Abstract
Flavonoids are most commonly conjugated with various sugar moieties by UDP-sugar:glycosyltransferases (UGTs) in a lineage-specific manner. Generally, the phylogenetics and regiospecificity of flavonoid UGTs are correlated, indicating that the regiospecificity of UGT differentiated prior to speciation. By contrast, it is unclear how the sugar donor specificity of UGTs evolved. Here, we report the biochemical, homology-modeled, and phylogenetic characterization of flavonoid 7-O-glucuronosyltransferases (F7GAT), which is responsible for producing specialized metabolites in Lamiales plants. All of the Lamiales F7GATs were found to be members of the UGT88-related cluster and specifically used UDP-glucuronic acid (UDPGA). We identified an Arg residue that is specifically conserved in the PSPG box in the Lamiales F7GATs. Substitution of this Arg with Trp was sufficient to convert the sugar donor specificity of the Lamiales F7GATs from UDPGA to UDP-glucose. Homology modeling of the Lamiales F7GAT suggested that the Arg residue plays a critical role in the specific recognition of anionic carboxylate of the glucuronic acid moiety of UDPGA with its cationic guanidinium moiety. These results support the hypothesis that differentiation of sugar donor specificity of UGTs occurred locally, in specific plant lineages, after establishment of general regiospecificity for the sugar acceptor. Thus, the plasticity of sugar donor specificity explains, in part, the extraordinary structural diversification of phytochemicals.
INTRODUCTION
The vast structural diversity in plant secondary metabolites is considered to be the consequence of chemical adaptation by plants to specific ecological niches, since plants exploit secondary metabolites in defense responses against pathogens, and in symbiotic relationships, such as nitrogen fixation and pollinator attraction (Gershenzon and Dudareva, 2007; Macías et al., 2007). A particular plant lineage develops specialized metabolites by acquisition of novel functions of biosynthetic enzymes to increase fitness to its environment. Therefore, specialized metabolites are ascribed to structural changes associated with functional differentiation during the course of enzyme gene evolution (Pichersky and Gang, 2000). It is noteworthy that a small number of amino acid differences are responsible for the functional differentiation of plant enzymes. For example, a single amino acid change can alter the substrate preference of O-methyltransferases (Gang et al., 2002). In the case of sesquiterpene synthases, mutational swaps of only nine amino acids are sufficient to interconvert the product specificities of the encoded mutant proteins in the background of each of the parent enzymes (Greenhagen et al., 2006; O'Maille et al., 2008). Moreover, substitution of only a single residue can substantially alter the product specificity of Clarkia eugenol and isoeugenol synthases; both are members of the PIP reductase family (Koeduka et al., 2008). However, it is generally difficult to identify the critical amino acids responsible for substrate specificity without the structural information of enzymes, since enzymes accumulate many naturally occurring amino acid changes that do not influence their catalysis, per se.
Glycosylation, which involves sugar conjugation from a sugar donor to an acceptor, is a key mechanism that regulates the bioactivity and storage of phytochemicals as well as the detoxification of xenobiotics in plants (Mackenzie et al., 1997; Bowles et al., 2005). This reaction is catalyzed by a superfamily of enzymes, UDP-sugar:glycosyltransferase (UGT), and is classified based on the UDP-sugar donor that is transferred, for example, glucosylation (UDP-glucose), rhamnosylation (UDP-rhamnose), and glucuronosylation (UDP-glucuronic acid [UDPGA]) (Ross et al., 2001; Lim and Bowles, 2004). In mammals, glucuronosylation is well known to occur during Phase II conjugative drug metabolism, which is catalyzed by glucuronic acid transferases/glucuronosyltransferases (GATs). Mammalian GATs are generally considered to be physiological membrane-bound enzymes (Senafi et al., 1994; King et al., 2000; Mackenzie et al., 2003). By contrast, plants evolved various soluble UGTs that have intrinsic sugar donor specificity:glucosyltransferase (GlcT), rhamnosyltransferase (RhaT), galactosyltransferase (GalT), arabinosyltransferase (AraT), and GAT (Miller et al., 1999; Sawada et al., 2005; Yonekura-Sakakibara et al., 2007, 2008). However, the physiological roles and biochemical activities of most plant UGTs remain elusive because higher-plant genomes have a high copy number of UGT genes (107 and 240 UGTs in Arabidopsis thaliana and grapevine [Vitis vinifera], respectively) (Li et al., 2001; Lim et al., 2003; Paquette et al., 2003; The French-Italian Public Consortium for Grapevine Genome Characterization, 2007). Moreover, the molecular structures of the sugar acceptors are extremely diverse.
Flavonoids, a large class of phenylpropanoid-derived secondary metabolites, are mostly glycosylated by UGTs with one or more sugar groups (Harborne and Baxter, 1999). The increasing number of biochemically characterized flavonoid UGTs allowed for the examination of the phylogenetic relationship between UGT structure and function (Ford et al., 1998; Yamazaki et al., 1999; Fukuchi-Mizutani et al., 2003; Nagashima et al., 2004; Ogata et al., 2005; Tohge et al., 2005). Each flavonoid, 3-O-glycosyltransferase, 5-O-glycosyltransferase, and 7-O-glycosyltransferase (i.e., F3GlyT, F5GlyT, and F7GlyT, respectively), forms a unique phylogenic clade (cluster I, II, and III, respectively) that is not limited by species, suggesting that the regiospecificity of flavonoid UGTs for the sugar acceptor (i.e., the specificity of the glycosylation site on the sugar acceptor) was established prior to speciation. Moreover, recent identification of cluster IV UGTs, which are specifically involved in glycosylation of the sugar moieties of glycosides (Morita et al., 2005; Sawada et al., 2005; Noguchi et al., 2008), showed that regiospecificity for the sugar acceptor, but not the sugar donor specificity, is the basis for flavonoid UGT clusters.
Sugar donor preference in plant UGTs is generally very specific. As it significantly impacts the vast structural diversity of natural phytochemicals, it is also of evolutionary and biochemical interest (Gachon et al., 2005). Elucidation of the crystal structures of flavonoid GlcTs provided information that was very useful for understanding the molecular basis of UGT catalysis (Shao et al., 2005; Offen et al., 2006). The PSPG box, a highly conserved motif among plant UGTs, is thought to be involved in sugar donor binding. A recent work on UGT94B1 using homology modeling and biochemical analysis, however, successfully identified a critical Arg residue for sugar donor specificity at the N terminus, which is far from the PSPG box (Osmani et al., 2008). The mechanisms responsible for the sugar donor specificity of UGTs remain largely unknown, although several studies related to this issue have been reported (Kubo et al., 2004; Offen et al., 2006; Yonekura-Sakakibara et al., 2007, 2008).
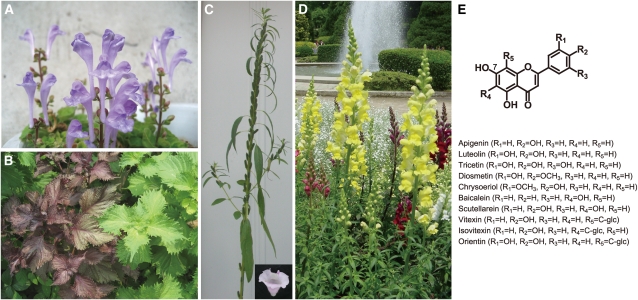
Generally, glucose is the most common sugar moiety in naturally occurring flavonoid glycosides. However, specific plant lineages have characteristic flavonoids conjugated with specific sugar moieties as their specialized metabolites. Flavonoid 7-O-glucuronide is commonly observed with flavonoid 7-O-glucoside in plants within the Lamiales order, such as Perilla frutescens (dietary herb), Antirrhinum majus (snapdragon flower), and Scutellaria baicalensis (a plant whose dried roots are used in Chinese natural medicine) (Figure 1) (Harborne, 1963; Asen et al., 1972; Subramanian and Nair, 1973; Abdalla et al., 1983; Tomimori et al., 1984; Kawasaki et al., 1988; Yoshida et al., 1993; Zhang et al., 1997; Hirotani et al., 1998; Harborne and Baxter, 1999; Huang et al., 1999; Yamazaki et al., 2003; Bremer et al., 2003; APG II, 2003, Wortley et al., 2005). Baicalin, a flavone 7-O-glucuronide, is predominantly accumulated in Scutellaria roots and has been well studied due to its beneficial effects on human health, such as its anti-allergic, antiviral, anti-inflammatory, and intestinal α-glucosidase inhibitory properties (Shibata and Hattori, 1931; Kida et al., 1982; Nishioka et al., 1998; Gao et al., 1999; Chou et al., 2003). The presence of this specialized flavonoid in Lamiales indicates that their parental plants had evolved a flavonoid 7-O-glucuronosyltransferase (F7GAT), which transfers glucuronic acid from UDPGA of sugar donor to the 7-OH group of flavonoids of sugar acceptor (Figure 2A). The first F7GAT, designated Sb UBGAT, was purified from S. baicalensis (Table 1) (Nagashima et al., 2000). Sb UBGAT specifically uses UDPGA as the sugar donor and glucuronosylates the 7-OH group of flavones with ortho-substituents, such as baicalein and scutellarein. However, flavonoids that lack substituents at the ortho-position of the 7-OH group, such as apigenin, luteolin, chrysin, quercetin, and daidzein, cannot be used as substrates for purified Sb UBGAT (Nagashima et al., 2000). The corresponding gene to Sb UBGAT/UGT88D1 has been predicted; however, it has yet to be certified experimentally.
Figure 1.
Lamiales Plants and Flavone Structures.
(A) Inflorescence of S. laeteviolaceae.
(B) P. frutescens var red (left) and green leaves (right) during the vegetative phase.
(C) Inflorescence of S. indicum. The inset shows the flower.
(D) Inflorescence of A. majus.
(E) Structures of the flavones used in the enzyme assays in this study.
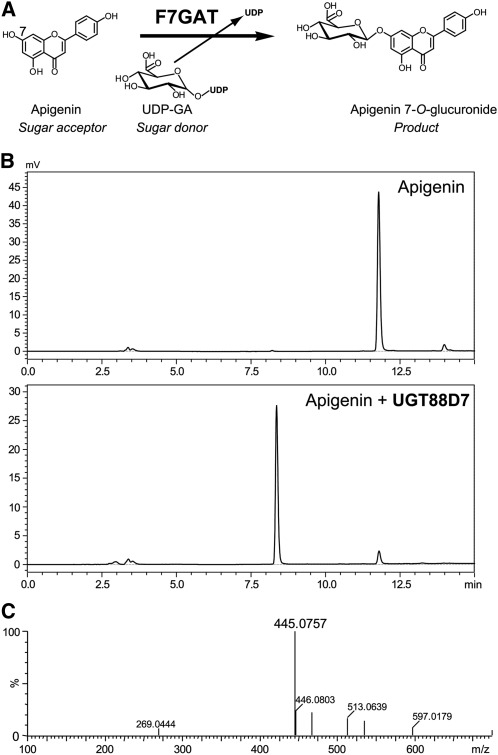
Figure 2.
Enzymatic Activity of P. frutescens F7GAT and UGT88D7.
(A) The reaction catalyzed by Lamiales F7GAT involves conjugation of glucuronic acid from the sugar donor, UDPGA, to the 7-O-position of acceptable flavonoids, such as apigenin.
(B) HPLC chromatograms of the reaction of UGT88D7 (bottom panel) or no enzyme (top panel) with apigenin and UDPGA. Chromatograms show the absorption at 350 nm.
(C) The MS spectrum of the product in the reaction of UGT88D7 with apigenin and UDPGA.
Table 1.
List of Lamiales F7GATs and Their Related UGTs
| Lamiales Order
|
Identity to Sb UBGAT (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Family | Species | Gene Product | UGT | Size (aa)b | MWa (kD) | Origin | Method | Reference | |
| Lamiaceae | S. baicalensis | Sb UBGAT | UGT88D1 | 442 | 48.650 | 100 | Root | Enzyme purification | Nagashima et al. (2000) |
| Lamiaceae | S. laeteviolaceae | Sl UGT1 | UGT88D5 | 455 | 50.156 | 78 | Root | RT-PCR | This work |
| Lamiaceae | P. frutescens | Pf UGT50 | UGT88D7 | 453 | 50.341 | 59 | Mature Leaf | cDNA library screening | This work |
| Lamiaceae | P. frutescens | Pf UGT57 | UGT88A7 | 472 | 51.886 | 41 | Mature Leaf | cDNA library screening | This work |
| Pedaliaceae | S. indicum | Si UGT23 | UGT88D6 | 457 | 50.295 | 60 | Immature seed | cDNA library screening | Noguchi et al. (2008) |
| Scrophulanaceae | A. majus | Am UGTcg10 | UGT88D4 | 457 | 50.566 | 55 | Floral bud | cDNA library screening | Ono et al. (2006) |
| Scrophulanaceae | A. majus | Am C4'GlcT | UGT88D3 | 457 | 50.853 | 53 | Floral bud | cDNA library screening | Ono et al. (2006) |
| Scrophulanaceae | L. vulgaris | Lv C4'GlcT | UGT88D2 | 454 | 50.389 | 50 | Petal | cDNA library screening | Ono and Nakayama (2007) |
MW, molecular weight.
aa, amino acids.
To examine how the sugar donor specificity of UGT might have emerged, we report the molecular identification and in vitro biochemical characterization of F7GATs from four different species of Lamiales. Using a combination of homology modeling and mutational analysis of F7GATs, we identified both a novel functional flavonoid UGT cluster, including Lamiales F7GATs, and also residues that are crucial for recognition of UDPGA. A sugar donor specificity shift of Lamiales F7GATs was achievable by only two amino acid substitutions proximal to the sugar donor in the substrate pocket. Phylogenetic analysis of Lamiales F7GATs suggests that local differentiation of sugar donor specificity often occurred after differentiation of general regiospecificity during the evolution of UGT in plants.
RESULTS
Molecular Cloning of Lamiales F7GAT Genes
A skullcap flower (Scutellaria laeteviolacea) is a popular floriculture phylogenetically related to S. baicalensis, the roots, but not leaves, of which also accumulate baicalin, as observed in the roots of S. baicalensis (Figure 1; see Supplemental Figure 1 online). To obtain candidate cDNAs encoding F7GATs, we performed RT-PCR cloning of cDNAs derived from the roots of S. laeteviolacea. We obtained an amplified fragment that was structurally similar to Sb UBGAT. The complete cDNA, designated Sl UGT1, showed 78% amino acid sequence identity to Sb UBGAT (Table 1).
To further obtain candidate cDNAs of Lamiales F7GATs, we screened 106 plaques of a P. frutescens red leaf cDNA library using a digoxigenin (DIG)-labeled Sl UGT1 probe (Yonekura-Sakakibara et al., 2000). We identified two candidate cDNAs, designated Pf UGT50 and Pf UGT57, that showed 63 and 44% amino acid sequence identity with Sl UGT1, respectively (Table 1). Furthermore, we explored Lamiales F7GAT candidates in silico based on homology with Sb UBGAT. We identified A. majus UGT contig10 (Am UGTcg10) and Sesamum indicum UGT23 (Si UGT23), which were previously isolated in our laboratory (Ono et al., 2006; Noguchi et al., 2008), as Lamiales F7GAT homologs. Am UGTcg10 and Si UGT23 showed 55 and 60% amino acid sequence identity with Sb UBGAT, respectively (Table 1).
As S. baicalensis F7GlcT (BAA83484) has regiospecificity toward the 7-position of the flavonoid in spite of its low primary sequence similarity to Sb UBGAT (Hirotani et al., 2000), we also explored Sb F7GlcT homologs from Lamiales plants for F7GAT. We isolated two cDNAs similar to the Sb F7GlcT gene using a Sb F7GlcT probe from the P. frutescens library. These homologs, designated Pf UGT2 and Pf UGT31, showed 38 and 72% amino acid sequence identity with Sb F7GlcT, respectively. Additionally, using an in silico search, we also determined that previously isolated A. majus UGT21, UGT36, and UGT38 are Lamiales F7GlcT homologs (Ono et al., 2006). A. majus UGT21, UGT36, and UGT38 showed 68, 40, and 39% amino acid sequence identity with S. baicalensis F7GlcT, respectively.
In summary, five F7GAT homologs and five F7GlcT homologs were identified from different species of Lamiales using different strategies. All the Lamiales F7GAT homologs, except Si UGT23, were isolated from each organ in which flavonoid 7-O-glucuronides accumulate (Table 1; see Supplemental Figures 1 and 2 online). The committee responsible for UDP-glucuronosyltransferase nomenclature (http://som.flinders.edu.au/FUSA/ClinPharm/UGT/) assigned Pf UGT57, Am UGTcg10, Sl UGT1, Si UGT23, and Pf UGT50 to UGT88A7, UGT88D4, UGT88D5, UGT88D6, and UGT88D7, respectively. The five F7GlcT homologs of Pf UGT2, Pf UGT31, Am UGT21, Am UGT36, and Am UGT38 correspond to UGT73A7, UGT73A13, UGT73A9, UGT73E2, and UGT73N1, respectively (Mackenzie et al., 1997). These Lamiales UGTs contain a PSPG box, which is a highly conserved domain among plant UGTs that is involved in secondary metabolism. In comparison with Lamiales F7GAT homologs based on the ClustalW multiple alignment, Sb UBGAT appeared to have a relatively short sequence that lacked several N-terminal amino acids (Table 1). To complete the sequence of Sb UBGAT, the lacking N-terminal region was obtained by amplifying the 5′-ends of the Sb UBGAT gene (see Supplemental Figure 3 online).
Characterization of Lamiales F7GATs
To evaluate the biochemical activities of the Lamiales F7GAT homologs, each recombinant protein fused with a His-tag was heterologously expressed under the control of a T7 lac promoter in Escherichia coli and purified using a nickel-affinity column. The purified F7GAT homologs were subjected to an enzymatic assay with possible flavonoid sugar acceptors and phenolic compounds. Subsequently, the reaction mixtures were analyzed using reversed-phase HPLC.
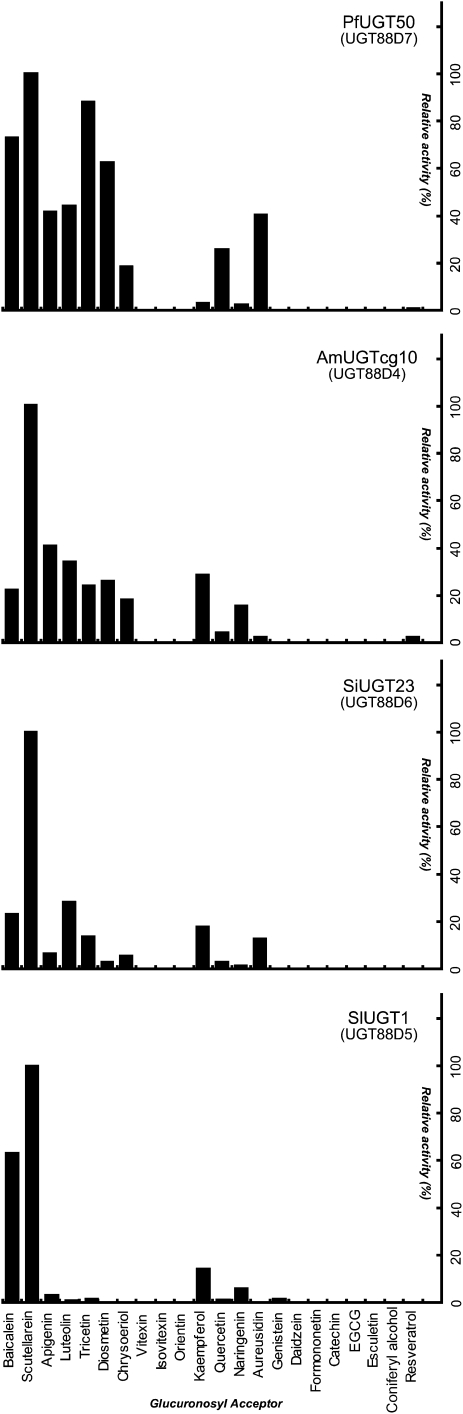
In the reaction with apigenin and UDPGA, the recombinant Pf UGT50/UGT88D7 protein yielded a product that was eluted with the same retention time as authentic apigenin 7-O-glucuronide (Figure 2B; see Supplemental Table 1 online). Liquid chromatography–mass spectrometry (LC-MS) analysis showed that the product exhibited a molecular ion at mass-to-charge ratio of 445.0757 [M − H]−, which was consistent with the mass calculation of apigenin 7-O-glucuronide (C21H17O11, 445.0771, err 1.4 mD) (Figure 2C). No product was observed in the reaction mixture without UDPGA, thus demonstrating that UGT88D7 is capable of glucuronosylation at the 7-position of apigenin. Each of the flavones tested, including scutellarein and baicalein, were good substrates for UGT88D7, showing its broad range of activity for sugar acceptors (Figure 3). Moreover, flavonols and aureusidin also served as substrates for UGT88D7. By contrast, none of the catechins, isoflavones, flavone C-glucosides at the ortho-position of 7-OH, or other phenolic compounds, such as stilbene and coumarin, served as substrates. The kinetic parameters calculated for the flavonoids showed that apigenin and scutellarein are more favorable substrates for UGT88D7 than other flavonoids tested (see Supplemental Table 2 online), consistent with the coaccumulation of their glucuronides and of transcripts of the UGT88D7 gene in leaves (see Supplemental Figure 2 online) (Yamazaki et al., 2003).
Figure 3.
Sugar Acceptor Specificity of Lamiales F7GATs.
The relative activities of Lamiales F7GATs toward 100 μM solutions of flavonoids and phenolic compounds were measured. The glucuronosylating activity toward scutellarein is taken to be 100%.
To determine the sugar donor specificity of UGT88D7, we examined several sugar donors for the catalysis of UGT88D7. UDPGA served as sugar donor, whereas neither UDP-glucose nor UDP-galactose did. These results confirmed that UGT88D7 is a P. frutescens F7GAT (Table 2). By contrast, another Sb UBGAT homolog, Pf UGT57/UGT88A7, was unable to use UDPGA but specifically used UDP-glucose instead (Table 2). In addition, P. frutescens UGT88D7 and UGT88A7 showed differential expression patterns (see Supplemental Figure 2 online).
Table 2.
Sugar Donor Specificity of Lamiales UGTs
| Relative Activity (%)
|
||||
|---|---|---|---|---|
| Name | UGT | UDP-Glc | UDP-Gal | UDP-GA |
| Pf UGT50 | UGT88D7 | 0 | 0 | 100 |
| Pf UGT57 | UGT88A7 | 100 | 0 | 0 |
| Am UGTcg10 | UGT88D4 | 0 | 0 | 100 |
| Sl UGT1 | UGT88D5 | 0 | 0 | 100 |
| Si UGT23 | UGT88D6 | 1 | 0 | 100 |
| At3g16520 | UGT88A1 | 100 | 3 | 2 |
| Am UGT21 | UGT73A9 | 100 | 7 | 2 |
| Am UGT36 | UGT73E2 | 100 | 0 | 0 |
| Am UGT38 | UGT73N1 | 100 | 2 | 0 |
| Pf UGT2 | UGT73A7 | 100 | 6 | 2 |
| Pf UGT31 | UGT73A13 | 100 | 6 | 0 |
| Pf UGT50 | UGT88D7(S127T) | 1 | 0 | 100 |
| UGT88D7(R350W) | 100 | 2 | 32 | |
| UGT88D7(S127T/R350W) | 100 | 3 | 4 | |
The glycosylating activity of each enzyme on three types of sugar donor (UDP-glucose, UGT-galactose, and UDP-glucuronic acid) was tested. Apigenin was used as sugar acceptor for evaluating the sugar donor specificity. Products were quantified based on peak area at A350. The highest activity in the three sugar donors is set as 100%.
Am UGTcg10/UGT88D4 and Si UGT23/UGT88D6 were identified based on their homology with Sb UBGAT; however, they showed higher sequence similarity with P. frutescens UGT88D7 than with Sb UBGAT. Both recombinant enzymes expressed in E. coli were able to catalyze flavonoid 7-O-glucuronosylation with sugar donor specificity exclusively for UDPGA (Table 2). These enzymes also tolerated flavonoids without substitutions at the ortho position of the 7-hydroxy group as sugar acceptors, as was the case for UGT88D7 (Figure 3). The kinetic parameters for the flavonoids differed between the enzymes. The most favorable substrate was apigenin for UGT88D6 in vitro (see Supplemental Table 2 online), which is consistent with the copresence of apigenin 7-O-glucuronide (see Supplemental Figure 1 online) and transcripts of UGT88D6 in S. indicum petals (see Supplemental Figure 2 online). Flavones were more favorable sugar acceptors than flavonols for UGT88D6, which was similar to the sugar acceptor preference of UGT88D7 (see Supplemental Table 2 online). By contrast, kaempferol was kinetically favored by UGT88D4, although flavone glucuronides are major metabolites in A. majus flowers (see Supplemental Figures 1 and 2 online) (Harborne, 1963). These results demonstrate that three Lamiales F7GATs, such as, P. frutescens UGT88D7, S. indicum UGT88D6, and A. majus UGT88D4, have broad specificities for sugar acceptors that are apparently distinct from that of Sb UBGAT.
Based on the highest sequence similarity with Sb UBGAT (Table 1), Sl UGT1/UGT88D5 was most likely the counterpart of Sb UBGAT in S. laeteviolacea. The recombinant UGT88D5 enzyme displayed F7GAT activity specific for baicalein and scutellarein, which are the major flavone aglycons in Scutellaria species (Figure 3; see Supplemental Table 2 and Supplemental Figure 1 online). Remarkably, flavones that lacked a substitution at the ortho position of the 7-hydroxy group, such as apigenin, were not substrates of UGT88D5. This strict specificity for the sugar acceptor was reminiscent of that of Sb UBGAT (Nagashima et al., 2000). The most favorable substrate was scutellarein for UGT88D5, whereas Sb UBGAT acted on baicalein more readily than on scutellarein (Nagashima et al., 2000). Based on their similar primary structures and enzymatic properties, we concluded that UGT88D5 was the S. laeteviolacea F7GAT corresponding to Sb UBGAT. None of the Lamiales F7GATs catalyzed glucuronosylation of the flavone C-glucosides, such as vitexin and orientin (Figure 3). Presumably, the glucose moiety at the ortho position of the 7-OH of flavones would sterically inhibit both sugar conjugation at the 7-position and recognition by F7GAT.
Identification of Residues Crucial to Sugar Donor Specificity of F7GAT
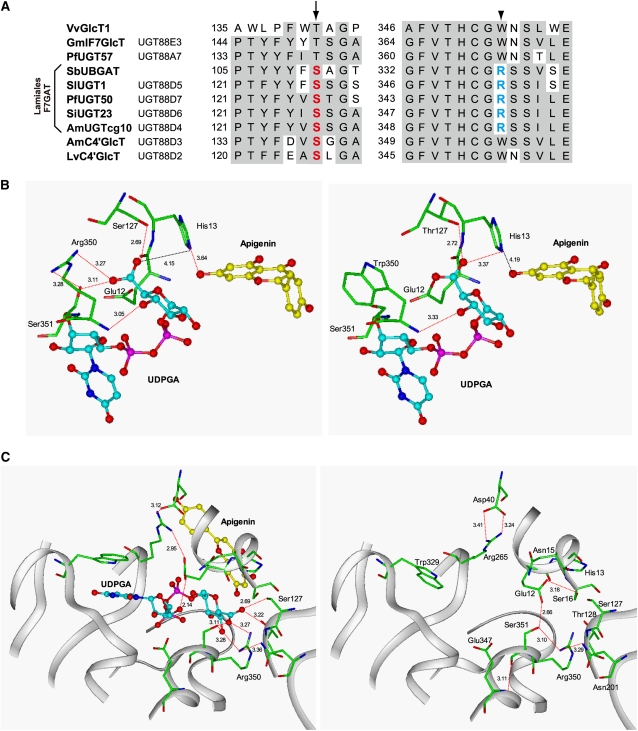
To examine the molecular basis for the sugar donor specificity of plant UGTs, we compared the amino acid sequences of Lamiales F7GATs with those of other F7GlcTs using ClustalW multiple alignment. Interestingly, a highly conserved Trp residue in the center of the PSPG box of the flavonoid GlyTs was replaced with Arg in all Lamiales F7GATs (Figure 4A). The main-chain amide hydrogen of the conserved Trp-353 apparently interacts with the O4 hydroxy group of glucose in crystallized V. vinifera F3GlcT (termed V. vinifera glycosyltransferase 1 [Vv GT1]); however, its role in sugar donor recognition is unknown (Offen et al., 2006). Another striking difference between the F7GlcTs and Lamiales F7GATs is the absence of a conserved Thr residue in the N-terminal domain of Lamiales F7GATs. The relevant Thr-141 in Vv GT1 is thought to participate in the recognition of the sugar donor, allowing an interaction with the O6 hydroxy group of glucose (Offen et al., 2006). Interestingly, all Lamiales F7GATs have a Ser residue at the equivalent position (Figure 4A).
Figure 4.
Homology Docking of UGT88D7 with UDP-GA and Apigenin.
(A) A ClustalW multiple alignment of UGT88 family enzymes and V. vinifera flavonoid 3-O-glucosyltransferase (Vv GT1), the crystal structure of which has been solved (Thompson et al., 1994; Offen et al., 2006). The arrows and arrowheads indicate the positions of the conserved Ser (red) and Arg (blue) residues in the Lamiales F7GATs, respectively.
(B) Homology models of UGT88D7 (left) and the S127T R350W double mutant (right) were constructed using Vv GlcT as a template (pdb code 2C1Z) and were subsequently used in docking experiments with UDPGA and apigenin. Plausible hydrogen bonds are shown by the red dotted lines (distance is within 4 Å). Carbon atoms are colored green for UGT amino acid residues, cyan for UDPGA, and yellow for apigenin. Oxygen atoms are red, nitrogen atoms are blue, and phosphorus atoms are magenta. Dotted lines in red depict the hydrogen bonds, and dotted lines in black depict diagnostic, interatomic distances (longer than 4 Å).
(C) A view of the homology model of UGT88D7 from another angle in the presence (left) or absence (right) of ligands. Ser-16 of UGT88D7 is located on the corresponding position of Arg-25 of Bp UGT94B1
To validate the importance of these unique residues in the sugar donor specificity of F7GAT, mutational analysis of PfUGT50/UGT88D7 was conducted. A UGT88D7-S127T mutant, in which Ser-127 was replaced with a Thr residue, displayed F7GAT activity similar to that of the wild type (Table 2). By contrast, the F7GAT activity of the R350W mutant was drastically compromised, while significant F7GlcT activity was retained. It is noteworthy that the S127T R350W double mutant exhibited negligible F7GAT activity but significant F7GlcT activity. Similarly, Thr and/or Trp residues were also introduced in Si UGT23/UGT88D6 and Am UGTcg10/UGT88D4 at positions corresponding to Ser-127 and Arg-350 of PfUGT50/UGT88D7, respectively. A sugar donor specificity shift was observed in both Lamiales mutants, in which Arg was replaced by Trp, which is consistent with the case of Pf UGT50/UGT88D7 mutants (see Supplemental Table 3 online). This alternative sugar donor specificity observed in these mutants of three Lamiales F7GAT enzymes demonstrated that both the Ser-127 and Arg-350 residues of UGT88D7 are required for recognition of UDPGA. To obtain further insight into the mechanism and importance of Ser-127 and Arg-350 in the sugar donor specificity of UGT88D7, detailed kinetic analyses were conducted (Table 3). The results showed that both the S127T and R350W single substitutions resulted in a decrease of the kcat and kcat/Km values in the glucuronosyl transfer reaction. The effects of the R350W substitution on kcat and kcat/Km values (727- and 239-fold diminutions, respectively) were much greater than those of the S127T substitution (8.8- and 33-fold diminutions, respectively). This illustrates the critical importance of Arg-350 in enabling the enzyme to catalyze the glucuronosyl transfer reaction. The double S127T R350W substitution markedly reduced kcat and kcat/Km (3520- and 3076-fold, respectively) without causing significant changes in Km values for apigenin (1.2-fold decrease) and UDP-GA (3-fold increase). Thus, the S127T substitution appeared to additively enhance the effects of the R350W substitution. By contrast, the double S127T R350W substitution caused an increase in kcat and kcat/Km glucosyl transfer reaction values (7- and 9-fold, respectively). The Km value for UDPGA was only slightly increased (2.6-fold) relative to the wild type, and that for UDP-Glc was somewhat decreased (1.4-fold). Again, R350W, rather than S127T, appeared to be primarily responsible for the observed increase in kcat and kcat/Km values, but simultaneous S127T substitution enhanced the effects of the R350W substitution. Judging from the ratios of the specificity constants [(kcat/Km)UDPGA/(kcat/Km)UDP-Glc], the specificity of UGT88D7 was completely shifted from that of GAT (ratio, 1143 for the wild type) to that of GlcT (ratio, 0.041 for the double mutant) by only two amino acid substitutions.
Table 3.
Kinetics of UGT88D7 Enzymes
| UGT88D7 | Substrate | Km (μM) | kcat (s−1) | kcat/Km (s−1·mM−1) |
|---|---|---|---|---|
| (A) | Donor | |||
| WT | UDP-GA | 11 ± 3 | 8.8 ± 0.5 | 800 |
| S127T | UDP-GA | 2.9 ± 0.2 | 0.27 ± 0.0005 | 91 |
| R350W | UDP-GA | 29 ± 1 | 0.031 ± 0.0005 | 1.1 |
| S127T R350W | UDP-GA | 9.4 ± 1.5 | 0.0025 ± 0.0001 | 0.26 |
| (A) | Donor | |||
| WT | UDP-Glc | 8.5 ± 1.9 | 0.0060 ± 0.0003 | 0.7 |
| S127T | UDP-Glc | 3.3 ± 0.3 | 0.0030 ± 0.00001 | 0.91 |
| R350W | UDP-Glc | 33 ± 5 | 0.13 ± 0.008 | 3.9 |
| S127T R350W | UDP-Glc | 6.7 ± 1.6 | 0.043 ± 0.003 | 6.4 |
| (B) | Acceptor | |||
| WT | Apigenin | 37 ± 2 | 240 | |
| S127T | Apigenin | 29 ± 2 | 9.2 | |
| R350W | Apigenin | 110 ± 10 | 0.29 | |
| S127T R350W | Apigenin | 98 ± 9 | 0.025 | |
| (C) | Acceptor | |||
| WT | Apigenin | 540 ± 20 | 0.011 | |
| S127T | Apigenin | 710 ± 70 | 0.0042 | |
| R350W | Apigenin | 410 ± 80 | 0.31 | |
| S127T R350W | Apigenin | 380 ± 60 | 0.11 |
Kinetic parameters for apigenin (A), UDP-GA (B), and UDP-Glc (C) were determined using wild-type enzymes and S127T, R350W, and S127T R350W mutant enzymes. As described in Methods, the kinetic parameters were calculated by fitting the initial velocity data to the Michaelis-Menten equation by nonlinear regression. In the case of apigenin (A), two types of UDP sugars were tested.
Identification of amino acid residues crucial for the recognition of UDPGA prompted us to examine their roles in GAT catalysis using three-dimensional structural homology modeling of F7GAT using the crystal structure of Vv GT1 as a template. The structural model of UGT88D7 docked with UDPGA and apigenin predicted that the cationic guanidinium moiety of Arg-350 would be in close proximity to the anionic carboxylate of the glucuronic acid moiety of UDPGA. The Arg residue indeed appears to play an important role in determining the sugar donor specificity of F7GAT catalysis (Figures 4B and 4C). The Ser-127 residue was also predicted to form a hydrogen bond with the carboxylate oxygen. The Thr-127 residue in the S127T mutant showed a contribution to the hydrogen bond in a similar manner of Ser-127, but its conformation appears to be slightly different from that of the Ser-127 residue due to the introduction of the methyl group at the Cβ position of the Ser residue. Thus, the conformation of the Thr-127 residue is preferable for a better hydrogen bond with the hydroxy group of the glucose moiety than a Ser residue does in this position.
Molecular Evolution of Lamiales F7GATs
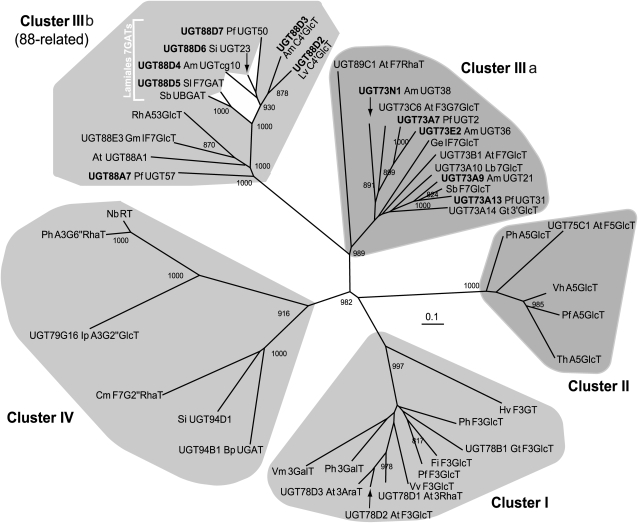
The identification of Lamiales F7GAT genes allowed us to compare the phylogenetic relationships among the F7GATs and other flavonoid UGTs at the amino acid level (Figure 5). The Lamiales F7GATs did not belong to any of previously known flavonoid UGT clusters (I, II, III, and IV). Rather, they formed a new cluster with other UGT88-related enzymes, such as Glycine max isoflavone 7-O-glucosyltransferase (Gm IF7GlcT/UGT88E3) and A. majus chalcone 4′-O-glucosyltransferase (Am C4′GlcT/UGT88D3, i.e., the 7-position of flavonoids corresponds to the 4′-position of chalcones) (Ono et al., 2006; Noguchi et al., 2007; Ono and Nakayama, 2007). P. frutescens UGT88A7 was also a member of this cluster; however, it was distinct from the F7GAT subclade (Figure 5; see Supplemental Data Set 1 online). The phylogenetic topology of UGT88-related genes was further validated using the maximum parsimony method (see Supplemental Figure 4 online). Notably, Lamiales F7GATs was structurally similar to Lamiales/Scrophulanaceae C4′GlcTs. These results reflect the regiospecificity of the sugar acceptor but do not indicate sugar donor specificity, which is consistent with the notion that regiospecificy is the criterion for clustering of flavonoid UGTs.
Figure 5.
A Phylogenetic Tree of Flavonoid UGTs.
An unrooted phylogenetic tree was constructed using TreeView version 1.6.6 software with the neighbor-joining method based on ClustalW multiple alignments of Lamiale F7GATs identified in this study and functionally characterized flavonoid UGTs. Number indicates >800 bootstrap value (1000 replicates). Bar = 0.1 amino acid substitutions per site. Functional clusters (I, II, IIIa, IIIb, and IV) of flavonoid UGTs are shaded gray. The UGT sequences are available in Supplemental Data Set 1 online.
Regarding sugar acceptor specificity, Lamiales 7GATs were further divided into two groups of Scutellaria with narrow specificity and the other Lamiales plants with broad specificity (Figure 3). These two groups were also phylogenetically distinguishable. Unexpectedly, codon usage corresponding to the Arg crucial to UDPGA recognition in the PSPG box differed between the Scutellaria (CGG) and other Lamiales F7GATs (AGG), whereas that for the Ser residue corresponding to the Ser-127 in UGT88D7 was often AGC among Lamiales F7GATs.
All Lamiales UGTs identified as Sb F7GlcT homologs in this study (Pf UGT2/UGT73A7, Pf UGT31/UGT73A13, Am UGT21/UGT73A9, Am UGT36/UGT73E2, and Am UGT38/UGT73N1) showed sugar donor specificity for UDP-glucose (Table 2). As expected, these UGTs were classified as belonging to cluster III, which is regarded as a functional group of UGT catalyzing the 7-O-glycosylation of flavonoids represented by Sb F7GlcT, Arabidopsis UGT73C6 and Allium cepa UGT73J1, although this phylogenetic cluster also contains UGTs involved in nonflavonoid and inducible xenobiotic metabolisms (Vogt and Jones, 2000; Hirotani et al., 2000; Jones et al., 2003; Kramer et al., 2003; Poppenberger et al., 2003, 2005; Achnine et al., 2005; Kim et al., 2006; Brazier-Hicks et al., 2007; Gandia-Herrero et al., 2008). The UGT88-related cluster and cluster III share <30% of their amino acids regardless of their similar region specificity on flavonoids. Considering both their functional similarity and structural diversity, we concluded that the UGT88-related cluster constituted a new functional group of UGTs with regiospecificity for the 7-position of flavonoids and designated this cluster as cluster IIIb of the flavonoid UGTs, whereas the previous cluster III was renamed IIIa (Figure 5).
DISCUSSION
Lamiales includes popular ornamental flowers and dietary herbs, such as snapdragon (A. majus), basil (Ocimum basilicum), and spearmint (Mentha spicata). In this study, we identified four UGT88D enzymes as F7GATs from different Lamiales species using both reverse-genetic and biochemical approaches based on the specialized metabolite. Most recently, we identified F7GAT-like genes from wishbone flower (Scrophulariaceae Torenia hybrid Th F7GATH), in which Ser and Arg residues corresponding to Ser-127 and Arg-350, respectively, in UGT88D7 are conserved (see Supplemental Figure 3 online). Thus, it is expected that the F7GAT gene is widespread among Lamiales plants. By contrast, Arabidopsis (Brassicaceae) is unlikely to possess the F7GAT gene, because no flavonoid 7-O-glucuronide derivatives have been reported in this species (Tohge et al., 2005). Moreover, the singlet UGT88 enzyme in the Arabidopsis genome, UGT88A1, has GlcT activity toward the 7-position of apigenin, but no GAT activity. The sugar acceptors of UGT88A1 in vivo remain unknown (Table 2) (Paquette et al., 2003; Lim et al., 2004). Therefore, F7GAT might have occurred locally in the lineage of specific plants. It should be noted that flavonoid 7-O-glucuronide is a specialized, but not unique, metabolite for Lamiales, since it is also found in the blue pigment complex of blue cornflowers (Asteraceae, Centaurea cyanus) and rye leaves (Poaceae, Secale cereale) (Schulz et al., 1985; Shiono et al., 2005). Therefore, it is of evolutionary interest to clarify whether or not F7GATs of cornflower and rye occurred independently of Lamiales F7GATs, from the chemotaxonomic viewpoint.
Given that glucose is the most common sugar moiety in flavonoid glycosides, and all functionally characterized members of cluster IIIb, including the singlet Arabidopsis UGT88A1, are GlcT (Lim et al., 2004; Ogata et al., 2005; Ono et al., 2006; Noguchi et al., 2007), it is reasonable to propose that extant Lamiales F7GATs differentiated from a parental cluster IIIb F7GlcT via a sugar donor specificity shift from UDP-glucose to UDPGA. The observation that distinct reversion of sugar donor specificity of UGT88D7 can result from only two mutations (i.e., S127T and R350W) supports this idea. It should be noted that A. majus and P. frutescens have multiple copies of UGT88 family genes, unlike Arabidopsis, which indicates that there is local gene duplication of UGT88 family genes in Lamiales after general establishment of regiospecificity of flavonoid UGT. The duplicate copies of the parental UGT88 gene could provide gene sources for subsequent functional differentiation of F7GAT (Ober, 2005).
Mutational analysis of UGT88D7 in vitro followed by homology modeling in silico successfully identified two amino acid residues that are crucial to the sugar donor specificity of UDPGA (Table 3, Figure 4). It is important to note that the Ser residue corresponding to Ser-127 of UGT88D7 is not unique to Lamieales F7GATs but is also present in Linaria vulgaris UGT88D2 and A. majus UGT88D3, which are Lamiales/Scrophulanaceae C4′GlcTs (Table 1, Figure 4). This result is consistent with the observation that S127T per se did not influence sugar donor specificity (Table 2). Codon usage for the Ser residue in Lamiales F7GATs, Lv C4′GlcT, and Am C4′GlcT were AGC, AGC, and AGT, respectively. The codons for the Thr residue used by Pf UGT57, At UGT88A1, and Gm IF7GlcT were ACC, ACC, and ACT, respectively. Thus, the Ser residue can be explained by the C-to-G transversion at the second position of the Thr codon and probably occurred in a common ancestral Lamiales UGT gene of F7GATs and C4′GlcTs prior to the acquisition of the Arg residue (Figure 5). Because S127T showed substantial additive effects on R350W in sugar donor specificity, the Ser residue could facilitate UDPGA recognition of F7GAT. In this context, acquisition of the Ser residue in Lamiales UGT88 enzymes may be regarded as a preadaptation for F7GAT. As another possible explanation, the Ser residue of Lamiales F7GATs and of Lamiales C4′GlcTs may have occurred independently. However, no F7GATs lacking the Ser residue have been found, suggesting that this hypothesis is much less likely. Interestingly, a red daisy cluster IV flavonoid GAT, UGT94B1, also has a Ser residue (Ser-143) corresponding to Ser-127 of UGT88D7 (see Supplemental Figure 3 online) (Sawada et al., 2005). Since Ser-143 is predicted to be able to form an H bond with UDPGA in the homology model (Osmani et al., 2008), it may also be involved in sugar donor specificity in a fashion similar to that proposed for Ser-127 in UGT88D7.
On the other hand, Arg-350, which is a unique residue in the PSPG-box of F7GATs, is the primary residue that determines the sugar donor specificity of F7GAT, as R350W causes a shift in sugar donor specificity from UDPGA to UDP-glucose. The cationic property of the Arg residue could allow for interaction with the anionic glucuronic acid of UDPGA (Figures 4B and 4C). Identification of a crucial Arg residue in the PSPG box of Lamiales F7GATs supports the previous notion that the PSPG box is involved in sugar donor binding.
Importantly, upon the double S127T R350W substitution, the observed change of sugar donor specificity in UGT88D7 arose mainly from changes in the kcat value but not in the Km value (Table 3). Additionally, the occurrence of Arg at position 350 should be of critical importance for enhancing the kcat value in the glucuronosyl transfer reaction (see above). In the model of the UGT88D7-S127T R350W double mutant docked with ligands (Figure 4B), due to the absence of Arg-350, the carboxylate of UDPGA is slightly shifted toward His-13, which corresponds to the catalytically important His-20 of Vv GT1 (Offen et al., 2006; see Supplemental Figure 3 online). This interaction takes His-13 away from the 7-OH group of apigenin and likely impairs the role of His-13. This appears to consistently account for why the R350W substitution results in a significant diminution in the kcat value of F7GAT activity but not in that of F7GlcT activity.
Results on the crucial role of the Arg-350 residue from cluster IIIb Lamiales F7GATs are comparable to those from cluster IV UGT94B1, where, through homology modeling, Arg-25 was predicted to be proximal to the carboxylate of UDPGA and was also demonstrated to be crucial for UDPGA specificity (Osmani et al., 2008, 2009). Although the corresponding Arg residues in UGT94B1 and Lamiales F7GATs are localized at completely different locations, being at the N terminus of UGT94B1 and at the PSPG box of Lamiales F7GATs, based on primary sequences (see Supplemental Figure 3 online), both Arg residues are predicted to be proximal to the carboxylate of UDPGA in each ternary structure model. Moreover, in mammalian GATs, an Arg residue proximal to UDPGA is also considered to be required for GAT activity (Zakim et al., 1983; Ouzzine et al., 2002; Miley et al., 2007). Thus, the results of studies of plant and animal GATs lead to the conclusion that an Arg residue proximal to the carboxylate of UDPGA is the biochemical basis of the GAT enzyme, and acquisition of the Arg residue is the most innovative event permitting differentiation to GAT. Interestingly, there is a single nucleotide polymorphism in the first position of the codon for the Arg residue in Lamiales F7GATs. Scutellaria F7GATs and the other Lamieales F7GATs have AGG and CGG for the Arg residue codons, respectively. The Trp residue that is highly conserved in the PSPG box of flavonoid UGTs is mostly encoded as TGG. Thus, it is likely that independent single nucleotide substitutions (T→A transversion and T→C transition) cause the critical replacement of Trp with Arg in the two Lamiales F7GAT lineages. Alternatively, an additional silent mutation might have occurred after differentiation of a common ancestral F7GAT; this possibility cannot be ruled out.
Identification of the important residues of Lamiales F7GATs for GAT activity allowed us to generate GAT by introducing the Ser and Arg residues in GlcTs of cluster IIIb. However, Pf UGT57(UGT88A7)-T139S W367R and Gm IF7GlcT(UGT88E3)-T150S W371R mutants did not show F7GAT activity. Given a similar result observed in the case of Sorgham bicolor UGT85B1 (Osmani et al., 2008), such unsuccessful GAT engineering indicates that the Arg residue is required but is not sufficient for GAT evolution. This in turn suggests the complexity of the sugar donor specificity of UGT. Considering the importance of an Arg residue proximal to the carboxylate group of UDPGA, putative important residues could also be in the vicinity of the predicted UDP-sugar binding pocket. Further consideration of homology modeling analysis could indicate other surrounding amino acids, some of which may also participate in GAT evolution. Ser-16, Tyr-125, Val-126, Ser-127, Thr-128, Gly-129, Thr-200, Asn-201, Gly-202, Leu-234, Cys-348, Gly-349, Ser-351, Ser-352, Ile-353, Leu-354, Glu-355, Glu-371, Gln-372, Asn-375, and Phe-378 are found to be close to the crucial Arg-350; within 5 Å in the model structure of UGT88D7 (Figure 4C). The multiple alignment of GATs and GlcTs in cluster IIIb showed that Ser-351 is conserved in all GATs, while most GlcTs, except for UGT88D3, have Asn at this position (Figure 4A; see Supplemental Figure 3 online). The other amino acid residues were mostly conserved between GlcT and GAT. Given that Ser-351 interacts with the carboxylate of UDPGA and the guanidinium group of Arg-350, it is also likely to contribute to the stabilization of the interaction between UDPGA and Arg-350 through hydrogen bonding. On the other hand, the bulky Asn residue in this position in UGT88A7-W367R mutants, and other related GlcT mutants, such as Gm IF7GlcT(UGT88E3)-T150S W371R, might sterically disturb the interaction.
It has been reported that the carboxylate group of UDPGA interacts with the guanidinium group of Arg-25 in UGT94B1 (Osmani et al., 2008). The conformation of the side chain in Arg-25 in the structural model of UGT94B1 indicated an extended form, while Arg-350 in UGT88D7 was a folded form. The structural model of UGT88D7 suggests that the folded conformation of Arg-350 is stabilized by interactions with Ser-351 and Asn-201, together with interactions with the carboxylate group of UDPGA. Thus, the side chain of Arg-350 would maintain a folded form during the enzyme reaction. In a ligand-free structural model of UGT88D7 (Figure 4), Ser-351 interacted with Glu-12 instead of the carboxylate of UDPGA, maintaining the hydrogen bond with Arg-350. Glu-12 would thus augment the role of Ser-351 in the stabilization of the folded form of Arg-350 in the absence of ligands. In the presence of ligands, the Glu-12 residue seems to interact with Arg-265, which is also highly conserved in Lamiales F7GATs. In the UGT88A7-T139S W367R mutant, Gly-16 and Asn-368 (corresponding to Glu-12 and Ser-351 in UGT88D7, respectively) might not contribute to the stabilization of the folded conformation of the introduced Arg-367, thereby resulting in unsuccessful GAT engineering. Future reciprocal biochemical studies using mutagenesis of these residues may clarify an evolutional path to GAT.
It should also be noted that the critical Arg-25 of UGT94B1 and Arg-350 of Lamiales F7GATs are unique residues both within and among clusters (Figures 4 and 5). Undoubtedly, Arg in the PSPG box of Lamiales F7GATs and in the N-terminal of UGT94B1 had occurred locally and independently. The positional plasticity of the crucial Arg residue not only highlights a convergent evolution of sugar donor specificity of UGT, but it also suggests that the GAT gene cannot be assigned based solely on the position of the Arg-25 and Arg-350. Thus, it is possible that another Arg residue may be present in unidentified GATs and that it is catalyzing the glucuronosylation of phytochemicals beneficial for human life (for example, the soybean's soyasapogenol, green tea's theasaponin, licorice's glycyrrhizin, and St. John's wort's miquelianin) (Butterweck et al., 2000; Kurosawa et al., 2002; Yoshikawa et al., 2007; Seki et al., 2008). A homology modeling strategy will help identify the crucial residue based on the biochemical basis of GAT.
The high copy number of plant UGTs should facilitate functional differentiation of plant UGTs. It is widely considered that differentiation of sugar donor specificity occurred after differentiation of general regiospecificity because flavonoid UGTs with the same regiospecificity are structurally conserved beyond species, irrespective of sugar donors, as observed in Arabidopsis F3RhaT/UGT78D1, F3GlcT/UGT78D2, and F3AraT/UGT78D3 in cluster I, petunia (Petunia hybrida) F3GalT and F3GlcT in cluster I, Arabidopsis F7GlcT/UGT73C6 and F7RhaT/UGT89C1 in cluster IIIa, and Am C4′GlcT/UGT88D3 and Am F7GAT/UGT88D4 in cluster IIIb (Figure 5) (Miller et al., 1999; Jones et al., 2003; Ono et al., 2006; Yonekura-Sakakibara et al., 2007, 2008). Notably, structural similarity among different sugar UGTs with the same regiospecificity is variable, probably reflecting the different timing of occurrence of new sugar donor specificity in each case. Considering the vast array of specialized phytochemical glycosides, it is not surprising that local differentiation of sugar donor specificity of UGTs occurred often in the plant kingdom. Lamiales F7GAT is one example of local differentiation of the UGT superfamily of enzymes, which is responsible for the chemotaxomoic feature.
In consideration of differential accumulation patterns in flavonoid 7-O-glucuronides and transcripts of F7GAT in Lamiales plants (in addition to diverse sugar acceptor specificities of Lamiales F7GAT, as shown in Figure 3; see Supplemental Figures 1 and 2 online), change would have occurred in both the biochemical activity and the spatial regulation of F7GAT. This is consistent with notions that duplicate genes, especially those involved in secondary metabolism, not only permit a single copy to differentiate functionally but also show elevated gene expression variation, probably due to low genetic constraint (Des Marais and Rausher, 2008; Kliebenstein, 2008). Thus, it is expected that flavonoid 7-O-glucuronides have different ecological/physiological roles in each plant, although it is difficult to understand the ecological/physiological roles of specialized metabolites, especially of the colorless and nonvolatile metabolites. In the cases of sesame and snapdragon F7GAT, products are observed in petals, suggesting that they may be exploited as copigments in flower coloration (Asen et al., 1972).
It is well known that mammalian UGTs efficiently metabolize most phytochemicals in dietary foods to their glucuronides. Recent pharmacological studies of drug metabolism reevaluated the belief that the sole function of glucuronosylation of phytochemicals, such as health-promoting flavonoids and pain-killing morphine, is inactivation of these compounds for excretion. Results of these studies demonstrate that glucuronsylation also impacts both the biological activity and localization of phytochemicals by altering the permeability and stability of aglycons (Kroemer and Klotz, 1992; Juergenliemk et al., 2003; O'Leary et al., 2003; Zhang et al., 2007). However, due to experimental cost and humanity, it is not practical to retrieve glucuronides from animals fed with phytochemicals. Our findings herein provide an efficient in vitro production system of flavonoid glucuronides for further verification of their in vivo effects.
This study on specialized metabolism of Lamiales sheds light on the molecular mechanism underlying the discrimination of sugar donors and offers a partial explanation for the fundamental question of why plants have been able to produce the great variety of specific enzyme functions necessary for the production of a vast array of specialized metabolites. The structure-guided approach should be one of the most powerful strategies for identifying critical residues of enzymes in future work (Zubieta et al., 2001, 2003; Gang et al., 2002; Osmani et al., 2008, 2009). As demonstrated here, collection of natural variants from a specific plant lineage based on specialized metabolites not only helps identification of candidate residues in determining the specificity of enzymes but also elucidates their possible molecular evolutional path. The local differentiation of sugar donor specificity of UGTs is a likely driving force for the large chemical diversity observed in nature.
METHODS
Plant Materials
Perilla frutescens var crispa red form was kindly provided by M. Maeda (Suntory Research Center). Scutellaria laeteviolacea cultivar Yakushimensis was obtained from a flower shop in Muko, Kyoto, Japan. Both plants were grown in a greenhouse under natural conditions. Antirrhinum majus cultivar Snap yellow was purchased at a flower shop in Shimamoto, Osaka, Japan. Sesamum indicum cultivar Masekin and Scutellaria baicalensis were kindly provided by M. Katsuta (National Institute of Crop Science, Ibaragi, Japan) and K. Ishiguro (Mukogawa Women's University, Hyogo, Japan), respectively.
Chemicals
Scutellarin was prepared as previously described (Yoshida et al., 1993). In brief, leaves of P. frutescnes (144.4 g/∼200 leaf blades) were pulverized in liquid N2, and the polyphenols were extracted overnight using 1.5 liters of 50% CH3CN containing 0.1% HCOOH (v/v). The extract was filtered and then concentrated by evaporation. The resulting extract was applied to a 600-mL DIAION CHP-20P column (Mitsubishi Chemical), and step-wise eluted using 600 mL each of 10, 20, 30, 40, and 50% CH3CN after washing with 1.2 liters of water. Due to the large amount of flavonoid glycosides in the 30% fraction, this fraction was further separated using HPLC with a Develosil C30-UG-5 column (20 × 250 mm; Nomura Chemical) and a linear gradient of 18 to 54% (v/v) CH3CN containing 0.1% (v/v) trifluoroacetic acid for 100 min, followed by 54% for 20 min at a flow rate of 6 mL/min. Flavonoids were monitored at A280 using an SPD-M10A photodiode array detector (Shimadzu). Scutellarin (16 mg) was isolated. Scutellarein was prepared by digesting 3 mg of scutellarin wtih β-glucuronidase/arylsulfatase (Roche) in 0.2 M CH3COONa buffer, pH 5.0, at 37°C for 2 h. The reaction mixture was subjected to a Sep-Pack-C18 cartridge (20cc; Waters) to wash out salts and enzymes by 5% (v/v) CH3CN and then eluted by 80% CH3CN containing 0.1% (v/v) trifluoroacetic acid. Finally, 0.4 mg of scutellarein was obtained after the reaction mixture was lyophilized.
Apigenin 7-O-glucuronide and aureusidin were prepared from petals of A. majus (cv butterfly yellow). The structure of apigenin 7-O-glucuronide was confirmed using NMR as described previously (Noguchi et al., 2008; see Supplemental Table 1 online). Tricetin was obtained from petals of a transgenic rose coexpressing Flavonoid 3′,5′-hydoroxylase (F3′5′H) and Flavone synthase (FNS) (Katsumoto et al., 2007; PCT/JP2008/61603). Baicalein, formononetin, resveratrol, UDP-galactose, UDP-glucose, and UDP-glucuronic acid were purchased from Sigma-Aldrich. Genistein and daizein were purchased from Fujicco. Apigenin, luteolin, diosmetin, chrysoeriol, vitexin, isovitexin, orientin, kaempferol, quercetin, naringenin, esculetin, and coniferylalcohol were purchased from Funakoshi. (+)-Catechin and (−)-epigallocatechin gallate (EGCG) were purchased from Nacalai tesque.
Molecular Cloning
Total RNA was extracted from the roots of S. laeteviolacea using an RNeasy plant mini kit (Qiagen). cDNAs were synthesized from 1 μg of total RNA using a first-strand synthesis system for RT-PCR (Invitrogen). To obtain cDNA of Sb UBGAT homologs, PCR with rTaq DNA polymerase (TaKaRa Bio) was run at 94°C for 3 min followed by 30 cycles at 94°C for 1 min, 54°C for 1 min, and 72°C for 2 min using the Sl UGT-Fw and Sl UGT-Rv primers (see Supplemental Table 4 online). An amplified fragment was subcloned into pCR-TOPO II (Invitrogen). Sequencing reactions were done using a BigDye-Terminator ver.3.1 cycle sequencing kit (Applied Biosystems) and then analyzed using a 3100 genetic analyzer (Applied Biosystems). The nucleotide sequence of Sl UGT1 was confirmed to be a Sb UBGAT homolog using ClustalW alignment in MACVECTOR software version 7.2.2 (Accelrys; Thompson et al., 1994).
Both the screening and detection of positive clones were performed with a DIG-DNA labeling and detection kit (Roche). A cDNA library derived from red leaves of P. frutescens that had been constructed previously was used to screen Pf F7GATs (Yonekura-Sakakibara et al., 2000). Approximately106 pfu of the cDNA library was screened with the Sl UGT1 probe, which was DIG labeled using PCR according to the manufacturer's instructions (Roche). Positive clones were detected under low-stringency hybridization conditions as described previously by Yonekura-Sakakibara et al. (2000). After the second screening, positive clones were excised into the pBK-CMV plasmid (Stratagene), and the nucleotide sequences were determined. Candidate cDNAs were selected based on homology with Sb UGAT, as determined using ClustalW alignment and BLASTx homology searches. Because Sl UGT1 had an incomplete open reading frame, the 5′- and 3′-ends were amplified with a GeneRacer kit (Invitrogen) and the following primer sets: GR-Sl UGT-Rv and Sl UGT-nest-Rv for 5′-rapid amplification of cDNA ends (RACE); and GR-Sl UGT-Fw and Sl UGT-nest-Fw for 3′-RACE. Full-length cDNAs of Pf UGT2, Pf UGT31, Pf UGT50, and Pf UGT57 were obtained by this screening. The nucleotide sequences of Am UGTcg10, Am UGT21, Am UGT36, Am UGT38, and Si UGT23 were previously determined and are available in the DNA data bank of Japan (DDBJ) database (Ono et al., 2006; Ono and Nakayama, 2007; Noguchi et al., 2008).
Since the lack of an N-terminal region in Sb UBGAT/UGT88D1 (accession number AB042277) was predicted by multiple alignment, the rapid amplification of 5′-RACE of Sb UBGAT was performed using GR-Sb 7GAT-Rv and Sb 7GAT-nest-Rv primers to obtain the full-length cDNA. The full-length cDNA of SbUBGAT was deposited in the DDBJ database. Th F7GATH-1 and Th F7GATH-2 are Torenia hybrid F7GAT homologs that were obtained by PCR using primers Am UGT88-Fw2 and Am UGT88-Rv3, followed by RACE, using specific primers GR-Th 7GATH-Fw, GR-Th 7GATH-Rv, Th 7GATH-nest-Fw, and Th 7GATH-nest-Rv, as shown in Supplemental Table 4 online.
Heterologous Expression
The coding sequences of Pf UGT50, Pf UGT57, Am UGTcg10, Sl UGT1, Si UGT23, Pf UGT2, Pf UGT31, Am UGT21, Am UGT36, Am UGT38, and At UGT88A1 were amplified by PCR using the specific primer sets shown in Supplemental Table 4 online. The amplified fragments were cloned into a pCR4Blunt-TOPO vector using a kit (Zero Blunt TOPO PCR cloning kit for sequencing; Invitrogen) and sequenced to confirm the absence of PCR errors. Plasmids of Pf UGT50, Am UGTcg10, Pf UGT2, Pf UGT31, Am UGT21, and Am UGT38 cDNAs were digested using NdeI and BclI for Pf UGT50, and NdeI and BamHI for Am UGTcg10, Pf UGT2, Pf UGT31, Si UGT23, Am UGT21, Am UGT38, and At UGT88A1. The resulting DNA fragments were ligated with a pET-15b vector (Novagen) that had previously been digested with NdeI and BamHI. The plasmids of PfGT57 (BamHI), Am UGT36 (BamHI), and SlUGT1 (Ndel) cDNAs were digested, and the resulting DNA fragments were ligated with a pET-15b vector that had previously been digested with NdeI and BamHI, respectively, to obtain the plasmids pET-15b-Pf UGT57 and pET-15b-Sl UGT1. The resultant plasmids were transformed into Escherichia coli BL21(DE3). The transformant cells were precultured at 37°C for 16 h in a Luria-Bertani broth containing 50 μg/mL ampicillin. Twenty milliliters of the preculture was then inoculated into 800 mL of the same Luria-Bertani medium. After cultivation at 37°C until the OD600 reached 0.5, isopropyl 1-R-d-thiogalactoside was added to the broth at a final concentration of 0.4 mM, which was then further cultivated at 22°C for 20 h. All subsequent operations were conducted at 0 to 4°C. The recombinant E. coli cells were harvested by centrifugation (7000g, 15 min), washed with distilled water, and resuspended in buffer A (20 mM sodium Pi, pH 7.4, containing 14 mM 2-mercaptoethanol and 0.5 M NaCl) containing 20 mM imidazole. The cells were disrupted at 4°C by five cycles of ultrasonication (where one cycle corresponds to 10 kHz for 1 min followed by an interval of 1 min). The cell debris was removed by centrifugation (7000g, 15 min). To the supernatant solution, polyethyleneimine was slowly added to a final concentration of 0.12% (v/v). The mixture was allowed to stand at 4°C for 30 min, followed by centrifugation (7000g, 15 min). The supernatant was applied to a HisTrap HP column (1 mL; GE Healthcare Bio-Science) that had been equilibrated with buffer A containing 20 mM imidazole. The column was washed with buffer A containing 20 mM imidazole, and the enzyme was eluted with buffer A containing 200 mM imidazole. The active column-bound fractions were concentrated and desalted using Vivaspin 30,000 MWCO (Vivascience), followed by substitution with buffer B (20 mM potassium Pi, pH 7.5, containing 14 mM 2-mercaptoethanol). The protein concentration was determined using the Bradford method (Bradford, 1976) with BSA as a standard. SDS-PAGE was performed according to the method of Laemmli (1970), and the proteins in the gels were visualized using Coomassie Brilliant Blue R 250 (see Supplemental Figure 5 online).
Site-Directed Mutagenesis
In vitro mutagenesis of the Pf UGT50, Pf UGT57, Si UGT23, Am UGTcg10, and Gm IF7GlcT genes was performed using recombinant PCR with the pET-15b vector containing each wild-type cDNA as the template and specific mutagenesis oligonucleotide primers (see Supplemental Table 4 online) to obtain the following site-directed mutants: Pf UGT50 (S127T, R350W, and the S127T R350W double mutant), Pf UGT57 (T139S, W367R, and the T139S W367R double mutant), Si UGT23 (S127T, R354W, and the S127T R354W double mutant), Am UGTcg10 (S127T, R355W, and the S127T R355W double mutant), and Gm IF7GlcT (T150S, W371R, and the T150S W371R double mutant). The amplified fragments were digested with BstBI and SacI for Pf UGT50 and its mutants, NdeI and BamHI for Si UGT23, Am UGTcg10, Gm IF7GlcT, and their mutants, and BstBI and SalI for Pf UGT57 and its mutants, and the resulting DNA fragments were ligated with pET-15b as described above. Individual mutations were verified by DNA sequencing of both strands.
Enzyme Assays and Kinetics
The standard reaction mixture (50 μL) consisted of 100 μM glycosyl acceptor, 2 mM glycosyl donor, 50 mM potassium phosphate buffer, pH 7.5, and enzyme. After a 10-min preincubation of the mixture without the enzyme at 30°C, the reaction was initiated by addition of the enzyme. After incubation at 30°C for 30 min, the reaction was stopped by the addition of 50 μL of CH3CN containing 0.5% (v/v) trifluoroacetic acid. The substrates and glycosylated products (except for esculetin, aureusidin, and their glycosides) were analyzed using reversed-phase HPLC on a Develosil C30-UG-5 column (4.6 × 150 mm; Nomura Chemical) with a linear gradient of 4.5 to 90% (v/v) CH3CN containing 0.1% (v/v) trifluoroacetic acid for 18 min at a flow rate of 1 mL/min. Esculetin, aureusidin, and the glycosides were separated using a linear gradient of 4.5 to 45% (v/v) CH3CN containing 0.1% (v/v) trifluoroacetic acid for 20 min at a flow rate of 1 mL/min. Naringenin, genistein, daidzein, formononetin, and their glycosides were detected at 280 nm, and the others were detected at 350 nm.
To determine the initial velocity of F7GATs, the assays were performed under steady state conditions using the standard assay system (see above) with various substrate concentrations. The apparent Km and Vmax values for the glucuronosyl donor and acceptor substrates in the presence of a saturating concentration of the counter substrate were determined by fitting the initial velocity data to the Michaelis-Menten equation using nonlinear regression analysis (Segel, 1975; Leatherbarrow, 1990).
LC-MS
LC-MS analysis was performed using a Q-TOF Premier mass spectrometer (Micromass) outfitted with an electrospray ion source operated in the V-Optics negative mode. A Develosil C30-UG-3 column (i.d. 2.0 × 150 mm, Nomura Chemical). was used for LC on a Shimadzu LC-20AD HPLC with a ternary solvent system composed of water (A), acetonitrile (B), and 10% formic acid in water (C). Each sample was eluted using a linear gradient of 50 to 90% solvent B in solvent A for 20 min at a flow rate of 0.2 mL min−1 and then was eluted with 90% solvent B for 5 min. The flow of solvent C was kept constant at 1%. The column eluent was coupled to the Lockspray electrospray ion source. In the reference channel of the Lockspray ion source, a 0.6 ng/μL solution of leucine enkephalin in 50% acetonitrile and 0.1% HCOOH was continuously delivered using a Jasco PV-2085 Plus semi-micro HPLC pump system at a flow rate of 10 μL/min to provide a lock mass ion at a mass-to-charge ratio of 554.2615. A 3-kV capillary voltage and a 58.0-kV cone voltage were applied for the production of the requisite molecular (precursor) ions in a negative ionization mode.
LC analysis (for Supplemental Figure 1 online) was performed as below. Flavonoids were extracted from pulverized plant tissue (1 g fresh weight) with 10 mL of 50% CH3CN. The resulting extract was filtered with Millex-LH (Millipore, and was further separated using HPLC with a Shim-pack FC-ODS column (i.d. 4.6 × 150 mm; Shimadzu). and a linear gradient of 18 to 63% (v/v) CH3CN containing 0.1% (v/v) trifluoroacetic acid for 10 min, followed by 63% for 6 min at a flow rate of 0.6 mL/min. Flavonoids were monitored at A280 or A330 using an SPD-M20A photodiode array detector ranging from 250 to 400 nm (Shimadzu). In this condition, baicalin and apigenin 7-O-glucuronide are eluted at retention times of 8.33 and 7.44 min, respectively. As described in Methods, the procedure for MS analysis was performed in negative mode.
Homology Modeling
The crystal structure of Vv GT1 (pdb code 2C1Z) was used to construct a homology model of F7GAT using the Homology module installed in the Insight II molecular modeling system (Molecular Simulations). The glucose portion of the UDP-glucose bound in Vv GT1 was modified to the glucuronic acid moiety and was replaced with glucuronic acid in the binding site of the F7GAT model. Apigenin was docked into the substrate binding site, corresponding to the substrate (kaempferol) in Vv GT1. The conformational space for the Arg-350 residue was examined by rotating the side chain and was found at the site that is occupied by water molecules in the crystal structure. After covering the initial model structure of F7GAT with water molecules of 10 Å thick, additional structure optimization was performed using the molecular mechanics program, Discover3 (Molecular Simulations). The final complex structure showed that Arg-350 is located close to the carboxylate oxygen atom of the glucuronic acid portion of UDPGA; hydrogen bonds are formed there.
Phylogenetic Analysis
As shown in Figure 6, an unrooted tree was constructed using TreeView version 1.6.6 software (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) based on the ClustalW multiple alignment using the neighbor-joining method with the following settings: MATRIX blosum, GAPOPEN 10.0, GAPEXT 0.2, GAPDIST 8, MAXDIV 40, and 1000 bootstrap counts (Thompson et al., 1994).
RT-PCR
RT-PCR for Supplemental Figure 2A online was performed on cDNAs prepared from each organ of P. frutescens cv Aochirimen using specific primer sets (Pf 50-Fw and -Rv for Pf UGT50/UGT88D7, Pf 57-Fw and -Rv for Pf UGT57/UGT88A7, and Pf rRNA-Fw and -Rv for Pf rRNA, which are listed in Supplemental Table 4 online). Each PCR product was separated on a 1% agarose gel and detected by ethidium bromide staining. RT-PCR for Supplemental Figure 2B online was performed on cDNAs prepared from each organ of S. indicum cv Masekin using specific primer sets for Si UGT23/UGT88D6 (Si UGT23-Fw and -Rv). The primer set for Si rRNA, the definition of sesame seed stages, and the RT-PCR procedure were described in our previous work (Noguchi et al., 2008; Supplemental Data online). RT-PCR followed by DNA gel blotting for Supplemental Figure 2C online was performed on cDNAs prepared from each organ of A. majus cultivar butterfly yellow using specific primer sets for Am UGTcg10/UGT88D4 (Am UGTcg10-Fw and -Rv). The primer set for Am GAPDH, definition of snapdragon flower stages, and RT-PCR procedure were described in our previous work (Morita et al., 2005; Ono et al., 2006).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL/DDBJ databases under the following accession numbers: UGT88A7 (Pf UGT57), AB362992; UGT88D4 (Am UGTcg10), AB362988; UGT88D5 (Sl UGT1), AB362989; UGT88D6 (Si UGT23), AB362990; UGT88D7 (Pf UGT50), AB362991; UGT73A9 (Am UGT21), AB362993; UGT73A7 (Pf UGT2), AB362994; UGT73A13 (Pf UGT31), AB362995; UGT73E2 (Am UGT36), AB371297; UGT73N1 (Am UGT38), AB371298; Th F7GATH-1, AB477350; Th F7GATH-2, AB477351; full-length Sb UBGAT (UGT88D1), AB479151; Pf rRNA, AB185210; Si rRNA, AJ236041; and Am GAPDH, X59517. Vv GT1 structural information is available in Protein Data Bank (PDB) (pdb code 2C1Z).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Metabolite Profiles of Lamiales Plants.
Supplemental Figure 2. Expression Profile of Three Lamiales F7GAT Genes (Pf UGT50, Si UGT23, and Am UGTcg10).
Supplemental Figure 3. Amino Acid Alignment of UGT88-Related Enzymes.
Supplemental Figure 4. Evolutionary Relationships of Cluster IIIb of Flavonoid UGTs.
Supplemental Figure 5. SDS-PAGE after Affinity Purification of His-Tagged Lamiales F7GAT Proteins Expressed in E. coli.
Supplemental Table 1. NMR of Apigenin 7-O-Glucuronide
Supplemental Table 2. Kinetic Data of Lamiales F7GATs.
Supplemental Table 3. Sugar Donor Specificity of Si UGT23 and Am UGTcg10.
Supplemental Table 4. Primer Sequences.
Supplemental Data Set 1. Amino Acid Sequences of UGTs Used to Generate the Phylogenetic Tree in Figure 5.
Supplementary Material
Acknowledgments
We thank P.I. Mackenzie (Flinders University, Australia) for assigning UGT numbers and Y. Honma (Tohoku University), N. Tateishi, M. Nakao (Suntory), J. Murata (SUNBOR), S. Ikushiro (Toyama Prefectural University, Toyama, Japan), K. Ishiguro (Mukogawa Women's University, Hyogo, Japan), and M. Mizutani (Kobe University, Hyogo, Japan) for the experimental materials and productive discussions related to this study. We also thank M. Maeda, N. Kasajima, A. Ohgaki, K. Iwasa, A. Saito, and H. Toyonaga (Suntory) for their excellent technical support with the experimental procedures.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Eiichiro Ono (eiichiro_ono@suntory.co.jp).
Online version contains Web-only data.
References
- Abdalla, M.F., Saleh, N.A.M., Gabr, S., Abu-Eyta, A.M., and El-Said, H. (1983). Flavone glycosides of Salvia triloba. Phytochemistry 22 2057–2060. [Google Scholar]
- Achnine, L., Huhman, D.V., Farag, M.A., Sumner, L.W., Blount, J.W., and Dixon, R.A. (2005). Genomics-based selection and functional characterization of triterpene glycosyltransfererases from the model legume Medicago truncatula. Plant J. 41 875–887. [DOI] [PubMed] [Google Scholar]
- APG II (2003). An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG II. Bot. J. Linn. Soc. 141 399–436. [Google Scholar]
- Asen, S., Norris, K.H., and Stewart, R.N. (1972). Copigment of aurone and flavone from petals of Antirrhinum majus. Phytochemistry 11 2739–2741. [Google Scholar]
- Bowles, D., Isayenkova, J., Lim, E.-K., and Poppenberger, B. (2005). Glycosyltransferases: Managers of small molecules. Curr. Opin. Plant Biol. 8 254–263. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Brazier-Hicks, M., Offen, W.A., Gershater, M.C., Revett, T.J., Lim, E.-K., Bowles, D.J., Davies, G.J., and Edwards, R. (2007). Characterization and engineering of the bifunctional N- and O-glucosyltransferase involved in xenobiotic metabolism in plants. Proc. Natl. Acad. Sci. USA 104 20238–20243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer, B., Bremer, K., Chase, M.W., Reveal, J.L., Soltis, D.E., Soltis, P.S., and Stevens, P.F. (2003). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot. J. Linn. Soc. 141 399–436. [Google Scholar]
- Butterweck, V., Juergenliemk, G., Nahrstedt, A., and Winterhoff, H. (2000). Flavonoids from Hypericum perforatum show antidepressant activity in the forced swimming test. Planta Med. 66 3–6. [DOI] [PubMed] [Google Scholar]
- Chou, T.-C., Chang, L.-P., Li, C.-Y., Wong, C.-S., and Yang, S.-P. (2003). The anti-inflammatory and analgesic effects of baicalin in carrageenan-evoked thermal hyperalgesia. Anesth. Analg. 97 1724–1729. [DOI] [PubMed] [Google Scholar]
- Des Marais, D.L., and Rausher, M.D. (2008). Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature 454 762–765. [DOI] [PubMed] [Google Scholar]
- Ford, C.M., Boss, P.K., and Hoj, P.B. (1998). Cloning and characterization of Vitis vinifera UDP-glucose:flavonoid 3-O-glucosyltransferas, a homolog of the enzyme encoded by the maize Bronze-1 locus that may primarily serve to glucosylate anthocyanindins in vivo. J. Biol. Chem. 273 9224–9233. [DOI] [PubMed] [Google Scholar]
- Fukuchi-Mizutani, M., Okuhara, H., Fukui, Y., Nakao, M., Katsumoto, Y., Yonekura-Sakakibara, K., Kusumi, T., and Tanaka, Y. (2003). Biochemical and molecular characterization of a novel UDP-glucose:anthocyanin 3′-O-glucosyltransferase, a key enzyme for blue anthocyanin biosynthesis, from gentian. Plant Physiol. 132 1652–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon, C.M., Langlois-Meurinne, M., and Saindrenan, P. (2005). Plant secondary metabolism glycosyltransferases: The emerging functional analysis. Trends Plant Sci. 10 542–549. [DOI] [PubMed] [Google Scholar]
- Gandia-Herrero, F., Lorenz, A., Larson, T., Graham, I.A., Bowles, D.J., Rylott, E.L., and Bruce, N.C. (2008). Detoxification of the explosive 2,4,6-trinitrotoluene in Arabidopsis: Discovery of bifunctional O- and C-glucosyltransferases. Plant J. 56 963–974. [DOI] [PubMed] [Google Scholar]
- Gang, D.R., Lavid, N., Zubieta, C., Chen, F., Beuerle, T., Lewinsohn, E., Noel, J.P., and Pichersky, E. (2002). Characterization of phenylpropene O-methyltransferases from sweet basil: Facile change of substrate specificity and convergent evolution within a plant O-methyltransferase family. Plant Cell 14 505–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Z., Huang, K., Yang, X., and Xu, H. (1999). Free radical scavenging and antioxidant activities of flavonoids extracted from the radix of Scutellaria baicalensis Georgi. Biochim. Biophys. Acta 1472 643–650. [DOI] [PubMed] [Google Scholar]
- Gershenzon, J., and Dudareva, N. (2007). The function of terpene natural products in the natural world. Nat. Chem. Biol. 3 408–414. [DOI] [PubMed] [Google Scholar]
- Greenhagen, B.T., O'Maille, P.E., Noel, J.P., and Chappell, J. (2006). Identifying and manipulating structural determinates linking catalytic specificities in terpene synthases. Proc. Natl. Acad. Sci. USA 103 9826–9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborne, J.B. (1963). Plant polyphenols. X. Flavone and aurone glycosides of Antirrhinum. Phytochemistry 2 327–334. [Google Scholar]
- Harborne, J.B., and Baxter, H. (1999). The Handbook of Natural Flavonoids, Vol. 2. (New York: John Wiley & Sons).
- Hirotani, M., Kuroda, R., Suzuki, H., and Yoshikawa, T. (2000). Cloning and expression of UDP-glucose:flavonoid 7-O-glucosyltransferase from hairy root cultures of Scutellaria baicalensis. Planta 210 1006–1013. [DOI] [PubMed] [Google Scholar]
- Hirotani, M., Nagashima, S., and Yoshikawa, T. (1998). Baicalin and baicalein productions of cultured Scutellaria baicalensis cells. Nat. Med. 52 440–443. [Google Scholar]
- Huang, Y., De Bruyne, T., Apers, S., Ma, Y., Claeys, M., Pieters, L., and Vlietinck, A. (1999). Flavonoid glucuronides from Picria fel-terrae. Phytochemistry 52 1701–1703. [Google Scholar]
- Jones, P., Messner, B., Nakajima, J., Schaffner, A.R., and Saito, K. (2003). UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J. Biol. Chem. 278 43910–43918. [DOI] [PubMed] [Google Scholar]
- Juergenliemk, G., Boje, K., Huewel, S., Lohmann, C., Galla, H-J., and Nahrstedt, A. (2003). In Vitro studies indicate that miquelianin (quercetin 3-O-β-D-glucuronopyranoside) is able to reach the CNS from the small intestine. Plant. Med. 69 1013–1017. [DOI] [PubMed] [Google Scholar]
- Katsumoto, Y., et al. (2007). Engineering of the rose flavonoid biosynthetic pathway successfully generated blue-hued flowers accumulating delphinidin. Plant Cell Physiol. 48 1589–1600. [DOI] [PubMed] [Google Scholar]
- Kawasaki, M., Hayashi, T., Arisawa, M., Morita, N., and Berganza, L.H. (1988). 8-Hydroxytricetin 7-glucuronide, a β-glucuronidase inhibitor from Scoparia dulcis. Phytochemistry 27 3709–3711. [Google Scholar]
- Kida, A., Nishi, K., Nagai, H., Matsuura, N., and Tsuchiya, H. (1982). Anti-allergic actions of crude drugs and blended Chinese traditional medicines. Effects on type I and type IV. Yakugaku Zasshi 80 31–41. [PubMed]
- King, C.D., Rios, G.R., Green, M.D., and Tephly, T.R. (2000). UDP-glucuronosyltransferases. Curr. Drug Metab. 1 143–161. [DOI] [PubMed] [Google Scholar]
- Kim, J.H., Kim, B.G., Park, Y., Ko, J.H., Lim, C.E., Lim, J., Lim, Y., and Ahn, J.-H. (2006). Characterization of flavonoid 7-O-glucosyltransferase from Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 70 1471–1477. [DOI] [PubMed] [Google Scholar]
- Kliebenstein, D.J. (2008). A role for gene duplication and natural variation of gene expression in the evolution of metabolism. PLoS One 3 e1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeduka, T., Louie, G.V., Orlova, I., Kish, C.M., Ibdah, M., Wilkerson, C.G., Bowman, M.E., Baiga, T.J., Noel, J.P., Dudareva, N., and Pichersky, E. (2008). The multiple phenylpropene synthases in both Clarkia breweri and Petunia hybrida represent two distinct protein lineages. Plant J. 54 362–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, C.M., Prata, R.T.N., Willits, M.G., Luca, V.D., Steffens, J.C., and Gracer, G. (2003). Cloning and regiospecificity studies of two flavonoid glucosyltransferases from Allium cepa. Phytochemistry 64 1069–1076. [DOI] [PubMed] [Google Scholar]
- Kroemer, H.K., and Klotz, U. (1992). Glucuronidation of drugs. Clin. Pharmacokinet. 23 292–310. [DOI] [PubMed] [Google Scholar]
- Kubo, A., Arai, Y., Nagashima, S., and Yoshikawa, T. (2004). Alteration of sugar donor specificities of plant glycosyltransferases by a single point mutation. Arch. Biochem. Biophys. 429 198–203. [DOI] [PubMed] [Google Scholar]
- Kurosawa, Y., Takahara, H., and Shiraiwa, M. (2002). UDP-glucuronic acid:soyasapogenol glucuronosyltransferase involved in saponin biosynthesis in germinating soybean seeds. Planta 215 620–629. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Leatherbarrow, R.J. (1990). Using linear and non-linear regression to fit biochemical data. Trends Biochem. Sci. 15 455–458. [DOI] [PubMed] [Google Scholar]
- Li, Y., Baldauf, S., Lim, E.-K., and Bowles, D.J. (2001). Phylogenic analysis of the UDP-glycosyltranserase multigene family of Arabidopsis thaliana. J. Biol. Chem. 276 4338–4343. [DOI] [PubMed] [Google Scholar]
- Lim, E.-K., Ashford, D.A., Hou, B., Jackson, R.G., and Bowles, D.J. (2004). Arabidopsis glycosyltransferases as biocatalysts in fermentation for resioselective synthesis of diverse quercetin glucosides. Biotechnol. Bioeng. 87 623–631. [DOI] [PubMed] [Google Scholar]
- Lim, E.-K., Baldauf, S., Li, Y., Elias, L., Worrall, D., Spencer, S.P., Jackson, R.G., Taguchi, G., Ross, J., and Bowles, D.J. (2003). Evolution of substrate recognition across a multigene family of glycosyltransferases in Arabidopsis. Glycobiology 13 139–145. [DOI] [PubMed] [Google Scholar]
- Lim, E.-K., and Bowles, D.J. (2004). A class of plant glycosyltransferases involved in cellular homeostasis. EMBO J. 23 2915–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macías, F.A., Galindo, J.L., and Galindo, J.C. (2007). Evolution and current status of ecological phytochemistry. Phytochemistry 68 2917–2936. [DOI] [PubMed] [Google Scholar]
- Mackenzie, P., Little, J.M., and Radominska-Pandya, A. (2003). Glucosidation of hyodeoxycholic acid by UDP-glucuronosyltransferase 2B7. Biochem. Pharmacol. 65 417–421. [DOI] [PubMed] [Google Scholar]
- Mackenzie, P.I., et al. (1997). The UDP glycosyltransferase gene superfamily: Recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 7 255–269. [DOI] [PubMed] [Google Scholar]
- Miley, M.J., Zielinska, A.K., Keenan, J.E., Bratton, S.M., Radominska-Pandya, A., and Redinbo, M.R. (2007). Crystal structure of the cofactor-binding domain of the human phase II drug-metabolisi enzyme UDP-glucuronosyltransferase 2B7. J. Mol. Biol. 369 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, K.D., Guyon, V., Evans, J.N., Shuttleworth, W.A., and Taylor, L.P. (1999). Purification, cloning, and heterologous expression of a catalytically efficient flavonol 3-O-galactosyltransferase expressed in the male gametophyte of Petunia hybrida. J. Biol. Chem. 274 34011–34019. [DOI] [PubMed] [Google Scholar]
- Morita, Y., Hoshino, A., Kikuchi, Y., Okuhara, H., Ono, E., Tanaka, Y., Fukui, Y., Saito, N., Nitasaka, E., Noguchi, H., and Iida, S. (2005). Japanese morning glory dusky mutants displaying reddish-brown or purplish-gray flowers are deficient in a novel glycosylation enzyme for anthocyanin biosynthesis, UDP-glucose:anthocyanidin 3-O-glucoside-2''-O-glucosyltransferase, due to 4-bp insertions in the gene. Plant J. 42 353–363. [DOI] [PubMed] [Google Scholar]
- Nagashima, S., Hirotani, M., and Yoshikawa, T. (2000). Purification and characterization of UDP-glucuronate:baicalein 7-O-glucuronosyltransferase from Scutellaria baicalensis Georgi. cell suspension cultures. Phytochemistry 53 533–538. [DOI] [PubMed] [Google Scholar]
- Nagashima, S., Inagaki, R., Kubo, A., Hirotani, M., and Yoshikawa, T. (2004). cDNA cloning and expression of isoflavonoid-specific glucosyltransferase from Glycyrrhiza echinata cell-suspension cultures. Planta 218 456–459. [DOI] [PubMed] [Google Scholar]
- Nishioka, T., Kawabata, J., and Aoyama, Y. (1998). Baicalein, an α-glucosidase inhibitor from Scutellaria baicalensis. J. Nat. Prod. 61 1413–1415. [DOI] [PubMed] [Google Scholar]
- Noguchi, A., Fukui, Y., Iuchi-Okada, A., Kakutani, S., Satake, H., Iwashita, T., Nakao, M., Umezawa, T., and Ono, E. (2008). Sequential glucosylation of a furofuran lignan, (+)-sesaminol by Sesamum indicum UGT71A9 and UGT94D1 glucosyltransferases. Plant J. 54 415–427. [DOI] [PubMed] [Google Scholar]
- Noguchi, A., Saito, A., Homma, Y., Nakao, M., Sasaki, N., Nishino, T., Takahashi, S., and Nakayama, T. (2007). A UDP-glucose:isoflavone 7-O-glucoyaltransferase from the roots of soybean (Glycine max) seedling. J. Biol. Chem. 282 23581–23590. [DOI] [PubMed] [Google Scholar]
- Ober, D. (2005). Seeing double: Gene duplication and diversification in plant secondary metabolism. Trends Plant Sci. 10 444–449. [DOI] [PubMed] [Google Scholar]
- Offen, W., Martinez-Fleites, C., Yang, M., Lim, E.-K., Davis, B.G., Tarling, C.A., Ford, C.M., Bowles, D.J., and Davies, G.J. (2006). Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 25 1396–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata, J., Kanno, Y., Itoh, Y., Tsugawa, H., and Suzuki, M. (2005). Anthocyanin biosynthesis in roses. Nature 435 757–758. [DOI] [PubMed] [Google Scholar]
- O'Leary, K.A., Day, A.J., Needs, P.W., Mellon, F.A., O'Brien, N.M., and Williamson, G. (2003). Metabolism of Quercetin-7- and Quercetin-3-glucuronides by an in vitro hepatic model: The role of human β-glucuronidase, sulfotransferase, catechol-O-methyltransferase and multi-resistant protein 2 (MRP2) in flavonoid metabolism. Biochem. Pharmacol. 65 479–491. [DOI] [PubMed] [Google Scholar]
- O'Maille, P.E., Malone, A., Dellas, N., Andes Hess, B., Jr., Smentek, L., Sheehan, I., Greenhagen, B.T., Chappell, J., Manning, G., and Noel, J.P. (2008). Quantitative exploration of the catalytic landscape separating divergent plant sesquiterpene synthases. Nat. Chem. Biol. 4 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, E., Fukuchi-Mizutani, M., Nakamura, N., Fukui, Y., Yonekura-Sakakibara, K., Yamaguchi, M., Nakayama, T., Tanaka, T., Kusumi, T., and Tanaka, Y. (2006). Yellow flowers generated by expression of the aurone biosynthetic pathway. Proc. Natl. Acad. Sci. USA 103 11075–11080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, E., and Nakayama, T. (2007). Molecular breeding of novel yellow flowers by engineering the aurone biosynthetic pathway. Transgen. Plant J. 1 66–80. [Google Scholar]
- Osmani, S., Bak, S., Imberty, A., Olsen, C.E., and Møller, B.L. (2008). Catalytic key anmino acids and UDP-sugar donor specificity of a plant glucuronosyl Transferase, UGT94B1. Plant Physiol. 148 1295–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani, S.A., Bak, S., and Møller, B.L. (2009). Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry 70 325–347. [DOI] [PubMed] [Google Scholar]
- Ouzzine, M., Gulberti, S., Levoin, N., Netter, P., Magdalou, J., and Fournel-Gigleux, S. (2002). The donor substrate specificity of the human β1,3-glucuronosyltransferase I toward UDP-glucuronic acid is determined by two crucial histidine and arginine residues. J. Biol. Chem. 277 25439–25445. [DOI] [PubMed] [Google Scholar]
- Paquette, S., Moller, B.L., and Bak, S. (2003). On the origin of family 1 plant glycosyltransferases. Phytochemistry 62 399–413. [DOI] [PubMed] [Google Scholar]
- Pichersky, E., and Gang, D.R. (2000). Genetics and biochemistry of secondary metabolites in plants: An evolutionary perspective. Trends Plant Sci. 5 439–445. [DOI] [PubMed] [Google Scholar]
- Poppenberger, B., Berthiller, F., Lucyshyn, D., Sieberer, T., Schuhmacher, R., Krska, R., Kuchler, K., Glössl, J., Luschnig, C., and Adam, G. (2003). Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J. Biol. Chem. 278 47905–47914. [DOI] [PubMed] [Google Scholar]
- Poppenberger, B., Fujiokam, S., Soenom, K., Georgem, G.L., Vaistij, F.E., Hiranuma, S., Seto, H., Takatsuto, S., Adam, G., Yoshida, S., and Bowles, D. (2005). The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc. Natl. Acad. Sci. USA 102 15253–15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, J., Li, Y., Lim, E-K., and Bowles, D. J. (2001). Higher plant glycosyltransferases. Genome Biol. 2 reviews 3004.1–3004.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada, S., Suzuki, H., Ichimaida, F., Yamaguchi, M.A., Iwashita, T., Fukui, Y., Hemmi, H., Nishino, T., and Nakayama, T. (2005). UDP-glucuronic acid:anthocyanin glucuronosyltransferase from red daisy (Bellis perennis) flowers. Enzymology and phylogenetics of a novel glucuronosyltransferase involved in flower pigment biosynthesis. J. Biol. Chem. 280 899–906. [DOI] [PubMed] [Google Scholar]
- Schulz, M., Strack, D., Weissenböck, G., Markham, K.R., Dellamonica, G., and Chopin, J. (1985). Two luteolin O-glucuronides from primary leaves of Secale cereal. Phytochemistry 24 343–345. [Google Scholar]
- Segel, I.H. (1975). Enzyme Kinetics. Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. (New York: John Wiley & Sons).
- Seki, H., Ohyama, K., Sawai, S., Mizutani, M., Ohnishi, T., Sudo, H., Akashi, T., Aoki, T., Saito, K., and Muranaka, T. (2008). Licorice beta-amyrin 11-oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc. Natl. Acad. Sci. USA 105 14204–14209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senafi, S.B., Clarke, D.J., and Burchell, B. (1994). Investigation of the substrate specificity of a cloned expressed human bilirubin UDP-glucuronosyltransferase: UDP-sugar specificity and involvement in steroid and xenobiotic glucuronidation. Biochem. J. 303 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, H., He, X., Achnine, L., Blount, J.W., Dixon, R.A., and Wang, X. (2005). Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell 17 3141–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, S., and Hattori, S. (1931). Über die konstitution des baicalein und des wogonin. Yakugaku Zasshi 51 15–17. [Google Scholar]
- Shiono, M., Matsugaki, M., and Takeda, K. (2005). Structure of the blue cornflower pigment. Nature 436 791. [DOI] [PubMed] [Google Scholar]
- Subramanian, S.S., and Nair, A.G.R. (1973). Scutellarin and hispidulin-7-O-glucuronide from the leaves of Clerodendrum indicum and Clerodendron infortunatum. Phytochemistry 12 1195. [Google Scholar]
- The French-Italian Public Consortium for Grapevine Genome Characterization (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449 463–468. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge, T., et al. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42 218–235. [DOI] [PubMed] [Google Scholar]
- Tomimori, T., Miyaichi, Y., Imoto, Y., Kizu, H., and Tanabe, Y. (1984). Studies on the constituents of Scutellaria species III. On the flavonoid constituents of the root of Scutellaria baicalensis GEORGI. Yakugaku Zasshi 104 524–528. [DOI] [PubMed] [Google Scholar]
- Vogt, T., and Jones, P. (2000). Glycosyltransferases in plant natural product synthesis: Characterization of a supergene family. Trends Plant Sci. 5 380–386. [DOI] [PubMed] [Google Scholar]
- Wortley, A.H., Rudall, P.J., Harris, D.J., and Scotland, R.W. (2005). How much data are needed to resolve a difficult phylogeny? Case study in Lamiales. Syst. Biol. 54 697–709. [DOI] [PubMed] [Google Scholar]
- Yamazaki, M., Gong, Z., Fukuchi-Mizutani, M., Fukui, Y., Tanaka, Y., Kusumi, T., and Saito, K. (1999). Molecular cloning and biochemical characterization of a novel anthocyanin 5-O-glucosyltransferase by mRNA differential display for plant forms regarding anthocyanin. J. Biol. Chem. 274 7405–7411. [DOI] [PubMed] [Google Scholar]
- Yamazaki, M., Nakajima, J., Yamanashi, M., Sugiyama, M., Makita, Y., Springob, K., Awazuhara, M., and Saito, K. (2003). Metabolomics and differential gene expression in anthocyanin chemo-varietal forms of Perilla frutescens. Phytochemistry 62 987–995. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara, K., Tanaka, Y., Fukuchi-Mizutani, M., Fujiwara, H., Fukui, Y., Ashikari, T., Murakami, Y., Yamaguchi, M., and Kusumi, T. (2000). Molecular and biochemical characterization of a novel hydroxycinnamoyl-CoA: anthocyanin 3-O-glucoside-6”-O-acyltransferase from Perilla frutescens. Plant Cell Physiol. 41 495–502. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara, K., Tohge, T., Matsuda, F., Nakabayashi, R., Takayama, H., Niida, R., Watanabe-Takahashi, A., Inoue, E., and Saito, K. (2008). Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20 2160–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara, K., Tohge, T., Niida, R., and Saito, K. (2007). Identification of a flavonol 7-O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. J. Biol. Chem. 282 14932–14941. [DOI] [PubMed] [Google Scholar]
- Yoshida, K., Kameda, K., and Kondo, T. (1993). Digulucuronoflavones from purple leaves of Perilla ocimoides. Phytochemistry 33 917–919. [DOI] [PubMed] [Google Scholar]
- Yoshikawa, M., Morikawa, T., Nakamura, S., Li, N., Li, X., and Matsuda, H. (2007). Bioactive saponins and glycosides. XXV. Acylated oleanane-type triterpene saponins from the seeds of tea plant (Camellia sinensis). Chem. Pharm. Bull. (Tokyo) 55 57–63. [DOI] [PubMed] [Google Scholar]
- Zakim, D., Hochman, Y., and Kenny, W.C. (1983). Evidence for an active site arginine in UDP-glucuronosyltransferase. J. Biol. Chem. 258 6430–6434. [PubMed] [Google Scholar]
- Zhang, L., Lin, G., Kouacs, B., Jani, M., Krajcsi, P., and Zuo, Z. (2007). Mechanistic study on the intestinal absorption and disposition of baicalein. Eur. J. Pharm. Sci. 31 221–231. [DOI] [PubMed] [Google Scholar]
- Zhang, Y.-Y., Guo, Y.-Z., Ageta, H., Harigaya, Y., Onda, M., Hashimoto, K., Ikeya, Y., Okada, M., and Maruno, M. (1997). Studies on the constituents of roots of Scutellaria planipes. Planta Med. 63 536–539. [DOI] [PubMed] [Google Scholar]
- Zubieta, C., He, X.Z., Dixon, R.A., and Noel, J.P. (2001). Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat. Struct. Biol. 8 271–279. [DOI] [PubMed] [Google Scholar]
- Zubieta, C., Ross, J.R., Koscheski, P., Yang, Y., Pichersky, E., and Noel, J.P. (2003). Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell 15 1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.