Abstract
Gibberellins (GAs) play important roles in regulating reproductive development, especially anther development. Our previous studies revealed that the MYB transcriptional factor GAMYB, an important component of GA signaling in cereal aleurone cells, is also important for anther development. Here, we examined the physiological functions of GA during anther development through phenotypic analyses of rice (Oryza sativa) GA-deficient, GA-insensitive, and gamyb mutants. The mutants exhibited common defects in programmed cell death (PCD) of tapetal cells and formation of exine and Ubisch bodies. Microarray analysis using anther RNAs of these mutants revealed that rice GAMYB is involved in almost all instances of GA-regulated gene expression in anthers. Among the GA-regulated genes, we focused on two lipid metabolic genes, a cytochrome P450 hydroxylase CYP703A3 and β-ketoacyl reductase, both of which might be involved in providing a substrate for exine and Ubisch body. GAMYB specifically interacted with GAMYB binding motifs in the promoter regions in vitro, and mutation of these motifs in promoter-β-glucuronidase (GUS) transformants caused reduced GUS expression in anthers. Furthermore, a knockout mutant for CYP703A3 showed gamyb-like defects in exine and Ubisch body formation. Together, these results suggest that GA regulates exine formation and the PCD of tapetal cells and that direct activation of CYP703A3 by GAMYB is key to exine formation.
INTRODUCTION
Gibberellins (GAs), growth factors with a tetracyclic diterpenoid structure, are essential regulators of diverse growth and developmental processes throughout the life cycle of the plant (Fleet and Sun, 2005). Among various physiological events regulated by GA, reproductive development is one of the most important events (Pharis and King, 1985; King and Evans, 2003). For example, GA application accelerates flowering in the facultative long-day plant Arabidopsis thaliana, particularly under short-day conditions (Langridge, 1957; Bagnall, 1992). Similarly, GAs application promotes the development of male flowers in cucumber (Cucumis sativus), whereas in maize (Zea mays) and castor bean (Ricinus communis), GAs promote the development of female flowers (Pharis and King, 1985). Conversely, GA deficiency in some mutants causes failure or delay of flowering (Koornneef and van der Veen, 1980; Wilson et al., 1992; Sun and Kamiya, 1994; Thornsberry et al., 2001). Additionally, GA-deficient flowers of several plant species exhibit abnormal anther development, resulting in male sterility (Nester and Zeevaart, 1988; Goto and Pharis, 1999). For example, a tomato (Solanum lycopersicum) GA-deficient (gib-1) mutant is defective in pollen formation, with arrest of anther development at the premeiotic stage (Jacobsen and Olszewski, 1991). In Arabidopsis, a GA-deficient mutant, ga1, has poorly developed anthers containing nonviable pollen grains (Koornneef and van der Veen, 1980; Goto and Pharis, 1999). There is also some evidence that GAs are important for pollen germination and elongation (Viti et al., 1990; Singh et al., 2002; Chhun et al., 2007). All of these observations strongly suggest that GA is important for flowering and development of floral organs, especially of anthers. In spite of their importance, little is known about the molecular function of GA in these processes.
During the past decade, several factors essential for GA perception have been identified, mainly through screens for GA-signaling mutants of rice (Oryza sativa) and Arabidopsis. In particular, the identification of genes encoding the GA receptor GIBBERELLIN-INSENSITIVE DWARF1 (GID1) (Ueguchi-Tanaka et al., 2005, 2007b; Griffiths et al., 2006; Nakajima et al., 2006; Willige et al., 2007), DELLA proteins (Peng et al., 1997, 1999; Silverstone et al., 1998; Ikeda et al., 2001; Chandler et al., 2002; Itoh et al., 2002), and the F-box protein GID2/ SLEEPY1 (SLY1) in rice and Arabidopsis (McGinnis et al., 2003; Sasaki et al., 2003; Dill et al., 2004; Fu et al., 2004), and the physiological and biochemical analyses of these gene products, have enabled us to construct a model of GA perception (Ueguchi-Tanaka et al., 2007a). According to this model, when GA is present, the GID1 receptor binds GA. The GID1/GA complex then interacts with the negative regulator of GA action, the DELLA protein, which results in degradation of DELLA protein through the SCFGID2/SLY1 (for Skp1, Cullin, F-box) proteasome pathway. The degradation of DELLA protein allows GA action to occur. Although the earliest events of GA signaling, from GA perception by GID1 to DELLA protein degradation, are now better understood, the molecular mechanism downstream of DELLA degradation remains unclear. Recent studies have demonstrated that PHYTOCHROME INTERACTING FACTOR proteins can interact with DELLA proteins to regulate light-dependent hypocotyl growth (de Lucas et al., 2008; Feng et al., 2008). Another important factor, which has been known to function downstream of DELLA protein degradation, is GAMYB. GAMYB was originally isolated as a positive GA-signaling component that regulates the expression of most GA-inducible genes in cereal aleurone cells (Gubler et al., 1995, 1999; Tsuji et al., 2006). Recently, some reports have described evidence for a molecular function of GAMYB outside of the aleurone, indicating that GAMYB plays an important role in flower development, especially in anther development (Murray et al., 2003; Achard et al., 2004; Kaneko et al., 2004; Millar and Gubler, 2005). For example, loss-of-function mutations of the rice GAMYB gene result in defects in flower development, especially in anther and pollen development (Kaneko et al., 2004). Although at present there is no direct evidence that GAMYB functions as a GA signaling factor in anther development, studying the molecular function of GAMYB in anthers provides us with an opportunity to understand this GA signaling pathway.
In this article, we describe the physiological function of GA in anther development by phenotypic analyses of a comprehensive set of rice GA-deficient, GA-insensitive, and gamyb mutants. Our analyses showed that these mutants exhibit common defects in programmed cell death (PCD) of tapetal cells, exine formation, and Ubisch body formation. In addition, we investigated the mechanism of GA signaling in anther development, with a focus on the molecular function of GAMYB. Microarray analysis suggested that GAMYB functions as a dominant component in GA signaling during anther development. We also identified two target genes of GAMYB, a cytochrome P450 hydroxylase CYP703A3 and β-ketoacyl reductase (KAR), both of which encode lipid metabolic enzymes. A knockout mutant of CYP703A3 showed defects in the formation of exine and Ubisch bodies, which are the same defects seen in the gamyb mutants. Considering all of these observations, we conclude that GA functions in the anther to promote exine and Ubisch body formation by GAMYB-dependent induction of CYP703A3.
RESULTS
Phenotypic Analysis of Impaired Anthers in GA-Deficient and GA-Insensitive Mutants
Although there are some reports that GA-related mutants show abnormal floral phenotypes (Koornneef and van der Veen, 1980; Nester and Zeevaart, 1988; Jacobsen and Olszewski, 1991; Goto and Pharis, 1999), there has been no detailed study of the involvement of GA during flower development. Therefore, we first examined flower development in rice GA-deficient and GA-insensitive mutants (see Supplemental Figure 1 online). We used four mutants, gid1-4, gid1-7, gid2-5, and gamyb-2, as GA-insensitive mutants, and oscps1-1 as a GA-deficient mutant. gid1-4 and gid1-7 are null and intermediate alleles of the soluble GA receptor gene, GID1, respectively (Ueguchi-Tanaka et al., 2005). gid2-5 is a null allele of GID2, which encodes an F-box protein involved in the degradation of a rice DELLA protein, SLENDER RICE1 (SLR1) (Sasaki et al., 2003). oscps1-1 is a null allele of CPS1, which encodes ent-copanyl diphosphate synthase, the enzyme that catalyzes the first step of the GA biosynthesis pathway. oscps1-1 is one of the most severe GA-deficient mutants in rice (Sakamoto et al., 2004). All of these mutants showed dwarfism, although gid1-4, a knockout allele of GID1, had the most severe phenotype, even more severe than oscps1-1 (see Supplemental Figure 1A online). In contrast with the other GA-related mutants we tested, gamyb-2, a null allele of GAMYB, did not show any abnormal phenotypes during the vegetative stage, consistent with a previous report (Kaneko et al., 2004).
We then compared floral development in these mutants (see Supplemental Figure 1B online). Floral formation was significantly delayed in these GA-related mutants: after transfer from long-day (16 h) to short-day (10 h) conditions, flowering took ∼6 to 7 weeks in gid1-4 and 5 weeks in oscps1-1, gid2-5, and gamyb-2, compared with 4 weeks in the wild type. All of these mutants developed a complete set of floral organs (see Supplemental Figure 1B online); however, the mutants developed abnormal stamens with shrunken and whitened anthers (see Supplemental Figure 1C online). The phenotypes of the abnormal stamens of these mutants were similar, although the degree of severity differed among the mutants (see Supplemental Figure 1C online). The severity of the anther phenotype correlated with the severity of dwarfism, except in the case of gamyb-2, which had defective anthers but normal plant height. In contrast with the anthers, pistils in these mutants developed normally (see Supplemental Figure 1D online).
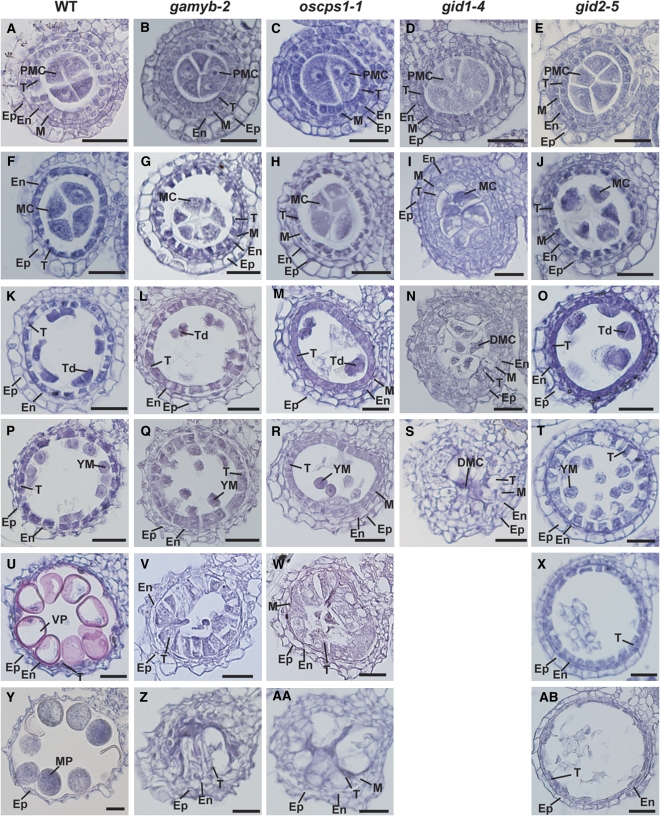
We then examined the developmental process of the anther through microscopy observation of transverse sections. Based on the cellular events visible under the light microscope, we divided the anther developmental process into six stages (Figure 1; see Supplemental Figure 2 online). At the pollen mother cell (PMC) stage, there was no sign of abnormal development or morphology in any of the mutants (Figures 1A to 1E; see Supplemental Figures 2A to 2E online). At the meiosis (MEI) stage, the PMCs of the wild type started meiosis to form tetrads (Figure 1F). The cytoplasm of the tapetal cells became deeply condensed at this stage, and the middle layer of cells was completely degraded in the wild type (Figure 1F; see Supplemental Figure 2F online). By contrast, the PMCs in mutant anthers at this stage were condensed, especially in gid1-4 (Figures 1G to 1J), and the middle layer of cells was not degraded (see Supplemental Figures 2G to 2J online). Deep condensation of the tapetal cells was also observed in the mutants, except for in gid1-4, which showed slightly enlarged tapetal cells devoid of condensation and a failure in epidermal cell expansion (Figures 1F to 1J; see Supplemental Figures 3G and 3H online). At the tetrad (TD) stage in the wild type, the PMCs completed meiosis to form tetrads, and tapetal cells began to degenerate (Figure 1K; see Supplemental Figure 2K online). Tetrads formed in all of the mutants except for gid1-4, but fewer tetrads formed than in the wild type (Figures 1K to 1O). The tapetal cells of these mutants started to expand (Figures 1L to 1O; see Supplemental Figures 2L to 2O online). This trend was most apparent in gid1-4, which had enlarged tapetal cells that nearly filled the locule and contained the degraded meiocyte (Figure 1N). At the young microspore (YM) stage, microspores in the wild type were released into the locules, and the tapetal layer became much thinner (Figure 1P). In gamyb-2, oscps1-1, and gid2-5, the enlargement of tapetal cells continued (Figures 1Q, 1R, and 1T). At this stage, in gid1-4, enlarged abnormal tapetal cells almost filled the locule (Figure 1S). At the vacuolated pollen (VP) stage, in the wild type, microspores were enlarged and the tapetal layer was almost gone (Figure 1U). In oscps1-1, the tapetal cells continued expanding, to the point of almost filling the locule space (Figure 1V), and a similar phenotype was also seen in gamyb-2, but to a lesser extent (Figure 1W). In gid2-5, the vacuolated tapetal cells began to degenerate, and the microspores were completely collapsed at this stage (Figure 1X). At the mature pollen (MP) stage, pollen in the wild type accumulated starch and completed its development (Figure 1Y). In gamyb-2 (Figure 1Z) and oscps1-1 (Figure 1AA), vacuolated tapetal cells completely filled the locule space. By contrast, the tapetal cells in gid2-5 continued to degenerate but were still visible at this stage (Figure 1AB). The microscopy analyses revealed that the abnormal developmental processes observed in gamyb-2 and oscps1-1 are nearly identical, with both mutants having abnormal enlargement of tapetal cells and collapse of microspores. These same abnormal processes were observed in gid1-4 but they proceeded more severely and quickly. In gid2-5, collapse of microspores and partial enlargement of tapetal cells also occurred at the YM stage, but occupation of the locule by enlarged tapetal cells was not observed. We consider that such phenotypic differences may be due to the slight differences in GA response defects among GA-related mutants, as supported by some recent reports (Ariizumi et al., 2008; Ueguchi-Tanaka et al., 2008).
Figure 1.
Histological Analysis of Anther Development in Wild-Type and GA-Related Mutants.
Anther development in wild-type and GA-related mutants was compared at the six different stages: PMC, MEI, TD, YM, VP, and MP. Ep, epidermal cell layer; En, endothecial cell layer; M, middle layer; T, tapetal layer; MC, meiocyte; DMC, degraded meiocyte; Td, tetrad. Bars = 25 μm.
(A) to (E) Transverse sections of anthers at the PMC stage in the wild type (A), gamyb-2 (B), oscps1-1 (C), gid1-4 (D), and gid2-5 (E).
(F) to (J) Transverse sections of anthers at the MEI stage in wild type (F), gamyb-2 (G), oscps1-1 (H), gid1-4 (I), and gid2-5 (J).
(K) to (O) Transverse sections of anthers at the TD stage in the wild type (K), gamyb-2 (L), oscps1-1 (M), gid1-4 (N), and gid2-5 (O).
(P) to (T) Transverse sections of anthers at the YM stage in the wild type (P), gamyb-2 (Q), oscps1-1 (R), gid1-4 (S), and gid2-5 (T).
(U) to (X) Transverse sections of anthers at the VP stage in the wild type (U), gamyb-2 (V), oscps1-1 (W), and gid2-5 (X).
(Y) to (AB) Transverse sections of anthers at the MP stage in the wild type (Y), gamyb-2 (Z), oscps1-1 (AA), and gid2-5 (AB).
Defect in PCD of Tapetal Cells in GA-Related Mutants
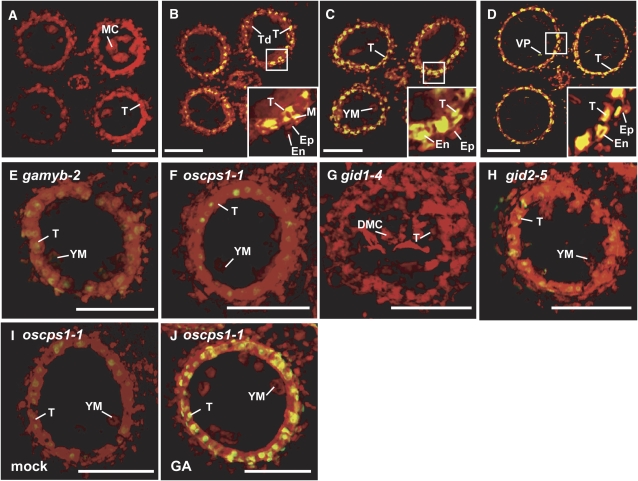
The degeneration of tapetal cells is thought to be caused by PCD (Li et al., 2006). To examine the PCD process of the enlarged tapetal cells observed in GA-related mutants, we performed a TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling) assay of tapetal cells of the wild type and mutants. At the MEI stage, no signal was detected in wild-type anthers (Figure 2A). At the TD stage, some of the tapetal and middle layer cells in the wild type showed a TUNEL-positive signal (Figure 2B, inset), indicating that PCD of these cells starts at this stage. At the YM stage, almost all the tapetal cells in the wild type showed strong TUNEL-positive signals (Figure 2C). At the VP stage, strong signals were still visible in degenerating tapetal cells and also appeared in the outer cell layers (Figure 2D, inset). This staining pattern of the TUNEL assay corresponds closely with the degradation process of tapetal cells (cf. with Figure 1). By contrast, no clear TUNEL-positive signal was seen in tapetal cells of gamyb-2, oscps1-1, or gid1-4 mutants (Figures 2E to 2G), whereas some faint signals were observed in gid2-5 at the YM stage (Figure 2H). The results of the TUNEL assay are consistent with the abnormal tapetal cells without degeneration in gamyb-2, oscps1-1, and gid1-4 and with their retardation of degeneration in gid2-5 (Figure 1), suggesting that the failure of tapetal cell degradation in these mutants is due to a defect in their PCD.
Figure 2.
TUNEL Assay of Anthers in Wild-Type and GA-Related Mutants.
(A) MEI stage in the wild type.
(B) TD stage in the wild type.
(C) and (E) to (H) YM stage in the wild type (C), gamyb-2 (E), oscps1-1 (F), gid1-4 (G), and gid2-5 (H).
(D) VP stage in the wild type.
(I) Mock-treated anther at the YM stage in oscps1-1.
(J) GA3-treated anther at the YM stage in oscps1-1.
The box in each panel outlines the portion of the anther cell layers shown at higher magnification in the inset. Nuclei were stained with propidium iodide (red), while yellow fluorescence is a TUNEL-positive signal. Ep, epidermal cell layer; En, endothecial cell layer; M, middle layer; T, tapetal layer; MC, meiocyte; DMC, degraded meiocyte; Td, tetrad. Bars = 50 μm.
To confirm that GA is responsible for PCD of tapetal cells, we treated these mutants with GA3 at the panicle primordium stage (before the PMC stage). GA treatment rescued the defect in anther development of oscps1-1 and was accompanied by the degradation of tapetal cells and the occurrence of TUNEL-positive signals (Figures 2I and 2J; see Supplemental Figures 4C and 4D online). By contrast, no phenotypic rescue by GA treatment was seen in the GA-insensitive mutants gamyb-2, gid1-4, and gid2-5 (see Supplemental Figures 4A, 4B, and 4E to 4H online). These observations confirm that GA is essential for PCD of tapetal cells and is involved in triggering the processes leading to their degradation.
Defects in Ubisch Body and Exine Formation in GA-Related Mutants
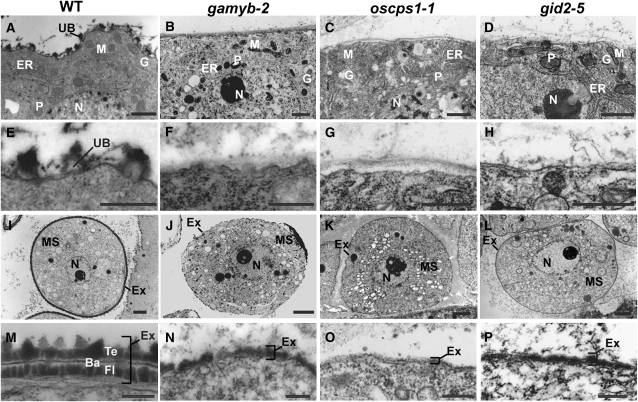
We observed cross sections of mutant anthers at the YM stage using transmission electron microscopy (TEM). In the cytoplasm of tapetal cells, there was no difference between the wild type and the mutants (Figures 3A to 3D). However, there were apparent differences on the surfaces of the tapetal cells. The wild-type tapetal cells produced numerous densely stained Ubisch bodies (Figure 3E), which consist of sporopollenin with a complex of fatty acid derivatives and phenolic constituents (Ahlers et al., 1999, 2000; Bubert et al., 2002; Wang et al., 2003). By contrast, such dense staining was not seen in the mutants (Figures 3F to 3H). The TEM observation also revealed some abnormal morphologies in other anther wall layers of the mutants, namely, the retardation of middle layer degradation in all mutants and incompletely developed vacuoles in the epidermal cells of all mutants except for gid2-5 (see Supplemental Figures 3A to 3J online). This suggests that GA action is not limited to the tapetal layer, but extends to the anther wall layers, including the tapetal layer. Inside the microspores, no abnormal structures were observed in these mutants (Figures 3I to 3L; see Supplemental Figures 5A to 5D online). The surfaces of wild-type microspores were surrounded by exine consisting of three layers (tectum, bacula, and foot layer) (Figure 3M), while microspores in the mutants were surrounded by a defective exine consisting of a single layer (Figures 3N to 3P). The impaired formation of exine in microspores might be affected by the defect in Ubisch bodies seen in the tapetal cells, since Ubisch bodies supply proteins and sporopollenin to microspores for development of exine (Piffanelli et al., 1998; Wang et al., 2003; Jung et al., 2006).
Figure 3.
Ultrastructural Analysis of Ubisch Bodies and Exine in Wild-Type and GA-Related Mutants by TEM.
(A) to (D) The cytoplasm of tapetal cells in the wild type (A), gamyb-2 (B), oscps1-1 (C), and gid2-5 (D). N, nucleus; P, plastid; M, mitochondria; ER, endoplasmic reticulum; G, Golgi body; UB, Ubisch body. Bars = 1 μm.
(E) to (H) The plasma membrane of tapetal cells in the wild type (E), gamyb-2 (F), oscps1-1 (G), and gid2-5 (H). Bars = 500 nm.
(I) to (L) The cytoplasm of microspores in the wild type (I), gamyb-2 (J), oscps1-1 (K), and gid2-5 (L). MS, microspore; N, nucleus; Ex, exine. Bars = 2 μm.
(M) to (P) Microspore coat of the wild type (M), gamyb-2 (N), oscps1-1 (O), and gid2-5 (P). Ba, bacula; Te, tectum; Fl, foot layer. Bars = 250 nm.
Most GA-Regulated Genes in the Rice Anther Are Modulated by GAMYB
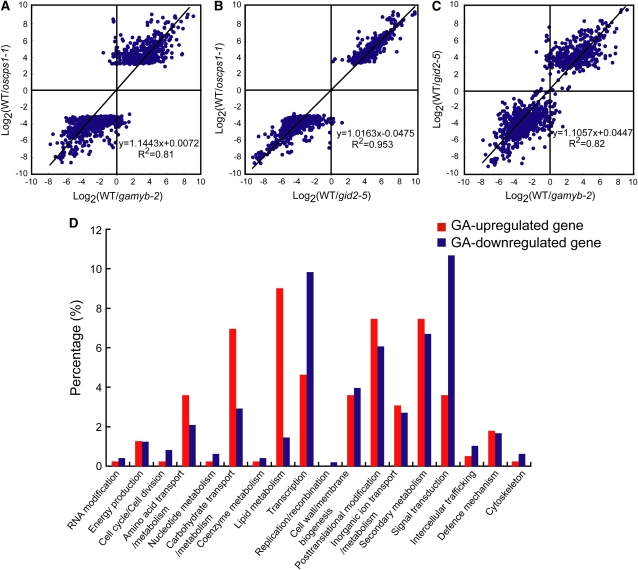
Our phenotypic analyses showed that the abnormal anther phenotype of gamyb-2 is quite similar to that of other GA-related mutants, having enlarged tapetal cells that do not undergo PCD, collapsed microspores, and failure of exine formation. These findings suggest that GAMYB plays a significant role in the GA signaling pathway in anthers. To test this hypothesis, we compared the expression profile of gamyb-2 with profiles of the GA-related mutants oscps1-1, gid1-7, and gid2-5 by two-color microarray analysis using RNAs from anthers at the YM stage. The microarray slides contained ∼44,000 60-mer oligonucleotides corresponding to rice transcriptional units. The microarray experiment was repeated twice using independently isolated RNA, and each experiment included a dye-swapping technical replicate for each biological replicate. To identify differentially expressed genes, the Bioconductor Rank Prod package was conducted on log2 normalized signal ratio at P < 0.0001. The multiple testing in this method was taken into account by the use of percentage of false prediction (pfp) at the condition of pfp < 0.01. Under these conditions, we identified 1114 probes (870 genes) that differed in expression between oscps1-1 and the wild type: 469 probes (390 genes) were identified as GA upregulated (repressed in oscps1-1), and 645 probes (480 genes) were identified as GA downregulated (induced in oscps1-1) (see Supplemental Data Set 1 online).
Using these 1114 probes, we compared the expression pattern between oscps1-1 and gamyb-2. The average log2 values of these 1114 probes were plotted against those calculated from the gamyb-2/wild type analysis (Figure 4A). In this figure, the x axis represents the average log2 values of the signal ratio of the wild type versus gamyb-2, the y axis represents that of the wild type versus oscps1-1, and each spot represents one probe. The analysis revealed that the expression patterns of oscps1-1 and gamyb-2 are very similar. The correlation coefficient between the wild type/oscps1-1 and wild type/gamyb-2 expression ratio was 0.81, and the slope of the regression line was 1.1. This indicates that the expression levels of most of the probes selected during the wild type/oscps1-1 microarray analysis were changed to a similar degree in gamyb-2, supporting the hypothesis that most GA-regulated genes in the anther are affected to a similar extent by GAMYB.
Figure 4.
Microarray Analysis of Genes Regulated by GA, GAMYB, and GID2.
Axes show the log2 value of the ratios of signal intensities observed in the indicated plants. Each spot represents one probe.
(A) Scatterplot analysis to compare the expression of genes regulated by GA (oscps1-1 background) and GAMYB in the anther.
(B) Scatterplot analysis to compare the expression of genes regulated by GA (oscps1-1 background) and GID2 in the anther.
(C) Scatterplot analysis to compare the expression of genes regulated by GAMYB and GID2 in the anther.
(D) Functional classification of GA-regulated genes identified by the comparison of genes differentially expressed between the wild type and oscps1-1.
We next compared the expression patterns of these 1114 probes between oscps1-1 and gid2-5 anthers (Figure 4B). Almost all of these 1114 probes identified in oscps1-1 were similarly regulated in gid2-5. The correlation coefficient of these analyses was quite high (0.95), and the slope of the regression line was 1.0, demonstrating a strong positive correlation between Os CPS1- and GID2-dependent regulations. These results also indicate that the gene expression regulated by GA in rice anthers is almost completely governed by GID2 function and therefore by the GID1-DELLA–dependent GA signaling system (see Discussion). We then compared the effects of gamyb-2 and gid2-5 mutations on the expression of 1114 probes. The spots in the scatterplot were located around Y = X, although the correlation between gid2-5 and gamyb-2 (0.82) was lower than that between gid2-5 and oscps1-1 (0.95) (Figure 4C). This result indicates that, although most GA-regulated genes are modulated by both GID2 and GAMYB, the modulation by GID2 was more tightly correlated to GA-mediated change than was the modulation by GAMYB. We also attempted to examine the correlation of gene expression profiles between the GA-deficient mutant oscps1-1 and the GA receptor mutant gid1-4. However, it was difficult to compare the expression profiles between gid1-4 and oscps1-1 at the YM stage because the phenotype of the gid1-4 anther is much more severe than that of oscps1-1, and the biological conditions of the two mutant anther types was too different to compare their expression patterns (cf. Figures 1R and 1S). Therefore, we used a weak allele of gid1, gid1-7, which is able to develop vacuolated pollen. For analyzing the data from gid1-7, we further selected 830 probes from the 1114 probes, using the significance level calculated in the wild type/gid1-7 microarray analysis (P < 0.05). Even though gid1-7 shows a much milder phenotype than oscps1-1, the correlation coefficient between oscps1-1 and gid1-7 was calculated as being 0.68 (see Supplemental Figure 6 online), indicating an apparent correlation of gene expression between oscps1-1 and gid1-7. The slope of the regression line was calculated as being 1.4, which might reflect the difference in severity of the two mutants.
Some GA-Regulated Genes in Anthers Have GAMYB Binding-Like Motifs in Their Promoter Regions
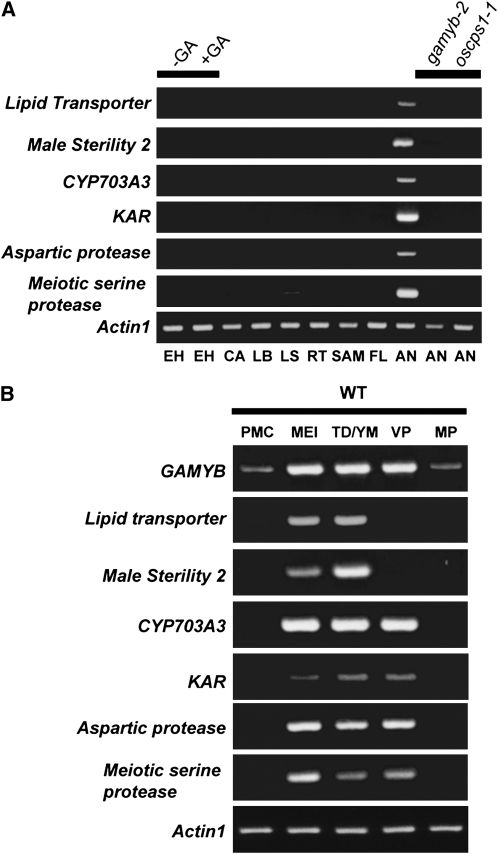
The 870 GA-regulated genes selected in the microarray analysis were categorized into 17 functional groups using Clusters of Orthologous Groups (COG) analysis at http://www.ncbi.nlm.nih.gov/COG/, with manual adjustment when necessary (Figure 4D; see Supplemental Data Set 2 online). Genes implicated in lipid metabolism, posttranslational modification, and secondary metabolism were more numerous in the GA-upregulated group, whereas those involved in transcription and signal transduction were more numerous in the GA-downregulated group. Based on previous results seen for barley (Hordeum vulgare) GAMYB in aleurone cells (Gubler et al., 1995, 1999), we expected that rice GAMYB would directly enhance the expression of some of GA-upregulated genes in the anther. Because 8-bp DNA sequences with a C/TAAC core preferentially interacting with barley GAMYB have been identified (GAMYB binding motifs) (Gubler et al., 1999), we searched for such sequences in the 5′-flanking region of all GA-regulated genes, which are registered in the Rice Annotation Project Database (RAP-DB; http://rapdb.dna.affrc.go.jp/). We found that 182 out of the 390 GA-upregulated genes (47%) contain GAMYB binding motifs in their 5′-flanking regions, whereas 190 out of 480 GA-downregulated genes (40%) contain such sequences in their 5′-flanking region (see Supplemental Data Set 3 online). The frequency of GAMYB binding sites in the GA-upregulated genes was also enriched relative to the overall 50,259 genes (40%). The statistical analysis confirmed that the frequency of the GAMYB binding motif in the 5′-flanking region of GA-upregulated genes is significantly higher (at the 5% level) than the frequencies of GA-downregulated and overall genes, upon performing the χ2 test. This supports the hypothesis that some GA-upregulated genes might be regulated by a direct interaction between GAMYB and GAMYB binding motifs. We then selected six GA-upregulated genes categorized as having roles in posttranslational modification and lipid metabolism and containing GAMYB binding-like sequences with a perfect match or a single mismatch from GAMYB binding motifs. RT-PCR analysis revealed that these genes were preferentially expressed in anthers but not in other organs tested, including GA-treated embryoless half-seeds (Figure 5A), where GAMYB actively functions in GA signaling. Consistent with the results of the microarray experiments, the expression of these genes was not detected in oscps1-1 and gamyb-2 anthers (Figure 5A).
Figure 5.
Expression Analysis of GA-Regulated Genes during Anther Development.
(A) Spatial expression of GA-regulated genes in various tissues by RT-PCR analysis. EH, embryoless half-seed; CA, callus; LB, leaf blade; LS, leaf sheath; RT, root; VM, vegetative meristem; FL, mature flower (lemma > 8 mm); AN, anther; SAM, shoot apical meristem. Embryoless half-seeds were treated with (+) or without (–) 10−5 M GA3 for 3 d. Rice Actin1 was used as a control. Results presented are representative of three independent experiments.
(B) Temporal expression of GA-regulated genes in developing anthers at five stages of anther development by RT-PCR analysis. Each developmental stage was scored according to the length of the lemma: PMC stage (lemma 2 mm), MEI stage (2 to 3 mm), TD/YM stage (3 to 5 mm), VP stage (5 to 8 mm), and MP stage (> 8 mm). Rice Actin1 was used as a control. Results presented are representative of three independent experiments.
We also examined the expression pattern of these six genes during anther development and compared their expression pattern with that of GAMYB (Figure 5B). The expression of the GA-upregulated genes in anthers was very low at the PMC stage, started to increase during the MEI stage, and peaked at the TD/YM stage or VP stage (Figure 5B). At the MP stage, their transcript levels were hardly detectable (Figure 5B). The expression pattern of GAMYB was essentially the same as these GA-upregulated genes with lower expression at the PMC and MP stages (Figure 5B). This overlapping expression of GA-upregulated genes and GAMYB also supports the hypothesis that GAMYB positively regulates the expression of these genes via the GA signaling pathway during anther development.
GAMYB Interacts with the GAMYB Binding-Like Motifs in GA-Upregulated Genes of Anthers and Regulates Their Expression
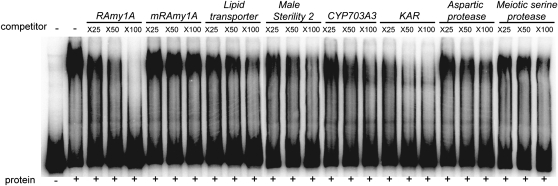
To examine the direct interaction between GAMYB and the GAMYB binding-like motifs found in the GA-upregulated genes, we performed a competitive gel-shift assay using the authentic GAMYB binding sequence derived from the rice α-amylase1A gene, RAmy1A, as a probe (Figure 6). The retarded band formed by GAMYB and its binding sequence of RAmy1A was eliminated by addition of an excess amount of the same nonlabeled sequence in a dose-dependent manner, but not by the mutated sequence (Figure 6, RAmy1A and mRAmy1A). The fragments containing GAMYB binding-like motifs of the six GA-upregulated genes inhibited the formation of its retarded band with varying efficiencies, but more effectively than mRAmy1A (Figure 6). These results indicate that GAMYB interacts specifically with the GAMYB binding-like motifs found in these GA-upregulated genes.
Figure 6.
Competitive Gel-Shift Assay.
DNA fragments containing the GAMYB binding-like motifs of six GA-upregulated genes inhibited the interaction between GAMYB and the 32P-labeled RAmy1A probe (−380 to −85). Competition experiments were performed using increasing molar amounts (×25, ×50, and ×100) of the indicated unlabeled fragment; lanes 3 to 5, RAmy1A (−380 to −85); lanes 6 to 8, mutated RAmy1A (mRAmy1A) (−380 to −85); lanes 9 to 11, Lipid transporter (−501 to −250); lanes 12 to 14, Male Sterility 2 (−425 to −123); lanes 15 to 17, CYP703A3 (−510 to +40); lanes 18 to 20, KAR (−332 to −4); Lanes 21 to 23, Aspartic protease (−495 to −126); and Lanes 24 to 26, Meiotic serine protease (−397 to −142). Numbers in parentheses indicate the distance in base pairs from the transcriptional initiation site.
CYP703A3 and KAR Genes Are Candidate Targets for GAMYB in Exine Formation
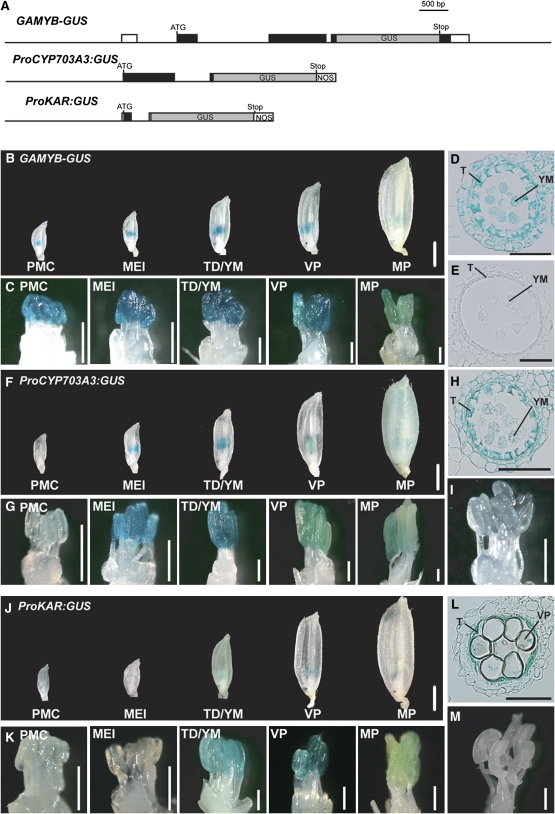
To further study whether GAMYB modulates the expression of GA-upregulated genes in vivo by interacting with their GAMYB binding-like motifs, we focused on two genes implicated in lipid metabolism (Figure 4D), CYP703A3 and KAR (see Supplemental Figures 7 and 8 and Supplemental Data Sets 4 and 5 online). As fatty alcohols are essential for exine and Ubisch body formation as a precursor of sporopollenin, we expected that these genes might be responsible for GA-mediated exine and Ubisch body formation under the regulation of GAMYB. We produced promoter-β-glucuronidase (GUS) reporter constructs, ProCYP703A3:GUS and ProKAR:GUS (Figure 7A), and transformed these constructs into wild-type plants. We also introduced a GAMYB-GUS translational fusion to compare its expression pattern to that of the two lipid metabolism genes (Figure 7A). In the presence of the complete GAMYB gene, GUS expression was detected in the anthers of developing flowers but not in other floral organs (Figures 7B and 7C). GUS expression was observed at every stage, with lowest expression at the MP stage and highest at the TD/YM stage (Figure 7C). This GUS expression pattern is consistent with the GAMYB expression pattern observed by RT-PCR analysis (Figure 5B). We also examined the cellular localization of GAMYB-GUS expression at the TD/YM stage and found strong GUS activity in the tapetal cells and faint GUS activity in the other anther wall layers and microspores (Figure 7D), while no staining was seen in the vector control plant (Figure 7E). Faint GUS expression in the microspore and anther wall layers is consistent with the abnormal phenotype of these cells, indicating that GAMYB may function in the development of these cells.
Figure 7.
GUS Expression under the Control of the Promoters of GAMYB, CYP703A3, and KAR during Anther Development.
(A) Schematic representation of the GAMYB-GUS, ProCYP703A3:GUS, and ProKAR:GUS constructs. Untranscribed regions and introns are shown as thin lines, untranslated regions as open boxes, and coding regions as closed boxes. GUS reporter regions are indicated in gray boxes. ATG, ATG start codon; Stop, stop codon; NOS, nos terminator.
(B) to (D) GUS expression of GAMYB-GUS (line #5) in the wild type. We analyzed 13 T0-independent lines.
(B) Whole flowers at each developmental stage. Bar = 1 mm.
(C) Close-up view of stamens. Bars = 400 μm.
(D) Cross section of anther at the TD/YM stage. Bar = 50 μm.
(E) Anther cross section of a transgenic line at the TD/YM stage carrying the vector control. Bar = 50 μm.
(F) to (H) GUS expression of ProCYP703A3:GUS in the wild type (line #4). We analyzed 10 T0-independent lines.
(F) Whole flowers at each developmental stage. Bar = 1 mm.
(G) Close-up view of stamens. Bars = 400 μm.
(H) Cross section of an anther at the TD/YM stage. Bar = 50 μm.
(I) GUS expression of ProCYP703A3:GUS in gamyb-2 (line #3). Bar = 400 μm. We analyzed 12 T0-independent lines.
(J) to (L) GUS expression of ProKAR:GUS in the wild type (line #7). We analyzed 10 T0-independent lines.
(J) Whole flowers at each developmental stage. Bar = 1 mm.
(K) Close-up view of stamens. Bars = 400 μm.
(L) Cross section of an anther at the VP stage. Bar = 50 μm.
(M) GUS expression of ProKAR:GUS in gamyb-2 (line #2). We analyzed 13 T0-independent lines. Bar = 400 μm. T, tapetal layer; YM, young microspore; VP, vacuolated pollen.
In ProCYP703A3:GUS transgenic plants, GUS expression was similar to that in GAMYB-GUS plants and was preferentially observed during the MEI to TD/YM stages (Figures 7F and 7G). The cross sections confirmed that the strong GUS activity at the TD/YM stage was localized in tapetal cells and that the weak GUS activity was in the other anther wall layers and microspores, similar to the pattern seen for GAMYB-GUS expression (Figures 7E and 7H). When we introduced the same construct into the gamyb-2 plants, no GUS expression was observed (Figure 7I), which is consistent with the expression analysis by RT-PCR (Figure 5A). In ProKAR:GUS transgenic plants, GUS activity was preferentially observed at the VP stage (Figures 7J and 7K), mainly in tapetal cells (Figures 7E and 7L). Again, consistent with the RT-PCR results shown in Figure 5A, GUS activity was not detected in the anthers of gamyb-2 (Figure 7M). The expression analysis by RT-PCR indicated that KAR was expressed during a longer period in the anther developmental process (Figure 5B) than was seen in the promoter-GUS experiment (Figure 7K). This apparent inconsistency might be caused by the detection sensitivity of RT-PCR being much higher than that of the promoter-GUS experiment: GUS activity was faint even in anthers showing the strongest GUS activity in these transformants (Figure 7K).
We also examined the direct regulation of CYP703A3 and KAR expression by the GAMYB function. For this experiment, we introduced a chimeric construct ProAct1:mOs GAMYB-GR, which consisted of the full-length GAMYB cDNA with nine synonymous nucleotide substitutions at the miRNA159 target site and the rat glucocorticoid receptor (GR), under the control of a rice constitutive promoter, Actin1 (see Supplemental Figure 9A online). The mRNA levels for CYP703A3 and KAR in the gamyb-2 flowers carrying ProAct1:mOs GAMYB-GR were quite low, consistent with the GUS staining patterns in Figures 7I and 7M, whereas the dexamethasone (DEX) treatment for 4 h caused more than ∼10- and ∼3-fold CYP703A3 and KAR mRNA levels, respectively (see Supplemental Figures 9B and 9C online). Such an increase in mRNA levels was not inhibited by cotreatment with the protein synthesis inhibitor cycloheximide (CHX), indicating that GAMYB directly induces expression of CYP703A3 and KAR. In the case of KAR, the DEX+CHX treatment caused hyperinduction of its mRNA, suggesting the existence of an unknown repressor for KAR expression, which is constantly produced and rapidly degraded.
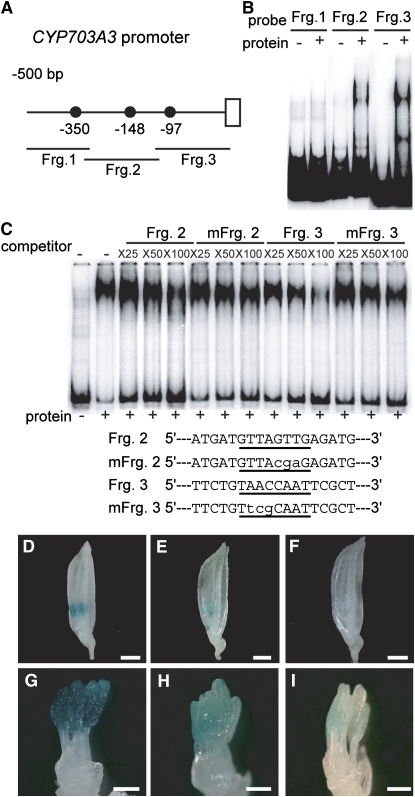
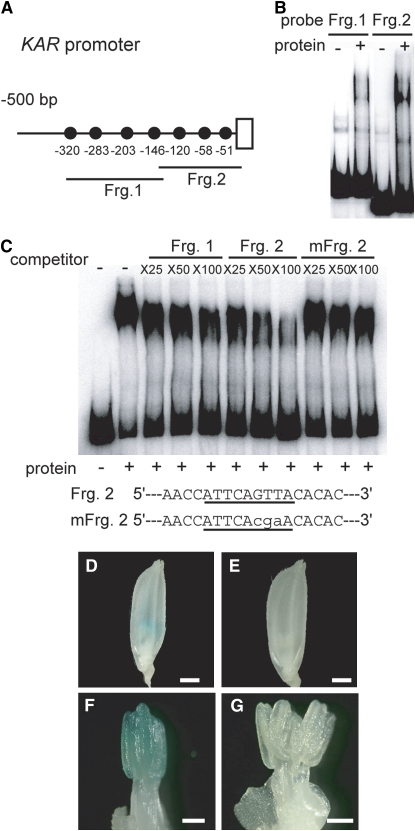
As there are three and seven GAMYB binding-like motifs in the promoter regions of the CYP703A3 and KAR genes, respectively (Figures 8A and 9A), we attempted to identify which GAMYB binding like motifs specifically interact with GAMYB by a gel-shift assay using fragmented CYP703A3 and KAR promoter sequences containing these motifs (closed circles in Figures 8A and 9A). The recombinant GAMYB protein bound to Fragments 2 and 3 of the CYP703A3 promoter and to Fragments 1 and 2 of the KAR promoter (Figures 8B and 9B, respectively). Next, we used the GAMYB binding sequence of RAmy1A as a probe and performed a competition experiment with fragments of the CYP703A3 and KAR promoters. Formation of a complex of GAMYB and the RAmy1A target sequence was inhibited by Fragments 2 and 3 of the CYP703A3 promoter in a dose-dependent manner, with Fragment 3 inhibiting formation of this complex more effectively (Figure 8C). The fragments containing mutated GAMYB binding-like motifs (mFrg. 2 and mFrg. 3) did not effectively inhibit the formation of the complex (Figure 8C). In the case of the KAR promoter, Fragment 2 inhibited formation of the complex more effectively than Fragment 1, while mFrg. 2, which is mutated at the GAMYB binding-like motif (−120 bp), did not compete (Figure 9C). Other GAMYB binding-like motifs found in the KAR promoter did not exhibit effective competitive activity.
Figure 8.
In Vivo Function of GAMYB Binding-Like Motifs in CYP703A3.
(A) Schematic representation of the 5′-flanking region of CYP703A3. The closed circles show GAMYB binding-like motifs. Fragments 1, 2, and 3 (Frg. 1, bp −510 to −308; Frg. 2, bp −328 to −112; Frg. 3, bp −131 to +40) were used as probes or competitors for gel-shift assays in (B) and (C).
(B) Gel-shift assay with the recombinant GAMYB protein and 32P-labeled Frg. 1 to 3 presented in (A).
(C) Competitive gel-shift assay. The interaction between GAMYB and 32P-labeled RAmy1A probe was efficiently competed by Fragment 3 but not by Fragment 2, whereas mutagenized Fragment 2 or 3 (mFrg. 2 and 3), which contain displaced nucleotides at the GAMYB binding site, did not show effective competitive activity. Competition experiments were performed using increasing molar amounts (×25, ×50, and ×100) of the indicated unlabeled fragment. The sequences of the oligonucleotides used as competitor fragments are shown at the bottom of the panel. The putative binding site is underlined, and the mutations are indicated in lowercase letters.
(D) to (F) Whole flowers at the TD/YM stage. Bars = 1 mm.
(G) to (I) Close-up view of stamens at the TD/YM stage. Bars = 300 μm.
(D) and (G) Flower from a plant transformed with the intact ProCYP703A3:GUS construct (line #6).
(E) and (H) Flower from a plant transformed with the mutagenized ProCYP703A3 (mFrg. 2):GUS construct (line #12).
(F) and (I) Flower from a plant transformed with the mutagenized ProCYP703A3 construct (mFrg. 3):GUS (line #9). We analyzed 10 T0-independent lines for ProCYP703A3:GUS, 12 lines for mFrg. 2, and 18 lines for mFrg. 3.
Figure 9.
In Vivo Function of a GAMYB Binding-Like Motif in KAR.
(A) Schematic representation of the 5′-flanking region of KAR. The closed circles show GAMYB binding-like motifs. Fragments 1 and 2 (Frg. 1, bp −332 to −123; Frg. 2, bp −143 to −4) were used as probes or competitors for gel-shift assays in (B) and (C), respectively.
(B) Gel-shift assay with the recombinant GAMYB protein and 32P-labeled Fragments 1 and 2 presented in (A).
(C) Competitive gel-shift assay. The interaction between GAMYB and 32P-labeled RAmy1A probe was efficiently competed by Fragment 2 but not by Fragment 1, whereas mutagenized Fragment 2 (mFrg. 2), which contains displaced nucleotides at the GAMYB binding-like motif, did not show effective competitive activity. Competition experiments were performed using increasing molar amounts (×25, ×50, and ×100) of the indicated unlabeled fragment. The sequences of the oligonucleotides used as Frg. 2 and mFrg.2 are shown at the bottom of the panel. The putative binding site is underlined, and the mutations are indicated in lowercase letters.
(D) and (E) Whole flowers at the VP stage. Bars = 1 mm.
(F) and (G) Close-up view of stamens at the VP stage. Bars = 300 μm.
(D) and (F) Flower transformed with the intact ProKAR:GUS (line #8).
(E) and (G) Flower transformed with the mutagenized ProKAR (mFrg. 2):GUS (line #12). We analyzed 10 T0-independent lines for ProKAR:GUS and 12 lines for mFrg. 2.
Next, we examined whether these GAMYB binding sites function as cis-elements for the GAMYB-dependent expression of these genes. We generated mutated promoter-GUS constructs carrying mutated GAMYB binding sites and transformed these into wild-type plants. As described in Figure 7, GUS expression occurs in anthers when GUS is driven by the native CYP703A3 or KAR promoter (Figures 8D, 8G, 9D, and 9F). Among transgenic plants containing mutated CYP703A3 promoters, lower GUS expression was observed in almost all T0 transgenic plants carrying the mFrg.2 promoter (10 of 12 lines), whereas no or very low activity was observed in all of the mFrg.3 T0 lines (18 of 18 lines) (Figures 8D to 8I; see Supplemental Figures 10A to 10C online). Similarly, all T0 transgenic plants containing the mFrg.2 promoter of KAR showed very low GUS activity (12 of 12 lines) (Figures 9D to 9G; see Supplemental Figures 11A and 11B online). These results show that the GAMYB binding sites in the promoters of CYP703A3 and KAR genes are necessary for their expression in developing anthers.
Mutation of CYP703A3 Causes a Defect in Exine and Ubisch Body Formation
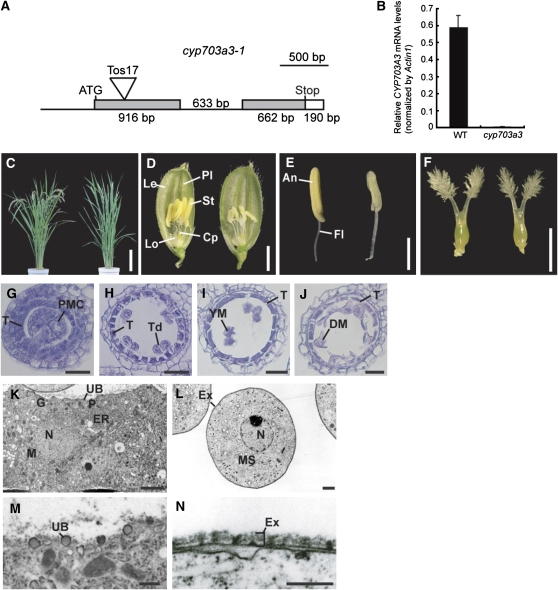
The above observations suggest that GAMYB is involved in exine and Ubisch body formation by regulating the expression of target genes, such as CYP703A and KAR. To test this model, we obtained a retrotransposon insertion mutant of CYP703A3 from the Rice Tos17 Insertion Mutant Database (http://tos.nias.affrc.go.jp/∼miyao/pub/tos17/index.html.en, NG6024). DNA sequence analysis revealed that a retrotransposon, Tos17, was inserted into exon 1 of CYP703A3 (Figure 10A). Quantitative RT-PCR analysis detected a very low level of CYP703A3 transcript in flowers of the cyp703a3 mutant, whereas some level of transcript was detected in the wild type (Figure 10B). The introduction of the wild-type allele of CYP703A3 into this mutant rescued the abnormal phenotype mentioned (see Supplemental Figure 12 online). These results demonstrate that this mutation is a loss-of-function mutant of CYP703A3.
Figure 10.
Characterization of a Loss-of-Function Mutant of CYP703A3.
(A) Site of Tos17 insertion in the CYP703A3 gene. Untranscribed and intron regions are represented as lines. Coding and untranslated regions are represented as gray and open boxes, respectively. The insertion site of Tos17 is indicated as a triangle. Bar = 500 bp.
(B) Quantitative RT-PCR analysis of CYP703A3 in flowers. Total RNA was extracted from wild-type and cyp703a3-1 flowers. The transcript was assayed by real-time PCR relative to an internal Actin1 control. Data are means ± sd from three replicates.
(C) Gross morphology of the wild type (left) and the cyp703a3-1 mutant (right) at the ripening stage. Bar = 20 cm.
(D) Flowers of the wild type (left) and cyp703a3-1 (right). Cp, carpel; Le, lemma; Lo, lodicule; Pl, palea; St, stamen. Bar = 2 mm.
(E) Stamens of the wild type (left) and cyp703a3-1 (right). An, anther; Fl, filament. Bar = 1 mm.
(F) Pistils of the wild type (left) and cyp703a3-1 (right). Bar = 1 mm.
(G) to (J) Transverse sections of cyp703a3-1 anthers at the PMC stage (G), TD stage (H), YM stage (I), and MP stage (J). T, tapetal layer; Td, tetrad; DM, degraded microspore. Bars = 25 μm.
(K) to (N) Ultrastructural analysis of cyp703a3-1 anthers by TEM.
(K) The cytoplasm of tapetal cell in cyp703a3-1. N, nucleus; P, plastid; M, mitochondria; ER, endoplasmic reticulum; G, Golgi body; UB, Ubisch body. Bar = 1 μm.
(L) The cytoplasm of a cyp703a3-1 microspore. MS, microspore; N, nucleus; Ex, exine. Bar = 2 μm.
(M) Defective Ubisch body on the plasma membrane of tapetal cells in cyp703a3-1. Bar = 500 nm.
(N) Defective exine on the microspore coat in cyp703a3-1. Bar = 250 nm.
No abnormal phenotype was observed in the cyp703a3 mutant at the vegetative stage (Figure 10C). The mutant developed flowers with normal organs (Figures 10D and 10F), except stamens, which contained shrunken and whitened anthers (Figure 10E). We observed cross sections of mutant anthers at different developmental stages. At the PMC or TD stage, no abnormal phenotype was observed in the mutant anthers, and PMCs underwent normal meiosis to form tetrads of haploid microspores (Figures 10G and 10H). At the YM stage, microspores released from tetrads were irregularly shaped in the mutant (Figure 10I). At the MP stage, collapse of microspores occurred as in GA-related mutant anthers, while degeneration of tapetal cells occurred normally in this mutant, as in the wild type (Figure 10J). We also observed the ultrastructure of tapetal and microspore cells by TEM. There was no abnormal phenotype in the cytoplasm of tapetal or microspore cells (Figures 10K and 10L; see Supplemental Figure 5E online). However, apparent abnormal phenotypes were seen on the surfaces of these cells (Figures 10M and 10N). Ubisch bodies at the tapetal cell surface of the mutant showed incomplete development, with smaller, less-dense particles than those in the wild type (cf. Figure 10M [mutant] and Figure 3E [wild type]). The exine of the mutant microspores consisted of a single layer (Figure 10N), while the wild-type microspores contained a normal three-layered exine (Figure 3M). These observations indicate that loss of function of the CYP703A3 gene impairs exine and Ubisch body formation. Such phenotypes of impaired exine and Ubisch body formation are essentially the same as those of GA-related mutants, while the degeneration of tapetal cells occurred in the cyp703a mutant as in the wild type (see Discussion).
DISCUSSION
GA Is Important for Formation of Exine and Ubisch Bodies and for PCD of Tapetal Cells
It has been reported that GA deficiency or insensitivity in several plant species causes abnormal development of anthers, which often leads to male sterility. For example, Izhaki et al. (2002) reported that petunia (Petunia hybrida) plants treated with a GA synthesis inhibitor, paclobutrazol, or overexpressing a GA signal repressor, SPY, developed immature pollen grains and degenerated tapetal cells. The tomato GA-deficient mutants gib-1 and gib-2 show defects in the meiosis of PMCs (Nester and Zeevaart, 1988; Jacobsen and Olszewski, 1991). In addition, the tapetal cells of gib-2 are developmentally arrested, resulting in their abnormal degradation (Nester and Zeevaart, 1988). In the Arabidopsis GA-deficient mutant ga1-3, microspore and tapetal cell development appears to be arrested prior to pollen mitosis, thus preventing the formation of mature pollen (Cheng et al., 2004). All these studies have shown that GA is essential for normal development of pollen and tapetal cells, although whether the defect of pollen development in these mutants is a direct effect of GA deficiency or a secondary effect of arrested tapetal cell development has been unclear (Cheng et al., 2004).
The results presented here indicate that the primary target of GA action during anther development is the tapetal cell. Microscopic observations revealed that the first abnormal event during anther development in all GA-related mutants tested is enlargement of tapetal cells (Figure 1; see Supplemental Figure 2 online). Furthermore, GA deficiency or insensitivity causes defective PCD of tapetal cells (Figure 2). During development, these enlarged tapetal cells eventually occupied the entire locule, taking up the space where pollen development normally occurs. Based on the observations of a rice male sterile mutant, tdr, Li et al. (2006) came to a similar conclusion, that is, that production of functional pollen grains depends on the timely death and degeneration of tapetal cells. On the other hand, GA deficiency or insensitivity also causes defective Ubisch body formation in tapetal cells, seriously affecting the formation of exine (Figure 3). The exine provides a physiological resistance to the internal pressure of pollen and various types of biological attack and dehydration (Ku et al., 2003; Edlund et al., 2004). Sporopollenin, a major component of both exine and Ubisch bodies, is synthesized in the tapetal cell and is carried from the tapetal cell via Ubisch bodies to the pollen wall, where it contributes to the formation of the exine structure (Piffanelli et al., 1998; Wang et al., 2003; Jung et al., 2006). All these results strongly support the idea that failure in pollen development in GA-related mutants is a secondary effect of abnormal tapetal cell development and defective Ubisch body formation in tapetal cells.
The Role of GAMYB in the GA-Mediated Gene Expression System in Anther
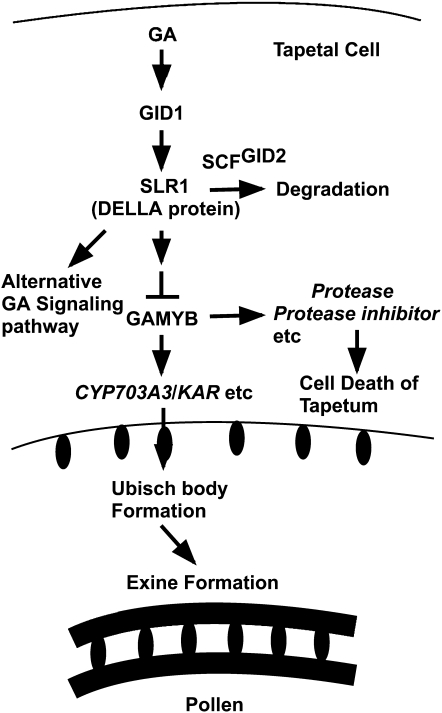
Microarray analysis showed a similar expression pattern between gamyb-2 and a GA-deficient mutant, oscps1-1, and also between gamyb-2 and an F-box–deficient mutant, gid2-5 (Figure 4). This strongly suggests that expression of most GA-regulated genes in anthers is modulated by GAMYB, which appears to function as a major player in GA-mediated gene expression downstream of the GID1/DELLA GA perception system (Figure 11). Microscopy analyses of anthers revealed that all of the GA-related mutants examined have a similar abnormal anther development phenotype as that of gamyb (Figures 1 to 3), supporting the idea that GAMYB functions as a major player in GA signaling in anther development.
Figure 11.
Model for GA Signaling in the Anther.
GA signaling predominantly works in tapetal cells through perception by the GID1/DELLA (SLR1) system that leads to degradation of DELLA protein by SCFGID2 proteasome pathway and downstream action of GAMYB. GAMYB activates the expression of CYP703A3, KAR, and other genes involved in the synthesis of sporopollenin, which is an essential component of the Ubisch body in the tapetal cell and exine in the pollen coat and also activates some GA-upregulated genes, such as Protease and Protease inhibitor that are probably involved in cell death of tapetal cells. See text for details.
The microarray analysis also showed very high correlation (coefficient value = 0.95) between gene expression patterns in the oscps1-1 and gid2-5 mutants (Figure 4). This indicates that the GID1/DELLA system is the predominant mechanism of GA perception in the rice anther, as in other organs, although it has been reported that there is a DELLA-independent or partially dependent component of GA-dependent gene regulation in the Arabidopsis flower (Cao et al., 2006). It is noteworthy that when we compared the correlation coefficient among the GA-related mutants, the correlation coefficient between oscps1-1 and gid2-5 (0.95) was highest, whereas the correlation coefficient between oscps1-1 and gamyb-2 (0.81) was similar to that between gid2-5 and gamyb-2 (0.82) (Figure 4). These differences in correlation coefficients may reflect the distance between these components in the signaling pathway, and there may be alternative GA signaling pathways independent from GAMYB in the anther (Figure 11). However, as we could not find any differences in phenotypes between gamyb-2 and other GA mutants in the anther, such alternative pathway(s) may be involved in other unknown event(s) in the anther.
In this study, we focused on GA-upregulated genes identified by the microarray analysis because GAMYB functions as a positive regulator of GA signaling in cereal aleurone cells (Gubler et al., 1995, 1999). Among the GA-upregulated genes, the frequency of genes implicated in lipid metabolism and posttranslational modification, which are logical candidates to be related to the function of tapetal cells (see below), was much higher than that of genes categorized into other functions (Figure 4D). Furthermore, GAMYB binding motifs were found with higher frequency in the 5′-flanking regions of GA-upregulated genes than in GA-downregulated genes (see Supplemental Data Set 3 online), suggesting that GAMYB directly and positively regulates the expression of some GA-upregulated genes in anthers, although the set of genes upregulated by GA in the anther does not overlap with that in aleurone cells, as previously reported (Tsuji et al., 2006). Which specific set of GAMYB-regulated genes is functional in these tissues probably depends on which transcription factors are cooperating with GAMYB, which has a tissue-specific pattern of activity, as previously discussed by Tsuji et al. (2006).
Direct interaction between GAMYB and GAMYB binding-like motifs in the 5′-flanking regions of six GA-upregulated genes was confirmed by a competitive gel-shift assay (Figure 6). Among these six genes, we confirmed by several approaches that GAMYB regulates the expression of CYP703A3 and KAR, which are involved in lipid biosynthesis, through its interaction with their GAMYB binding-like motifs at their 5′-flanking regions (Figures 5 and 7 to 9; see Supplemental Figure 9 online). Studies on an Arabidopsis ortholog of CYP703A3, CYP703A2, have shown that this enzyme functions as a lauric acid in-chain hydroxylase that provides medium-chain hydroxy fatty acids as essential building blocks during sporopollenin biosynthesis, and its loss of function causes impaired exine development (Morant et al., 2007). Similarly, in this study, we showed that a loss-of-function mutant for CYP703A3 exhibited abnormal exine and Ubisch body formation similar to that seen in gamyb-2 and GA-related mutants (Figure 10). These results demonstrate that the CYP703A family members function as sporopollenin biosynthetic enzymes that play an important role in exine formation in both Arabidopsis and rice. On the other hand, KAR encodes β-ketoacyl reductase, a key enzyme required for the first reductive step in de novo synthesis of fatty acids (Toomey and Wakil, 1966), which is a component of sporopollenin (Wang et al., 2003). The importance of fatty acid synthesis for sporopollenin synthesis, and consequently for exine and Ubisch body formation, is supported by the following observation: in the tapetum, the reduction of acetyl-CoA (a substrate for fatty acid synthesis) by expression of antisense pyruvate dehydrogenase E1α driven by a tapetum-specific promoter exhibits aberrant exine formation and premature Ubisch body formation (Yui et al., 2003).
Considering all of these results, we can propose a simple model: GA activates GAMYB function via the GID1/DELLA perception pathway, resulting in upregulation of CYP703A and KAR, which are involved in the synthesis of sporopollenin components, and consequently contributes to Ubisch body formation in tapetal cells and exine formation in pollen (Figure 11). It has been noted that Arabidopsis has a unique secretory tapetum that does not involve Ubisch bodies, whereas most plant species (including rice) have a secretory tapetum that uses Ubisch bodies as a carrier material (Huysmans et al., 1998; Piffanelli et al., 1998; Wang et al., 2003; Jung et al., 2006). Therefore, we have to be careful when proposing to adapt this model to anther development in Arabidopsis. However, we believe that similar GA signaling might function to regulate the expression of CYP703A2 even in the Arabidopsis tapetum because the Arabidopsis CYP703A2 gene has a GAMYB binding motif in its 5′-flanking region (−78 bp from the transcriptional initiation site), similar to the one in the rice CYP703A3 gene. To test this hypothesis, it would be necessary to establish whether GA regulates the developmental process of pollen exine formation via direct GAMYB (MYB33/65) regulation of their target genes in Arabidopsis.
Our microarray analysis revealed that a high frequency of genes implicated in posttranslational modification were under the positive regulation of GAMYB (see Supplemental Data Set 2 online). This group contains many protease genes, including subtilisin-like protease, meiotic Ser proteinase, and aspartic protease. Furthermore, we confirmed that GAMYB directly binds to GAMYB binding-like motifs of some of these genes in vitro (Figure 6). There are reports that some proteases play an important role in PCD (Beers et al., 2000, 2004), and the result of the TUNEL assay demonstrated that the tapetal cells in all GA-related mutants we examined failed to degenerate (Figure 2).
Recently, Li et al. (2006) reported that the rice TDR gene, which encodes a putative basic helix-loop-helix protein, is required for tapetal degradation through the upregulation of a Cys protease, CP1, and a protease inhibitor, Osc6. According to our microarray data, the expression level of TDR is decreased by ∼3.6-fold in gamyb-2, suggesting that TDR might function downstream of GAMYB, as proposed by Wijeratne et al. (2007), but its expression was not decreased by the same degree in oscps1-1 (67%) or gid2-5 (71%) relative to the level in the wild type. Similarly, the expression of CP1 is slightly downregulated in oscps1-1 (48%), gid2-5 (42%), and gamyb-2 (71%) in comparison to in the wild type. On the other hand, the expression of Osc6 is dramatically decreased (by >500-fold) in these three mutants. Furthermore, we identified a GAMYB binding-like motif in the Osc6 promoter region, and the DNA sequence containing the motif (base pair −388 to −301) inhibited the interaction between the RAmy1A probe and GAMYB (see Supplemental Figure 13 online). This indicates that GAMYB directly interacts with the GAMYB binding-like motif found in the Osc6 promoter region. Taken together, these results indicate that the TDR-regulated pathway for tapetal PCD might partially overlap with the pathway that is regulated by GA signaling via GAMYB. Further study on these protease and protease inhibitor genes will provide the basis for understanding the GA-dependent PCD of tapetal cells.
METHODS
Plant Materials and Chemical Treatment
The rice cultivar Oryza sativa cv Nipponbare (wild type); GA-deficient mutant oscps1-1 (mutant of ent-copalyl diphosphate synthase; Sakamoto et al., 2004); GA-insensitive mutants gid1-4 and gid1-7 (mutants of GA receptor GID1; Ueguchi-Tanaka et al., 2005), gid2-5 (mutant of F-box protein; Sasaki et al., 2003) and gamyb-2 (mutant of GAMYB protein; Kaneko et al., 2004); and cyp703a3-1 were used in this study. The seeds of all plants were immersed in water for 2 d, grown for 1 month in a greenhouse, and then transplanted to the field. We identified homozygous plants of oscps1-1, gid1-4, gid1-7, and gid2-5 from the seedlings derived from heterozygous plants based on their dwarfism. Genotyping to identify gamyb-2 plants was performed by PCR amplification using two sets of primers, GAMYB12-U and LTR4A, and GAMYB12-U and GAMYB11-L. Genotyping for cyp703a3-1 was also performed by PCR using two sets of primers, AK106843-19L and LTR4A, and AK106843-19U and AK106843-19L. Primer sequences used in this study are listed in Supplemental Table 1 online.
For the GA treatment experiment, plants at the panicle primordium stage were sprayed with 10−5 M GA3 or 0.1% ethanol (mock) and then harvested after 2 weeks. For DEX treatment, the detached flowers were placed in a solution of 150 mM sucrose containing 10 μM DEX (Sigma-Aldrich) or ethanol for 4 h at 22°C. For inhibition of protein synthesis, the flowers were pretreated in a solution of 10 μM CHX (Sigma-Aldrich) and then incubated for 4 h in a solution containing 10 μM CHX and 10 μM DEX, or 10 μM CHX and ethanol as a control.
Plasmid Construction and Rice Transformation
PCR amplifications for all constructs were performed using high-fidelity PrimeStar DNA polymerase (Takara). Primer sequences used in this study are listed in Supplemental Table 1 online. The PCR-amplified fragments of all constructs were sequenced to ensure that no mutations were introduced.
For the ProCYP703A3:GUS construct, the CYP703A3 promoter region (position −2000 to +1584 from the transcriptional initiation site) was amplified from genomic DNA using CYP703A3-SacIU and CYP703A3-SmaIL primers. The resulting PCR fragment contained a SacI linker sequence at its 5′ end and a SmaI linker at the 3′ end. This amplified PCR fragment was then cloned into the pCR4 Blunt-TOPO vector (Invitrogen) to give ProCYP703A3-pCR4. ProCYP703A3-pCR4 was digested with SacI, blunted with T4 DNA polymerase (Takara), and then digested with SmaI. The 3.5-kb blunted SacI-SmaI fragment was ligated into the SmaI site of the pBI-Hm2 vector (kindly provided by Kenzo Nakamura, Nagoya University) in the correct orientation.
For the ProKAR:GUS construct, the KAR promoter region (position −2000 to +501) was amplified from a genomic subclone using KAR-SpeIU and KAR-EcoRVL primers. The resulting fragment contained a SpeI linker site at its 5′ site and an EcoRV linker site at its 3′ site. This fragment was then introduced into the pCR4 Blunt-TOPO vector, generating the plasmid ProKAR-pCR4. ProKAR-pCR4 was then digested with SpeI and EcoRV, and inserted into the XbaI-SmaI site of the pBI-Hm2 vector.
To produce the mutagenized construct of ProCYP703A3:GUS and ProKAR:GUS, PCR was performed on the plasmid ProCYP703A3-pCR4 or ProKAR-pCR4 with one set of mutagenized primers corresponding to each mutation, as previously reported (Ueguchi-Tanaka et al., 2007b). Each of the mutated constructs of ProCYP703A3-pCR4 and ProKAR-pCR4 was introduced into the pBI-Hm2 vector, as described above.
To create the GAMYB-GUS translational fusion, the GUS coding sequence was amplified from the vector pBI121 (Toyobo) using GUS-ClaIU and GUS-ClaIL primers. The resulting fragment contained a ClaIU linker site at each end. This fragment was then introduced into the pCR4 Blunt-TOPO vector, generating the plasmid GUS-pCR4. The GUS fragment from GUS-pCR4 (cut out with ClaI) and StuI-ClaI and MluI-ClaI fragments from the GAMYB genomic clone GAMYB-pBS (previously made in pBluescript II SK+ vector [Stratagene]; Kaneko et al., 2004) were inserted in the StuI-MluI site of GAMYB-pBS in the correct orientation, resulting in GAMYB-GUS-pBS. GAMYB-GUS-pBS was digested with SalI and then blunted with T4 DNA polymerase (Takara). The resulting fragment was ligated into the SmaI site of the pBI-Hm12 vector (kindly provided by H. Hirano) in the correct orientation.
To produce the GAMYB cDNA-GR mutagenized construct, the full-length GAMYB cDNA was synthesized by PCR using GAMYB-XbaIU and GAMYB-SmaIL primers. This fragment was introduced into the pCR4 Blunt-TOPO vector, and PCR mutagenesis was then used to exchange the miRNA159 target site. The mutated constructs of GAMYB cDNA were constructed in pAct-GR-Hm2.
These chimeric constructs were introduced into Agrobacterium tumefaciens strain EHA101 and used to infect rice callus according to Hiei et al. (1994). Transformed cells and plants were selected by hygromycin, and then regenerants were grown to maturity in pots in a greenhouse.
Microscopy Analysis
For light microscopy, tissues were fixed in FAA (5% formaldehyde, 5% glacial acetic acid, and 63% ethanol) and dehydrated in a graded ethanol series. The samples were embedded in Paraplast Plus (Sherwood Medical). Microtome sections of 10 μm in thickness were applied to silane-coated glass slides (Matsunami Glass). The sections were deparaffinized in xylene, dehydrated through a graded ethanol series, and dried overnight before staining with hematoxylin.
TUNEL Assay
Deparaffinized sections were washed in PBS (160 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, and 1.5 mM KH2PO4) for 5 min and incubated in 20 μg/mL proteinase K in protease K buffer (100 mM TrisHCl, pH 8.0, and 50 mM EDTA) for 20 min at 37°C. The sections were washed with PBS for 5 min and fixed in 4% (w/v) paraformaldehyde in PBS for 10 min and again washed with PBS twice for 5 min. Detection of nuclear DNA fragmentation was performed using a TUNEL apoptosis detection kit (DeadEnd Fluorometric TUNEL system; Promega) according to the manufacturer's instructions. For nuclei observation, samples were counterstained with propidium iodide solution. Samples were observed using a fluorescence confocal scanning microscope (Olympus).
Ultrastructural Analysis with TEM
The anthers were vacuum infiltrated three times for 10 min with fixative (4% paraformaldehyde and 1% glutaraldehyde in 0.05 M cacodylate buffer, pH 7.4). Procedures for ultrastructural studies were essentially the same as described previously (Hara-Nishimura et al., 1993).
Microarray Analysis
Agilent 44K rice oligoarrays (Agilent Technologies), which contain 44,000 features, were used for two-color oligoarrays in this study. Each feature consists of a 60-mer oligonucleotide corresponding to a full-length cDNA of rice. Total RNA was extracted from anthers of plants with the predicted genotypes using the RNeasy plant mini kit (Qiagen). Each microarray experiment was performed twice using RNA samples that had been isolated independently (two biological replicates). All microarray experiments were performed as described by Tsuji et al. (2006). Feature extraction software (Agilent Technologies) was used to delineate and measure the signal intensity of each spot in the array and to normalize intensities using the Lowess method (Cleveland, 1979). Each experiment included a dye-swapping test as a technical replicate for each biological replicate. For microarray data quality control, we examined both dye-dependent effects and distribution of the ratio after normalization using dye-swapping tests in all microarray experiments and confirmed that all microarray experiments in this study generated high-quality data without significant dye-dependent effects and skewness of ratio distribution. To identify differentially expressed genes, the values of log2 normalized signal ratio in each experiment were statistically analyzed using rank product statistics as described by Breitling et al. (2004). A list of GA-regulated genes with statistically significant changes in expression between the wild type and oscps1-1 anthers was generated using the RankProduct method (Gentleman et al., 2004; Hong et al., 2006) of BioConductor (P value < 0.0001). As a check measure for the multiple testing problem, the multiple testing in this method was taken into account using the percentage of false prediction at the condition of pfp < 0.01 (Breitling et al., 2004; Hong et al., 2006). Under these conditions, we obtained 1114 spots that corresponded to GA-regulated genes in anthers. For the comparison between gid1-7 and oscps1-1, we selected a subset of 830 spots from the original 1114 spots using a significance level of P value < 0.05 calculated in the wild type/gid1-7 duplicate microarray experiments.
Promoter Analysis and Biological Theme Representation Analysis
From the rice genomic sequences registered in RAP-DB (http://rapdb.dna.affrc.go.jp/), 0.5-kb 5′-flanking sequences of all 870 GA-regulated genes and 50,259 overall genes were obtained using Perl scripts developed by us. For promoter analysis in the GA-regulated genes, we used 42 sets of 8-bp GAMYB binding motif to construct a BLAST database, based on their preferential interaction with barley (Hordeum vulgare) GAMYB (Gubler et al., 1999). Similarity searches of the upstream sequences were performed using the BLASTN program against the constructed database.
Functional categories of the GA-regulated genes were classified by sequence similarities between protein sequences for each probe and those in the COG database (http://www.ncbi.nlm.nih.gov/COG/), with manual adjustment according to Gene Ontology information obtained from The Institute for Genomic Research database (http://www.tigr.org/) when necessary.
RNA Isolation and RT-PCR Analysis
Total RNA was isolated from tissue samples using TRIzol reagent (Invitrogen). The isolation of total RNA was performed according to the manufacturer's instructions. For the classification of anther developmental stage, we divided the process of rice anther development into five stages, according to the length of lemma (Itoh et al., 2005): PMC stage (lemma 2 mm), meiosis stage (2 to 3 mm), tetrad and young microspore stage (3 to 5 mm), vacuolated pollen stage (5 to 8 mm), and mature pollen stage (>8 mm). The tetrad and young microspore stages were collected together because this classification system could not distinguish between these stages. The first strand of cDNA was synthesized from 1 μg of total RNA using an Omniscript reverse transcription kit (Qiagen). Transcripts were quantified by RT-PCR or real-time RT-PCR analyses using one-twentieth of the resulting cDNA as template. Real-time RT-PCR was performed with the LightCycler system (Roche) with the SYBR Green PCR kit (Qiagen). For RT-PCR analysis, an adequate volume of each cDNA sample was used for PCR amplification with different numbers of cycles (20 to 35 cycles) to confirm the linear range of PCR amplification for each gene. The results were confirmed using two or three independent biological replicates. The Actin1 gene from rice was used as an internal standard for normalizing cDNA concentration variations. Primer sequences used in this study are listed in Supplemental Table 1 online.
Gel-Shift Assay
To produce a recombinant GAMYB protein, Escherichia coli BL21 (DE3) pLysS Rossetta-gami 2 (Novagen) was transformed with an expression plasmid, the full-length GAMYB cDNA-pET32a, previously made in the pET32a vector (Novagen) (Tsuji et al., 2006). Then, 5 mL of precultured cells were added to 500 mL of Luria-Bertani medium in a 2-liter flask and cultured at 37°C until reaching an OD600 of 0.4 to 0.6. Recombinant proteins were induced by the addition of 0.4 mM isopropyl-β-d-thiogalactopyranoside and further incubated at 15°C for 18 h. The purification of recombinant proteins was performed as previously described (Ueguchi-Tanaka et al., 2007b).
The DNA fragments were generated by PCR using genomic DNA and the specific primers. The resulting DNA fragments were cloned into the pCR4 Blunt-TOPO vector. Nucleotide substitutions in RAmy1A, CYP703A3, and KAR promoter fragments were introduced by PCR as previously reported (Ueguchi-Tanaka et al., 2007b). The RAmy1A, CYP703A3, and KAR promoter fragments, amplified using M13F and M13R primers, were digested with EcoRI. The resulting fragments were labeled with 32P-dATP using the Klenow fragment and purified on the NICK columns (GE Healthcare). DNA binding reactions were performed as previously reported (Tsuji et al., 2006). Primer sequences used in this study are listed in Supplemental Table 1 online.
Phylogenetic Analysis
Multiple sequence alignments were produced using ClustalX 2.0 (with default parameters; e.g., amino acid substitution matrix Gonnet250, slow pairwise alignments, gap opening penalty 10, gap extension penalty 0.1 for pairwise, and 0.2 for multiple alignment) (Larkin et al., 2007) and then manually adjusted to optimize alignments (available in Supplemental Data Sets 4 and 5 online). Phylogenetic trees were constructed using MEGA 4.0 with the neighbor-joining method, JTT matrix, and complete deletion of gaps (Tamura et al., 2007). Bootstrap values calculated from 1000 replicates are shown.
Histochemical Analysis of GUS Activity
For GUS staining, tissues were transferred to microcentrifuge tubes containing a solution of 50 mM sodium phosphate, pH 7.0, 7% methanol, and 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide for 12 h at 37°C. Stained tissues were destained with an ethanol series.
Accession Numbers
Sequence data from this article can be found in the Rice Annotation Project Database (http://rapdb.dna.affrc.go.jp/) under the following accession numbers: CYP703A3 (Os08g0131100), KAR (Os12g0242700), Lipid transporter (Os08g0546300), Male Sterility2 (Os03g0167600), Aspartic protease (Os09g0452800), and Meiotic serine protease (Os04g0543700).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phenotypic Analysis of Reproductive Organs in Wild-Type and GA-Related Mutants.
Supplemental Figure 2. Higher Magnification of Transverse Section from Wild-Type and GA-Related Mutants.
Supplemental Figure 3. Ultrastructural Analysis of the Anther Wall Layers in Wild-Type and GA-Related Mutants by TEM.
Supplemental Figure 4. Histological Analysis of GA3-Treated Anthers in GA-Related Mutants.
Supplemental Figure 5. The Cytoplasm of Microspores in the Wild Type, GA-Related Mutants, and cyp703a3-1 by TEM.
Supplemental Figure 6. Scatterplot Analysis to Compare Genes Regulated by GA (oscps1-1 Background) and GID1 in Anther.
Supplemental Figure 7. Structure of CYP703A3.
Supplemental Figure 8. Structure of KAR.
Supplemental Figure 9. Expression Analysis for CYP703A3 and KAR Genes Using the DEX-Inducible mOs GAMYB-GR Transgenic gamyb-2 Plant.
Supplemental Figure 10. GUS Activity of Stamens in All Transgenic Plants Carrying the Native or Mutagenized ProCYP703A3:GUS Construct.
Supplemental Figure 11. GUS Activity of Stamens in All Transgenic Plants Carrying the Native or Mutagenized ProKAR:GUS Construct.
Supplemental Figure 12. Complementation Test of cyp703a3-1.
Supplemental Figure 13. Competitive Gel-Shift Assay with Osc6 Promoter Region.
Supplemental Table 1. PCR Primers Used in This Study.
Supplemental Data Set 1. Microarray Analysis of 1114 GA-Regulated Probes.
Supplemental Data Set 2. Functional Classification of 870 GA-Regulated Genes.
Supplemental Data Set 3. GAMYB Binding Motif in the Promoter of 870 GA-Regulated Genes.
Supplemental Data Set 4. Protein Sequences Used to Generate the Dendogram in Supplemental Figure 7A.
Supplemental Data Set 5. Protein Sequences Used to Generate the Dendogram in Supplemental Figure 8A.
Supplementary Material
Acknowledgments
We thank E. Koketsu, M. Kawamura, and H. Ohmiya (Nagoya University) for technical assistance. We also thank Y. Nagamura and R. Motoyama (Rice Genome Resource Center of the National Institute of Agrobiological Sciences, Japan) for the use of the rice microarray analysis system and technical support. The mutant line for CYP703A3 gene described in this article was obtained from the Rice Genome Resource Center of the National Institute of Agrobiological Sciences, Japan (http://www.nias.affrc.go.jp/). This study was supported by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (19570037 to M.U.-T., 19043015 to K.Y., and 18107001 and 18075006 to M.M.), the Ministry of Agriculture, Forest, and Fisheries of Japan (Green Technology Project IPG0003; M.M.), Grants-in-Aid for Exploratory Research from the Japan Society for Promotion of Science (19651084 to K.Y.), and research fellowships from the Japan Society for the Promotion of Science for Young Scientists (K.A.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Makoto Matsuoka (makoto@agr.nagoya-u.ac.jp).
Online version contains Web-only data.
References
- Achard, P., Herr, A., Baulcombe, D.C., and Harberd, N.P. (2004). Modulation of floral development by a gibberellin-regulated microRNA. Development 131 3357–3365. [DOI] [PubMed] [Google Scholar]
- Ahlers, F., Bubert, H., Steuernagel, S., and Wiermann, R. (2000). The nature of oxygen in sporopollenin from the pollen of Typha angustifolia L. Z. Naturforsch. [C] 55 129–136. [DOI] [PubMed] [Google Scholar]
- Ahlers, F., Thom, I., Lambert, J., Kuckuk, R., and Wiermann, R. (1999). 1H NMR analysis of sporopollenin from Typha angustifolia. Phytochemistry 50 1095–1098. [Google Scholar]
- Ariizumi, T., Murase, K., Sun, T.-P., and Steber, C.M. (2008). Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. Plant Cell 20 2447–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnall, D.J. (1992). Control of flowering in Arabidopsis thaliana by light, vernalisation and gibberellins. Aust. J. Plant Physiol. 19 401–409. [Google Scholar]
- Beers, E.P., Jones, A.M., and Dickerman, A.W. (2004). The S8 serine, C1A cysteine and A1 aspartic protease families in Arabidopsis. Phytochemistry 65 43–58. [DOI] [PubMed] [Google Scholar]
- Beers, E.P., Woffenden, B.J., and Zhao, C. (2000). Plant proteolytic enzymes: possible roles during programmed cell death. Plant Mol. Biol. 44 399–415. [DOI] [PubMed] [Google Scholar]
- Breitling, R., Armengaud, P., Amtmann, A., and Herzyk, P. (2004). Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573 83–92. [DOI] [PubMed] [Google Scholar]
- Bubert, H., Lambert, J., Steuernagel, S., Ahlers, F., and Wiermann, R. (2002). Continuous decomposition of sporopollenin from pollen of Typha angustifolia L. by acidic methanolysis. Z. Naturforsch. [C] 57 1035–1041. [DOI] [PubMed] [Google Scholar]
- Cao, D., Cheng, H., Wu, W., Soo, H.M., and Peng, J. (2006). Gibberellin mobilizes distinct DELLA-dependent transcriptomes to regulate seed germination and floral development in Arabidopsis. Plant Physiol. 142 509–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, P.M., Marion-Poll, A., Ellis, M., and Gubler, F. (2002). Mutants at the Slender1 locus of barley cv Himalaya: Molecular and physiological characterization. Plant Physiol. 129 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, H., Qin, L., Lee, S., Fu, X., Richards, D.E., Cao, D., Luo, D., Harberd, N.P., and Peng, J. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131 1055–1064. [DOI] [PubMed] [Google Scholar]
- Chhun, T., Aya, K., Asano, K., Yamamoto, E., Morinaka, Y., Watanabe, M., Kitano, H., Ashikari, M., Matsuoka, M., and Ueguchi-Tanaka, M. (2007). Gibberellin regulates pollen viability and pollen tube growth in rice. Plant Cell 19 3876–3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland, W. (1979). Robust locally weighted regression and smoothing scatter plots. J. Am. Stat. Assoc. 74 829–836. [Google Scholar]
- de Lucas, M., Davière, J.M., Rodríguez-Falcón, M., Pontin, M., Iglesias-Pedraz, J.M., Lorrain, S., Fankhauser, C., Blázquez, M.A., Titarenko, E., and Prat, S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451 480–484. [DOI] [PubMed] [Google Scholar]
- Dill, A., Thomas, S.G., Hu, J., Steber, C.M., and Sun, T.-P. (2004). The Arabidopsis F-box protein SLEEPY1 targets GA signaling repressors for GA-induced degradation. Plant Cell 16 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund, A.F., Swanson, R., and Preuss, D. (2004). Pollen and stigma structure and function: The role of diversity in pollination. Plant Cell 16 (suppl.): S84–S97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet, C.M., and Sun, T.-P. (2005). A DELLAcate balance: The role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 8 77–85. [DOI] [PubMed] [Google Scholar]
- Fu, X., Richards, D.E., Fleck, B., Xie, D., Burton, N., and Harberd, N.P. (2004). The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16 1406–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman, R.C., et al. (2004). Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5 R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, N., and Pharis, R.P. (1999). Role of gibberellin in the development of floral organs of the gibberellin-deficient mutant, ga1-1, of Arabidopsis thaliana. Can. J. Bot. 77 944–954. [Google Scholar]
- Griffiths, J., Murase, K., Rieu, I., Zentella, R., Zhang, Z.L., Powers, S.J., Gong, F., Phillips, A.L., Hedden, P., Sun, T.-P., and Thomas, S.G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18 3399–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., Kalla, R., Roberts, J.K., and Jacobsen, J.V. (1995). Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pI alpha-amylase gene promoter. Plant Cell 7 1879–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., Raventos, D., Keys, M., Watts, R., Mundy, J., and Jacobsen, J.V. (1999). Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J. 17 1–9. [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura, I., Takeuchi, Y., and Nishimura, M. (1993). Molecular characterization of a vacuolar processing enzyme related to a putative cysteine proteinase of Schistosoma mansoni. Plant Cell 5 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei, Y., Ohta, S., Komari, T., and Kumashiro, T. (1994). Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6 271–282. [DOI] [PubMed] [Google Scholar]
- Hong, F., Breitling, R., McEntee, C.W., Wittner, B.S., Nemhauser, J.L., and Chory, J. (2006). RankProd: A bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22 2825–2827. [DOI] [PubMed] [Google Scholar]
- Huysmans, S., El-Ghazaly, G., and Smets, E. (1998). Orbicules in angiosperms: Morphology, function, distribution, and relation with tapetum types. Bot. Rev. 64 240–272. [Google Scholar]
- Ikeda, A., Ueguchi-Tanaka, M., Sonoda, Y., Kitano, H., Koshioka, M., Koshioka, M., Futsuhara, Y., Matsuoka, M., and Yamaguchi, J. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, J., Nonomura, K., Ikeda, K., Yamaki, S., Inukai, Y., Yamagishi, H., Kitano, H., and Nagato, Y. (2005). Rice plant development: from zygote to spikelet. Plant Cell Physiol. 46 23–47. [DOI] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M., and Matsuoka, M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izhaki, A., Borochov, A., Zamski, E., and Weiss, D. (2002). Gibberellin regulates post-microsporogenesis processes in petunia anthers. Physiol. Plant. 115 442–447. [DOI] [PubMed] [Google Scholar]
- Jacobsen, S.E., and Olszewski, N.E. (1991). Characterization of the arrest in anther development associated with gibberellin deficiency of the gib-1 mutant of tomato. Plant Physiol. 97 409–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, K.H., Han, M.J., Lee, D.Y., Lee, Y.S., Schreiber, L., Franke, R., Faust, A., Yephremov, A., Saedler, H., Kim, Y.W., Hwang, I., and An, G. (2006). Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell 18 3015–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko, M., Inukai, Y., Ueguchi-Tanaka, M., Itoh, H., Izawa, T., Kobayashi, Y., Hattori, T., Miyao, A., Hirochika, H., Ashikari, M., and Matsuoka, M. (2004). Loss-of-function mutations of the rice GAMYB gene impair alpha-amylase expression in aleurone and flower development. Plant Cell 16 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, R.W., and Evans, L.T. (2003). Gibberellins and flowering of grasses and cereals: Prizing open the lid of the “florigen” black box. Annu. Rev. Plant Biol. 54 307–328. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., and van der Veen, J.H. (1980). Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.). Theor. Appl. Genet. 58 257–263. [DOI] [PubMed] [Google Scholar]
- Ku, S., Yoon, H., Suh, H.S., and Chung, Y.Y. (2003). Male-sterility of thermosensitive genic male-sterile rice is associated with premature programmed cell death of the tapetum. Planta 217 559–565. [DOI] [PubMed] [Google Scholar]
- Langridge, J. (1957). Effect of day-length and gibberellic acid on the flowering of Arabidopsis. Nature 180 36–37. [Google Scholar]
- Larkin, M.A., Blackshields, G., Brown, N.P., Chenna, R., McGettigan, P.A., McWilliam, H., Valentin, F., Wallace, I.M., Wilm, A., Lopez, R., and Thompson, J.D. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23 2947–2948. [DOI] [PubMed] [Google Scholar]
- Li, N., et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18 2999–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis, K.M., Thomas, S.G., Soule, F.D., Strader, L.C., Zale, J.M., Sun, T.-P., and Steber, C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.A., and Gubler, F. (2005). The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17 705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morant, M., Jørgensen, K., Schaller, H., Pinot, F., Møller, B.L., Werck-Reichhart, D., and Bak, S. (2007). CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19 1473–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, F., Kalla, R., Jacobsen, J., and Gubler, F. (2003). A role for HvGAMYB in anther development. Plant J. 33 481–491. [DOI] [PubMed] [Google Scholar]
- Nakajima, M., et al. (2006). Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46 880–889. [DOI] [PubMed] [Google Scholar]
- Nester, J.E., and Zeevaart, J.A.D. (1988). Flower development in normal tomato and a gibberellin-deficient (ga-2) mutant. Am. J. Bot. 75 45–55. [Google Scholar]
- Peng, J., Carol, P., Richards, D.E., King, K.E., Cowling, R.J., Murphy, G.P., and Harberd, N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., Richards, D.E., Hartley, N.M., Murphy, G.P., Devos, K.M., Flintham, J.E., Beales, J., Fish, L.J., Worland, A.J., Pelica, F., Sudhakar, D., and Christou, P. (1999). 'Green revolution' genes encode mutant gibberellin response modulators. Nature 400 256–261. [DOI] [PubMed] [Google Scholar]
- Pharis, R.P., and King, R.W. (1985). Gibberellins and reproductive development in seed plants. Annu. Rev. Plant Physiol. 36 517–568. [Google Scholar]
- Piffanelli, P., Ross, J.H.E., and Murphy, D.J. (1998). Biogenesis and function of the lipidic structures of pollen grains. Sex. Plant Reprod. 11 65–80. [Google Scholar]
- Sakamoto, T., Miura, K., Itoh, H., Tatsumi, T., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., Agrawal, G.K., Takeda, S., Abe, K., Miyao, A., and Hirochika, H. (2004). An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 134 1642–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, A., Itoh, H., Gomi, K., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., Jeong, D.H., An, G., Kitano, H., Ashikari, M., and Matsuoka, M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299 1896–1898. [DOI] [PubMed] [Google Scholar]
- Silverstone, A.L., Ciampaglio, C.N., and Sun, T.-P. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, D.P., Jermakow, A.M., and Swain, S.M. (2002). Gibberellins are required for seed development and pollen tube growth in Arabidopsis. Plant Cell 14 3133–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T.-P., and Kamiya, Y. (1994). The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell 6 1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24 1596–1599. [DOI] [PubMed] [Google Scholar]
- Thornsberry, J.M., Goodman, M.M., Doebley, J., Kresovich, S., Nielsen, D., and Buckler IV, E.S. (2001). Dwarf8 polymorphisms associate with variation in flowering time. Nat. Genet. 28 286–289. [DOI] [PubMed] [Google Scholar]
- Toomey, R.E., and Wakil, S.J. (1966). Studies on the mechanism of fatty acid synthesis XV. Preparation and general properties of β-ketoacyl acyl carrier protein reductase from Escherichia coli. Biochim. Biophys. Acta 116 189–197. [DOI] [PubMed] [Google Scholar]
- Tsuji, H., Aya, K., Ueguchi-Tanaka, M., Shimada, Y., Nakazono, M., Watanabe, R., Nishizawa, N.K., Gomi, K., Shimada, A., Kitano, H., Ashikari, M., and Matsuoka, M. (2006). GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant J. 47 427–444. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., Chow, T.-Y., Hsing, Y.C., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437 693–698. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Hirano, K., Hasegawa, Y., Kitano, H., and Matsuoka, M. (2008). Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant. Plant Cell 20 2437–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Nakajima, M., Ashikari, M., and Matsuoka, M. (2007. a). Gibberellin receptor and its role in gibberellin signaling in plants. Annu. Rev. Plant Biol. 58 183–198. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Nakajima, M., Katoh, E., Ohmiya, H., Asano, K., Saji, S., Hongyu, X., Ashikari, M., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2007. b). Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19 2140–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viti, R., Bartolini, S., and Vitagliano, C. (1990). Growth regulators on pollen germination in olive. Acta Hortic. 286 227–230. [Google Scholar]
- Wang, A., Xia, Q., Xie, W., Datla, R., and Selvaraj, G. (2003). The classical Ubisch bodies carry a sporophytically produced structural protein (RAFTIN) that is essential for pollen development. Proc. Natl. Acad. Sci. USA 100 14487–14492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijeratne, A.J., Zhang, W., Sun, Y., Liu, W., Albert, R., Zheng, Z., Oppenheimer, D.G., Zhao, D., and Ma, H. (2007). Differential gene expression in Arabidopsis wild-type and mutant anthers: Insights into anther cell differentiation and regulatory networks. Plant J. 52 14–29. [DOI] [PubMed] [Google Scholar]
- Willige, B.C., Ghosh, S., Nill, C., Zourelidou, M., Dohmann, E.M., Maier, A., and Schwechheimer, C. (2007). The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R., Heckman, J.W., and Somerville, C. (1992). Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 100 403–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui, R., Iketani, S., Mikami, T., and Kubo, T. (2003). Antisense inhibition of mitochondrial pyruvate dehydrogenase E1α subunit in anther tapetum causes male sterility. Plant J. 34 57–66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.