculate that jasmonate may interact with auxin to regulate LR formation. Our characterization of the jdl1/asa1-1 mutant, which showed defective LR formation in the presence of exogenous MeJA (Figure 1), strongly supported this speculation and provided insights into the action mechanisms of jasmonate to regulate LR formation. While MeJA promoted LR formation in the wild type, it repressed LR formation in jdl1/asa1-1 (Figure 1). The fact that the repressive effect of MeJA on LR formation can only be observed in the jdl1/asa1-1 mutant implies that, in the wild type, the repressive effect of MeJA on LR formation is masked by the JDL1 function. Therefore, elucidation of the biochemical function of the JDL1 gene product is critical to understand the molecular mechanisms underlying the distinct effect of MeJA on LR formation between the wild type and jdl1/asa1-1.
Gene cloning studies indicated that JDL1 encodes the well-characterized auxin biosynthetic gene ASA1 (Figure 2). Early studies indicated that the expression of ASA1 was induced by wounding and pathogen infections (Niyogi and Fink, 1992), suggesting a potential role for this gene in plant responses to abiotic or biotic stresses. Indeed, ASA1 was also demonstrated recently to be required for ethylene-induced auxin production and therefore plays a role in ethylene-mediated regulation of root growth inhibition (Stepanova et al., 2005). In this context, it is reasonable to propose that ASA1 may represent an integration node through which jasmonate exerts its effect on auxin biosynthesis and, subsequently, regulates LR formation. Supporting evidence for this hypothesis first came from our feeding experiments showing that anthranilate, Trp, and auxin can rescue or partially rescue the LR-deficient phenotype of the jdl1/asa1-1 mutant (Figure 3).
Furthermore, gene expression analyses indicated that MeJA activates the expression of ASA1 in a COI1-dependent manner (Figures 4B and 4C). In addition to ASA1, MeJA also induces the expression of a list of other genes known to be related to auxin biosynthesis (see Supplemental Figure 2 online). Finally, our measurement of free IAA levels demonstrated the role of ASA1 in mediating jasmonate-induced auxin biosynthesis (Figure 4H). Collectively, these results support a hypothesis that jasmonate activates the transcriptional expression of ASA1 and leads to increased free IAA levels, which eventually contribute to MeJA-induced promotion of LR formation in wild-type plants. It is worth noting that MeJA can increase free IAA levels (by up to 50%) even in the jdl1/asa1-1 mutant seedlings, although not as much as in wild-type seedlings (Figure 4H), indicating the existence of other targets of MeJA in IAA biosynthesis. Indeed, our qRT-PCR assays indicated that MeJA can induce the expression of the putative IAA conjugate hydrolase gene ILL5 (for ILR1-like5) (see Supplemental Figure 2C online). However, it remains to be determined whether the ILL5 gene product is an active IAA amidohydrolase (Davies et al., 1999; Woodward and Bartel, 2005).
Characterization of the jdl1/asa1-1 Mutant Reveals a Masked Role of Jasmonate in Regulation of Auxin Transport during LR Formation
However, the above oversimplified scenario cannot explain the phenomenon that MeJA represses LR formation in the jdl1/asa1-1 mutant. Extensive studies have together demonstrated that the phytohormone auxin serves as an instructive signal for LRP initiation and later development (Casimiro et al., 2001; Bhalerao et al., 2002; Benková et al., 2003; Laplaze et al., 2007; Swarup et al., 2008; Dubrovsky et al., 2008). Notably, a recent elegant observation proposed that LR positioning is regulated in a spatiotemporal manner by auxin accumulation in the root basal meristem (Ishikawa and Evans, 1995; Beemster et al., 2003; De Smet et al., 2007). Under this paradigm, the jasmonate-induced LR formation defect of jdl1/asa1-1 may result from suboptimal auxin accumulation in the root basal meristem. The employment of the auxin-responsive reporter proIAA2:GUS enabled us to inspect, at cellular resolution, local auxin accumulation in the root basal meristem. Our observations indicated that, in the wild type, the promotional effect of MeJA on LR formation correlated with increased proIAA2:GUS expression in the root basal meristem (Figures 5A and 5B). In the jdl1/asa1-1 mutant, the repressive effect of MeJA on LR formation correlated with reduced proIAA2:GUS in the root basal meristem (Figures 5E and 5F).
The basal meristem has been proposed to recycle the major auxin transport flows in the root, including the phloem-based shoot-derived auxin transport flow and the local PAT flows (Swarup et al., 2001, 2005; Marchant et al., 2002; Blilou et al., 2005; Leyser, 2005); therefore, root basal meristem exhibits an auxin accumulation maximum that is essential for LR initiation (De Smet et al., 2007). In this context, our observations of proIAA2:GUS expression in wild-type and mutant roots point to a possibility that, in addition to activate ASA1-dependent IAA biosynthesis, MeJA may also affect, directly or indirectly, the auxin transport flows that are important for local auxin accumulation in the root basal meristem. The ability of the aux1-22 mutant to reduce MeJA-induced proIAA2:GUS expression in the root basal meristem (Figures 5G and 5H) provided supporting evidence to this hypothesis, since AUX1 was reported to be involved in the two physiologically distinct auxin transport pathways (i.e., phloem-based transport and local PAT) in the root basal meristem (Swarup et al., 2001; Marchant et al., 2002; De Smet et al., 2007). Furthermore, we found that jasmonate exhibits a complex regulation of the expression of several auxin influx and efflux facilitators at both transcriptional and translational levels (Figure 6). Notably, in MeJA-treated wild-type roots, PIN1 and PIN2 protein levels were decreased (Figures 6H and 6I). In MeJA-treated jdl1/asa1-1 roots, the reduction was more severe than that in wild-type roots (Figures 6J and 6K). These results suggested that the ASA1 function (i.e., jasmonate-induced auxin biosynthesis) is important for JA-mediated regulation of auxin transport components.
Previous characterization of independent mutant alleles of ASA1, including trp5-2wvc1 (Rutherford et al., 1998) and tir7 (Ruegger et al., 1997; Ljung et al., 2005), which were isolated based on altered root waving phenotype and resistance to auxin transport inhibitors, respectively, provided evidence that mutation of the ASA1 function might affect auxin transport. Together with our observations in this work, these results raised the interesting question of how jasmonate regulates the expression of auxin transport components. Given that jasmonate promotes ASA1-dependent auxin biosynthesis and auxin itself can upregulate the expression of the PIN genes at the transcription level (Vieten et al., 2005), it is possible that the MeJA-induced upregulation of the transcripts of auxin transport-related genes is achieved indirectly through the auxin pathway (i.e., MeJA induces the expression of ASA1 that leads to increased auxin synthesis that, in turn, activates the transcriptional expression of these genes). Our data that the jdl1/asa1-1 mutation affects the jasmonate-regulated expression levels of AUX1, PIN1, and PIN2 (Figures 6A to 6G) lend support to this possibility.
Considering that MeJA positively regulates the biosynthesis of flavonoids (Dombrecht et al., 2007), which act as negative regulators of auxin transport (Brown et al., 2001; Buer and Muday, 2004; Peer et al., 2004; Besseau et al., 2007), it is also possible that the MeJA-mediated regulation of auxin transport-related genes is achieved through the upregulation of flavonoid biosynthesis by MeJA. In line with this idea, recent elegant work using flavonoid mutants with altered auxin transport indicated that flavonoids modulate the expression of the PIN genes at both mRNA and protein levels in a tissue-specific manner (Peer et al., 2004). Further studies are required to elucidate the regulatory mechanisms of jasmonate on the expression of auxin transport-related genes.
Jasmonate Mediates a Fine-Tuned Regulation of Auxin Accumulation in the Root Basal Meristem That Is Critical for LR Formation
Based on the results described in this study, we propose a model (Figure 8) to explain the distinct effect of jasmonate on LR formation between the wild type and jdl1/asa1-1. In wild-type plants, MeJA activates the transcriptional expression of ASA1, which leads to increased IAA biosynthesis in shoot and root tissues. IAA synthesized de novo by the ASA1-dependent pathway in the aerial part is delivered through phloem-based transport and/or long-distance PAT to the root basal meristem. IAA synthesized locally in the root by the ASA1-dependent pathway is also transported through PAT to the root basal meristem. It has been shown that defect of the ASA1 gene harbored by the tir7-1 mutation allele has less impact on root-specific IAA synthesis than on synthesis in the aerial portion of the seedling (Ljung et al., 2005), suggesting that MeJA-induced IAA accumulation in the root basal meristem is mainly derived from ASA1-dependent synthesis in shoot tissues. Considering the proposed dominant role of phloem-based transport in long-distance auxin distribution (Tanaka et al., 2006), it is likely that the majority of IAA produced in the aerial part by ASA1 is transported to the root basal meristem through phloem. In addition to activating ASA1-dependent auxin biosynthesis, MeJA also directly or indirectly decreases PIN1 and PIN2 protein levels, which results in a negative effect on PAT-mediated local auxin accumulation in the root basal meristem. The combinational effect of MeJA on IAA biosynthesis and transport leads to increased local auxin accumulation in the root basal meristem and, as a result, promotes LR formation.
Figure 8.
Model Showing Jasmonate Modulation of Auxin Accumulation in the Root Basal Meristem.
(A) In the wild type, JA stimulates the ASA1-dependent auxin synthesis in shoot and root tissues, which are transported to the root basal meristem through phloem-based and/or polar transport pathways. On the other hand, jasmonate reduces the protein levels of auxin efflux carriers (PIN1 and PIN2), which leads to decreased auxin transport capacities to the root basal meristem. The net effect of jasmonate is to increase local auxin accumulation in the root basal meristem and promote LR formation.
(B) In the jdl/asa1-1 mutant, ASA1-dependent auxin synthesis in the shoot and root tissues is blocked. Jasmonate still represses auxin transport to the root basal meristem through reducing the levels of PIN1 and PIN2 proteins. The net effect of jasmonate in the mutant is to decrease auxin accumulation in the root basal meristem and therefore suppresses LR formation. Arrows represent auxin transport flows, ovals in red represent ASA1-dependent auxin synthesis in shoot and root, and dashed lines represent the jdl1/asa1-1 mutation blocks ASA1-dependent auxin synthesis.
In jdl1/asa1-1 mutants, the jasmonate-induced auxin biosynthesis via ASA1 is blocked, which leads to defective auxin sources (both shoot and root tissues) for jasmonate-induced auxin accumulation in the root basal meristem. Jasmonate still exerts its negative effect on auxin transport capacities through reducing PIN1 and PIN2 protein levels. These effects likely cause the root basal meristem to be deprived of auxin and, as a consequence, repress LR formation. It should be noted that the repressive effect of jasmonate on local auxin accumulation in the root basal meristem (as well as LR formation) can only be observed in jdl1/asa1-1 but not in the wild type, suggesting that the ASA1 function (i.e., jasmonate-induced auxin biosynthesis) counteracts the repressive effect of jasmonate on auxin transport. Our results not only highlight the importance of ASA1 in mediating jasmonate-induced auxin biosynthesis, but also reveal a masked repressive role of jasmonate on auxin transport. The two aspects of jasmonate actions may represent a fine-tuned regulation of local auxin accumulation in the root basal meristem that is critical for LR formation. Given the established role of jasmonate in plant defense, the jasmonate-mediated fine-tuned regulation of LR formation may have adaptive significance under conditions in which jasmonate levels are elevated.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana mutants and/or transgenic lines described are in the Col-0 background. Some of the plant materials used in this study were previously described: wei2-2 (Stepanova et al., 2005), coi1-1 (Xie et al., 1998), coi1-2 (Xu et al., 2002), etr1-1 (Chang et al., 1993), ein2-5 (Alonso et al., 1999), proCYCB1;1:GUS (Colón-Carmona et al., 1999), DR5:GUS (Ulmasov et al., 1997), proYUC2:GUS (Cheng et al., 2006), proNIT3:GUS (Kutz et al., 2002), proIAA2:GUS and proIAA2:GUS/aux1-22 (Swarup et al., 2007), DR5rev:GFP (Benková et al., 2003), aux1-22 (Swarup et al., 2007), axr4-2 (Dharmasiri et al., 2006), lax3 aux1-21 (Swarup et al., 2008), proPIN1,2:GUS (Vieten et al., 2005) and proAUX1:GUS (Marchant et al., 2002). Seeds of Salk_040353 were obtained from the ABRC (Alonso et al., 2003).
Seeds were surface-sterilized for 15 min in 10% bleach, washed four times with sterile water, and plated on half-strength Murashige and Skoog (1962) medium. Plants were stratified at 4°C for 2 d in darkness and then transferred to a phytotron set at 22°C with a 16-h-light/8-h-dark photoperiod (light intensity 120 μmol m−2 s−1).
Isolation of the jdl1/asa1-1 Mutant
Ethyl methanesulfonate–mutagenized M2 seeds of the wild-type accession, Col-0, were kindly provided by Jianru Zuo (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing). M2 seeds were germinated and grown vertically on medium containing 20 μM MeJA for 10 d. Seedlings with reduced LRs were selected as jdl mutant candidates and directly transplanted to soil. Resulting M3 seeds from the putative mutants were confirmed on medium with 20 μM MeJA by LR phenotype. The original jdl1/asa1-1 mutant was backcrossed to Col-0 for three generations, and the resulting homozygous progenies were used in this study.
Map-Based Cloning of the JDL1 Gene
For mapping, the jdl1/asa1-1 mutant in the Col-0 background was crossed to the polymorphic ecotype Landsberg erecta (Ler), and the resulting F1 plants were self-pollinated to yield an F2 population segregating for the jdl1/asa1-1 mutant phenotypes. Simple sequence length polymorphism markers were used for linkage analysis using a previously described procedure (Lukowitz et al., 2000). In the fine-mapping process, we developed several PCR-based molecular markers. The primers for the simple sequence length polymorphism marker CER455774 are 5′-CTAATAGAAGTGGAAAGTAAGA-3′ and 5′-GGGGGAGTGATCAATGGTAT-3′. The following cleaved amplified polymorphic sequence markers were designed: for MJJ-1, the primers 5′-TAGAGGCCAACATGCACATATC-3′ and 5′-TGTTTTACCCTGCTCTTGCTTT-3′ were used, and the PCR product from Ler can be digested by AflIII (198 and 145 bp); for CER438063, the primers 5′-TACTTGTTCTTGGCTAAATCC-3′ and 5′-GACCCACCTCTAATAAATCTACTA-3′ were used, and the PCR product from Ler can be digested by EcoRV (241 and 391 bp).
The T-DNA insertion line Salk_040353 contains a T-DNA inserted in the ninth exon of ASA1 (At5g05730). Homozygous T-DNA insertion plants were identified by diagnostic PCR with the gene-specific primers 5′-TGATCGAGTGGAAAAAGGTTG-3′ and 5′-ATGAAAAGCAATGTGCTGAGC-3′. The homozygous Salk_040353 mutant was crossed with jdl1/asa1-1 for allelic testing.
To generate the DR5:GUS reporter line in the jdl1/asa1-1 mutant background, homozygous jdl1/asa1-1 plants were crossed to a transgenic line harboring the DR5:GUS construct (Ulmasov et al., 1997) to produce an F2 population. Putative DR5:GUS/jdl1/asa1-1 plants, which were homozygous for the jdl1/asa1-1 mutation as well as the DR5:GUS construct, were identified in the F2 population and then retested in the F3 population (i.e., in the F3 population, 100% of seedlings showed no LR formation in the presence of 20 μM MeJA; 100% of seedlings showed uniform staining for GUS). Similarly, the DR5rev:GFP (Benková et al., 2003), proCYCB1;1:GUS (Colón-Carmona et al., 1999), and proIAA2:GUS (Swarup et al., 2007) reporters were also introduced into the jdl1/asa1-1 mutant background. The proIAA2:GUS and DR5rev:GFP constructs were also transferred into the coi1-1 mutant (Xie et al., 1998) using a similar approach.
Plasmid Construction and Plant Transformation
All molecular manipulations were performed according to standard methods (Sambrook and Russell, 2001). A 2.0-kb genomic fragment upstream of the ASA1 translation start codon was amplified by PCR and cloned into the SalI/BamHI sites of the binary vector pCAMBIA1391-Z (CAMBIA), resulting in the transcriptional fusion of the ASA1 promoter with the GUS coding region. Similarly, a 2.0-kb promoter region of ASB1 was also fused with the GUS coding region in the binary vector pCAMBIA1391-Z. Primers used for PCR amplification are as follows: 5′-CGCGTCGACCTAGAATATGTTATGCTTC-3′ (SalI) and 5′-CTAGGATCCTGTAACGGCTAAGAACTC-3′ (BamHI) for ASA1 promoter; 5′-ATCAAGCTTTTCGGGCAGAGATCGCAGAGC-3′ (HindIII) and 5′-GCCGGATCCTGTCTGAGAAGCAAAGATTCCT-3′ (BamHI) for ASB1 promoter.
The above constructs were then transformed into Agrobacterium tumefaciens strain GV3101 (pMP90), which was used for transformation of Arabidopsis plants by vacuum infiltration (Bechtold and Pelletier, 1998).
Analysis of GUS Activity
Histochemical staining for GUS activity in transgenic plants was performed as described previously (Jefferson et al., 1987). Briefly, whole seedlings or various tissues were immersed in 1 mM 5-bromo-4-chloro-3-indolyl-β-glucuronic acid solution in 100 mM sodium phosphate, pH 7.0, 0.1 mM EDTA, 0.5 mM ferricyanide, 0.5 mM ferrocyanide, and 0.1% Triton X-100, after applying vacuum for 2 min, and were incubated at 37°C from 20 min to overnight. Chlorophyll was cleared from plant tissues by immersing them in 70% ethanol. Individual representative seedlings were photographed.
Observation of Plants
Seedlings of DR5:GUS/Col-0, DR5:GUS/jdl1/asa1-1, proIAA2:GUS/Col-0, proIAA2:GUS/jdl1/asa1-1, proIAA2:GUS/coi1-1, proIAA2:GUS/aux1-22, proASA1:GUS/Col-0, proASB1:GUS/Col-0, proPIN1,2:GUS/Col-0, proAUX1:GUS/Col-0, proYUC2:GUS/Col-0, and proNIT3:GUS/Col-0 reporter lines were photographed using the Leica Microsystems DM5000B microscope and DFC490 CCD camera. To observe the LRP and emerged LR, roots were first cleared using HCG solution (chloroacetaldehyde:water:glycerol = 8:3:1) for 30 s (Sabatini et al., 1999). The numbers of LRPs and emerged LRs were counted with the Interference Discrepancy Contrast Microscope system (Leica DM5000B microscope). Roots of DR5rev:GFP/Col-0, DR5rev:GFP/jdl1/asa1-1, and DR5rev:GFP/coi1-1 plants in different genetic backgrounds were mounted in a 10 μM propidium iodide solution for 3 to 5 min for confocal scanning. The fluorescence excitation and image acquisitions were monitored by a laser scanning confocal microscope (LSM 510; Zeiss).
Free IAA Measurement
To measure the influence of jasmonate on auxin levels, seeds of Col-0 and jdl1/asa1-1 were surface-sterilized. Approximately 30 mL of medium containing half-strength Murashige and Skoog salts (Caisson Laboratories), 1% sucrose, and 0.5% phytagel (Sigma-Aldrich), pH 5.7 was poured into each square Petri dish (100 × 100 × 15 mm), and 19 seeds were placed in a line, 1 cm from the top edge. The Petri dishes were incubated vertically at 22°C in a phytotron. Four milliliters of 100 μM MeJA (Bedoukian Research) was added to the plates in the horizontal position onto the Petri dish to cover the seedlings for 10 min. Petri dishes with MeJA or control solution were resealed and placed vertically in the growth chamber for another 2 d. Ethanol was used to make the 100 mM MeJA stock solution, resulting in 0.1% ethanol in the final 100 μM treatment solution, and the same amount of ethanol in water was used as the control treatment. Fresh whole seedlings (25 to 50 mg) were harvested, weighed, and then frozen in liquid nitrogen. After adding 3 ng of [13C6]IAA as internal standard, each sample was extracted, purified, methylated, and then analyzed using selected ion monitoring gas chromatography–mass spectrometry as described (Barkawi et al., 2008). At least 10 biological replicates were analyzed for each treatment.
Immunolocalization Assay for PIN Proteins
The PIN1 and PIN2 immunolocalization assays were performed using the InsituPro robot (Friml et al., 2002). The following antibodies and dilutions were used: anti-PIN1 (1:50), anti-PIN2 (1:400), and Alexa Fluor 488 or Alexa Fluor 555 (1:500) secondary antibodies (Molecular Probes). Fluorescent samples were inspected by the Zeiss Axioplan 2 confocal laser scanning microscope and Zeiss LSM 510 Image Browser software.
Gene Expression Analysis
For RNA gel blot analysis, total RNA was prepared by a guanidine thiocyanate extraction method, and RNA gel blot analysis was performed as described previously (Zheng et al., 2006). Ten micrograms of total RNA was separated in an agarose gel containing 10% formaldehyde, blotted onto Hybond N+ membrane (Amersham), and probed with 32P-labeled DNA fragment for ASA1. Primers used to amplify the ASA1 probe are 5′-GGCCGCCACCGAGTTCTTAG-3′ and 5′-GCAAATGTTCGCCGCTCAAA-3′.
For qRT-PCR analysis, 12-d-old Arabidopsis seedlings (Col-0 and jdl1/asa1-1) grown vertically on medium plates were treated with 50 μM MeJA for 3 h or untreated as a control. Whole seedlings or different tissues were frozen in liquid nitrogen for RNA extraction. RNA was extracted with the RNeasy kit (Qiagen). Poly(dT) cDNA was prepared from 10 μg of total RNA with Superscript III reverse transcriptase (Invitrogen) and quantified with an cycler apparatus (Bio-Rad) with the RealMasterMix (SYBR Green) kit (Tiangen) according to the manufacturer's instructions. PCR was performed in 96-well optical reaction plates heated for 5 min at 95°C to activate hot start Taq DNA polymerase, followed by 40 cycles of denaturation for 30 s at 95°C, annealing for 30 s at 59°C, and extension for 20 s at 68°C. Expression levels of target genes were normalized to ACTIN2 or ACTIN7 expression levels. The statistical significance was evaluated by t test analysis. Primers used to quantify gene expression levels are listed in Supplemental Table 1 online.
Accession Numbers
The Arabidopsis Genome Initiative locus identifiers for the genes characterized in this study are as follows: ACTIN2 (AT3g18780), ACTIN7 (AT5G09810), ASA1 (AT5G05730), ASB1 (AT1G25220), AUX1 (AT2G38120), CYCB1;1 (AT4G37490), IAA2 (AT3G23030), PIN1 (AT1G73590), PIN2 (AT5G57090), NIT3 (AT3G44320), and YUCCA2 (AT4G13260). Germplasm identification numbers for the seeds in this study are as follows: asa1-2 (SALK_040353), arf7-1 arf19-2 (CS24630), aux1-22 (AUX1-22), axr4-2 (CS8019), coi1-1 (coi1-1), coi1-2 (coi1-2), ein2-5 (ein2-5), etr1-1 (CS237), jdl1/asa1-1(this study), lax3 aux1-21 (lax3 aux1-21), slr1 (slr-1), and wei2-2/asa1-3 (SALK_ 017444).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Morphological Comparison of LRPs in the Wild Type and jdl1/asa1-1.
Supplemental Figure 2. MeJA-Induced Expression of Auxin Biosynthesis-Related Genes.
Supplemental Figure 3. MeJA Fails to Promote LR Formation in slr1 and arf7-1 arf19-2 Double Mutants.
Supplemental Figure 4. MeJA Differentially Regulates the Expression of DR5 Reporters in Wild-Type, coi1-1, and jdl1/asa1-1 Root Tips.
Supplemental Figure 5. MeJA-Regulated DR5:GUS Expression along the Primary Root Length of the Wild Type and jdl1/asa1-1.
Supplemental Table 1. DNA Primers Used for qRT-PCR Assays in This Study.
Supplementary Material
Acknowledgments
We thank the ABRC and Mark Estelle, Malcolm J. Bennett, Jose M. Alonso, Bannie Bartel, Yunde Zhao, Jianru Zuo, Hongwei Guo, and Daoxin Xie for kindly providing seeds used in this study. We thank Jennifer Normanly for critical reading of the manuscript. This work was supported by grants from the National Natural Science Foundation of China (90717007 and 30530440), the Ministry of Science and Technology of China (2007CB948200, 2006AA10A116, and 2006CB910604), and the Chinese Academy of Sciences (KSCX2-YW-N-045 and KSCX2-YW-N-015). Work in the laboratory of Jerry D. Cohen was supported by grants from the U.S. National Science Foundation (MCB0725149 and DBI 0606666) and the USDA, National Research Initiative (2005-35318-16197).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Chuanyou Li (cyli@genetics.ac.cn).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso, J.M., Hirayama, T., Roman, G., Nourizadeh, S., and Ecker, J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284 2148–2152. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Barkawi, L.S., Tam, Y.Y., Tillman, J.A., Pederson, B., Calio, J., Al-Amier, H., Emerick, M., Normanly, J., and Cohen, J.D. (2008). A high-throughput method for the quantitative analysis of indole-3-acetic acid and other auxins from plant tissue. Anal. Biochem. 372 177–188. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82 259–266. [DOI] [PubMed] [Google Scholar]
- Beemster, G.T.S., Fiorani, F., and Inzé, D. (2003). Cell cycle: The key to plant growth control? Trends Plant Sci. 8 154–158. [DOI] [PubMed] [Google Scholar]
- Benková, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertova, D., Jurgens, G., and Friml, J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602. [DOI] [PubMed] [Google Scholar]
- Besseau, S., Hoffmann, L., Geoffroy, P., Lapierre, C., Pollet, B., and Legrand, M. (2007). Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19 148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalerao, R.P., Eklof, J., Ljung, K., Marchant, A., Bennett, M., and Sandberg, G. (2002). Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J. 29 325–332. [DOI] [PubMed] [Google Scholar]
- Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I., Friml, J., Heidstra, R., Aida, M., Palme, K., and Scheres, B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433 39–44. [DOI] [PubMed] [Google Scholar]
- Brown, D.E., Rashotte, A.M., Murphy, A.S., Normanly, J., Tague, B.W., Peer, W.A., Taiz, L., and Muday, G.K. (2001). Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 126 524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse, J. (2005). Jasmonate: An oxylipin signal with many roles in plants. Vitam. Horm. 72 431–456. [DOI] [PubMed] [Google Scholar]
- Buer, C.S., and Muday, G.K. (2004). The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16 1191–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro, I., Beeckman, T., Graham, N., Bhalerao, R., Zhang, H., Casero, P., Sandberg, G., and Bennett, M.J. (2003). Dissecting Arabidopsis lateral root development. Trends Plant Sci. 8 165–171. [DOI] [PubMed] [Google Scholar]
- Casimiro, I., Marchant, A., Bhalerao, R.P., Beeckman, T., Dhooge, S., Swarup, R., Graham, N., Inze, D., Sandberg, G., Casero, P.J., and Bennett, M. (2001). Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza, J.L., Jr., Grisafi, P.L., and Fink, G.R. (1995). A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 9 2131–2142. [DOI] [PubMed] [Google Scholar]
- Chang, C., Kwok, S.F., Bleecker, A.B., and Meyerowitz, E.M. (1993). Arabidopsis ethylene-response gene ETR1: Similarity of product to two-component regulators. Science 262 539–544. [DOI] [PubMed] [Google Scholar]
- Cheng, Y., Dai, X., and Zhao, Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colón-Carmona, A., You, R., Haimovitch-Gal, T., and Doerner, P. (1999). Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20 503–508. [DOI] [PubMed] [Google Scholar]
- Creelman, R.A., and Mullet, J.E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 355–381. [DOI] [PubMed] [Google Scholar]
- Davies, R.T., Goetz, D.H., Lasswell, J., Anderson, M.N., and Bartel, B. (1999). IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet, I., et al. (2007). Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134 681–690. [DOI] [PubMed] [Google Scholar]
- De Smet, I., Vanneste, S., Inzé, D., and Beeckman, T. (2006). Lateral root initiation or the birth of a new meristem. Plant Mol. Biol. 60 871–887. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, S., Swarup, R., Mockaitis, K., Dharmasiri, N., Singh, S.K., Kowalchyk, M., Marchant, A., Mills, S., Sandberg, G., Bennett, M.J., and Estelle, M. (2006). AXR4 is required for localization of the auxin influx facilitator AUX1. Science 312 1218–1220. [DOI] [PubMed] [Google Scholar]
- Dombrecht, B., Xue, G.P., Sprague, S.J., Kirkegaard, J.A., Ross, J.J., Reid, J.B., Fitt, G.P., Sewelam, N., Schenk, P.M., Manners, J.M., and Kazan, K. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky, J.G., Rost, T.L., Colón-Carmona, A., and Doerner, P. (2001). Early primordium morphogenesis during lateral root initiation in Arabidopsis thaliana. Planta 214 30–36. [DOI] [PubMed] [Google Scholar]
- Dubrovsky, J.G., Sauer, M., Napsucialy-Mendivil, S., Ivanchenko, M.G., Friml, J., Shishkova, S., Celenza, J., and Benkova, E. (2008). Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl. Acad. Sci. USA 105 8790–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml, J., Benková, E., Blilou, I., Wisniewska, J., Hamann, T., Ljung, K., Woody, S., Sandberg, G., Scheres, B., Jürgens, G., and Palme, K. (2002). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108 661–673. [DOI] [PubMed] [Google Scholar]
- Fukaki, H., Okushima, Y., and Tasaka, M. (2007). Auxin-mediated lateral root formation in higher plants. Int. Rev. Cytol. 256 111–137. [DOI] [PubMed] [Google Scholar]
- Fukaki, H., Tameda, S., Masuda, H., and Tasaka, M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29 153–168. [DOI] [PubMed] [Google Scholar]
- Fukaki, H., and Tasaka, M. (2009). Hormone interactions during lateral root formation. Plant Mol. Biol. 69 383–396. [DOI] [PubMed] [Google Scholar]
- Himanen, K., Boucheron, E., Vanneste, S., de Almeida Engler, J., Inze, D., and Beeckman, T. (2002). Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14 2339–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, G.A., and Jander, G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59 41–66. [DOI] [PubMed] [Google Scholar]
- Ishikawa, H., and Evans, M.L. (1995). Specialized zones of development in roots. Plant Physiol. 109 725–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanchenko, M.G., Muday, G.K., and Dubrovsky, J.G. (2008). Ethylene-auxin interactions regulate lateral root initiation and emergence in Arabidopsis thaliana. Plant J. 55 335–347. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan, K., and Manners, J.M. (2008). Jasmonate signaling: toward an integrated view. Plant Physiol. 146 1459–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutz, A., Muller, A., Hennig, P., Kaiser, W.M., Piotrowski, M., and Weiler, E.W. (2002). A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana. Plant J. 30 95–106. [DOI] [PubMed] [Google Scholar]
- Laplaze, L., et al. (2007). Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19 3889–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser, O. (2005). Auxin distribution and plant pattern formation: How many angels can dance on the point of PIN? Cell 121 819–822. [DOI] [PubMed] [Google Scholar]
- Ljung, K., Hull, A.K., Celenza, J., Yamada, M., Estelle, M., Normanly, J., and Sandberg, G. (2005). Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17 1090–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz, W., Gillmor, C.S., and Scheible, W.R. (2000). Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 123 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy, J.E. (2005). Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ. 28 67–77. [DOI] [PubMed] [Google Scholar]
- Malamy, J.E., and Benfey, P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124 33–44. [DOI] [PubMed] [Google Scholar]
- Marchant, A., Bhalerao, R., Casimiro, I., Eklof, J., Casero, P.J., Bennett, M., and Sandberg, G. (2002). AUX1 promotes lateral root formation by facilitating indole-3-acetic acid distribution between sink and source tissues in the Arabidopsis seedling. Plant Cell 14 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz, M., et al. (2007). Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130 1044–1056. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15 473–497. [Google Scholar]
- Negi, S., Ivanchenko, M.G., and Muday, G.K. (2008). Ethylene regulates lateral root formation and auxin transport in Arabidopsis thaliana. Plant J. 55 175–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibau, C., Gibbs, D.J., and Coates, J.C. (2008). Branching out in new directions: The control of root architecture by lateral root formation. New Phytol. 179 595–614. [DOI] [PubMed] [Google Scholar]
- Niyogi, K.K., and Fink, G.R. (1992). Two anthranilate synthase genes in Arabidopsis: Defense-related regulation of the tryptophan pathway. Plant Cell 4 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima, Y., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottenschläger, I., Wolff, P., Wolverton, C., Bhalerao, R.P., Sandberg, G., Ishikawa, H., Evans, M., and Palme, K. (2003). Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc. Natl. Acad. Sci. USA 100 2987–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer, W.A., Bandyopadhyay, A., Blakeslee, J.J., Makam, S.N., Chen, R.J., Masson, P.H., and Murphy, A.S. (2004). Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16 1898–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwanski, E.R., and Last, R.L. (1995). Tryptophan biosynthesis and metabolism: Biochemical and molecular genetics. Plant Cell 7 921–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, A., Amakawa, T., Goto, N., and Tsurumi, S. (2001). Auxin is a positive regulator for ethylene-mediated response in the growth of Arabidopsis roots. Plant Cell Physiol. 42 301–307. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Hobbie, L., Brown, D., Bernasconi, P., Turner, J., Muday, G., and Estelle, M. (1997). Reduced naphthylphthalamic acid binding in the tir3 mutant of Arabidopsis is associated with a reduction in polar auxin transport and diverse morphological defects. Plant Cell 9 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford, R., Gallois, P., and Masson, P.H. (1998). Mutations in Arabidopsis thaliana genes involved in the tryptophan biosynthesis pathway affect root waving on tilted agar surfaces. Plant J. 16 145–154. [DOI] [PubMed] [Google Scholar]
- Ruzicka, K., Ljung, K., Vanneste, S., Podhorska, R., Beeckman, T., Friml, J., and Benkova, E. (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19 2197–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini, S., Beis, D., Wolkenfelt, H., Murfett, J., Guilfoyle, T., Malamy, J., Benfey, P., Leyser, O., Bechtold, N., Weisbeek, P., and Scheres, B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99 463–472. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning, 3rd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Staswick, P.E., Su, W., and Howell, S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova, A.N., Hoyt, J.M., Hamilton, A.A., and Alonso, J.M. (2005). A Link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17 2230–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup, K., et al. (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10 946–954. [DOI] [PubMed] [Google Scholar]
- Swarup, R., Friml, J., Marchant, A., Ljung, K., Sandberg, G., Palme, K., and Bennett, M. (2001). Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 15 2648–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup, R., Kramer, E.M., Perry, P., Knox, K., Leyser, H.M.O., Haseloff, J., Beemster, G.T.S., Bhalerao, R., and Bennett, M.J. (2005). Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat. Cell Biol. 7 1057–1065. [DOI] [PubMed] [Google Scholar]
- Swarup, R., Perry, P., Hagenbeek, D., Van Der Straeten, D., Beemster, G.T., Sandberg, G., Bhalerao, R., Ljung, K., and Bennett, M.J. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19 2186–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, H., Dhonukshe, P., Brewer, P.B., and Friml, J. (2006). Spatiotemporal asymmetric auxin distribution: A means to coordinate plant development. Cell. Mol. Life Sci. 63 2738–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiryaki, I., and Staswick, P.E. (2002). An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol. 130 887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, J.G., Ellis, C., and Devoto, A. (2002). The jasmonate signal pathway. Plant Cell 14(Suppl): S153–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, J., and Kato, J. (1980). Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.). Plant Physiol. 66 246–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten, A., Vanneste, S., Wisniewska, J., Benkova, E., Benjamins, R., Beeckman, T., Luschnig, C., and Friml, J. (2005). Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132 4521–4531. [DOI] [PubMed] [Google Scholar]
- Wasternack, C. (2007). Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. (Lond.) 100 681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward, A.W., and Bartel, B. (2005). Auxin: Regulation, action, and interaction. Ann. Bot. (Lond.) 95 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu, L., Liu, F., Lechner, E., Genschik, P., Crosby, W.L., Ma, H., Peng, W., Huang, D., and Xie, D. (2002). The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, W., Zhai, Q., Sun, J., Li, C.B., Zhang, L., Li, H., Zhang, X., Li, S., Xu, Y., Jiang, H., Wu, X., and Li, C. (2006). Bestatin, an inhibitor of aminopeptidases, provides a chemical genetics approach to dissect jasmonate signaling in Arabidopsis. Plant Physiol. 141 1400–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.