Abstract
At the onset of flowering, the Arabidopsis thaliana primary inflorescence meristem starts to produce flower meristems on its flank. Determination of floral fate is associated with changes in the growth pattern and expression of meristem identity genes and suppression of a subtending leaf called a bract. Here, we show a role in floral fate determination and bract suppression for the PUCHI gene, an AP2/EREBP family gene that has previously been reported to play roles in lateral root morphogenesis. Mutations in PUCHI cause partial conversion of flowers to inflorescences, indicating that PUCHI is required for flower meristem identity. PUCHI is transiently expressed in the early flower meristem and accelerates meristem bulging while it prevents the growth of the bract primordium. The function of PUCHI in floral fate determination and bract suppression overlaps that of the BLADE-ON-PETIOLE1 (BOP1) and BOP2 genes, which encode a pair of redundant regulatory proteins involved in various developmental processes, including leaf morphogenesis and flower patterning. We also show that PUCHI acts together with BOP1 and BOP2 to promote expression of LEAFY and APETALA1, two central regulators of floral meristem identity. Expression patterns of the PUCHI and BOP genes point to a role in spatial control of flower-specific activation of these meristem identity genes.
INTRODUCTION
Most aerial parts of a plant are generated postembryonically by the activity of the shoot meristem, a group of mitotically active cells that continuously add new structures at the shoot apex throughout the life cycle (Steeves and Sussex, 1989; Poethig, 2003). The shoot meristem initially produces vegetative leaves at its periphery and then produces flowers as it enters the reproductive phase. Plants show a wide variety of inflorescence morphologies, and the pattern of any particular inflorescence form is highly dependent on when and where flower primordia arise in the shoot meristem (Coen and Nugent, 1994; Benlloch et al., 2007; Prusinkiewicz et al., 2007).
In Arabidopsis thaliana, the primary inflorescence produces lateral meristems that develop into either secondary inflorescences or flowers (Schultz and Haughn, 1991). Secondary inflorescences, or branches, are produced immediately after the transition from the vegetative to the reproductive phase and show an indeterminate growth pattern that reiterates the pattern of the primary inflorescence. After several rounds of branch production, the primary inflorescence meristem begins to produce determinate floral meristems, which generate a fixed number of floral organs. The conversion of meristem identity from secondary inflorescence to flower is largely dependent on endogenous and environmental cues, which eventually converge on the expression of the floral meristem identity genes LEAFY (LFY) and APETALA1 (AP1), both encoding transcription factors (Weigel et al., 1992; Mandel et al., 1992; Weigel and Meyerowitz, 1993; Blázquez et al., 2006; Baurle and Dean, 2006; Kobayashi and Weigel, 2007).
Mutations in LFY and AP1 cause partial conversion of flowers into branch-like structures, whereas constitutive expression of either gene is sufficient to convert branches into flowers, indicating that these genes are critical factors for specifying floral meristem identity (Weigel et al., 1992; Bowman et al., 1993; Mandel and Yanofsky, 1995; Weigel and Nilsson, 1995). Expression of LFY is weak in leaf primordia during the vegetative phase but is strongly activated in floral meristems at the onset of flowering (Weigel et al., 1992; Blázquez et al., 1997; Hempel et al., 1997). The LFY protein directly activates transcription of AP1 and its redundant homolog CAULIFLOWER (CAL) in the floral meristem (Parcy et al., 1998; Wagner et al., 1999; William et al., 2004). AP1 and CAL in turn maintain LFY expression to ensure correct floral identity (Bowman et al., 1993; Liljegren et al., 1999).
The above results indicate that the attainment of the high level of LFY expression is a key step for floral meristem specification. Although many factors besides AP1 and CAL have been reported to promote LFY expression, none are expressed specifically in floral meristems but rather in a broader region (Blázquez et al., 1998; Blázquez and Weigel, 2000; Lee et al., 2000; Samach et al., 2000; Yu et al., 2002; Michaels et al., 2003; Smith et al., 2004; Abe et al., 2005; Wigge et al., 2005; Kanrar et al., 2008; Lee et al., 2008), raising the question of how local activation of LFY expression is regulated.
In Arabidopsis, an important feature that distinguishes a flower from a secondary inflorescence is the absence of subtending leaves or bracts in the flower. Whereas the secondary inflorescence meristem is initiated in the axil of a primordium that develops into a subtending leaf, the floral meristem is initiated as an adaxial subdomain of a flower primordium that also contains the abaxial cryptic bract domain. Subsequent development of the cryptic bract is strongly suppressed by an unidentified signal derived from the floral meristem (Nilsson et al., 1998; Long and Barton, 2000), resulting in the formation of a flower that lacks a visible subtending bract. Both LFY and its coregulator UNUSUAL FLORAL ORGANS (UFO) are involved in this process (Schultz and Haughn, 1991; Hepworth et al., 2006). Besides these, the two paralogous genes BLADE-ON-PETIOLE1 (BOP1) and BOP2, which encode proteins related to the disease resistance regulatory protein NONEXPRESSOR OF PR1 (NPR1), are redundantly required for suppression of the bract (Hepworth et al., 2005; Norberg et al., 2005). Although BOP1 and BOP2 have been suggested to participate in the transition from vegetative to reproductive development, their precise role in flower development is not yet clear.
Arabidopsis PUCHI, which is required for lateral root morphogenesis (Hirota et al., 2007), is another factor that is potentially involved in meristem identity and bract suppression. Mutations in this gene cause ectopic cell proliferation at the base of lateral root primordia, indicating that PUCHI is involved in cell division control during lateral root formation. In the shoot, on the other hand, puchi mutants produce characteristic ectopic tissue that is reminiscent of a bract although its exact identity remains unclear. The PUCHI protein belongs to the AP2/ethylene-responsive element binding protein family and is highly homologous to the maize (Zea mays) protein BRANCHED SILKLESS1 (BD1) and the rice (Oryza sativa) protein FRIZZY PANICLE (FZP), both of which affect floral meristem identity (Chuck et al., 2002; Komatsu et al., 2003).
Here, we provide evidence that, in addition to its role in lateral root development, PUCHI is involved in the determination of floral meristem identity and suppression of bract growth. PUCHI is expressed on the adaxial side of early floral primordium and is required for proper conversion of secondary inflorescences to flowers. The puchi mutations cause a prolonged phase of bract primordium growth while they delay the bulging of the floral meristem. We also show that PUCHI has an overlapping function with BOP1 and BOP2 in controlling floral meristem identity and that these genes together promote expression of LFY and AP1. The expression domains of PUCHI and BOP are restricted to lateral meristems and may provide a positional cue for flower-specific activation of these meristem identity genes.
RESULTS
puchi Mutations Affect Floral Meristem Identity
Floral transition in Arabidopsis is regulated by multiple endogenous and environmental factors, including daylength (Baurle and Dean, 2006; Kobayashi and Weigel, 2007). To investigate PUCHI gene function in flower development, we characterized two recessive alleles, puchi-1 and puchi-2 (Hirota et al., 2007), under continuous-light and short-day conditions. The timing of the meristem identity transition is commonly measured by counting the number of secondary inflorescences produced on the bolting stem prior to flower formation (Ratcliffe et al., 1998). We also counted rosette leaf number, which correlates well with flowering time (Koornneef et al., 1991).
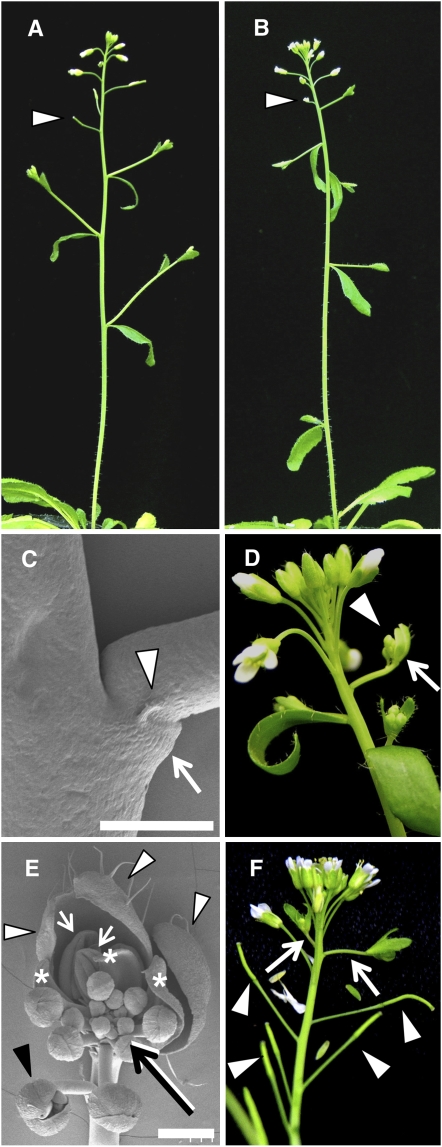
Under continuous-light conditions, both puchi-1 and puchi-2 mutants showed a small but significant increase in the number of secondary inflorescences compared with that of the wild type, whereas the number of rosette leaves was unaffected (Figures 1A and 1B, Table 1; see Supplemental Table 1 online). This phenotype was interpreted as very early arising flowers being completely transformed into secondary inflorescences. In addition, 20% (10 of 50) of puchi plants lacked a subtending cauline leaf in the uppermost secondary inflorescence and instead formed a flat leaf-like structure flanked by a pair of pin-shaped projections (Figure 1C; see Supplemental Figure 1A online). These solitary branches lacking normal cauline leaves occasionally (4%: 2 of 50 inflorescences) showed a mosaic of inflorescence and flower phenotypes (Figure 1D; see Supplemental Figure 1B online). The apex of the mosaic structures consisted of three sepal-like organs in the first whorl, a few petals and stamens inside them, and an indeterminate shoot at the center (Figure 1E). In addition, an extra flower often formed from the pedicel of these mosaic structures (Figure 1E, black arrowhead). These phenotypes appeared to represent an incomplete conversion of a flower to an inflorescence.
Figure 1.
Inflorescence Phenotypes of the Wild Type and puchi-1.
(A) to (E) Inflorescence of wild type (A) and puchi-1 ([B] to [E]) grown under continuous light conditions.
(A) Inflorescence of a wild-type plant.
(B) Inflorescence of a puchi-1 plant. Compared with the wild type, the number of nodes with secondary inflorescences is increased in puchi-1. Arrowheads indicate the first flower formed after the transition from secondary inflorescences to flowers.
(C) Scanning electron micrograph of a puchi-1 branch that lacks a subtending cauline leaf but instead has a flat leaf-like organ (arrow) flanked by a pair of pin-shaped projections (only one of them is apparent in this image; arrowhead). Bar = 500 μm.
(D) Primary inflorescence of puchi-1, showing a mosaic branch consisting of a flower (arrow) and an inflorescence (arrowhead).
(E) Scanning electron micrograph of a mosaic branch of puchi-1. White arrowheads indicate sepal-like organs in the first whorl. Asterisks and white arrows indicate petal- and stamen-like organs, respectively. The black arrow indicates the inflorescence-like shoot, and the black arrowhead indicates an extra flower produced from the pedicel. Bar = 500 μm.
(F) Primary inflorescence of puchi-1 grown under short-day conditions, showing ectopic secondary inflorescences (arrows). The ectopic secondary inflorescences are produced after six to nine flowers/siliques (arrowheads) have arisen on the primary inflorescence.
[See online article for color version of this figure.]
Table 1.
Inflorescence Architecture of puchi, bop1 bop2, and bop1 bop2 puchi Mutants Grown under Continuous-Light and Short-Day Conditions
| Condition | Genotype | SI with Cauline Leaf | SI without Cauline Leaf | Total SI | Plants Scored |
|---|---|---|---|---|---|
| CL | Col | 3.33 ± 0.11 | 0.0 ± 0.0 | 3.33 ± 0.11 | 30 |
| puchi-1 | 4.60 ± 0.10** | 0.20 ± 0.07** | 4.80 ± 0.11** | 30 | |
| puchi-2 | 4.43 ± 0.15** | 0.20 ± 0.07** | 4.63 ± 0.18** | 30 | |
| bop1 bop2 | 4.7 ± 0.14** | 0.30 ± 0.09** | 5.0 ± 0.18** | 30 | |
| bop1 bop2 puchi | 4.67 ± 0.11** | 19.60 ± 0.45** | 24.27 ± 0.46** | 30 | |
| SD | Col | 10.05 ± 0.32 | 0.0 ± 0.0 | 10.05 ± 0.0 | 20 |
| puchi-1 | 12.15 ± 0.32** | 3.35 ± 0.49** | 15.50 ± 0.57** | 20 | |
| puchi-2 | 10.90 ± 0.25* | 3.4 ± 0.41** | 14.30 ± 0.33** | 20 | |
| bop1 bop2 | 32.44 ± 0.96** | 2.67 ± 0.94** | 35.11 ± 0.86** | 9 | |
| bop1 bop2 puchi | 49.55 ± 1.27** | 0.0 ± 0.0 | 49.55 ± 1.27** | 9 |
The number of secondary inflorescences (SI) produced on the primary bolting stem was scored for each genotype. Values are mean ± se. Differences between wild-type and mutant plants are significant at the 0.05 > P > 0.01 (*) or the P < 0.01 (**) levels. Under short-day (SD) conditions, SI that formed at positions below and above the lowermost flower were both scored. In continuous light (CL), none of the genotypes produced any ectopic branches.
Under short-day conditions, puchi mutants clearly possessed more secondary inflorescences than did the wild type (Table 1). In addition, puchi plants produced ectopic secondary inflorescences after six to nine flowers had arisen on the primary inflorescence (Figure 1F; see Supplemental Figure 1C online). Typically, in such cases, one to three ectopic inflorescences were produced sequentially; these phases of ectopic inflorescence production could occur up to four times during inflorescence development, with each phase being separated by the formation of 1 to 10 flowers. These ectopic inflorescences reiterated the process of primary inflorescence (see Supplemental Figure 1D online), suggesting that the transformation of flowers to secondary inflorescences was complete.
Taken together, these results show that mutations in PUCHI caused partial conversion of flowers into inflorescences in the two photoperiod conditions examined and indicate that PUCHI controls the fate of lateral meristems. The puchi-1 and puchi-2 mutants gave essentially the same phenotypes in all aspects of shoot development, and we chose the puchi-1 allele for further analyses.
puchi Mutant Flowers Have Rudimentary Bracts
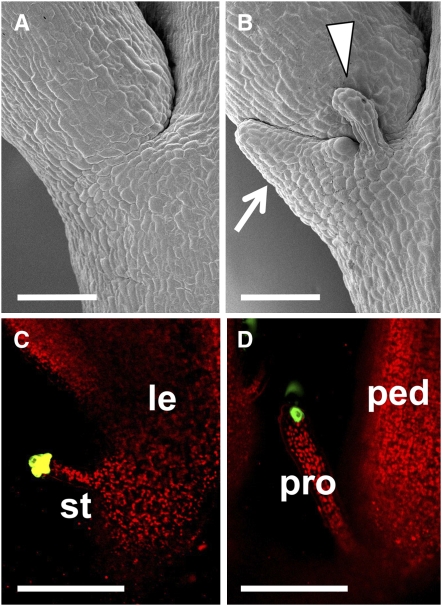
We next examined flower phenotypes in the puchi mutant. All puchi flowers had ectopic structures at the base of each pedicel, whereas wild-type flowers showed a smooth surface at the same position (Figures 2A and 2B; Hirota et al., 2007). These ectopic structures consisted of a flat leaf-like part and a pair of pin-shaped projections, similar to those observed at the base of the solitary secondary inflorescences (Figures 1C and 2B). The pin-shaped projections were morphologically similar to stipules formed at the base of the leaves, although their size was somewhat larger than normal stipules. Based on these observations, we speculated that these structures were a rudimentary bract associated with a pair of stipules. To test this prediction, we analyzed expression of green fluorescent protein (GFP) in the enhancer trap line E1238 (http://enhancertraps.bio.upenn.edu/default.html), in which the signal is detected in the stipules of leaves (Figure 2C). The GFP signal was detected in the pin-shaped projections of puchi mutant flowers (Figure 2D). Furthermore, mutation of the PRESSED FLOWER (PRS) gene, which is required for stipule formation in the leaf (Matsumoto and Okada, 2001; Nardmann et al., 2004), resulted in the loss of the pin-shaped projections when combined with the puchi mutation (see Supplemental Figures 2A to 2D online). These results indicate that the ectopic structures at the base of puchi pedicels comprise a rudimentary bract associated with a pair of stipules and that PUCHI is involved in the suppression of bract growth in flowers. Given that the absence of a subtending leaf is one of the characters that discriminate flowers from secondary inflorescences in Arabidopsis, the failure of bract suppression in the mutant may be explained as a partial conversion of flowers into secondary inflorescences.
Figure 2.
puchi Mutant Flowers Have Rudimentary Bracts at the Base of Their Pedicels.
(A) Scanning electron micrograph showing the base of the wild-type pedicel.
(B) Scanning electron micrograph showing the base of the puchi-1 pedicel. puchi produces a flat leaf-like organ (arrow) flanked by a pair of pin-shaped projections (only one is visible in this image; arrowhead).
(C) GFP expression of the enhancer trap line E1238 is detected in the stipule of the wild-type cauline leaf.
(D) GFP expression of the enhancer trap line E1238 is detected in the pin-shaped projection of the puchi-1 flower.
le, leaf; st, stipule; ped, pedicel; pro, pin-shaped projection. Bars = 100 μm.
In contrast with the phenotype in the morphology of the flower base, we could not detect any obvious abnormalities in the identity or the number of individual floral organs of puchi (see Supplemental Figures 3A to 3D and Supplemental Table 2 online), indicating that PUCHI is not involved in the specification or patterning of flower organs.
Early Flower Primordium Development in puchi
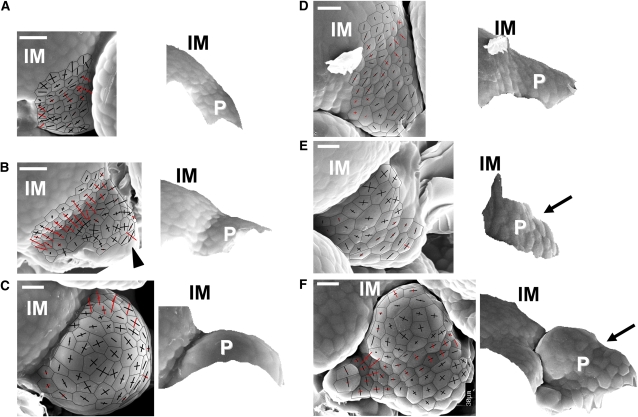
To investigate how PUCHI affects bract growth, we sought to examine early flower development in the puchi mutant in detail. To this end, we used a sensitive, noninvasive replica method combined with a three-dimensional reconstruction algorithm, which can reliably detect a cryptic bract in early flower primordia (Kwiatkowska, 2006).
Curvature plots on wild-type inflorescence apices enable the definition of four consecutive stages in flower primordium development (Kwiatkowska, 2006; Szczesny et al., 2009). The first floral stage is an initial bulging that leads to the formation of a shallow crease between the primordium protrusion and the primary inflorescence meristem (Figure 3A). This region is concave in the meridional direction (red curvature cross arms in Figure 3A) and convex in the latitudinal (black arms). The next stage is a second bulging, at which a convex region (marked by curvature crosses with both arms black) appears at the bottom of the shallow crease (Figure 3B; see also Kwiatkowska, 2006; Szczesny et al., 2009). The convex region corresponds to the floral meristem proper, while the concave region at the distal end (arrowhead in Figure 3B) corresponds to the bract primordium. In the third stage of bulge formation (Figure 3C), the temporarily apparent bract primordium disappears. During the final stage, the sepal primordia are formed (P5 in Supplemental Figure 4A online).
Figure 3.
Early Flower Primordium Development in puchi.
(A) The wild-type primordium at the initial bulging stage.
(B) The wild-type primordium at the second bulging stage.
(C) The wild-type primordium at the bulge stage.
(D) The puchi-1 primordium at the initial bulging stage.
(E) The puchi-1 primordium at the second bulging stage.
(F) The puchi-1 primordium at the bulge stage.
In each panel, scanning electron micrographs on which curvature crosses are overlaid (left), and side views of the reconstructed surface (right), were obtained from replicas of inflorescence shoot apices. Curvature cross-arms are aligned with the direction of curvature. The length of each cross-arm is proportional to the degree of curvature. Arms appear in red if the surface is concave in this direction and in black if it is convex. Three of the four consecutive stages, initial bulging, second bulging, and bulge, are shown. The arrowhead in (B) indicates the concave region at the distal end. Arrows in (E) and (F) point to the boundary between flower meristem proper and bract. IM, primary inflorescence meristem; P, flower primordium. Bars = 10 μm.
In the puchi mutant, we also recognized four similar consecutive stages, although both their geometry and their timing were different from those in the wild type. During the first stage of initial bulging, the mutant primordium was indistinguishable from that of the wild type at the beginning (data not shown) but then protruded further from the shoot axis than it did in the wild type (cf. Figures 3A and 3D). The upper surface of the primordium was largely flat or only slightly concave, forming a shelf-like shape (Figure 3D). The duration of initial bulging leading to formation of this shelf-like primordium, measured as the mean number of plastochrons, was longer than the equivalent stage in the wild type (mean of 3.89 plastochrons ± 0.11 se in puchi versus 3.16 ± 0.09 se in the wild type; n = 9 and 16, respectively). During the stage of the second bulging in puchi, a convex region (where both curvature cross arms are depicted in black) appeared within the shelf-like region, similar to the wild type. Unlike the wild type, however, the mutant bract did not disappear; the shelf-like primordium was partitioned into the floral meristem proper and the bract (Figure 3E).
The major difference between puchi and the wild type in the third stage was the permanent presence in puchi of a bract with stipules (cf. Figure 3C with 3F). Moreover, the duration of this stage was extended in the mutant. The mutant floral meristem proper grew into a finger-like structure devoid of sepals (Figure 3F) until it overgrew the primary inflorescence meristem (P7 in Supplemental Figure 4B online). At this stage, the cells of the floral meristem proper appeared enlarged both in the wild type and in puchi (cf. Figure 3C with 3A and Figure 3F with 3D). A prolonged duration of the bulge stage in the mutant was manifested in delayed sepal formation (cf. P5 in Supplemental Figure 4A with P9 in Supplemental Figure 4C online, in which the youngest sepal primordia are indicated by asterisks). This stage in the wild type begins at 6.69 plastochrons (mean ± 0.17 se; n = 16), but in puchi at 8.94 plastochrons (± 0.49 se; n = 9).
In summary, early flower development of puchi is characterized by a prolonged period of the initial bulging stage, leading to the formation of a shelf-like bract primordium instead of a shallow crease as in the wild type. Consequently, initiation of the second bulging that forms the floral meristem proper is delayed. These results point to a role for PUCHI in regulating the early phase of floral meristem development in the axil of a cryptic bract.
PUCHI Is Expressed in Lateral Meristems Developing at the Periphery of the Primary Meristem
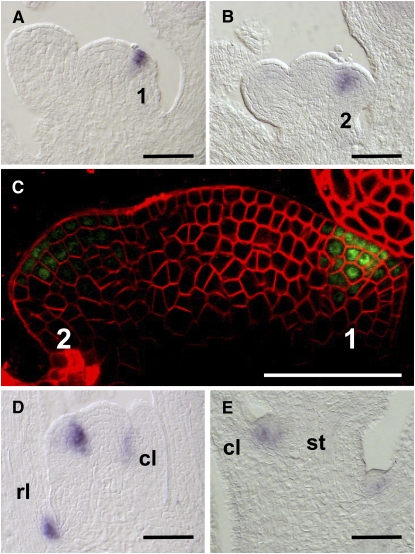
To investigate how PUCHI gene expression correlates with the mutant phenotypes, we monitored PUCHI mRNA by in situ hybridization. In the inflorescence apex, PUCHI expression was first detected in cells that had apparently begun to emerge from the inflorescence meristem as a buttress, which was morphologically equivalent to the stage 1 floral meristem (Figure 4A; Smyth et al., 1990). PUCHI expression continued until early stage 2 (Figure 4B) and disappeared before the initiation of sepal primordia. Accumulation of PUCHI mRNA was restricted to the adaxial side of floral primordia. The duration of PUCHI expression roughly corresponded to the first two stages in the analysis of surface morphology described above (i.e., the initial and second bulging stages).
Figure 4.
Expression Patterns of PUCHI.
(A) to (C) Wild-type inflorescence apices.
(A) PUCHI mRNA is detected on the adaxial side of the stage 1 flower primordium.
(B) PUCHI mRNA is detected on the adaxial side of the stage 2 flower primordum.
(C) Localization of GFP-PUCHI fusion protein expressed under the cis-regulatory elements of the PUCHI gene. The fusion protein is detected on the adaxial side of stages 1 and 2 floral meristem proper.
(D) Fourteen-day-old wild-type seedling apex that has just begun bolting. PUCHI mRNA is detected in the rosette and cauline leaf axils.
(E) PUCHI mRNA is detected in the axillary meristems of cauline leaves. Numbers indicate stages of flower development (Smyth et al., 1990).
rl, rosette leaf; cl, cauline leaf; st, stem. Bars = 50 μm.
Although PUCHI mRNA was detected on the adaxial side of the floral meristem, the puchi mutant displayed ectopic bract formation on the abaxial side. To localize the site of PUCHI action more precisely, we examined localization of a GFP-PUCHI fusion protein driven by the regulatory elements of PUCHI in the puchi-1 mutant background (genomic GFP-PUCHI; Hirota et al., 2007). The GFP signal was localized to the adaxial side of floral meristems at stages 1 and 2 (Figure 4C), corresponding well with the pattern found in the in situ hybridization experiments. Furthermore, the genomic GFP-PUCHI construct fully complemented the rudimentary bract phenotype of puchi. These results indicate that adaxial localization of PUCHI protein is sufficient to suppress bract outgrowth on the abaxial side.
Because PUCHI is involved in the determination of floral meristem identity, we examined whether PUCHI expression was restricted to the floral meristem or was also present in other types of lateral meristems. During vegetative development, PUCHI transcript accumulation was not detected in the shoot apex (data not shown). Shortly after the onset of flowering, however, PUCHI mRNA accumulated in the axillary meristems of rosette and cauline leaves (Figures 4D and 4E). These results suggest that PUCHI is expressed in all lateral meristems after the transition from the vegetative to the reproductive phase.
PUCHI and BOP Have Overlapping Functions
The paralogous genes BOP1 and BOP2 are redundantly required for various processes of shoot organ development, such as leaf formation, flower patterning, and formation of floral organ abscission zone (Ha et al., 2003; Hepworth et al., 2005; Norberg et al., 2005; McKim et al., 2008). Notably, bop1 bop2 mutant flowers are subtended by ectopic bracts (Norberg et al., 2005). We reexamined the bop1-4 bop2-11 double mutant and found that the bracts that subtended early arising flowers were rudimentary and thus were not readily visible (Figure 5C), whereas those formed in late arising flowers were much larger and showed more complete leaf-like features (Figures 5C, inset, and 5L, left; Norberg et al., 2005). In addition, bop1 bop2 showed significantly more secondary inflorescences than the wild type (Table 1), raising the possibility that BOP1 and BOP2 are involved in the determination of floral meristem identity. Because PUCHI, BOP1, and BOP2 affect similar processes of flower development, we tested for a possible interaction between the PUCHI and BOP genes.
Figure 5.
Genetic Interaction between puchi and bop Mutants.
(A) to (D) Forty-day-old primary inflorescences.
(A) The wild type.
(B) puchi-1.
(C) bop1-4 bop2-11. The inset shows an older (55 to 60 d old) inflorescence, producing visible bracts on the flower pedicels (arrowheads).
(D) puchi-1 bop1-4 bop2-11. Flowers are transformed into secondary inflorescence-like structures (arrows).
(E) to (H) Close-up view of inflorescence apices photographed when the inflorescences were ∼10 mm in length.
(E) The wild type.
(F) puchi-1.
(G) bop1-4 bop2-1.
(H) puchi-1 bop1-4 bop2-11.Unlike puchi and bop1 bop2 flowers (arrows in [F] and [G]), the triple mutant produces secondary inflorescences (arrowheads in [H]).
(I) A secondary inflorescence of puchi-1 bop1-4 bop2-11 in a position normally occupied by a flower in the wild type. Arrows indicate the formation of tertiary shoots in the leaf axils.
(J) Scanning electron micrograph showing a puchi-1 bop1-4 bop2-11 inflorescence apex at a similar stage to (H). Primordia produced by the primary inflorescence meristem (1°) behave like inflorescence meristems rather than like flower meristems (e.g., primordia indicated with 2°).
(K) Sixty-day-old inflorescence of a puchi-1 bop1-4 bop2-11 triple mutant showing formation of flowers subtended by well-developed bracts (arrowheads).
(L) Typical bracts of bop1-4 bop2-11 (left) and puchi-1 bop1-4 bop2-11 (right). Note that puchi mutant flowers have rudimentary bracts that are much smaller than those in double and triple mutants (cf. [L] with Figure 2B; see Supplemental Figure 2A online).
(M) Flowers of a bop1-4 bop2-11 double mutant.
(N) Flowers of a puchi-1 bop1-4 bop2-11 triple mutant.Arrows in (M) and (N) indicate sepal-petal hybrid organs in the first whorl.
Bars = 1 mm in (E) to (H) and (L) to (N) and 100 μm in (J).
[See online article for color version of this figure.]
We first generated a puchi-1 bop1-4 bop2-11 triple mutant, which displayed significantly enhanced phenotypes compared with the parental mutants with regard to both the determination of meristem identity and bract suppression (Figures 5A to 5L). The most striking feature of puchi bop1 bop2 plants was their altered inflorescence structure, which was characterized by the presence of a much higher number of secondary inflorescences: up to six- to sevenfold more than in either of the parental mutants (Table 1). Each of these secondary branches typically had nodes with associated axillary shoots and showed indeterminate growth (Figure 5I). Such branches were always subtended by well-developed cauline leaves when they were produced at the basal nodes, whereas the upper branches were not (see Supplemental Figure 5 online). Thus, the transition from secondary inflorescence meristems to floral meristems in this triple mutant was more severely impaired that in either of the parent mutants. Scanning electron microscopy of the primary inflorescence apex of this triple mutant confirmed that, during the initial stages of inflorescence development, the primary inflorescence meristem yielded secondary meristems that produced lateral organs in a spiral arrangement that is typical of a branch, rather than a whorled pattern as in the floral meristem (Figure 5J).
After ∼24 branches had appeared (Table 1), the primary shoot of the triple mutant started to produce flowers. However, these flowers were subtended by well-developed bracts that were much larger than those in puchi or bop1 bop2 mutants (Figures 5K and 5L). These results show that the activity of bract formation is also enhanced in the triple mutant. It has been reported that bop1 bop2 mutants produce flowers with abnormal morphology, such as increased numbers of floral organs and the presence of sepal-petal hybrid organs on the abaxial side of the first whorl (Hepworth et al., 2005; Norberg et al., 2005). The puchi mutation, however, did not exacerbate the floral phenotypes of bop1 bop2 (Figures 5M and 5N), again suggesting that PUCHI does not play a role in floral organ patterning. When puchi bop1 bop2 plants were grown under short-day conditions, they showed even more extreme phenotypes than when they were grown under continuous light (Table 1), indicating that the effects of these mutations and short-day photoperiod are additive.
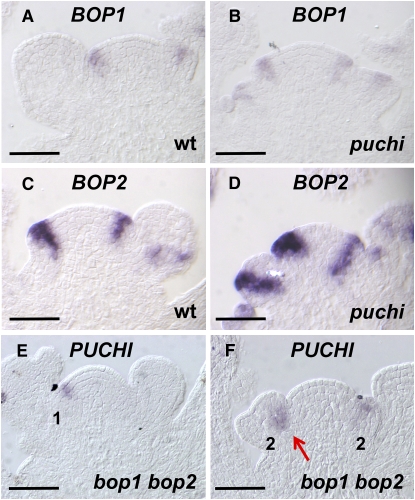
We next compared expression patterns of BOP1 and BOP2 between wild type and puchi mutant backgrounds. In the wild type, the earliest expression of both genes was found in the floral anlagen, from which a flower primordium will arise (Figures 6A and 6C; Hepworth et al., 2005; Norberg et al., 2005). Their expression persisted throughout early stage 1 and 2 floral meristems (Figures 6A and 6C). In the stage 2 primordium, BOP1 and BOP2 expression was detected in a central region that roughly corresponded to the zone between the floral meristem and the cryptic bract (Figures 6A and 6C; Long and Barton, 2000; Dinneny et al., 2004). In puchi, expression of BOP1 and BOP2 was similar to that in the wild type except that the signal was somewhat broader (Figures 6B and 6D). PUCHI expression in the bop1 bop2 mutant inflorescence apex was also analyzed and was generally similar to that in the wild type, although the signal was localized more internally in puchi mutant primordia at late stage 2 (Figures 6F and 6F). Collectively, these results suggest that the PUCHI and BOP genes are not related to each other in a hierarchical order of transcriptional control.
Figure 6.
BOP and PUCHI Are Expressed Independently of Each Other.
(A) BOP1 mRNA in longitudinal section of the wild-type inflorescence apex.
(B) BOP1 mRNA in longitudinal section of the puchi-1 inflorescence apex.
(C) BOP2 mRNA in longitudinal section of the wild-type inflorescence apex.
(D) BOP2 mRNA in longitudinal section of the puchi-1 inflorescence apex.
(E) PUCHI mRNA in longitudinal section of bop1-4 bop2-11 the inflorescence apex, showing expression in the stage 1 primordium.
(F) PUCHI mRNA in longitudinal section of the bop1-4 bop2-11 inflorescence apex, showing expression in the stage 2 primordia. PUCHI expression is unaffected (cf. [E] with Figure 4A) except in the late stage 2 flower, in which it tends to localize to the inner cells (cf. [F], arrow, with Figure 4B).
Bars = 50 μm.
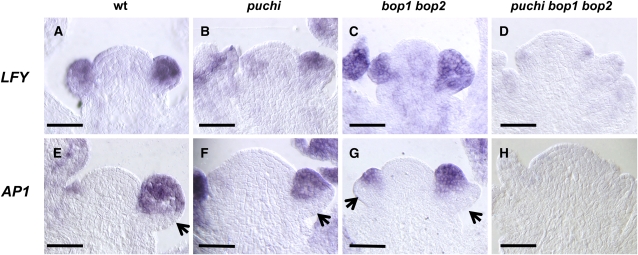
The PUCHI and BOP Genes Are All Required for LFY and AP1 Expression
The perturbation in floral meristem specification in puchi bop1 bop2 suggested that other genes responsible for floral meristem specification, such as LFY and AP1, might be inactive in this triple mutant. We therefore tested whether the puchi and bop mutations had any effect on LFY and AP1 expression.
In the wild-type inflorescence apex, LFY mRNA is first detected at a low level in the floral anlagen (Figure 7A; Weigel et al., 1992; Blázquez et al., 1997). LFY was uniformly expressed at a higher level throughout stage 1 and 2 flower primordia (Figure 7A). LFY expression was normal in puchi and bop1 bop2 mutant backgrounds (Figures 7B and 7C) but was markedly reduced in the puchi bop1 bop2 triple mutant inflorescence apex (Figure 7D). These results indicate that the PUCHI and BOP genes redundantly promote LFY expression during inflorescence development.
Figure 7.
Expression of the Floral Meristem Identity Genes LFY and AP1 in the puchi bop1 bop2 Mutant.
(A) to (H) Longitudinal sections hybridized with either a LFY or an AP1 probe.
(A) The wild-type inflorescence apex hybridized with the LFY probe.
(B) The puchi-1 inflorescence apex hybridized with the LFY probe.
(C) The bop1-4 bop2-11 inflorescence apex hybridized with the LFY probe.
(D) The puchi-1 bop1-4 bop2-11 inflorescence apex hybridized with the LFY probe. Note that compared with the wild type (A), LFY expression does not change in puchi single (B) or bop1 bop2 double (C) mutants but is markedly reduced in the puchi bop1 bop2 triple mutant (D).
(E) The wild-type inflorescence apex hybridized with the AP1 probe.
(F) The puchi-1 inflorescence apex hybridized with the AP1 probe.
(G) The bop1-4 bop2-11 inflorescence apex hybridized with the AP1 probe.
(H) The puchi-1 bop1-4 bop2-11 inflorescence apex hybridized with the AP1 probe. Note that compared with the wild type (E), AP1 expression is detected in a much smaller proportion of the adaxial cells of young flower primordia of puchi single (F) and bop1 bop2 double (G) mutants and is almost undetectable in the puchi bop1 bop2 triple mutant (H).
Arrows indicate the absence of AP1 expression on the abaxial side. Bars = 50 μm.
Next, we examined AP1 expression patterns. In the wild type, AP1 mRNA was detected at a high level in the adaxial cells of stage 1 and 2 floral primordia (Figure 7E; Mandel et al., 1992). A small group of abaxial cells in these early floral primordia did not express AP1 (Figure 7E); these cells correspond to the cryptic bract region of floral primordia. In the puchi and bop1 bop2 mutants, AP1 expression was detected in a much smaller proportion of the adaxial cells of young floral primordia (Figures 7F and 7G), consistent with ectopic bract formation in these backgrounds, and was almost undetectable in the inflorescence of puchi bop1 bop2 (Figure 7H). Thus, the severe inflorescence phenotype in the triple mutant correlates with a drastic reduction in the expression of genes involved in floral meristem specification.
DISCUSSION
PUCHI Is Required for Floral Meristem Identity
In this study, we have shown that puchi mutations affect inflorescence architecture in two ways. First, the number of secondary inflorescences is increased, indicating a conversion of early arising flowers into branches. Second, mutant flowers are subtended by rudimentary bracts, partially displaying the character of secondary inflorescences, which normally bear a subtending leaf. These results show that PUCHI is required for proper conversion of secondary inflorescences to flowers.
PUCHI is orthologous to maize BD1 and rice FZP, both of which also affect inflorescence architecture (Chuck et al., 2002; Komatsu et al., 2003; Hirota et al., 2007). The inflorescences of grasses show a unique type of lateral meristem called spikelet meristems, from which floral meristems arise (Thompson and Hake, 2009). Spikelet meristems initially produce bract-like organs called glumes. In bd1 and fzp mutants, spikelet meristems are replaced by indeterminate branch-like structures, indicating some functional similarity between the grass genes and PUCHI in the control of meristem identity. Several observations, however, point to important differences (this article; Chuck et al., 2002; Komatsu et al., 2003). First, expression of PUCHI is detected in the floral meristem proper (Figures 4A to 4C), whereas BD1 and FZP are expressed in the axil of glumes but not in the spikelet meristem itself. Second, both bd1 and fzp mutants display ectopic meristem formation in the axil of glumes, whereas no corresponding phenotype is observed in the puchi mutant. Third, the puchi mutation affects bract suppression, but neither bd1 nor fzp mutations affect this process. These results together suggest that Arabidopsis has adopted this type of gene to its own fate determination process in a different way to the grass species.
Another difference between PUCHI and BD1/FZP lies in the strength of the mutant phenotypes: the inflorescence phenotype of puchi is much more subtle than that of bd1 or fzp. It is possible that other Arabidopsis proteins function redundantly with PUCHI and partially mask the effects of the puchi single mutation. A good candidate is LEAFY PETIOLE, which is most closely related to PUCHI and shares 95% amino acid identity within its AP2 domain (van der Graaff et al., 2000; Hirota et al., 2007).
Relationship between the PUCHI and BOP Genes
Our analysis demonstrates that the PUCHI and BOP genes have overlapping functions and indicates that the relationship between these genes does not involve mutual transcriptional control. The strong phenotype in the puchi bop1 bop2 triple mutant reveals the critical roles played by the PUCHI and BOP genes in the control of meristem identity and bract suppression, although the molecular mechanism underlying this synergistic phenotype is currently unknown. BOP1 and BOP2 encode proteins with a BTP/POZ domain and ankyrin repeats, both of which are involved in protein–protein interactions. Their homolog NPR1 regulates pathogen-inducible gene expression by interacting with TGACG sequence-specific binding transcription factors (TGAs) in the nucleus. BOP1 and BOP2 have also been shown to interact with a TGA protein, PERIANTHIA (PAN), to regulate floral organ patterning (Hepworth et al., 2005). Because our analysis shows that the expression domains of PUCHI and BOP genes overlap in lateral meristems, at least partially, it will be important to test whether PUCHI interacts directly with BOP proteins.
The PUCHI and BOP Genes May Provide a Positional Cue for Activation of LFY and AP1 Expression
LFY and AP1 expression was greatly reduced in lateral meristems of the puchi bop1 bop2 triple mutant inflorescence apex (Figures 7D and 7H), demonstrating critical roles for the PUCHI and BOP genes in activating expression of these meristem identity genes. Because expression of AP1 requires LFY (Liljegren et al., 1999; Ratcliffe et al., 1999; Wagner et al., 1999), the loss of AP1 expression in the triple mutant is most simply explained by the loss of LFY activation. A threshold level of LFY expression is required to confer flower identity on the lateral primordia during the transition from vegetative to reproductive phase (Blázquez et al., 1997). Expression of LFY is regulated by multiple inputs, including SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), AGAMOUS-LIKE24 (AGL24), FLOWERING LOCUS T (FT), and gibberellins (Blázquez and Weigel, 2000; Yu et al., 2002; Schmid et al., 2003; Moon et al., 2005). Among these, the precise distribution of FT and gibberellins in the shoot apex remains unclear.
On the other hand, the two MADS transcription factors SOC1 and AGL24, which together form a complex and bind directly to the LFY promoter, are expressed throughout the shoot apex (Lee et al., 2000; Samach et al., 2000; Yu et al., 2002; Michaels et al., 2003), raising the possibility that other unknown factors are involved in floral meristem–specific activation of LFY (Lee et al., 2008). PUCHI and BOP genes are candidates for this effect because their expression is specific to lateral meristems. PUCHI and BOP genes are required for specification of floral meristem identity under both continuous-light and short-day conditions, suggesting that their actions are largely independent of these environmental cues. Our analysis thus suggests that PUCHI and the two BOP genes provide a positional cue for LFY and AP1 to be expressed in lateral meristems and perhaps act in concert with other flower-promoting signals, such as photoperiod.
Interestingly, expression of PUCHI is not restricted only to floral meristems but also occurs in secondary inflorescence meristems (Figures 4D and 4E), which normally maintain low levels of LFY expression (Ratcliffe et al., 1999). This result suggests that activation of LFY by PUCHI may require additional factors that are expressed in the floral meristems but not in secondary inflorescence meristems. It is also possible that some negative factor(s), such as TERMINAL FLOWER1, which is known to limit LFY expression to the floral meristem, may repress PUCHI function in the secondary inflorescence meristem.
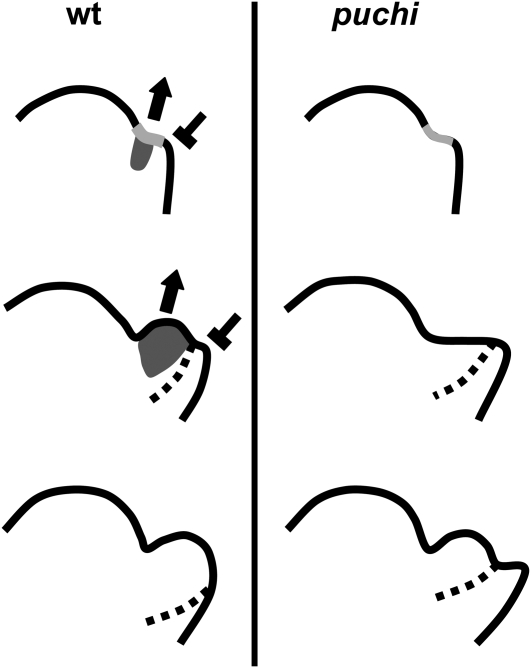
PUCHI Is a Novel Regulator for Shaping the Flower Primordium
Studies using molecular markers (Long and Barton, 2000) and, more recently, a sensitive method for surface morphology (Kwiatkowska, 2006) have indicated that the floral meristem of Arabidopsis is initiated in the axil of the cryptic bract, whose development is later suppressed by a signal derived from the floral meristem (Nilsson et al., 1998). We have shown here that PUCHI mRNA is transiently detected on the adaxial side of early floral primordia. Moreover, expression of GFP-PUCHI, in the same domain and at the same time, is sufficient to suppress the puchi phenotype. We propose that the domain of PUCHI expression corresponds to the floral meristem proper in the axil of a cryptic bract (Figure 8). Accumulation of the PUCHI protein in this domain accelerates the second bulging of the floral meristem proper and suppresses the growth of the shelf-like cryptic bract primordium. The expression domain of PUCHI does not overlap the cryptic bract, raising the possibility that PUCHI acts non-cell-autonomously in bract suppression. PUCHI may promote expression of a signaling molecule that can move from the floral meristem proper toward the abaxial cryptic bract region. Another possibility is that promotion of the second bulging of the floral meristem proper by PUCHI may indirectly affect growth of the cryptic bract, either by changing the distribution of mechanical stress across the flower primordium (Hamant et al., 2008) or by incorporating cells that would otherwise become a part of the bract into the floral meristem proper. The proposed function for PUCHI in flower primordium development is very similar to that suggested for UFO, whose mutation causes a delay in the development of the floral meristem proper relative to bract development (Hepworth et al., 2006). It will thus be important to determine whether these two genes interact in early flower development.
Figure 8.
A Model for Control of Morphogenesis by PUCHI in the Early Flower Primordium.
Flower primordium formation in the wild type (left) and puchi (right). The expression domain of PUCHI (dark gray) is deduced from Figures 4A to 4C. Dotted lines represent a putative boundary between the floral meristem proper and the cryptic bract, deduced from the AP1 expression domain (Figures 7E and 7F). Top: Initial bulging leading to the appearance of a shallow crease (light gray). In the wild type, PUCHI promotes the second bulging of the floral meristem (arrow) and represses the cryptic bract (T bar). Middle: The second bulging occurs from the shallow crease in the wild type primordium (left), whereas puchi forms a shelf-like primordium because the second bulging is delayed (right). Bottom: The bulge completes in the wild type and morphological signs of the cryptic bract disappear. In puchi, the second bulging now occurs and the cryptic bract remains morphologically apparent.
Previous analysis has shown that PUCHI is involved in morphogenesis of early lateral root primordia (Hirota et al., 2007). Initiation of a lateral root begins with anticlinal cell divisions of one or two pericycle cells and subsequent periclinal and anticlinal divisions result in bulging of a primordium with a restricted size along the radial dimension (Malamy and Benfey, 1997; Dubrovsky et al., 2001). Expression of PUCHI begins in cells throughout the early lateral root primordium and is later downregulated in the center (Hirota et al., 2007). By affecting the frequency of anticlinal relative to periclinal divisions, the puchi mutation causes ectopic cell proliferation in the periphery of the primordium, resulting in the formation of a wider and flatter lateral root primordium with a less prominent central dome (Hirota et al., 2007). These results indicate that PUCHI prevents cell proliferation in the periphery through the control of cell divisions. This phenotype in early lateral root formation is reminiscent of the ectopic bract growth observed in early flower formation in the puchi mutant. Although flowers and roots are very different in their anatomy, their developmental origins, and the regulatory genes involved in the fate specification process, further detailed analysis of PUCHI function may lead to the identification of a common mechanism that regulates morphogenesis of early lateral primordia both in the shoot and in the root.
METHODS
Plant Materials and Growth Conditions
All mutants were in the Arabidopsis thaliana cv Columbia (Col) background unless otherwise noted. The puchi-1 and puchi-2 mutants, both of which are the Col background, have been described previously (Hirota et al., 2007) and were backcrossed three times to Col before phenotypic analyses. The GAL4-GFP enhancer trap line E1238 (http://enhancertraps.bio.upenn.edu/default.html) was obtained from the ABRC (Ohio State University, Columbus, OH; stock number CS70083). bop1-4 and bop2-11 are null alleles (Ha et al., 2004, 2007) and were kindly provided by J.C. Fletcher and C.M. Ha. The prs mutant, which is in the Landsberg erecta background, was kindly provided by K. Okada. Seeds were imbibed, surface sterilized, and incubated at 4°C for 3 d. They were then sown and germinated on soil and grown at 23°C under continuous-light or short-day (8 h light/16 h dark) conditions unless otherwise noted. Light intensity was 15 μmol m−2 s−1 and 28 μmol m−2 s−1 in continuous light and short-day conditions, respectively.
Phenotypic Analyses
The number of rosette leaves was counted at bolting, and the number of secondary inflorescences was counted after formation of the first flower. Ectopic secondary inflorescences were counted shortly before senescence. Leaves on the primary bolting stem were considered as cauline leaves if they bear indeterminate secondary inflorescences. Leaves or rudimentary leaf-like structures subtending flowers were regarded as bracts (Dinneny et al., 2004). To estimate the number of secondary inflorescences, all plants were grown at the same time, in the same growth chamber, and at the same density per pot. These precautions were particularly important when counting the number of secondary inflorescences because the phenotype appeared sensitive to small fluctuations in growing conditions, such as temperature, humidity, or nutrients.
Photography and Microscopy
Photographs were taken with a digital camera (Velbon; Nikon). A Keyence VHX-900 digital microscope was used to take close-up images. Scanning electron microscopy of plant material was performed as described previously (Aida et al., 1999). To detect expression of GFP-PUCHI, inflorescence apices were fixed in 5% agarose (Gibco BRL) and incubated at 4°C for 20 min. Longitudinal sections of 100 μm were made using a vibrating-blade microtome (Microm International). Samples were stained with 50 μg/mL FM4-64 (Invitrogen), and fluorescence images were obtained using an FV1000 confocal laser scanning microscope (Olympus). GFP fluorescence was detected with the spectral settings at 490 to 540 nm for emission and 488 nm for excitation. FM4-64 fluorescence was detected with the spectral settings at 590 to 690 nm for emission and 543 nm for excitation.
Sequential Replicas and Quantitative Analysis of Primordium Shape
Inflorescence shoot apices of nine puchi plants were studied with the aid of the nondestructive sequential replica method (Williams and Green, 1988), in which dental polymer molds were taken from individual apices at 12-h intervals. Epoxy resin casts prepared from these molds were observed by scanning electron microscopy (sputter-coated; LEO435VP microscope). Replicas were taken from plants at 7 to 8 weeks after germination (11 h light/13 h dark), when the inflorescence axis length was between 2 and 10 mm and before the oldest flower bud had opened. Sequences of replicas obtained from puchi apices were compared with previously studied wild-type Col apices (Kwiatkowska, 2006).
For each cast, two micrographs were taken, one tilted at 10° with respect to the other, and used for stereoscopic reconstruction (Routier-Kierzkowska and Kwiatkowska, 2008) and geometric quantitation (Dumais and Kwiatkowska, 2002). Computer programs used for this analysis were written in Matlab (The Mathworks). Reconstructed surfaces of flower primordia were rotated so that the side views of the primordia could be compared. The shape of each primordium was quantified by means of principal curvature directions (i.e., the directions in which the curvature attained either maximal or minimal values).
In Situ Hybridization
For in situ hybridization, inflorescence apices were collected and fixed shortly after bolting, when the inflorescences were <10 mm in length. In situ hybridization was performed according to Takada et al. (2001). The BOP1 probe has been described by Ha et al. (2004). The LFY probe was transcribed using T3 RNA polymerase (Promega) from pDW124 (a gift from D. Weigel) linearized with BamHI. AP1 and BOP2 probes were transcribed using T3 RNA polymerase from RAFL22-60-H11 and RAFL15-22-D12 (provided by RIKEN) linearized with EcoRI. To synthesize the PUCHI probe, a cDNA fragment was amplified using PUCHI_F (5′-CTCCACAGTTTGTCATCGATC-3′) and PUCHI_R (5′-GACTGAGTAGAAGCCTGTAG-3′) primers, which excluded the AP2 domain to avoid cross-hybridization, and the blunt PCR product was cloned into pCR-Blunt II-TOPO (Invitrogen). The plasmid was linearized with SpeI and transcribed using T7 RNA polymerase (Promega). Hybridization was performed at 45°C. Western Blue (Promega) was used as the substrate for signal detection.
Accession Numbers
Sequence data from this article can be found in the GenBank and/or The Arabidopsis Information Resource data libraries under the following accession numbers: PUCHI (NP_197357/At5g18560), PRS (NP_180429/At2g28610), BOP1 (NP_191272/At3g57130), BOP2 (NP_181668/At2g41370), LFY (NP_200993/At5g61850), AP1 (NP_177074/At1g69120), BD1 (NP_001105200), and FZP (AB103120).
Supplemental Data
Supplemental Figure 1. Inflorescence Phenotypes of the puchi-2 Mutant.
Supplemental Figure 2. Formation of Pin-Shaped Projections in puchi Is Dependent on the PRESSED FLOWER Gene.
Supplemental Figure 3. puchi Flower Phenotypes.
Supplemental Figure 4. Sepal Formation Is Delayed in the puchi Mutant.
Supplemental Figure 5. Fifty-Day-Old Primary Inflorescence of the puchi bop1 bop2 Mutant Grown under Continuous-Light Conditions.
Supplementary Material
Acknowledgments
We thank Jennifer Fletcher and Chan Man Ha for bop1-4 and bop2-11 seeds, and Detlef Weigel and Kiyotaka Okada for pDW124 plasmid and prs seeds, respectively. We also thank the ABRC and RIKEN for providing materials. We thank Ian Smith and Seiji Takeda for their critical reading of the manuscript. This work was partly supported by the Ministry of Education, Culture, Sports, Science, and Technology, through Grant-in-Aid for Scientific Research on Priority Areas (14036222) to M.T., Grant-in-Aid for Young Scientists (18770036), and Grant-in-Aid for Scientific Research on Priority Areas (21027027) to M.A.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Mitsuhiro Aida (m-aida@bs.naist.jp).
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., Ichinoki, H., Notaguchi, M., Goto, K., and Araki, T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309 1052–1056. [DOI] [PubMed] [Google Scholar]
- Aida, M., Ishida, T., and Tasaka, M. (1999). Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126 1563–1570. [DOI] [PubMed] [Google Scholar]
- Baurle, I., and Dean, C. (2006). The timing of developmental transitions in plants. Cell 125 655–664. [DOI] [PubMed] [Google Scholar]
- Benlloch, R., Berbel, A., Serrano-Mislata, A., and Madueno, F. (2007). Floral initiation and inflorescence architecture: A comparative view. Ann. Bot. (Lond.) 100 659–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez, M.A., Ferrandiz, C.F., Madueno, F., and Parcy, F. (2006). How flower meristems are formed. Plant Mol. Biol. 60 855–870. [DOI] [PubMed] [Google Scholar]
- Blázquez, M.A., Green, R., Nilsson, O., Sussman, M.R., and Weigel, D. (1998). Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell 10 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez, M.A., Soowal, L.N., and Weigel, D. (1997). LEAFY expression and flower initiation in Arabidopsis. Development 124 3835–3844. [DOI] [PubMed] [Google Scholar]
- Blázquez, M.A., and Weigel, D. (2000). Integration of floral inductive signals in Arabidopsis. Nature 404 889–892. [DOI] [PubMed] [Google Scholar]
- Bowman, J.L., Alvarez, J., Weigel, D., Meyerowitz, E.M., and Smyth, D.R. (1993). Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119 721–743. [Google Scholar]
- Chuck, G., Muszynski, M., Kellogg, E., Hake, S., and Schmidt, R.J. (2002). The control of spikelet meristem identity by the branched silkless1 gene in maize. Science 298 1238–1241. [DOI] [PubMed] [Google Scholar]
- Coen, E.S., and Nugent, J.M. (1994). Evolution of flowers and inflorescences. Development 120(suppl.): 107–116. [Google Scholar]
- Dinneny, J.R., Yadegari, R., Fischer, R.L., Yanofsky, M.F., and Weigel, D. (2004). The role of JAGGED in shaping lateral organs. Development 131 1101–1110. [DOI] [PubMed] [Google Scholar]
- Dubrovsky, J.G., Rost, T.L., Colon-Carmona, A., and Doerner, P. (2001). Early primordium morphogenesis during lateral root initiation in Arabidopsis thaliana. Planta 214 30–36. [DOI] [PubMed] [Google Scholar]
- Dumais, J., and Kwiatkowska, D. (2002). Analysis of surface growth in shoot apices. Plant J. 31 229–241. [DOI] [PubMed] [Google Scholar]
- Ha, C.M., Jun, J.H., Nam, H.G., and Fletcher, J.C. (2004). BLADE-ON-PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 45 1361–1370. [DOI] [PubMed] [Google Scholar]
- Ha, C.M., Jun, J.H., Nam, H.G., and Fletcher, J.C. (2007). BLADE-ON-PETIOLE1 and 2 control Arabidopsis lateral organ fate through regulation of LOB domain and adaxial-abaxial polarity genes. Plant Cell 19 1809–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, C.M., Kim, G.T., Kim, B.C., Jun, J.H., Soh, M.S., Ueno, Y., Machida, Y., Tsukaya, H., and Nam, H.G. (2003). The BLADE-ON-PETIOLE1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development 130 161–172. [DOI] [PubMed] [Google Scholar]
- Hamant, O., Heisler, M.G., Jonsson, H., Krupinski, P., Uyttewaal, M., Bokov, P., Corson, F., Sahlin, P., Boudaoud, A., Meyerowitz, E.M., Couder, Y., and Traas, J. (2008). Developmental patterning by mechanical signals in Arabidopsis. Science 322 1650–1655. [DOI] [PubMed] [Google Scholar]
- Hempel, F.D., Weigel, D., Mandel, M.A., Ditta, G., Zambryski, P.C., Feldman, L.J., and Yanofsky, M.F. (1997). Floral determination and expression of floral regulatory genes in Arabidopsis. Development 124 3845–3853. [DOI] [PubMed] [Google Scholar]
- Hepworth, S.R., Klenz, J.E., and Haughn, G.W. (2006). UFO in the Arabidopsis inflorescence apex is required for floral-meristem identity and bract suppression. Planta 223 769–778. [DOI] [PubMed] [Google Scholar]
- Hepworth, S.R., Zhang, Y., McKim, S., Li, X., and Haughn, G.W. (2005). BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17 1434–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota, A., Kato, T., Fukaki, H., Aida, M., and Tasaka, M. (2007). The auxin-regulated AP2/EREBP gene PUCHI is required for morphogenesis in the early lateral root primordium of Arabidopsis. Plant Cell 19 2156–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanrar, S., Bhattacharya, M., Arthur, B., Courtier, J., and Smith, H.M.S. (2008). Regulatory networks that function to specify flower meristems require the function of homeobox genes PENNYWISE and POUND-FOOLISH in Arabidopsis. Plant J. 54 924–937. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., and Weigel, D. (2007). Move on up, its time for change—Mobile signals controlling photoperiod-dependent flowering. Genes Dev. 21 2371–2384. [DOI] [PubMed] [Google Scholar]
- Komatsu, M., Chujo, A., Nagato, Y., Shimamoto, K., and Kyozuka, J. (2003). FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130 3841–3850. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Hanhart, C.J., and van der Veen, J.H. (1991). A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229 57–66. [DOI] [PubMed] [Google Scholar]
- Kwiatkowska, D. (2006). Flower primordium formation at the Arabidopsis shoot apex: quantitative analysis of surface geometry and growth. J. Exp. Bot. 57 571–580. [DOI] [PubMed] [Google Scholar]
- Lee, H., Suh, S.S., Park, E., Cho, E., Ahn, J.H., Kim, S.G., Lee, J.S., Kwon, Y.M., and Lee, I. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., Oh, M., Park, H., and Lee, I. (2008). SOC1 translocated to the nucleus by interaction with AGL24 directly regulates LEAFY. Plant J. 55 832–843. [DOI] [PubMed] [Google Scholar]
- Liljegren, S.J., Gustafson-Brown, C., Pinyopich, A., Ditta, G.S., and Yanofsky, M.F. (1999). Interactions among APETALA1, LEAFY, and TERMINAL FLOWER1 specify meristem fate. Plant Cell 11 1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, J., and Barton, M.K. (2000). Initiation of axillary and floral meristems in Arabidopsis. Dev. Biol. 218 341–353. [DOI] [PubMed] [Google Scholar]
- Malamy, J.E., and Benfey, P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124 33–44. [DOI] [PubMed] [Google Scholar]
- Mandel, M.A., Gustafson-Brown, C., Savidge, B., and Yanofsky, M.F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360 273–277. [DOI] [PubMed] [Google Scholar]
- Mandel, M.A., and Yanofsky, M.F. (1995). A gene triggering flower development in Arabidopsis. Nature 377 522–524. [DOI] [PubMed] [Google Scholar]
- Matsumoto, N., and Okada, K. (2001). A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers. Genes Dev. 15 3355–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim, S.M., Stenvik, G.E., Butenko, M.A., Kristiansen, W., Cho, S.K., Hepworth, S.R., Aalen, R.B., and Haughn, G.W. (2008). The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development 135 1537–1546. [DOI] [PubMed] [Google Scholar]
- Michaels, S.D., Ditta, G., Gustafon-Brown, C., Pelaz, S., Yanofsky, M., and Amasino, R.M. (2003). AGL24 acts as a promoter of flowering in Arabidopsis is positively regulated by vernalization. Plant J. 33 867–874. [DOI] [PubMed] [Google Scholar]
- Moon, J., Lee, H., Kim, M., and Lee, I. (2005). Analysis of flowering pathway integrators in Arabidopsis. Plant Cell Physiol. 46 292–299. [DOI] [PubMed] [Google Scholar]
- Nardmann, J., Ji, J., Werr, W., and Scanlon, M.J. (2004). The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in lateral domain of shoot apical meristems. Development 131 2827–2839. [DOI] [PubMed] [Google Scholar]
- Nilsson, O., Wu, E., Wolfe, D.S., and Weigel, D. (1998). Genetic ablation of flowers in transgenic Arabidopsis. Plant J. 15 799–804. [DOI] [PubMed] [Google Scholar]
- Norberg, M., Holmlund, M., and Nilsson, O. (2005). The BLADE-ON-PETIOLE genes act redundantly to control the growth and development of lateral organs. Development 132 2203–2213. [DOI] [PubMed] [Google Scholar]
- Parcy, F., Nilsson, O., Busch, M.A., Lee, I., and Weigel, D. (1998). A genetic framework for floral patterning. Nature 395 561–566. [DOI] [PubMed] [Google Scholar]
- Poethig, R.S. (2003). Phase change and the regulation of developmental timing in plants. Science 301 334–336. [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz, P., Erasmus, Y., Lane, B., Hareder, L.D., and Coen, E. (2007). Evolution and development of inflorescence architectures. Science 316 1452–1456. [DOI] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Amaya, I., Vincent, C.A., Rothstein, S., Carpenter, R., Coen, E.S., and Bradley, D.J. (1998). A common mechanism controls the life cycle and architecture of plants. Development 125 1609–1615. [DOI] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Bradley, D.J., and Coen, E.S. (1999). Separation of shoot and floral identity in Arabidopsis. Development 126 1109–1120. [DOI] [PubMed] [Google Scholar]
- Routier-Kierzkowska, A.L., and Kwiatkowska, D. (2008). New stereoscopic reconstruction protocol for scanning electron microscope images and its application to in vivo replicas of the shoot apical meristem. Funct. Plant Biol. 35 1034–1046. [DOI] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288 1613–1616. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Uhlenhaut, N.H., Godard, F., Demar, M., Bressan, R., Weigel, D., and Lohmann, J.U. (2003). Dissection of floral induction pathways using global expression analysis. Development 130 6001–6012. [DOI] [PubMed] [Google Scholar]
- Schultz, E.A., and Haughn, G.W. (1991). LEAFY, a homeotic gene that regulates inflorescence development in Arabidopsis. Plant Cell 3 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, H.M., Campbell, B.C., and Hake, S. (2004). Competence to respond to floral inductive signals requires the homeobox genes PENNYWISE and POUND-FOOLISH. Curr. Biol. 14 812–817. [DOI] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves, T.A., and Sussex, I.M. (1989). Patterns in Plant Development, 2nd ed. (Cambridge, UK: Cambridge University Press).
- Szczesny, T., Routier-Kierzkowska, A.-L., and Kwiatkowska, D. (2009). Influence of clavata3–2 mutation on early flower development in Arabidopsis thaliana: Quantitative analysis of changing geometry. J. Exp. Bot. 60 679–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada, S., Hibara, K., Ishida, T., and Tasaka, M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128 1127–1135. [DOI] [PubMed] [Google Scholar]
- Thompson, B.E., and Hake, S. (2009). Translational biology: From Arabidopsis flowers to grass inflorescence architecture. Plant Physiol. 149 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff, E., Dulk-Ras, A.D., Hooykaas, P.J., and Keller, B. (2000). Activation tagging of the LEAFY PETIOLE gene affects leaf petiole development in Arabidopsis thaliana. Development 127 4971–4980. [DOI] [PubMed] [Google Scholar]
- Wagner, D., Sablowski, R.W.M., and Meyerowitz, E.M. (1999). Transcriptional activation of APETALA1 by LEAFY. Science 285 582–584. [DOI] [PubMed] [Google Scholar]
- Weigel, D., Alvarez, J., Smyth, D.R., Yanofsky, M.F., and Meyerowitz, E.M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69 843–859. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Meyerowitz, E.M. (1993). Activation of floral homeotic genes in Arabidopsis. Science 261 1723–1726. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Nilsson, O. (1995). A developmental switch sufficient for flower initiation in diverse plants. Nature 377 495–500. [DOI] [PubMed] [Google Scholar]
- Wigge, P.A., Kim, M.C., Jaeger, K.E., Busch, W., Schmid, M., Lohmann, J.U., and Weigel, D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309 1056–1059. [DOI] [PubMed] [Google Scholar]
- William, D.A., Su, Y., Smith, M.R., Lu, M., Baldwin, D.A., and Wagner, D. (2004). Genomic identification of direct target genes of LEAFY. Proc. Natl. Acad. Sci. USA 101 1775–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, M.H., and Green, P.B. (1988). Sequential scanning electron microscopy of a growing plant meristem. Protoplasma 147 77–79. [Google Scholar]
- Yu, H., Xu, Y., Tan, E.L., and Kumar, P.P. (2002). AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc. Natl. Acad. Sci. USA 99 16336–16341. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.