Abstract
The phytohormone gibberellin (GA) has long been known to regulate the growth, development, and life cycle progression of flowering plants. However, the molecular GA-GID1-DELLA mechanism that enables plants to respond to GA has only recently been discovered. In addition, studies published in the last few years have highlighted previously unsuspected roles for the GA-GID1-DELLA mechanism in regulating growth response to environmental variables. Here, we review these advances within a general plant biology context and speculate on the answers to some remaining questions. We also discuss the hypothesis that the GA-GID1-DELLA mechanism enables flowering plants to maintain transient growth arrest, giving them the flexibility to survive periods of adversity.
INTRODUCTION
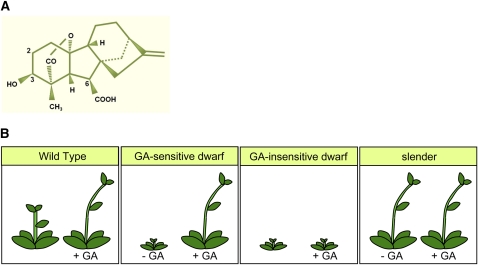
The gibberellins (GAs) are diterpenoid compounds found in plants, fungi, and bacteria, of which only a few actively regulate plant growth (are bioactive) (see http://www.plant-hormones.info/gibberellins.htm; Figure 1A). There has been much recent progress in understanding of the regulation of the biosynthesis and inactivation of bioactive GAs (reviewed in Yamaguchi, 2008). The GAs are derived from a basic diterpenoid carboxylic acid skeleton (Figure 1A). Among features crucial to bioactivity are the hydroxyl group on C3 and the carboxyl group on C6 (Figure 1A), lack of which cause loss of activity. In addition, hydroxylation on C2 causes inactivation and is an important mechanism for growth regulation in angiosperms (Yamaguchi, 2008). The slight structural differences between bioactive and inactive GAs is indicative of the tight fit of bioactive GAs in a specific pocket feature of the GA receptor. This has important implications that are explored further below.
Figure 1.
GA Structure and Response Mutant Categories.
(A) The structure of a bioactive GA (GA4), showing carboxylic (C6) and hydroxyl (C3) groups that are essential for biological activity, and the C2 site, hydroxylation of which abolishes biological activity.
(B) Schematic representation of plants in GA-related mutant categories. Normal (wild type) plants respond to exogenous GA (+GA) by increased growth. GA-sensitive dwarf mutants are GA-deficient (–GA) and grow in response to exogenous GA. GA-insensitive dwarf mutants do not grow in response to exogenous GA. Finally, slender mutant growth mimics that of GA-treated normal plants, even when additional mutations or chemical growth retardants cause GA deficiency.
The importance of GAs to angiosperm growth regulation is exemplified by the phenotype of GA-deficient mutants. For example, the GA-deficient Arabidopsis thaliana ga1-3 mutant lacks ent-kaurene synthetase A, an enzyme in the GA biosynthesis pathway, and this mutant exhibits a characteristic severe dwarf phenotype. In addition, ga1-3 mutant seed do not germinate; ga1-3 mutant shoots bear leaves that are shorter and darker green than the wild type and flower late in long-day photoperiods (and not at all in short-day photoperiods); and ga1-3 mutant flowers exhibit impaired petal and stamen development and are male sterile. All of these aspects of the GA-deficient phenotype can be corrected by exogenous GA (reviewed in Richards et al., 2001). Mutants such as ga1-3 are GA-sensitive dwarf mutants (Figure 1B) that are known in a number of different plant species and typically carry recessive mutations that reduce the activity of GA biosynthesis enzymes (Yamaguchi, 2008).
The molecular characterization of various GA response mutants led to the discovery of the GID1 and DELLA proteins, key components of the molecular GA-GID1-DELLA mechanism that enables plants to respond to GA. Here, we trace the history of these discoveries and discuss recent developments that point to a fundamental role for the GA-GID1-DELLA mechanism in regulating the growth response of flowering plants to environmental variables.
GA-OPPOSABLE PLANT GROWTH INHIBITORS
There are several categories of GA response mutants. Initial interest in such mutants was provoked by the possibility that they might provide clues as to how plant cells perceive and respond to the GA signal. For example, the phenotype of GA-insensitive dwarf mutants is not restored to normal by GA treatments (Figure 1B). Genetic studies of gibberellic acid-insensitive (gai; Arabidopsis), D8 (maize [Zea mays]), and Rht-B1b/Rht-D1b (wheat [Triticum aestivum]) mutants (Koornneef et al., 1985; Harberd and Freeling, 1989; Peng and Harberd, 1993; Peng et al., 1997, 1999a) identified common properties: the various mutant alleles involved act in a genetically dominant fashion and encode active (altered function) mutant products that confer dwarfism and reduced GA response. Further groupings of recessive (rather than dominant) mutations conferring GA-insensitive dwarfism have more recently been characterized (e.g., McGinnis et al., 2003; Sasaki et al., 2003; Ueguchi-Tanaka et al., 2005). Finally, recessive slender mutants exhibit exaggerated growth: tall and slim, they resemble wild-type plants treated with saturating levels of GA and remain tall even when GA deficient (e.g., Potts et al., 1985; Figure 1B).
Studies of some of the above categories of mutant enabled formal genetic definition of the mechanism via which GA promotes growth, long before the molecular basis of this mechanism was apparent. First, analyses of pea (Pisum sativum) slender mutants led to the proposal that GA works as an “inhibitor of an inhibitor” (Brian, 1957). This proposal was based on the observation that slender mutants grow tall irrespective of GA content (e.g., Potts et al., 1985). According to the inhibitor of an inhibitor hypothesis, plants contain an endogenous factor that inhibits growth, and GA promotes growth by overcoming this factor. Lack of the endogenous growth-inhibiting factor in slender mutants causes them to grow tall even when GA deficient. Second, genetic analysis of GA-insensitive maize dwarfing mutations led to an elaboration of the inhibitor of an inhibitor hypothesis. According to this elaboration, dominant GA-insensitive dwarfing mutations confer mutant forms of the growth-inhibiting factor that retain the capacity to inhibit growth but have lost the capacity to be overcome by GA (Harberd and Freeling, 1989). Thus, the recessive slender versus dominant GA-insensitive dwarf mutants could be seen as opposing mutational faces of the same coin: alternative loss-of-function or altered function outcomes of mutational change to an endogenous plant growth inhibitory factor whose molecular identity remained unknown.
As described below, molecular identification of the genes affected in GA-insensitive dwarf and slender mutants enabled the discovery of three of the major components of what is now known as the GA-GID1-DELLA mechanism of GA response regulation: the DELLA growth inhibitors; the F-box protein component of an E3 ubiquitin ligase that specifically targets the DELLAs for destruction in the proteasome in response to GA; and the GID1 GA receptor.
MOLECULAR DISCOVERY OF GA-OPPOSABLE PLANT GROWTH INHIBITORS: THE DELLA PROTEINS
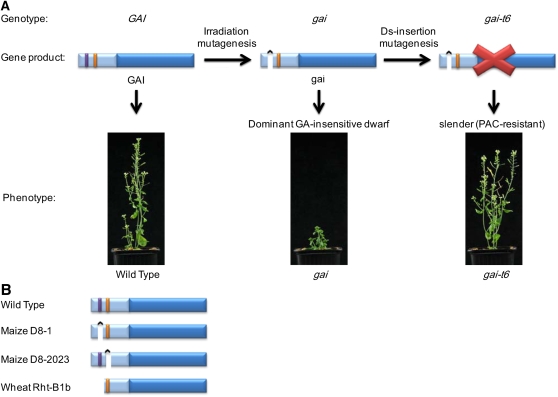
Discovery of the molecular identity of the endogenous plant GA-opposable growth inhibitory factor resulted from the molecular cloning of genes encoding what are now known as the DELLA proteins (or DELLAs), beginning with GAI. The Arabidopsis gai mutation confers dominant, GA-insensitive dwarfism (Koornneef et al., 1985; Peng and Harberd, 1993). An insertional mutagenesis approach enabled the molecular cloning of gai via isolation of a Ds transposon inactivated allele (gai-t6; Peng et al., 1997; Figure 2A). Initial analysis of the amino acid sequence of GAI gave little insight into how this protein might regulate GA response. However, approximately two-thirds of the GAI molecule (at the C-terminal end) was found to be related in sequence to the transcriptional regulator SCARECROW (SCR), suggesting that GAI might also be a transcriptional regulator (Peng et al., 1997). More intriguing findings came from the comparison of the DNA sequences of the GAI and mutant gai alleles. This comparison showed that the gai open reading frame carries a small in-frame deletion mutation and thus encodes an altered product, a mutant gai protein that lacks a 17–amino acid segment (now known as the DELLA domain, named after its first five amino acids; Peng et al., 1997; Figure 2A). This finding was particularly significant because it identified the molecular basis of the altered function previously ascribed to the products of dominant GA-insensitive dwarfing alleles. It is the lack of a functional DELLA domain in gai that causes the reduced GA response of gai mutant plants. Further experiments showed that the gai-t6 (loss-of-function) allele confers partial resistance to the dwarfing effects of the GA biosynthesis inhibitor paclobutrazol (PAC; Peng et al., 1997), a characteristic property of slender alleles (e.g., Potts et al., 1985). Thus, lack of GAI (in gai-t6) causes PAC resistance (slender phenotype), while lack of the DELLA domain (in gai) causes a constitutively active mutant growth inhibitor (gai) whose genetically dominant action can no longer be opposed by GA (Figure 2A).
Figure 2.
Wild-Type and Mutant DELLAs.
(A) The Arabidopsis mutant gai allele confers dominant GA-insensitive dwarfism and was derived from the wild-type allele (GAI) via irradiation mutagenesis (see Koornneef et al., 1985; Peng et al., 1997). gai encodes a mutant gai protein that lacks the DELLA domain (purple section in the GAI protein). It is the lack of the DELLA domain that causes the altered function of gai, making it a constitutive growth inhibitor whose activity is not opposed by GA. Subsequent Ds insertion mutagenesis experiments yielded the gai-t6 allele. This knockout allele does not encode a functional GAI (or gai) protein and confers a tall, slender (PAC-resistant) phenotype.
(B) The mutant maize D8-1, D8-2023, and wheat Rht-B1b proteins confer dominant GA-insensitive dwarfism. The maize and wheat genomes encode DELLA proteins (d8 and Rht-B1a, respectively). Maize mutant alleles encode mutant D8-1 (lacks the DELLA domain, domain I, in purple), mutant D8-2023 (lacks the TVHYNPS domain, domain II, in orange), and wheat mutant Rht-B1b lacks domain I (see also Peng et al., 1999a).
Arabidopsis RGA was identified by loss-of-function mutations conferring partial suppression of the ga1-3 phenotype (a characteristic property of slender alleles; Silverstone et al., 1997) and shown to encode a protein (RGA) closely related to GAI (Silverstone et al., 1998). Subsequently, it became clear that the Arabidopsis genome contains three further GAI/RGA-related genes: RGL1, RGL2, and RGL3 (e.g., Lee et al., 2002). The gene products GAI, RGA, RGL1, RGL2, and RGL3 comprise the full Arabidopsis complement of what are now known collectively as the DELLAs.
These findings, together with further analysis, confirmed that the DELLAs are indeed the previously inferred endogenous GA-opposable plant growth inhibitors. For example, the lack of such inhibitors would be predicted to suppress the GA-deficient mutant phenotype of ga1-3. Accordingly, lack of GAI and RGA suppresses the dwarfed shoot phenotype normally conferred by ga1-3 (Dill and Sun, 2001; King et al., 2001), lack of RGL2 permits GA-independent germination of ga1-3 seed (Lee et al., 2002; Tyler et al., 2004), while lack of RGA, RGL1, and RGL2 permits normal stamen and petal growth in ga1-3 flowers (Cheng et al., 2004; Tyler et al., 2004). In effect, progressive reduction of DELLA function tends toward complete suppression of all visible GA-deficient phenotypes exhibited by ga1-3 (Cheng et al., 2004; Tyler et al., 2004). In addition, the Arabidopsis DELLAs display both overlapping (e.g., GAI and RGA in stem elongation) and relatively discrete (e.g., RGL2 in seed germination) GA response regulation functions. The correspondence between DELLAs and the inferred GA-opposable growth inhibitor was confirmed by the demonstration that slender mutants in species other than Arabidopsis carry loss-of-function mutations in GAI/RGA orthologs (e.g., Ikeda et al., 2001; Chandler et al., 2002) and that La and Cry, mutant alleles of which confer the pea slender phenotype upon which the original inhibitor of an inhibitor hypothesis was based (Brian, 1957), correspond to DELLA-encoding genes (Weston et al., 2008).
The molecular characterization of the Arabidopsis gai mutation highlighted the importance of the DELLA domain (Figure 2A; Peng et al., 1997). This importance was further emphasized in studies of the GA insensitivity of dwarfing mutants of maize (D8-1; D8-2023) and wheat (Rht-B1b). As with gai, the GA insensitivity of these mutants is conferred by mutant DELLAs that lack functional conserved N-terminal domains, termed DELLA (domain I) or TVHYNPS (domain II) (Figure 2B; Peng et al., 1999a). Further studies identified domain I or domain II mutations in GA-insensitive barley (Hordeum vulgare) and rice (Oryza sativa) mutants (Chandler et al., 2002; Asano et al., 2009). In addition, deletion of domain I of Arabidopsis RGA (as conferred by the rga-Δ17 transgene) results in a mutant RGA protein (rga-Δ17) that, similarly to gai, confers GA-insensitive dwarfism (Dill et al., 2001). Thus, in a general sense, mutation of domain I or II of DELLAs can prevent GA opposability and confers GA-insensitive dwarfism. We will return to the biological functions of domains I and II later in this review.
The wheat Rht-B1b (and Rht-D1b) alleles are important in modern agriculture, in that they confer the increased grain yields of the green revolution varieties containing them (Peng et al., 1999a). In attempts to further expand the agricultural utility of domain I mutant DELLAs, the Arabidopsis gai gene was shown to confer dwarfism when expressed in basmati rice (Peng et al., 1999a; Fu et al., 2001), and a switchable form of gai was developed that can be used to induce growth restraint flexibly as and when required (Ait-ali et al., 2003).
In summary, wild-type DELLAs were identified as GA-opposable plant growth inhibitors that can be specifically mutated (in domains I and/or II) to become growth inhibitors that are resistant to the effects of GA. However, the mechanism via which GA overcomes the growth inhibitory effects of wild-type DELLAs was unknown.
GA PROMOTES PROTEASOME-DEPENDENT DESTRUCTION OF DELLAs
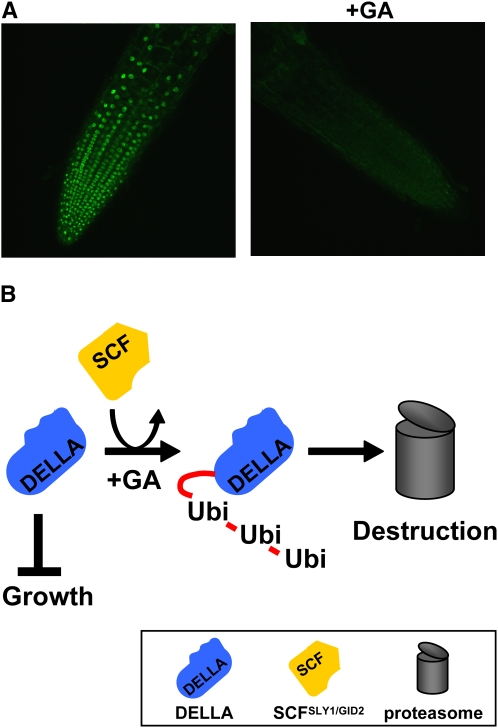
The stage was now set for in-depth molecular analyses of how GA overcomes the growth inhibiting effects of the DELLAs. The first breakthrough came from studies of DELLAs fused to the green fluorescent protein (GFP), in particular GFP-RGA. GFP-RGA was shown to accumulate in root cell nuclei and to disappear from those nuclei within a few hours of GA treatment (Silverstone et al., 2001; Figure 3A). Further studies showed that GA causes disappearance of cereal DELLAs (e.g., barley SLN1 and rice SLR1; Gubler et al., 2002; Itoh et al., 2002). These observations suggest that GA overcomes the growth inhibitory effects of DELLAs by promoting their disappearance. Intriguingly, GA does not promote the disappearance of DELLA proteins lacking domain I (Dill et al., 2001; Fu et al., 2004), thus showing that these mutant proteins, whose growth-inhibiting properties are resistant to GA, are also resistant to GA-promoted disappearance.
Figure 3.
GA Promotes the Proteasome-Dependent Destruction of DELLAs.
(A) GA treatment causes disappearance of GFP-RGA from Arabidopsis root cell nuclei. Right, GA-treated (4 h) pRGA:GFP-RGA roots; left, control (see also Silverstone et al., 2001).
(B) Schematic representation of the DELLA-SCFSLY1/GID2 interaction. The DELLA protein (blue) inhibits growth. GA stimulates an interaction between the DELLA protein and the E3 ubiquitin ligase SCFSLY1/GID2, resulting in polyubiquitination of DELLA. Polyubiquitination targets the DELLA protein for destruction by the proteasome.
How then does GA cause the disappearance of DELLAs? One possibility was that DELLAs are destroyed by the proteasome (Sullivan et al., 2003). Initial experiments showed that the specific proteasome inhibitor MG132 inhibits the GA-promoted disappearance of the barley DELLA SLN1 (Fu et al., 2002). Targeting of proteins to the proteasome occurs via polyubiquitination: the attachment of a polymeric chain consisting of ubiquitin protein residues. Polyubiquitination is achieved by E3 ubiquitin ligases, among which is the so-called SCF complex class (Sullivan et al., 2003). It is the F-box protein component of the SCF complex that provides specificity, with each F-box protein having specific affinity for a particular set of target proteins. Interaction between SCF and the target protein promotes polyubiquitination and subsequent destruction of the target by the proteasome. Molecular genetic analysis of GA-insensitive dwarf mutants identified an F-box protein (rice GID2/Arabidopsis SLY1) that is part of a DELLA-interacting E3 ubiquitin ligase that interacts with a C-terminal region of the DELLA protein (McGinnis et al., 2003; Sasaki et al., 2003; Dill et al., 2004; Fu et al., 2004) and that targets DELLAs for destruction by the proteasome (Figure 3B).
These discoveries enabled a molecular understanding of how GA overcomes DELLA function and hence promotes growth: the growth-inhibiting DELLAs are targeted for destruction by the proteasome in response to GA following polyubiquitination by a specific E3 ubiquitin ligase of the SCF class (SCFSLY1/GID2), thus relieving DELLA-mediated growth inhibition. Nevertheless, the question of how GA promotes the DELLA- SCFSLY1/GID2 interaction remained, and it was only with the discovery of the GA receptor that it became clear how this occurs.
THE GA RECEPTOR INTERACTS WITH DOMAINS I AND II OF THE DELLA PROTEIN
The discovery of the GA receptor (Ueguchi-Tanaka et al., 2005) was a major advance in understanding of the GA-GID1-DELLA mechanism. A rice gene (GID1) initially identified by loss-of-function gid1 alleles (conferring GA-insensitive dwarfism) was shown to encode a nuclear GA receptor protein (GID1; Ueguchi-Tanaka et al., 2005). GID1 is localized (at least predominantly) in the nucleus and acts there as a soluble GA receptor, having high affinity for bioactive GAs and low or nonexistent affinity for inactive GAs. Remarkably, GID1 binds specifically with the rice DELLA SLR1 when both proteins are expressed in yeast in the presence of bioactive GA. Unlike the case in Arabidopsis, whose genome encodes multiple DELLAs, SLR1 is the sole DELLA protein encoded by the rice genome. Accordingly, lack of SLR1 (in a slr1-1 loss-of-function mutant) suppresses the dwarfism conferred by a gid1 loss-of-function allele, and SLR1 accumulates in the gid1-1 loss-of-function mutant. Taken together, these observations suggested that GA potentiates an in planta interaction between the GID1 GA receptor and DELLAs and that this interaction stimulates the GA-dependent destruction of DELLAs, thus promoting growth (Ueguchi-Tanaka et al., 2005).
Genes encoding multiple GID1-related GA receptors were discovered in Arabidopsis, and mutants multiply homozygous for loss-of-function mutations in these genes were shown to exhibit GA-insensitive dwarfism (Griffiths et al., 2006; Iuchi et al., 2007; Willige et al., 2007). Further analysis showed that the GID1–DELLA interaction specifically involves the conserved N-terminal domains I and II of the DELLA protein (Peng et al., 1999a; Griffiths et al., 2006; Ueguchi-Tanaka et al., 2007; Willige et al., 2007; Feng et al., 2008), thus finally explaining why mutant DELLA proteins lacking these domains confer GA insensitivity. In addition, the GA-stimulated interaction between GID1 and DELLA itself increases the affinity of DELLA for the SCFSLY1/GID2 E3 ubiquitin ligase (Griffiths et al., 2006), presumably causing increased polyubiquitination of DELLA, subsequent destruction of DELLA by the proteasome, and the promotion of growth.
Recent crystal structure studies have deepened our understanding of the molecular spatial relationships governing the interactions between GID1, GA, and DELLA. Essentially, the GID1 protein possesses a central pocket that accommodates bioactive GA. Polar groups in the bioactive GA molecule interact directly with GID1 and with water molecules via hydrogen bonding, resulting in tight specificity of fit of bioactive GA within the pocket. For example, the C3-hydroxyl group (Figure 1A), which is essential for GA activity, is hydrogen bonded to a specific Tyr residue in the internal surface of the GID1 pocket via a bridging water molecule (Murase et al., 2008; Shimada et al., 2008). Conversely, addition of a 2-hydroxyl group, an in planta mechanism for the inactivation of bioactive GAs in angiosperms (Yamaguchi, 2008), is predicted to cause steric interference and conformational changes that will not favor receptor binding (Murase et al., 2008).
Binding of GA causes an allosteric change to GID1, which results in the N terminus forming a lid to the pocket (Murase et al., 2008; Shimada et al., 2008). Once in place, the upper surface of the lid binds with the DELLA protein, specifically with the N-terminal region defined by domains I and II (Murase et al., 2008). The formation of the GA-GID1-DELLA complex is thought to induce a conformational change in a C-terminal domain of the DELLA protein that stimulates substrate recognition by the SCFSLY1/GID2 E3 ubiquitin ligase, proteasomic destruction of DELLA, and the consequent promotion of growth.
Intriguing recent evidence suggests that overcoming DELLA-mediated growth inhibition does not equate solely with the destruction of DELLA. Experiments using mutant plants lacking the F-box component of the SCFSLY1/GID2 E3 ubiquitin ligase have indicated that the GA-dependent formation of the GID1-DELLA complex itself (independently of subsequent protein destruction) reduces the growth-repressing effects of the DELLAs (Ariizumi et al., 2008; Ueguchi-Tanaka et al., 2008). Given that DELLAs inhibit growth by interacting with specific growth-promoting transcription factors (see below), it is likely that formation of the GID1-DELLA complex reduces the availability of DELLAs for interaction with those transcription factors, thus reducing the resultant growth inhibitory effects. Thus, the GA-stimulated GID1–DELLA interaction inherently reduces DELLA-mediated growth repression as well as stimulating DELLA destruction.
HOW DELLAs INHIBIT PLANT GROWTH
The above advances define the molecular mechanism via which GA opposes the growth inhibitory properties of the DELLAs, but they do not explain how DELLAs inhibit growth. Because the C-terminal domain of DELLAs is closely related to that of SCR and other transcriptional regulators, it seemed possible that the DELLAs associate with DNA. However, there are no reports of direct DELLA–DNA associations. In addition, the moderate degree of promoter enrichment obtained in chromatin immunoprecipitation studies (Zentella et al., 2007) suggested that the association of RGA with gene promoters might involve additional transcriptional regulators.
Further advances in understanding of how DELLAs inhibit growth have come from studies of plant light responses. Plant growth is strongly influenced by the light environment. For example, the growth of the Arabidopsis seedling hypocotyl (expansion of the embryonic stem during seed germination and seedling establishment) is inhibited by light. A number of studies had previously implicated GA regulation as a component of light-mediated hypocotyl growth regulation (e.g., Peng and Harberd, 1997), but it is only recently that the DELLAs have been shown to contribute to this process. For example, DELLA-deficient Arabidopsis seedling hypocotyl growth is partially insensitive to light-mediated growth inhibition (Achard et al., 2007a), and DELLA deficiency also compromises the shade avoidance response (whereby plants sense and respond to competing neighbors; Djakovic-Petrovic et al., 2007). Thus, DELLA-mediated growth inhibition is a component of light-mediated growth inhibition.
The light signals that elicit hypocotyl growth regulation are first perceived by photoreceptors and subsequently communicated to signal-transducing transcription factors, such as PHYTOCHROME INTERACTING FACTOR3 (PIF3) and PIF4. Recent studies identified a physical interaction between a conserved Leu-heptad repeat in the DELLAs and the DNA recognition domain of the PIF factors (de Lucas et al., 2008; Feng et al., 2008). This interaction inhibits the binding of PIF4 to gene promoter target recognition sequences (de Lucas et al., 2008), suggesting that DELLAs restrain growth by sequestering PIFs into inactive protein complexes, thus inhibiting their ability to promote growth via gene transcriptional activation. These observations provide a possible general framework for understanding how DELLAs inhibit plant growth. According to this framework, DELLAs inhibit growth by interfering with the activity of growth-promoting transcription factors.
ALTERNATIVE (NON-GA) ROUTES TO DELLA-DEPENDENT GROWTH CONTROL
The above described advances provide a molecular framework for understanding how GA quantitatively regulates plant growth. However, it is becoming clear that there are additional, GA-independent factors that can modulate the growth inhibitory function of the DELLAs. One such factor is the putative O-GlcNAc transferase encoded by SPY (reviewed in Richards et al., 2001). Loss-of-function spy alleles partially suppress the dwarf phenotypes conferred by ga1-3, gai, and rga-Δ17 (Peng et al., 1997, 1999b; Silverstone et al., 1997, 2007). O-GlcNAc transferases typically catalyze O-GlcNAc modification of target Ser residues in regulatory proteins. A reduction in SPY activity (e.g., as conferred by spy alleles) causes an increase in DELLA protein abundance (as detected antigenically or via GFP-DELLA fusion), rather than the reduction that might a priori have been expected for a mutation that causes a reduction in DELLA-mediated growth inhibition (Shimada et al., 2006; Silverstone et al., 2007). One plausible explanation for these observations (as yet unverified) is that O-GlcNAc modification of DELLAs activates (or enhances) the growth inhibition function of DELLAs. Modulation of O-GlcNAc transferase activity via environmental or developmental variables thus provides a potential non-GA route for growth control via modulation of DELLA activity.
THE GA-GID1-DELLA PLANT GROWTH REGULATORY MECHANISM
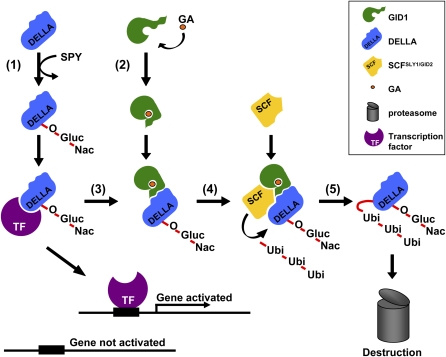
The advances in understanding outlined in the preceding paragraphs can be summarized as follows (Figure 4): (1) DELLAs are growth inhibitors that act (at least in part) by interfering with the activity of growth-promoting transcription factors. In addition, the growth inhibitory properties of DELLAs may be enhanced by O-GlcNAc modification due to SPY. (2) GA binds within the GA binding pocket of the GID1 GA receptor, causing the folding of a lid structure to which the N-terminal region of the DELLAs binds. (3) The GA-GID1-DELLA interaction reduces the growth-repressing effect of DELLAs, perhaps by reducing their capacity for interacting with growth-promoting transcription factors. (4) The GA-GID1-DELLA interaction also stimulates the binding of DELLA to the SCFSLY1/GID2 E3 ubiquitin ligase. (5) Polyubiquitination of DELLA by SCFSLY1/GID2 targets DELLA for destruction by the proteasome, thus finally removing the agent of DELLA-mediated growth inhibition (Figure 4).
Figure 4.
The GA-GID1-DELLA Mechanism of Angiosperm Growth Regulation.
For step-by-step guide to points (1) to (5) in the mechanism, see main text. Growth results from activation of growth-promoting genes (gene activated). Formation of the GA-GID1-DELLA complex frees transcription factors from sequestration by DELLAs, enabling previously inactive growth-promoting genes to become activated.
The GA-GID1-DELLA growth regulatory mechanism thus summarized operates in angiosperms and appears to be a relatively recent innovation in plant evolution, having arisen sometime between the divergences of the bryophytes and the lycophytes from the land plant lineage (Hirano et al., 2007; Yasumura et al., 2007). The most prominent function of the angiosperm GA-GID1-DELLA mechanism is regulation of the growth of organs following their definition in shoot and root apical meristems. For example, the Arabidopsis ga1-3 mutant has both a dwarfed shoot (Richards et al., 2001) and a short root (Fu and Harberd, 2003), in part because GA regulates postmeristematic growth of these organs. In fact, the GA-GID1-DELLA mechanism plays a major role in the regulation of organ extension growth of shoot and root and does so via interaction with the auxin signaling pathway: auxin regulates growth (at least in part) by the modulation of both GA biosynthesis and GA responsiveness (Ross et al., 2000; Fu and Harberd, 2003). Recent evidence indicates that GA-GID1-DELLA regulation of root extension growth is not generally distributed throughout root cell layers, but operates primarily in the endodermis (Ubeda-Tomás et al., 2008).
In addition to its major role in modulating extension growth of plant organs subsequent to their definition in apical meristems, the GA-GID1-DELLA mechanism also determines some aspects of developmental patterning. For example, maintenance of pluripotency in the cells of the vegetative shoot apical meristem (the group of cells from which arise the aerial parts of angiosperms) is in part dependent on dampening of GA-GID1-DELLA signaling mediated by KNOX transcription factors (Hay et al., 2002; Jasinski et al., 2005). Conversely, growing (postmeristematic) cells characteristically have activated GA signaling.
THE GA-GID1-DELLA MECHANISM ENABLES A FLEXIBLE GROWTH REGULATION RESPONSE TO ENVIRONMENTAL VARIABILITY
Possible reasons why angiosperms might have evolved the GA-GID1-DELLA mechanism of growth regulation become clear when one considers the plant life strategy. Plants are sessile organisms that need to be able to respond appropriately and in situ to the biotic and abiotic environmental challenges with which they are faced. There is a particular need to regulate growth in response to environmental variability, in part because growth is energetically demanding and in part because growth-driven increases in body volume/surface area tend to increase vulnerability. Recent studies show that the GA-GID1-DELLA mechanism is highly integrated within the overall angiosperm informational signaling and response system. We have already touched upon the relationship between GA-GID1-DELLA and light signaling in growth regulation. Additional examples come from studies of the relationship between GA-GID1-DELLA, the phytohormone ethylene, and plant stress responses. A general idea emerging from these studies is that the GA-GID1-DELLA mechanism enables plants to maintain transient growth arrest and thus to survive periods of adversity.
Ethylene is perceived by the ETR1 family of ethylene receptors, thus causing inactivation of CTR1 (a Raf kinase–related repressor of ethylene signaling) and resultant accumulation of EIN3 and EIN3-like ethylene-signaling transcription factors (Guo and Ecker, 2004). Initial investigations of the relationship between ethylene signaling and the GA-GID1-DELLA mechanism showed that ethylene inhibits DELLA-deficient mutant Arabidopsis seedling root growth less than that of the wild type and that ethylene inhibits the GA-induced disappearance of GFP-RGA via CTR1-dependent signaling (Achard et al., 2003). In addition, the maintenance of the exaggerated apical hook structure typical of dark-grown ethylene-treated seedlings was shown to be dependent on loss of DELLA-mediated growth inhibition (Achard et al., 2003; Vriezen et al., 2004). Thus, there is a connection between ethylene response and the GA-GID1-DELLA mechanism and a correlation between ethylene-mediated growth inhibition and DELLA accumulation. The conclusion from these studies is that ethylene inhibits growth (at least in part) via a DELLA-dependent mechanism. However, there are circumstances in which ethylene promotes (rather than inhibits) the growth of light-grown hypocotyls, and in this case, promotion interestingly is also accompanied by an accumulation (rather than a depletion) of GFP-RGA in hypocotyl nuclei (Vandenbussche et al., 2007). It seems that in this case ethylene still promotes GFP-RGA accumulation, but this accumulation is not translated into growth inhibition. Thus, the frequently observed negative correlation between growth and DELLA accumulation is not absolute and can be broken in specific circumstances. As described above in the case of the spy mutant phenotype, these observations are indicative of a physiological role for regulation of inherent growth inhibitory activity of DELLAs that is independent of DELLA accumulation.
The close relationships between ethylene signaling and the GA-GID1-DELLA mechanism continue through stages of the plant life cycle beyond seedling emergence. For example, recent studies show that ethylene regulates the transition from vegetative to reproductive growth, and does so, at least in part, via interaction with the GA-GID1-DELLA mechanism. The activation of ethylene signaling results in a reduction in GA levels, and this delays floral induction via a DELLA-dependent mechanism. In addition, a lack of DELLAs suppresses the delayed flowering characteristic of the constitutive ethylene response phenotype conferred by ctr1 loss-of-function alleles (Achard et al., 2007b). Further analysis indicates that ethylene likely affects DELLAs downstream of the EIN3 transcriptional regulator and delays flowering by repressing the activity of floral meristem identity genes (LEAFY and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1; Achard et al., 2007b). These observations identify the GA-GID1-DELLA mechanism as a previously unknown bridge between the ethylene signaling pathway and floral meristem identity genes in the regulation of the transition between vegetative to reproductive growth, probably providing a mechanism via which environmental stress regulates that transition.
Several studies have directly implicated the GA-GID1-DELLA mechanism in regulating the response of plants to environmental stress. For example, high salinity slows the growth of plants. Studies with DELLA-deficient Arabidopsis mutants showed that salt slows growth via a partially DELLA-dependent mechanism (Achard et al., 2006). Further studies revealed that salt causes a reduction in in planta bioactive GA levels (Achard et al., 2006) and that this reduction can be explained, at least in part, by activation of genes encoding GA-2oxidase, an enzyme that deactivates bioactive GAs by addition of a hydroxyl group to the 2C position (Figure 1A; Magome et al., 2008). Thus, salt inhibits plant growth by reducing bioactive GA abundance, in turn causing accumulation of DELLAs and consequent growth inhibition. Salt-activated DELLA-dependent growth inhibition presumably has adaptive significance because plants lacking DELLAs are more susceptible, whereas plants in which DELLAs accumulate (e.g., ga1-3) are more resistant to the lethal effects of extreme salt (Achard et al., 2006). These observations indicate that the GA-GID1-DELLA mechanism provides plants with a means of regulating growth appropriate to environmental conditions, enabling a slowing of growth and reduced energetic commitment during periods of environmental adversity. Achard et al. (2008a) identified a reduction in reactive oxygen species levels as a possible mechanism for the restraint of growth and the concomitant promotion of survival of adverse salinity.
A link between ethylene and the GA-GID1-DELLA mechanism has also been implicated in the growth responses of rice to flooding. Flood-tolerant rice survives temporary submergence via a transient inhibition of growth and carbohydrate consumption that is controlled by an ethylene-responsive factor ERF-type transcription factor encoded by the Sub1A gene. Submergence causes ethylene production and the consequent activation of Sub1A. In turn, Sub1A inhibits elongation growth by causing increased accumulation of the rice DELLA SLR1 and of a related protein, SLRL1 (Fukao and Bailey-Serres, 2008). SLRL1 is a protein that inhibits growth but lacks functional domains I and II (see above) and thus inhibits growth in a GA-resistant fashion. Therefore, SLRL1 is functionally analogous to the mutant DELLA proteins encoded by the mutant gai, D8-1, D8-2023, and Rht-B1b alleles described above and is a protein that rice has recruited to function in flooding response. Thus, as shown previously in Arabidopsis (Achard et al., 2003), ethylene can inhibit rice growth using a GA-GID1-DELLA–dependent mechanism (via SLR1) and additionally using a DELLA-related non-GA opposable growth inhibitory protein (SLRL1). Activation of SLR1 and SLRL1 by Sub1A suppresses energy-consuming processes, resulting in reduced carbohydrate consumption and inhibited growth in response to flooding in rice (Fukao and Bailey-Serres, 2008), just as DELLAs inhibit growth in response to salt stress in Arabidopsis (Achard et al., 2006). These observations suggest that the GA-GID1-DELLA mechanism provides a general mechanism for inhibition of growth and associated resource consumption in response to environmental adversity.
The likelihood that the GA-GID1-DELLA mechanism is indeed a general mechanism for modulating growth response to the environment is increased by recent discoveries that it regulates growth in response to change in a wide variety of environmental variables. For example, the effects of cold temperatures on plant growth signaled by the cold-induced CBF1 factor-dependent pathway are mediated via effects on GA metabolism (and consequently on DELLAs; Achard et al., 2008b), and growth and developmental responses to phosphate starvation are also modulated by the GA-GID1-DELLA pathway (Jiang et al., 2007).
An unexpected recent development has been the discovery that the GA-GID1-DELLA mechanism plays a role in the regulation of plant–pathogen interactions. Studies of the pathogen responses of DELLA-deficient Arabidopsis plants showed that DELLA accumulation differentially affects response to different classes of pathogen. Thus, presence of DELLAs is associated with susceptibility to virulent biotrophs and with resistance to necrotrophs, properties that are related to an effect of the GA-GID1-DELLA pathway on the balance between jasmonic acid and salicylic acid signaling (Navarro et al., 2008). Although concordant with previous examples of the GA-GID1-DELLA mechanism acting as a mediating bridge between different signaling pathways, these observations were unexpected because they represent identification of a GA-GID1-DELLA–regulated phenomenon that is not strictly growth related (the pathogenicity assays used in these experiments involved relatively mature leaves that were no longer in the growing phase). It is possible that there are additional non-growth aspects of GA-GID1-DELLA biology that are yet to be discovered.
DOES THE GA-GID1-DELLA MECHANISM HAVE ADAPTIVE SIGNIFICANCE?
Given that the GA-GID1-DELLA mechanism plays such a prominent role in the response of plants to environmental variables, it might be expected that possession of this mechanism is selectively advantageous and that variant forms of it might have adaptive significance. Recent studies monitored the effects of artificial selection on an experimental wheat population segregating for Rht-B1a and Rht-B1b (wheat DELLA-encoding alleles conferring tall and dwarf plant phenotypes, respectively; Peng et al., 1999a; Raquin et al., 2008). Mean plant height increased steadily with number of generations post the founder population generation, as did the relative frequency of the RhtB1a allele, indicating that Rht-B1a was under strong positive selection throughout the 17 generations of the experiment (Raquin et al., 2008). At first sight this may seem paradoxical given the well-documented increases in grain yield conferred by Rht-B1b (see above), the very increases that were responsible for the use of the Rht-B1b allele in green revolution wheat varieties. However, taller plants presumably compete more effectively for light and nutrients when grown in heterogeneous populations (as opposed to the homogeneous populations typical of modern wheat fields). In addition, the strength of selection against Rht-B1b indicates that alleles conferring DELLAs resistant to opposition by GA will be at a competitive disadvantage in many natural environments, perhaps because the impaired GA opposability imposes a clamp on the flexibility with which plants can modulate growth in response to environmental change.
THE GA-GID1-DELLA MECHANISM: OUTSTANDING ISSUES AND QUESTIONS
As reviewed above, recent research has shown that the DELLAs are the GA-opposable endogenous plant growth inhibitors whose existence was first postulated in the late 1950s. We have summarized the molecular nature of the GA-GID1-DELLA mechanism, the mechanism via which plants respond to GA, and argued that this mechanism enables plasticity of growth in response to environmental variability. We now point out a number of outstanding issues and questions that are likely fertile areas for future research.
First, as described above, the recent demonstration that DELLAs interact with PIF3/PIF4 transcription factors provides a potential general model for understanding how DELLAs inhibit growth. However, the degree of specificity of this interaction remains unclear. The PIFs are a subset of the basic helix-loop-helix (bHLH) family of transcription factors. Perhaps DELLAs interact with a wider spectrum of bHLHs than those defined as PIFs, thus sequestering various bHLHs away from the gene promoters that they activate. Such a scenario might explain, at least in part, the pervasive roles that DELLAs appear to play in angiosperm biology. In addition, since the bHLH transcription factor PIL5 regulates the expression of DELLA-encoding genes (Oh et al., 2007), it is possible that DELLAs feedback regulate their own expression via interaction with bHLH transcription factors.
Second, this review has shown how GA regulates the growth inhibition function of DELLAs. However, there are increasing indications that DELLA function can also be modified via routes that do not directly involve GA. Among these possible routes are transcriptional regulation of genes encoding DELLAs (e.g., Oh et al., 2007; Fukao and Bailey-Serres, 2008), posttranslational activation of DELLAs, and non-GA routes for modulating DELLA abundance. One possible route for posttranslational activation of DELLAs is the likely O-GlcNAc transferase activity encoded by SPY (discussed above), although modulation of this activity has not yet been shown to regulate DELLA activity differentially in response to physiological or environmental variables. If O-GlcNAc modification indeed enhances the growth inhibitory properties of DELLAs, it is possible that this modification promotes the interaction between DELLAs and PIF3/4 (or other bHLHs). A possible point of non-GA regulation of DELLA abundance is the rate of polyubiquitination (and hence destruction) of DELLAs by the SCFSLY1/GID2 E3 ubiquitin ligase. For example, transgenic overexpression of SLY1 (presumably causing increased SLY1 availability) promotes growth by increasing DELLA destruction (Fu et al., 2004). Thus, SLY1 availability can be a limiting factor in plant growth regulation, and differential regulation of SLY1 expression is potentially a mechanism via which different plant signaling pathways may (independently of GA) regulate plant growth by reducing DELLA-mediated growth inhibition. Future studies will determine the contributions of these (and other) non-GA routes to the modulation of DELLA growth inhibitory function.
Another likely important area of future investigation concerns the role of DELLAs as integrators of growth responses to a variety of signals. It has been suggested that the GA-GID1-DELLA mechanism is nodal and provides a mechanism for integrating multiple input signals into a single growth output (Alvey and Harberd, 2005). However, many questions remain, not least with respect to the unraveling of the relative extents to which other signals (including other hormones) impact upon GA-GID1-DELLA activity through regulation of bioactive GA biosynthesis and inactivation, regulation of DELLA susceptibility to GA-mediated opposition of DELLA activity (interaction with the GA receptor and/or subsequent degradation), or modulation of DELLA activity by other means. Furthermore, it is clear that the GA-GID1-DELLA mechanism is not the sole mechanism for integration of growth regulation. For example, DELLAs inhibit hypocotyl growth in response to light, but a DELLA-deficient hypocotyl is still partially photomorphogenetic (is longer than the wild type in light but not as long as a dark-grown wild-type hypocotyl; Achard et al., 2007a; Feng et al., 2008). Thus, there must be additional response integrators that work in conjunction with the DELLAs to inhibit hypocotyl growth in response to light. By extension, it is likely that there are additional integrators that coordinate the regulation of growth in response to environmental variables in general, and determining their relationship with the DELLAs is an important task for the future.
An additional emerging idea concerns the possible relationship between DELLA-mediated growth inhibition and resource allocation. Essentially, growth inhibition might have adaptive significance when environmental impacts (both biotic and abiotic) threaten resource limitation. Prioritization of resource allocation may result in resources being diverted away from growth in favor of defense against pathogens or in the adoption of a strategy that involves reduced resource consumption during a period of wait for improved environmental conditions. Thus, the GA-GID1-DELLA mechanism may enable the robust adaptation of various aspects of angiosperm biology to environmental threat.
Finally, it is possible that consideration of the meaning of “growth inhibition” at the cellular level will prove fruitful. The intriguing potential role of reactive oxygen species in GA-GID1-DELLA–mediated growth regulation is described above, and this is likely to be an important area of further investigation. In addition, plant cell expansion is often characterized as being the product of opposing forces of (growth-promoting) intracellular turgor pressure and the (growth-inhibiting) turgor-resisting forces of the cell wall. This view has parallels with the proposal that growth is regulated by the opposing forces of GA and a GA-opposable growth inhibitor, the inhibitor of an inhibitor proposal with which this review began. Could it be that an increase in DELLA-mediated inhibition of growth equates ultimately to an increase in the resistance of the cell wall?
Acknowledgments
Work in our laboratory is funded by the UK Biotechnology and Biological Sciences Research Council (response modes Grant BB/F020759/1).
References
- Achard, P., Baghour, M., Chapple, A., Hedden, P., Van Der Straeten, D., Genschik, P., Moritz, T., and Harberd, N.P. (2007. b). The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. USA 104 6484–6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard, P., Cheng, H., De Grauwe, L., Decat, J., Schoutteten, H., Moritz, T., Van Der Straeten, D., Peng, J., and Harberd, N.P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311 91–94. [DOI] [PubMed] [Google Scholar]
- Achard, P., Gong, F., Cheminant, S., Alioua, M., Hedden, P., and Genschik, P. (2008. b). The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20 2117–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard, P., Liao, L., Jiang, C., Desnos, T., Bartlett, J., Fu, X., and Harberd, N.P. (2007. a). DELLAs contribute to plant photomorphogenesis. Plant Physiol. 143 1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard, P., Renou, J.-P., Berthomé, R., Harberd, N.P., and Genschik, P. (2008. a). Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 18 656–660. [DOI] [PubMed] [Google Scholar]
- Achard, P., Vriezen, W.H., Van Der Straeten, D., and Harberd, N.P. (2003). Ethylene regulates Arabidopsis development via modulation of DELLA protein growth repressor function. Plant Cell 15 2816–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait-ali, T., Rands, C., and Harberd, N.P. (2003). Flexible control of plant architecture and yield via switchable expression of Arabidopsis gai. Plant Biotechnol. J. 1 337–343. [DOI] [PubMed] [Google Scholar]
- Alvey, L., and Harberd, N.P. (2005). DELLA proteins: Integrators of multiple plant growth regulatory inputs? Physiol. Plant. 123 153–160. [Google Scholar]
- Ariizumi, T., Murase, K., Sun, T.-p., and Steber, C.M. (2008). Proteolysis-independent downregulation of DELLA repression in Arabidopsis by the gibberellin receptor GIBBERELLIN INSENSITIVE DWARF1. Plant Cell 20 2447–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano, K., Hirano, K., Ueguchi-Tanaka, M., Angeles-Shim, R.B., Komura, T., Satoh, H., Kitano, H., Matsuoka, M., and Ashikari, M. (2009). Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice. Mol. Genet. Genomics 281 223–231. [DOI] [PubMed] [Google Scholar]
- Brian, P.W. (1957). The effects of some microbial metabolic products on plant growth. Symp. Soc. Exp. Biol. 11 166–182. [PubMed] [Google Scholar]
- Chandler, P.M., Marion-Poll, A., Ellis, M., and Gubler, F. (2002). Mutants at the Slender1 locus of barley cv Himalaya. Molecular and physiological characterisation. Plant Physiol. 129 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, H., Qin, L., Lee, S., Fu, X., Richards, D.E., Cao, D., Luo, D., Harberd, N.P., and Peng, J. (2004). Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131 1055–1064. [DOI] [PubMed] [Google Scholar]
- de Lucas, M., Davierre, J.M., Rodriguez-Falcon, M., Pontin, M., Iglesias-Pedraz, J.M., Lorrain, S., Fankhauser, C., Blazquez, M.A., Titarenko, E., and Prat, S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451 480–484. [DOI] [PubMed] [Google Scholar]
- Dill, A., Jung, H.-S., and Sun, T.-p. (2001). The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. USA 98 14162–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill, A., and Sun, T.-p. (2001). Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill, A., Thomas, S.G., Hu, J., Steber, C.M., and Sun, T.-p. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djakovic-Petrovic, T., de Wit, M., Voesenek, L.A.C.J., and Pierik, R. (2007). DELLA protein function in growth responses to canopy signals. Plant J. 51 117–126. [DOI] [PubMed] [Google Scholar]
- Feng, S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X., and Harberd, N.P. (2003). Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421 740–743. [DOI] [PubMed] [Google Scholar]
- Fu, X., Richards, D.E., Ait-ali, T., Hynes, L.W., Ougham, H., Peng, J., and Harberd, N.P. (2002). Gibberellin-mediated proteasome-dependent degradation of the barley DELLA-protein SLN1 repressor. Plant Cell 14 3191–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X., Richards, D.E., Fleck, B., Xie, D., Burton, N., and Harberd, N.P. (2004). The Arabidopsis mutant sly1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell 16 1406–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X., Sudhakar, D., Peng, J., Richards, D.E., Christou, P., and Harberd, N.P. (2001). Expression of Arabidopsis GAI in transgenic rice represses multiple gibberellin responses. Plant Cell 13 1791–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao, T., and Bailey-Serres, J. (2008). Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. USA 105 16814–16819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths, J., Murase, K., Rieu, I., Zentella, R., Zhang, Z.L., Powers, S.J., Gong, F., Phillips, A.L., Hedden, P., Sun, T.P., and Thomas, S.G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18 3399–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., Chandler, P.M., White, R.G., Llewellyn, D.J., and Jacobsen, J.V. (2002). Gibberellin signaling in barley aleurone cells. Control of SLN1 and GAMYB expression. Plant Physiol. 129 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., and Ecker, J.R. (2004). The ethylene signaling pathway: New insights. Curr. Opin. Plant Biol. 7 40–49. [DOI] [PubMed] [Google Scholar]
- Harberd, N.P., and Freeling, M. (1989). Genetics of dominant gibberellin-insensitive dwarfism in maize. Genetics 121 827–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, A., Kaur, H., Phillips, A., Hedden, P., Hake, S., and Tsiantis, M. (2002). The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. 12 1557–1565. [DOI] [PubMed] [Google Scholar]
- Hirano, K., et al. (2007). The GID1-mediated gibberellin perception mechanism is conserved in the lycophyte Selaginella moellendorfii but not in the bryophyte Physcomitrella patens. Plant Cell 19 3058–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, A., Ueguchi-Tanaka, M., Sonoda, Y., Kitano, H., Koshioka, M., Futsuhara, Y., Matsuoka, M., and Yamaguchi, J. (2001). Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M., and Matsuoka, M. (2002). The gibberellin signalling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi, S., Suzuki, H., Kim, Y.C., Iuchi, A., Kuromori, T., Ueguchi-Tanaka, M., Asami, T., Yamaguchi, I., Matsuoka, M., Kobayashi, M., and Nakajima, M. (2007). Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. Plant J. 50 958–966. [DOI] [PubMed] [Google Scholar]
- Jasinski, S., Piazza, P., Craft, J., Hay, A., Woolley, L., Rieu, I., Phillips, A., Hedden, P., and Tsiantis, M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15 1560–1565. [DOI] [PubMed] [Google Scholar]
- Jiang, C., Gao, X., Liao, L., Harberd, N.P., and Fu, X. (2007). Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signalling pathway in Arabidopsis. Plant Physiol. 145 1460–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, K.E., Moritz, T., and Harberd, N.P. (2001). Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159 767–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Elgersma, A., Hanhart, C.J., van Loenen-Martinet, E.P., van Rign, L., and Zeevaart, J.A.D. (1985). A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol. Plant. 65 33–39. [Google Scholar]
- Lee, S., Cheng, H., King, K.E., Wang, W., Husssain, A., Lo, J., Harberd, N.P., and Peng, J. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16 646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magome, H., Yamaguchi, S., Hanada, A., Kamiya, Y., and Oda, K. (2008). The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J. 56 613–626. [DOI] [PubMed] [Google Scholar]
- McGinnis, K.M., Thomas, S.G., Soule, F.D., Strader, L.C., Zale, J.M., Sun, T.-p., and Steber, C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase, K., Hirano, Y., Sun, T.-p., and Hakoshima, T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456 459–463. [DOI] [PubMed] [Google Scholar]
- Navarro, L., Bari, R., Achard, P., Lisón, P., Nemri, A., Harberd, N.P., and Jones, J.D.G. (2008). DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr. Biol. 18 650–655. [DOI] [PubMed] [Google Scholar]
- Oh, E., Yamaguchi, S., Hu, J., Yusuke, J., Jung, B., Paik, I., Lee, H.S., Sun, T.-p., Kamiya, Y., and Choi, G. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19 1192–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., Carol, P., Richards, D.E., King, K.E., Cowling, R.J., Murphy, G.P., and Harberd, N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., and Harberd, N.P. (1993). Derivative alleles of the Arabidopsis gibberellin-insensitive (gai) mutation confer a wild-type phenotype. Plant Cell 5 351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., and Harberd, N.P. (1997). Gibberellin deficiency and response mutations suppress the stem elongation phenotype of phytochrome deficient mutants of Arabidopsis. Plant Physiol. 113 1051–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., et al. (1999. a). ‘Green Revolution’ genes encode mutant gibberellin response modulators. Nature 400 256–261. [DOI] [PubMed] [Google Scholar]
- Peng, J., Richards, D.E., Moritz, T., Caño-Delgado, A., and Harberd, N.P. (1999. b). Extragenic suppressors of the Arabidopsis gai mutation alter the dose–response relationship of diverse gibberellin responses. Plant Physiol. 119 1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts, W.C., Reid, J.B., and Murfet, I.C. (1985). Internode length in Pisum. Gibberellins and the slender phenotype. Physiol. Plant. 63 357–364. [Google Scholar]
- Raquin, A.-L., Brabant, P., Rhoné, B., Balfourier, F., Leroy, P., and Goldringer, I. (2008). Soft selective sweep near a gene that increases plant height in wheat. Mol. Ecol. 17 741–756. [DOI] [PubMed] [Google Scholar]
- Richards, D.E., King, K.E., Ait-ali, T., and Harberd, N.P. (2001). How gibberellin regulates plant growth and development: A molecular genetic analysis of gibberellin signalling. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 67–88. [DOI] [PubMed] [Google Scholar]
- Ross, J.J., O'Neill, D.P., Smith, J.J., Kerckhoffs, L.H.J., and Elliott, R.C. (2000). Evidence that auxin promotes gibberellin A1 biosynthesis in pea. Plant J. 21 547–552. [DOI] [PubMed] [Google Scholar]
- Sasaki, A., Itoh, H., Gomi, K., Ueguchi-Tanaka, M., Ishiyama, K., Kobayashi, M., Jeong, D.H., An, G., Kitano, H., Ashikari, M., and Matsuoka, M. (2003). Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299 1896–1898. [DOI] [PubMed] [Google Scholar]
- Shimada, A., Ueguchi-Tanaka, M., Nakatsu, T., Nakajima, M., Naoe, Y., Ohmiya, H., Kato, H., and Matsuoka, M. (2008). Structural basis for gibberellin recognition by its receptor GID1. Nature 456 520–523. [DOI] [PubMed] [Google Scholar]
- Shimada, A., Ueguchi-Tanaka, M., Sakamoto, T., Fujioka, S., Takatsuto, S., Yoshida, S., Sazuka, T., Ashikari, M., and Matsuoka, M. (2006). The rice SPINDLY gene functions as a negative regulator of gibberellin signalling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J. 48 390–402. [DOI] [PubMed] [Google Scholar]
- Silverstone, A.L., Ciampaglio, C.N., and Sun, T.-p. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Jung, H.S., Dill, A., Kawaide, H., Kamiya, Y., and Sun, T.-p. (2001). Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13 1555–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Mak, P.Y.A., Martinez, E.C., and Sun, T.-p. (1997). The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146 1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Tseng, T.S., Swain, S.M., Dill, A., Jeong, S.Y., Olszewski, N.E., and Sun, T.-p. (2007). Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol. 143 987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, J.A., Shirasu, K., and Deng, X.-W. (2003). The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nat. Rev. Genet. 4 948–958. [DOI] [PubMed] [Google Scholar]
- Tyler, L., Thomas, S.G., Hu, J.H., Dill, A., Alonso, J.M., Ecker, J.R., and Sun, T.-p. (2004). DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomás, S., Swarup, R., Coates, J., Swarup, K., Laplaze, L., Beemster, G.T.S., Hedden, P., Bhalerao, R., and Bennett, M.J. (2008). Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat. Cell Biol. 10 625–628. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Ashikari, M., Nakajima, M., Itoh, H., Katoh, E., Kobayashi, M., Chow, T.Y., Hsing, Y.I., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437 693–698. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Hirano, K., Hasegawa, Y., Kitano, H., and Matsuoka, M. (2008). Release of the repressive activity of rice DELLA protein SLR1 by gibberellin does not require SLR1 degradation in the gid2 mutant. Plant Cell 20 2437–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka, M., Nakajima, M., Katoh, E., Ohmiya, H., Asano, K., Saji, S., Hongyu, X., Ashikari, M., Kitano, H., Yamaguchi, I., and Matsuoka, M. (2007). Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19 2140–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche, F., Vancompernolle, B., Rieu, I., Ahmad, M., Phillips, A., Moritz, T., Hedden, P., and Van Der Straeten, D. (2007). Ethylene-induced Arabidopsis hypocotyl elongation is dependent on but not mediated by gibberellins. J. Exp. Bot. 58 4269–4281. [DOI] [PubMed] [Google Scholar]
- Vriezen, W.H., Achard, P., Harberd, N.P., and Van Der Straeten, D. (2004). Ethylene-mediated enhancement of apical hook formation in etiolated Arabidopsis thaliana seedlings is gibberellin dependent. Plant J. 37 505–516. [DOI] [PubMed] [Google Scholar]
- Weston, D.E., Elliot, R.C., Lester, D.R., Rameau, C., Reid, J.B., Murfet, I.C., and Ross, J.J. (2008). The pea DELLA proteins LA and CRY are important regulators of gibberellin synthesis and root growth. Plant Physiol. 147 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige, B.C., Ghosh, S., Nill, C., Zourelidou, M., Dohmann, E.M.N., Maier, A., and Schwechheimer, C. (2007). The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59 225–253. [DOI] [PubMed] [Google Scholar]
- Yasumura, Y., Crumpton-Taylor, M., Fuentes, S., and Harberd, N.P. (2007). Step-by-step acquisition of the gibberellin-DELLA growth-regulatory mechanism during land-plant evolution. Curr. Biol. 17 1225–1230. [DOI] [PubMed] [Google Scholar]
- Zentella, R., Zhang, Z.L., Park, M., Thomas, S.G., Endo, A., Murase, K., Fleet, C.M., Jikumaru, Y., Nambara, E., Kamiya, Y., and Sun, T.P. (2007). Global analysis of DELLA direct targets in early gibberellin signalling in Arabidopsis. Plant Cell 19 3037–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]