Abstract
Remorins (REMs) are proteins of unknown function specific to vascular plants. We have used imaging and biochemical approaches and in situ labeling to demonstrate that REM clusters at plasmodesmata and in ∼70-nm membrane domains, similar to lipid rafts, in the cytosolic leaflet of the plasma membrane. From a manipulation of REM levels in transgenic tomato (Solanum lycopersicum) plants, we show that Potato virus X (PVX) movement is inversely related to REM accumulation. We show that REM can interact physically with the movement protein TRIPLE GENE BLOCK PROTEIN1 from PVX. Based on the localization of REM and its impact on virus macromolecular trafficking, we discuss the potential for lipid rafts to act as functional components in plasmodesmata and the plasma membrane.
INTRODUCTION
Current models of the plasma membrane (PM) predict the existence of a patchwork of specialized and dynamic microdomains coordinating a variety of cellular functions. Among the well-characterized PM microdomains are the membrane rafts (Brown, 2006). Most of the evidence for the existence of membrane rafts relies on indirect methods, such as detergent extraction (Schroeder et al., 1994). However, a number of recent studies raised concerns regarding such methods (Heerklotz, 2002; Munro, 2003; Kenworthy, 2008; Roche et al., 2008). To face the controversy, various microscopy approaches have been developed to establish a link between biochemical and imaging techniques (for review, see Marguet et al., 2006). In the animal and yeast fields, a recent consensus defines membrane rafts as “small (10 to 200 nm), heterogeneous, highly dynamic, sterol- and sphingolipid-enriched domains that compartmentalize cellular processes” (Pike, 2006). Membrane rafts are thought to act as docking sites for specific proteins involved in many important cellular processes, including secretion, signal transduction, and the perception of pathogens (Rajendran and Simons, 2005). They also may be important in virus infection, including steps such as entry, assembly, and budding (Chazal and Gerlier, 2003). Specific proteins seem to be associated with raft microdomains. In animal cells, the best-characterized examples are caveolins, which form scaffolds for caveolae, small (50 to 100 nm) flask-shaped invaginations of the PM (Parton and Simons, 2007). Reggie proteins (also called flotillins) are also considered to be bona fide raft protein markers. They associate with the cytosolic leaflet of the PM via acylations (myristoylation and/or palmitoylation) where they form stable clusters readily observable by confocal and immunogold electron microscopy (Langhorst et al., 2005).
Evidence for the presence of rafts in higher plant cells is emerging. Simple biochemical techniques, such as low-temperature nonionic detergent treatments, yield detergent-insoluble membranes (DIMs) that are thought to be the biochemical counterpart of lipid rafts (Bhat and Panstruga, 2005). For example, in the alga Chlorella kessleri, the hexose-proton symporter HUP1 is present in a sterol-enriched DIM fraction, a typical property for raft protein. When fused to green fluorescent protein (GFP), this protein shows (1) a patchy distribution in the PM of algal cells, (2) a similar spotty distribution in the PM when expressed in yeast, and (3) homogenous distribution in yeast mutants lacking the typical raft lipids ergosterol and sphingolipids (Grossmann et al., 2006). This study, albeit in a heterologous system, showed that a plant membrane protein has the property of being concentrated in specific sterol- and sphingolipid-enriched raft membrane compartments. The antibiotic filipin, which binds to sterol, shows pronounced labeling at fungal pathogen entry sites, suggesting either an aggregation of a plant raft-like domain or the release of sterol-enriched fungal PM-derived material (Bhat and Panstruga, 2005). Recently, Kierszniowska et al. (2009) used an N14/N15 quantitative proteomic approach to distinguish between true sterol-dependent plant raft proteins, sterol-independent nonraft proteins, and copurifying contaminants in DIMs. Plant raft proteins were shown to comprise a variable set of signaling components and a core set of unknown cell wall–related proteins.
In plants, although a number of proteins enriched in DIMs have been reported, their specific localization to raft-like microdomains within the PM has not been demonstrated. Subsets of proteins have been found to copurify with Triton X-100 (TX100)–insoluble membranes (Bhat and Panstruga, 2005), but the validity of assigning these proteins to membrane subdomains has been criticized on the grounds of specificity and correspondence with in vivo data (Shaw, 2006).
The prototype remorin protein was discovered as a potato (Solanum tuberosum) PM protein differentially phosphorylated in presence of oligogalacturonides but was first designated phosphorylated protein 34 kD or pp34 (Jacinto et al., 1993). Later, characterization showed it to be very hydrophilic and was consequently named remorin (from remora, smaller fish adhering to larger fishes and ships) in relation to its hydrophilicity and membrane location (Reymond et al., 1996). Absence of any targeting signature in the primary sequence suggests that remorin is synthesized in the cytosol and targeted to the cytosolic leaflet of the PM by an as yet unknown mechanism (Reymond et al., 1996). Remorin was later identified from PM-derived DIMs from tobacco (Nicotiana tabacum) leaves (Mongrand et al., 2004), BY-2 cells (Morel et al., 2006), Medicago truncatula roots (Lefebvre et al., 2007), leek (Allium porrum) seedlings (Laloi et al., 2007), and Arabidopsis thaliana seedlings and calli (Bhat et al., 2005; Shahollari et al., 2005; Laloi et al., 2007). Subsequent genomic and EST sequencing revealed remorins as a tracheophytae-specific protein family with >100 members. Remorins found in DIMs belong to the group 1b, characterized by a high Pro content in the N-terminal region (Raffaele et al., 2007). In Solanaceae, remorins form a very coherent monophyletic clade (with 86.9 to 95.5% of amino acid sequence similarities; see Supplemental Figure 1A online).This clade includes the first discovered remorin from potato (Reymond et al., 1996) and remorins from tobacco (Mongrand et al., 2004; Morel et al., 2006) and tomato (Solanum lycopersicum; Bariola et al., 2004). This work is focused on this prototype clade of remorins from Solanaceae, further designated as REM here.
After almost two decades, the function of REM remains enigmatic. Its potential association with PM microdomains has relaunched the debate as to its function, but the tools required to visualize putative membrane domains in vivo have been limited. Here, we show that REM is highly enriched in sterol/sphingolipid-rich DIM fractions purified from the PM. In addition, REM is anchored to the cytosolic leaflet of the PM. We investigated the localization of REM using immunogold labeling coupled to electron microscopy (EM), allowing plant PM domains to be visualized on a nanometer scale. We show that REM displays a clear lateral segregation, forming cytosolic domains of ∼70 nm. These studies also revealed that in vivo, REM is not only located in PM microdomains, but also at plasmodesmata. The physiological function of REM was further analyzed in transgenic plants having different levels of REM expression. Our results show that REM interferes with cell-to-cell movement of a plant virus, the Potato virus X (PVX), and binds directly to the virus movement protein TGBp1.
RESULTS
REM Is a Biochemical Marker for PM Cytosolic Leaflet DIMs
To assist in the biochemical characterization of REM, we generated specific polyclonal antibodies to the full-length potato REM protein (St REM1.3) and to a consensus peptide (called anti-REM and anti-REMpep, respectively). These antibodies recognized a single band in crude leaf total protein extracts from Solanaceae (see Supplemental Figure 1A online). We also used anti-REMpp34 (Reymond et al., 1996) and anti-REMα130 (Bariola et al., 2004) antibodies. All the antibodies are specific for the REM protein (see Supplemental Figure 1B online).
REM Is Located in the Cytosolic Leaflet of the PM
To determine experimentally in which leaflet of the PM REM is located, we purified PM vesicles by phase partition, a method generating mostly sealed, right-side-out (RSO) vesicles (Larsson, 1988). Second, we obtained inside-out vesicles after subsequent freeze/thaw cycles from this starting material. Trypsin was used as a nonpermeating protease to determine the accessibility of REM to hydrolysis when exposed on the outside of the vesicle. Figure 1A shows that less REM was hydrolyzed when more RSO vesicles were present, indicating that REM is located in the cytosolic leaflet of the PM. Quantification of the protein blot signal and the proportion of RSO vesicles showed that most of REM is located in the cytosolic face (see calculation in Supplemental Methods online).
Figure 1.
REM Is Located in the Cytosolic Leaflet of the PM and Is a Biochemical Marker of PM DIMs.
(A) REM is found in the cytosolic leaflet of the PM. Representative protein gel blot (mean ± sd, n = 3) shows the detection of REM in phase partition-purified tobacco leaf PMs submitted to three subsequent cycles of freezing/thawing to progressively reverse the orientation of the RSO vesicles, followed by trypsin digestion.
(B) Top: REM is a biochemical marker for plant DIMs. Isolation of DIMs from the PM using a TX100-to-protein ratio of 15. Ten fractions of equal volume were collected from the top to the bottom of the gradient. Precipitated proteins corresponding to half the volume of each fraction were analyzed by protein gel blots with antibodies to REM and to PMA. Bottom: Representative histogram shows total protein distribution along the gradient after DIM preparation (n = 10).
(C) REM becomes TX100-soluble after mβCD treatment. PMs were treated or not with mβCD and submitted to the DIM isolation procedure. REM level was quantified in treated whole PMs (inset) and in the different fractions of the sucrose gradient by protein gel blots.
REM Is Highly Enriched in Sterol- and Sphingolipid-Enriched Detergent-Insoluble PM Fractions
To estimate the enrichment of REM in DIMs, PM preparations were subjected to TX100 treatment at a ratio TX100-to-protein of 15, suitable to obtain a maximum enrichment of both phytosterols and sphingolipids, lipid markers for membrane rafts (Mongrand et al., 2004). Sucrose gradient fractions were analyzed to determine the enrichment of REM in detergent-insoluble (top of the gradient) and detergent-soluble (bottom of the gradient) fractions. As described previously (e.g., Mongrand et al., 2004), ∼10% of total PM protein is present in the DIM fraction; thus, most of the proteins are in the detergent-soluble fraction in the bottom of the sucrose gradient (Figure 1B). Protein gel blot analysis clearly showed that in the PM, REM is almost exclusively present in DIMs and not in detergent-soluble membranes (DSMs) (Figure 1B). We also examined the distribution of the PM proton pump ATPases (PMAs) found to be present in plant DIM fractions from other plant species (Mongrand et al., 2004; Borner et al., 2005; Morel et al., 2006, Lefebvre et al., 2007). In contrast with REM, PMAs are present both in DIM and DSM fractions with only a minor proportion (<20%) in DIMs (Figure 1B). REM can therefore be considered as a biochemical protein marker of tobacco PM rafts.
We recently showed that phytosterol depletion, monitored by fluorescence spectroscopy, led to an increase in lipid acyl chain disorder, suggesting a diminution of the overall liquid-phase heterogeneity. This indicates that free sterols are crucial for the lateral structure of the tobacco PM and may be key components for the formation of lipid rafts (Roche et al., 2008). Reciprocally, the disruption of phytosterol-rich domains led to the complete removal of the DIM fraction obtained after TX100 extraction (Roche et al., 2008). To determine whether REM associated with DIMs is altered by removal of membrane-free sterols, methyl β-cyclodextrin (mβCD), a chelator of free sterols, was included in the DIM isolation procedure. Figure 1C shows that mβCD treatment depletes DIMs from the top of the sucrose density gradient (very little protein is detected in these fractions, ∼3% of total proteins; see Supplemental Figure 2 online) and causes REM relocation to DSM subfractions at the bottom of the gradient (Figure 1C). We checked that the phospholipid composition of the PM is not altered by the mβCD treatment, despite the ∼50% reduction in sterols (see Supplemental Figure 2 online; Roche et al., 2008). Importantly, mβCD treatment does not significantly affect the amount of PM-localized REM (Figure 1C, inset), suggesting a reorganization of this protein within the PM.
Remorin Clusters in Microdomains in the Cytosolic Leaflet of the PM
Immunogold Labeling of REM on Purified PM Coupled with Point Pattern Statistical Analysis
We designed an EM strategy to study the distribution of REM in membrane microdomains in an mβCD-dependent manner. We first checked by EM that treatment with mβCD does not significantly modify the morphology of tobacco PM vesicles (Figure 2A). Using an in-batch immunogold labeling procedure, we further observed that REM label is clustered in domains within the isolated PM (Figure 2A). To analyze these data quantitatively, we performed a point pattern analysis as described by Prior et al. (2003). We estimated a density function by computing statistical distances between the gold particles using Ripley's K-functions. Figure 2A shows the density (K) functions, and their first minimum derivative, indicated by an arrowhead. The only consistent minimum across a series of images pointed to the existence of ∼75-nm diameter clusters (Figure 2B). This result clearly shows that REM is clustered in the PM preparation fractions and forms membrane microdomains. By contrast, immunogold labeling of PM vesicles with antibodies to PMA shows no significant clustering of gold particles, but rather a homogeneous labeling (see Supplemental Figure 3A online). Importantly, a negative control without primary antibodies showed an 80% decrease of gold labeling for the same amount of PM vesicles. This result allows us to rule out the hypothesis of an unspecific clustering of the secondary antibodies (see Supplemental Figure 3B online).
Figure 2.
REM Locates in Membrane Domains in the Cytosolic Leaflet of the Tobacco Leaf PM.
(A) Left: Transmission electron micrographs of negatively stained tobacco PM vesicles treated or not with mβCD (20 mM, 30 min). Center: REM locates in membrane domains of ∼80 nm. PM vesicles were treated or not with mβCD then immunogold labeled in batch, detected by GAR10, and observed by EM. Arrows point to obvious areas of REM clustering. Right: Statistical point patterns show analyses of the whole images by Ripley's K function. First minimum (arrowhead) of the derivative of K function (dotted) indicates the average size of clusters seen in the picture.
(B) K-function analyses calculated as described in (A) are pooled and reported.
(C) REM is clustered in DIMs on one side of the bilayer. PMs and DIMs processed as described in (A) were embedded in resin, ultrathin cut, and observed by EM. The PM appeared mainly as vesicles (V); DIMs appeared as membrane sheets more or less parallel to the section plane. Arrows show the border of the lipid raft bilayer.
The clustering effect disappeared when the PM preparations were treated previously with mβCB (Figure 2A, bottom). These observations allow us to establish for REM a correlation between mβCB-dependent TX100 insolubility (Figure 1) and mβCB-dependent microdomain organization in the plant PM (Figure 2). Importantly, mβCD treatment did not change the total density (number of gold particles) of PM labeling (Figure 2B) but induced a change in REM distribution from clusters to random distribution. This result is consistent with mβCD treatment not modifying the amount of REM attached to the PM (as shown in Figure 1C, inset). For more detailed ultrastructural observations, ultrathin sections of the same in batch immunolabeled PM fractions were observed by EM. The PM mostly appears as vesicles with occasional clusters of REM label of ∼60 to 80 nm in diameter (Figure 2C).
In parallel, we also analyzed DIM fractions. As expected, immunogold-labeled DIMs appeared as a population of nonvesiculated bilayer membrane ribbons as described by Mongrand et al. (2004), heavily labeled for REM. Clusters of REM-rich domains were observed on only one side of the lipid bilayer, consistent with REM being located mostly in one side of the PM in vivo (i.e., the cytosolic leaflet of the PM; Figure 2C). It should be noted that the final overall size of REM-labeled domains in purified DIMs is overestimated because TX100 treatment is known to trigger aggregation of preexisting rafts (Chamberlain, 2004).
Translational GFP Fusions of Full-Length REM Show Discontinuous Labeling in the PM
To gain insight into REM behavior in vivo, we assessed the subcellular localization of full-length REM fusions to GFP or yellow fluorescent protein (YFP) reporters following expression under the control of the 35S cauliflower mosaic virus promoter. Constructs were expressed transiently in tobacco leaves or BY-2 cells using Agrobacterium tumefaciens–mediated transformation. Protein fusions were observed by confocal laser scanning microscopy. GFP fused to PMA4 stably expressed in BY-2 cells or transiently expressed in tobacco epidermis (Lefebvre et al., 2004) was used as a PM nonraft marker (Figures 3A and 3B). Median planes clearly showed YFP-REM colocalized with the PM marker (Figure 3A). Translational fusions of the fluorescent reporters YGP or GFP at either the C or N terminus of REM showed similar localization (see Supplemental Figure 4 online). Maximum projections (z series) of an expressing cell showed that no internal organelles were labeled (Figure 3B, two-dimensional maximum projection).
Figure 3.
Translational-GFP/YFP Fusions to REM Are Targeted to the PM.
(A) Tobacco epidermis cells expressing the PM H+ pump ATPase (PMA4) fused to GFP (GFP-PMA4) and YFP-REM; observation by confocal microscopy shows that REM localizes to the PM.
(B) Two-dimensional maximal projection of tobacco epidermis cell expressing GFP-REM showing labeling distribution through the whole cell; no labeling was observed in the cytoplasm (cytosol and organelles).
(C) Detailed comparison between GFP-PMA and GFP-REM labeling in tobacco epidermis and BY-2 cells through PM and secant confocal planes. By contrast with GFP-PMA4, GFP-REM shows a patchy localization both in the PM plane and in close-up view through secant plane.
A careful examination of z serial images showed that the GFP-REM labeling was not uniform, but exhibited a patchy distribution in the PM both in tobacco epidermis and BY-2 cells (Figure 3C). Analysis of images taken in the plane of the PM indicated that these fluorescent membrane domains were ∼600 nm in diameter and represented ∼25% of the surface of the PM (see Supplemental Figure 5 online). It should be noted that this size is likely overestimated due to fluorescence diffusion, as reviewed by Hanson and Kohler (2001). By contrast, PMA4-GFP fluorescence was more homogeneous in the plane of the PM both in tobacco epidermis and BY-2 cells (Figure 3C).
REM Is Located Both in PM Microdomains and in Plasmodesmata
REM has been proposed to have some of the properties of viral movement proteins and may interact with plasmodesmata (PD) (Reymond et al., 1996). Subcellular distribution of GFP-REM expressed in tobacco epidermis or BY-2 cells showed the fusion protein remaining with the retracted PM upon plasmolysis, although Hechtian strands connected to punctate labeling on the cell wall remained visible (Figure 4A). These results led us to ask whether REM could be detected within PD. Colocalization experiments between YFP-REM and GFP-PDLP1, a recently identified type I membrane protein targeted to PDs (Thomas et al., 2008), showed that colabeled punctuate structures were retained on the cell wall after plasmolysis (Figure 4B; see Supplemental Figure 6 online).
Figure 4.
Translational-GFP/YFP Fusions to REM Localize Both in the PM and PD.
(A) REM is located in adhesion sites (arrows) between the PM and cell wall in plasmolyzed calcofluor-stained (CaFl) BY-2 cells (left) or epidermal leaf tobacco cells (right) expressing GFP-REM.
(B) REM colocalized with the PD marker PDLP1. Plasmolyzed tobacco cells expressing YFP-REM and GFP-PDLP1. Arrowheads show colabeled spots retained in the cell wall after plasmolysis, and asterisks indicate attachment sites to the retracted protoplast (see Supplemental Figure 5 online).
To gain insight into the location of endogenous REM in plant tissues (without 35S-driven overexpression), we first performed indirect immunolabeling of tomato root cells, using primary antibodies to REM with secondary antibodies coupled to fluorescent probes. The labeling outlined the PM, and no labeling was within the cytoplasm. In the PM, REM was distributed with a patchy pattern in membrane domains of ∼600 nm (Figure 5A). As discussed for the GFP fusion, the size of domains is likely overestimated. Incorporation of REM into new PM, best seen on the division wall at mitosis, appeared only at late telophase when the phragmoplast is sealed to form the new division wall and accompanying PM (see Supplemental Figure 7 online).
Figure 5.
Endogenous REM Localizes Both in PM and PD in Vivo.
(A) Localization of REM by immunofluorescence in tomato root cells.
(B) Representative electron micrographs showing immunogold-labeled REM along the PM of high-pressure frozen tobacco root apices immunolabeled with antibodies to REMα130 and detected by GAR5 (arrowheads).
(C) Representative electron micrographs and corresponding drawings showing immunogold-labeled REM in PD. Left: High-pressure frozen tobacco root apex immunolabeled with antibodies to REMα130 and detected by GAR5 (arrows). Right: Chemically fixed tomato shoot apex immunolabeled with antibodies to REMpp34 and detected by GAR10; arrowheads show the necks of PD. CW, cell wall.
Immunogold labeling was also performed on tomato and tobacco root and shoot tissues subjected to either chemical fixation or to high-pressure freeze substitution. Thin sections were analyzed by indirect immunogold labeling using two different polyclonal antibodies raised against REM. REM labeling was observed along the PM in small clusters of gold particles (Figure 5B). In longitudinal sections through PDs, gold labeling was present proximal to the PD and along the length of the PD channel (Figure 5C).
Toward Biological Functions of REM
Transgenic Tomato Lines Misexpressing REM Do Not Reveal Any Striking Phenotype
Transgenic tomato lines containing sense or antisense REM cDNA sequences expressed from the 35S cauliflower mosaic virus promoter were generated. We selected four lines overaccumulating REM (O1, O2, O408, and O437) and two lines underaccumulating REM (u327 and u407) for the further studies. These plants contained between 3.3× (O437) to 0.5× (u407) the amount of REM present in wild-type plants (Figure 6A). Wild-type plants and tomato transformed with the empty vector (P21) served as controls. Importantly, in these plants, the transgenic protein was still targeted to its natural compartment (i.e., the PM; Figure 6A, inset). Compared with the wild type and P21, none of the transgenic plants revealed any significant growth abnormalities during the first 10 weeks. However, after this period, a slightly accelerated senescence could be observed in plants overexpressing REM (Figure 6B).
Figure 6.
Transgenic Tomato Lines with Altered REM Levels.
(A) REM transgenes significantly impact REM accumulation in 1-month-old transgenic tomatoes. Lines overaccumulating (O) or underaccumulating REM (u) were selected. Wild-type and transgenic plants transformed with the empty vector (P21) were also analyzed. Protein gel blot on crude total protein extract from T2 transgenic tomato leaves (further used in Figure 7 experiments; at least six plants per line) was quantified and expressed as percentage of signal in the wild type; error bars show sd. Inset: The PM was purified from some T2 transgenic tomato leaves, and REM level was measured by protein gel blotting
(B) Modulation of REM level in tomato causes no significant alteration in plant development except a slight early senescence in overaccumulating plants.
REM Misexpression in Tomato Lines Alters PVX Virus Propagation
The presence of REM in PDs (Figures 4 and 5) led us to focus on the physiological function generally associated with these cellular structures. PDs provide cell-to-cell connections that enable the symplastic transport of molecules from 1 to 60 kD (for review, see Oparka, 2004) . The paradigm for the latter process is the movement of viruses through PDs to achieve a local spreading infection and, ultimately, symplastic loading into the phloem to initiate systemic infection. Therefore, virus infections have provided effective monitors of the functionality of symplastic transport through PDs. To test the importance of REM in PDs trafficking, we used PVX. PVX infects tomato plants, and substantial knowledge is available with respect to its mode of cell-to-cell movement (Verchot-Lubicz et al., 2007).
REM Can Modulate PVX Spreading throughout the Plant
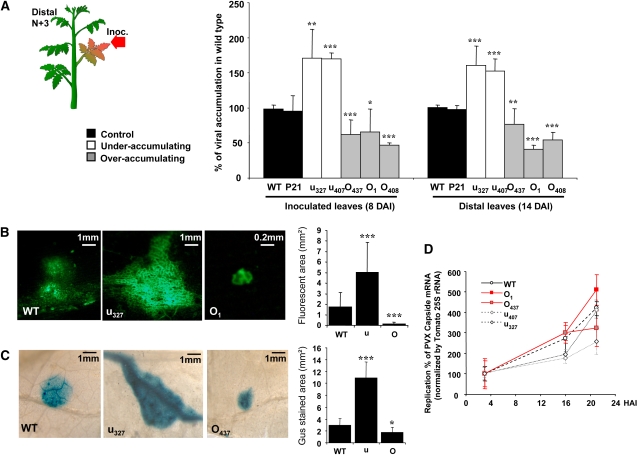
We assessed the impact of altered accumulation of REM on the ability of PVX to establish infections. Transgenic plants were inoculated with PVX, and viral accumulation in both inoculated leaves and systemically invaded leaves was evaluated using ELISA. At 8 d after inoculation (DAI), compared with wild-type and control plants, a significant increase in viral accumulation in the inoculated leaves of plants underaccumulating REM was observed (Figure 7A). At 14 DAI, the amount of virus in distal leaves, three nodes above the inoculated leaf, was also significantly increased in these plants. Consistently, reduced virus accumulation in both inoculated and distal leaves was observed in plants overexpressing REM (Figure 7A). These results indicate that REM is involved either directly or indirectly in movement of PVX through PD.
Figure 7.
REM Misexpression Alters PVX Virus Propagation.
(A) PVX viral charge is inversely correlated with REM level in 4-week-old transgenic tomatoes, both at the local and systemic levels. Viral charge was assayed by ELISA using antibodies to PVX coat protein on inoculated leaves (at 8 DAI) and distal (n+3) leaves (14 DAI). Three independent experiments were performed with seven to eight plants for each transgenic line and nontransgenic (WT) or empty vector control (P21).
(B) and (C) PVX spreading was inversely correlated with REM level in transgenic tomatoes. Four-week-old wild-type and transgenic tomato lines were inoculated with PVX-GFP (B) or PVX-GUS (C). Representative pictures of inoculated leaves were taken at 4 DAI. Graphs show average area of infection foci from ∼150 pictures of various lines of each genotype.
(D) Protoplasts of transgenic tomatoes misexpressing REM were inoculated with 4 μg of purified PVX RNA. PVX RNA accumulation was measured by qRT-PCR and normalized to 25S rRNA. HAI, hours postinoculation.
For graphs, error bars show sd, and significance is assessed by a Student's t test (*, P < 0.1; **, P < 0.05; ***, P < 0.001).
REM Is Involved in PVX Cell-to-Cell Movement
To test whether REM alters the cell-to-cell movement of PVX in inoculated leaves, wild-type and transgenic tomato plants were inoculated with PVX-GFP (Figure 7B) (Santa-Cruz et al., 1996). Results were also confirmed using PVX expressing the GUS reporter gene (Figure 7C) (Chapman et al., 1992). Analysis of the size of the infection foci with tagged virus at 4 DAI revealed differences in the spread of the virus between transgenic plants and the wild type. In plants with low REM levels, the mean diameter of the foci was threefold larger than in wild-type plants (Figure 7B). By contrast, infection foci in REM-overexpressing plants were up to eightfold smaller than in the wild type, and phloem loading leading to systemic infection was rare in these plants. Furthermore, small discrete infection foci (<30 cells) were frequently observed on inoculated leaves of REM sense lines (Figure 7B) but never observed in wild-type or antisense lines. These results indicate that overexpression of REM negatively impacts cell-to-cell movement of PVX in tomato.
REM Does Not Impair PVX Replication
The reduced accumulation and spread of PVX in overexpressing transgenic lines might be explained by reduced virus replication in single cells (i.e., the result of defective virus replication). Tomato protoplasts were prepared (Kohm et al., 1993) from transgenic lines and inoculated with PVX transcripts (Kavanagh et al., 1992). As shown in Figure 7D, the time course accumulation of viral RNA (monitored by quantitative RT-PCR [qRT-PCR]) was not significantly different in protoplasts from the various transgenic lines. This result indicates that the cell-to-cell movement of PVX, rather than its replication, is affected by REM.
REM Influences PVX Cell-to-Cell Movement by Directly Binding to TGBp1 Movement Protein
PVX requires three movement proteins, named triple gene block proteins (TGBp1, -2, and -3), and the viral coat protein (CP) to facilitate viral cell-to-cell transfer and vascular transport. To discover the mechanism by which REM could play a role in PVX propagation, we analyzed putative interactions between REM and TGBp1, -2, or -3, or CP using the split-ubiquitin assay (Deslandes et al., 2003). Our data clearly show a strong interaction between TGBp1 and REM in yeast (Figure 8A). To confirm the physical interaction of TGBp1 and REM, we performed a pull-down assay using glutathione S-transferase (GST)-tagged REM. We used Agrobacterium infiltration to express GFP-tagged viral proteins (TGBp1, TGBp2, or CP) in tobacco leaves. Pull-down assays were performed using glutathione-agarose beads alone (negative control) or beads coupled with GST-tagged REM. TGBp1-GFP was the only viral protein pulled down by REM in this assay (Figure 8B).
Figure 8.
REM Directly Binds PVX TGBp1 to Alter PVX Cell-to-Cell Transfer.
(A) REM directly interacts with TGBp1 in a yeast split ubiquitin assay. Split ubiquitin interaction between REM and PVX movement (TGBP-1, -2, and -3) and capsid (CP) proteins.
(B) REM directly interacts with TGBp1 in a pull-down assay. GFP-tagged TGP1-2 and CP were transiently expressed in tobacco leaves. Leaf crude extracts were analyzed by protein gel blots using antibodies to GFP. Proteins were extracted and incubated overnight with glutathione sepharose beads with or without GST-tagged REM. Pulled-down proteins were analyzed after three washes.
(C) YFP-REM and TGBp1-GFP colocalize when transiently expressed in tobacco leaves. Image shows two-dimensional maximal projections throughout the cell.
Localization studies performed on PVX movement proteins revealed that TGBp1 is crucial for PVX cell-to-cell movement (Krishnamurthy et al., 2002) and targets PD (Howard et al., 2004). We used tobacco leaves agroinfiltrated with TGBp1-GFP and REM-YFP to test the colocalization of these proteins. As expected, confocal microscopy confirmed the colocation of TGBp1-GFP and REM-YFP (Figure 8C).
DISCUSSION
Prototype REM Is a Marker for Plant PM Rafts
In this article, we show that REM is almost exclusively present in the DIM fraction (Figures 1B and 1C). The use of mβCD, a chelator of free sterols, led to the complete removal of the DIM fraction obtained after TX100 extraction and causes REM relocation to the TX100-soluble fractions. These results are in agreement with quantitative proteomics using differential treatment with mβCD, in which two members of the Arabidopsis remorin protein family (At2g45820 and At3g61260) were identified as mβCD-responsive DIM proteins (Kierszniowska et al., 2009) and are therefore primary candidates for an association with a sterol-rich membrane microenvironment, defined as lipid rafts.
In contrast with REM, the proton pump PMA, used in our experiments as a PM marker, is a non-mβCD-responsive raft protein (Kierszniowska et al., 2009). Although previous works showed that PMA is slightly enriched in DIMs versus total PM preparations when the same amount of proteins per lane was loaded (Mongrand et al., 2004; Borner et al., 2005; Shahollari et al., 2005; Morel et al., 2006; Laloi et al., 2007), this protein preferentially partitions into DSM fractions (Lefebvre et al., 2007; this work) and therefore does not constitute a marker for imaging plant rafts. Our localization data thus clearly show a characteristic difference in the patterns of distribution for REM and PMA in the PM.
Evidence for Sterol-Dependent Membrane Microdomains in the Cytosolic Leaflet of Plant PMs
Although estimates of the size of lipid rafts in animals and yeast may vary depending on the physiological state of the cell, reported diameters are ∼100 nm (Pike, 2006). In our work, immunogold labeling/EM coupled to statistical analysis showed that REM clusters in domains of ∼75 nm. Evidence that these clusters relate to lipid rafts was provided by the demonstration that the REM labeling pattern became random following mβCD treatment, likely due to free sterol depletion, which neither modifies the amount of REM attached to the PM (Figure 1C) nor changes the total density of PM labeling (number of gold particles, Figure 2B). The random distribution of the REM raft marker after mβCD treatment is also important, showing that preparation, fixation, and gold labeling of the PM vesicles does not induce clustering of the labeled protein per se, particularly considering the fact that REM can interact with itself (Bariola et al., 2004). Proteolytic analysis of RSO and inside out PM vesicles pointed to REM being located on the cytosolic face of the PM, a conclusion also supported by the immunolabeling of the PM (Figure 2C).
REM Is a PD-Associated Protein
Immunolabeling and colocalization with PDLP1 show that REM is associated with PDs. The presence of REM indicates that the PM lining PDs likely contains lipid raft membrane domains and may therefore contain other proteins associated with these structures. Proteins commonly assigned to lipid rafts are the Glycosyl PhosphatidylInositol-anchored proteins (Borner et al., 2005). Hence, it is interesting that a GPI-anchored glucanase (Levy et al., 2007) and a GPI-anchored callose binding protein (Simpson et al., 2009) are among the few identified PD proteins.
Because REM is widely distributed in the PM, and the PM between cells is contiguous through PDs, presence of REM in PDs need not implicate it directly in trafficking of macromolecules through PD. However, we showed that REM interferes with the cell-to-cell movement of PVX virus and interacts with TGBp1 movement protein. This virus protein has been shown previously to be targeted to PD (Howard et al., 2004) and to be responsible for virus-induced gene silencing, which operates as a systemic, sequence-specific defense system (Voinnet et al., 2000). Because plants overexpressing REM show an early senescence phenotype, it could be argued that the inhibition of virus movement could be an indirect consequence of the incurred stress (e.g., by callose deposition; Jongebloed et al., 2004; Benitez-Alfonso et al., 2009), perhaps associated with extraneous targeting of the overexpressed protein. However, the reverse effect on virus movement in plants with lower REM levels (with no apparent phenotype when compared with wild type) makes this unlikely. In addition, overexpressed REM-GFP fusion showed no cytoplasm labeling (cytosol or organelles), and anti-REM protein gel blots performed on PM preparations from REM overaccumulating plants show that overexpressed REM accumulates in the PM. Together, these results suggest that REM overexpression and reporter fusion do not significantly alter REM subcellular localization.
Toward the Understanding of REM Function at the PD
The precise mechanisms by which endogenous proteins or viral genomes traffic through PDs are poorly understood, but for processes not dependent upon molecular diffusion, specific regulators must be required. It is possible that such regulators might be directly associated with PDs where they could physically interact with the virus movement protein(s) or the translocating virus itself. The identification of REM, and lipid rafts, as structural features within the PD, raises the possibility that REM could act as such a regulator. The fact that PVX movement is inversely related to REM accumulation suggests that REM acts as a negative regulator of virus movement. We further demonstrated that REM is likely involved in altering PVX cell-to-cell movement by direct physical interaction with the virus protein TGBp1.
In animal cells, the lipid raft hypothesis proposes that the recruitment and aggregation of lipid rafts is an important driving force (Kusumi and Suzuki, 2005). For example, anchorage of HIV on cells leads to coaggregation of viral particles with surface nucleolin at membrane raft microdomains. (Nisole et al., 2002), and sphingomyelin and cholesterol promote the surface aggregation of HIV-1 gp41 monomer (Saez-Cirion et al., 2002). In plants, viral ribonucleoprotein complexes are thought to be trafficked to PD by their attachment to the cytosolic face of the endoplasmic reticulum (Oparka, 2004). Similarly, REM could titrate out TGBp1 and prevent TGBp1 from carrying out its role in virus movement. REM-containing lipid rafts could serve as a counteracting membrane platform for viral ribonucleoprotein docking to PD, leading to a reduced PVX movement.
An interesting parallel can be drawn with animal cell systems where PM rafts are used by viruses as entry sites, platforms for the assembly of viral components, and scaffolds for the budding of virus from infected cells (for review, see Chazal and Gerlier, 2003). Plant viruses are symplastically isolated, so the role of lipid rafts must be fundamentally different, but these structures could be involved in cell-to-cell movement of the virus.
More generally, it should be noted that REM was also found in the interactome of rice infected with rice yellow mottle virus (Brizard et al., 2006), and the expression of REM homologs in susceptible Arabidopsis accessions was changed following infection with potyviruses (Valérie Schurdi-Levraud, personal communication). On the other hand, Arabidopsis REMs were recently found to accumulate differentially and to undergo posttranslational modifications (i.e., phosphorylation in plants expressing the bacterial avrRpm1 effector; Widjaja et al., 2009). Therefore, REM involvement in plant–pathogen interactions does not seem to be restricted to viruses. Posttranslational modification of REM protein could be involved in activation of REM-based blocking of PVX cell-to-cell movement and in the host-pathogen relationship in general. This hypothesis is consistent with previously reported phosphorylation of REM, which could be a possible regulatory mechanism (Reymond et al., 1996). Additional experiments will be required to reveal the range of pathogens affected by REM and the associated mechanisms. Key questions emerging from this work also relate to the role of REM in the PM or whether REM has a more general role in macromolecular trafficking through PD.
METHODS
Plant Material, Inoculation, and Fluorescence Microscopy
Leaves were obtained from 8-week-old tobacco plants (Nicotiana tabacum cv Xanthi) grown in a growth chamber at 25°C under 16/8 h day/night conditions. Four-week-old tobacco plants were used for Agrobacterium tumefaciens (strain GV3101)–mediated transient expression (Batoko et al., 2000). N. tabacum cv Bright Yellow-2 (BY-2) suspension cultured cells were grown in a modified Murashige and Skoog medium as previously described (Couchy et al., 2003) and transformed as described by Nagata et al. (1992) with the constructs described in Supplemental Table 1 online. Four-day-old BY-2 tobacco cells were cocultivated with Agrobacterium strains during 2 d, and the REM transient expression was analyzed by confocal laser scanning microscopy. FM4-64 was used at the concentration of 4.25 μM and the calcofluor white at 0.01% (0.1 μM). Confocal imaging was performed 48 h after agroinfiltration using a Leica TCS SP2 confocal microscope with a ×63 oil immersion objective and LCS Lite Leica software. Laser and image settings were as described by Brandizzi et al. (2002), and GFP/YFP acquisitions were performed by sequential image recording between frames according to the manufacturer's instructions. Cells were plasmolyzed with 4% NaCl for 15 min.
Tomato plants (Solanum lycopersicum cv Ailsa Craig) were grown in a growth chamber at 150 μE/m2/s, 25 to 18°C under 16-8 h day/night, and 75% humidity conditions. Transgenic tomato lines were generated using GV3101Agrobacterium strains carrying the constructs according to Fillatti et al. (1987), except that kanamycin was removed during root regeneration. Transformed plants were screened by growth on 50 mg/mL kanamycin, by PCR, and by protein gel blot using antibody to REMpep. All experiments involving tomato trangenics were performed on T2 plants grown on Murashige and Skoog plates containing 50 mg/mL kanamycin, and each plant was assayed for its REM level by protein gel blot analysis.
Molecular Cloning, Transcription, and qRT-PCR
Standard and Gateway molecular techniques were used for cloning and subcloning, with vectors and primers given in Supplemental Tables 1 and 2 online. All the GFP fusion constructs were built using prototype StREM1.3. PVX cDNA transcription was performed with the Ambion mMESSAGE mMACHINE kit according to the manufacturer's instructions. qRT-PCR was performed as described by Joubes et al. (2008) with the primers shown in Supplemental Table 2 online.
Preparation of Highly Enriched PM Fractions and DIMs, and mβCD Treatment of PMs
Tobacco leaf PM was purified by phase partition as described by Mongrand et al. (2004). To perform sterol depletion, PM fractions were incubated with stirring for 30 min at room temperature with 20 mM mβCD (Cell Culture Tested; Sigma-Aldrich) in buffered conditions (20 mM Tris, pH 7.6, 150 mM NaCl, and 1 mM PMSF; see also Roche et al., 2008). PMs were further submitted to TX100 treatment (final concentration 1%, v/v) with a detergent-to-PM protein mass ratio of 15 at 4°C for 30 min. Treated membranes were brought to a final concentration of 52% sucrose (w/w), overlaid with successive 3-mL steps of 40, 35, and 30% sucrose in TBS buffer (w/w), and then centrifuged for 16 h at 200,000g in a TST41 rotor (Sorvall). DIMs could be recovered above the 30 to 35% layers as an opaque band. This fraction was washed in 4 mL of TBS buffer to remove residual sucrose. The protein concentration was determined with a BCA protein assay to avoid TX100 interference, using BSA as a protein standard. Proteins in the sucrose density gradient were precipitated in 10% cold trichloroacetic acid for 30 min at 4°C. After centrifugation, the pellet was first washed with 10% trichloroacetic acid in water to remove residual sucrose and finally with cold acetone before being resuspended in Laemli buffer for SDS-PAGE. To check the morphology of PM vesicles after mβCD treatment, vesicles were deposited on collodion-coated grids and negatively stained in 1% ammonium molybdate for 2 min and observed by TEM.
To analyze the repartition of REM in the cytosolic versus apoplastic leaflet of the PM, PM vesicles were purified by phase partition generating mostly sealed, RSO vesicles (Larsson, 1988). Orientation of tobacco PM vesicles obtained by phase partition was determined by measurement of Mg2+-dependent proton pump ATPase activity according to Serrano and Portillo (1990). Inside-out vesicles were obtained after subsequent freeze in liquid nitrogen and thaw cycles in ice from RSO PM vesicles. Thirty micrograms of vesicles were treated at 37°C for 30 min in the presence of 1 μL of trypsin (7500 U·mg−1; Sigma-Aldrich). Proteins were analyzed by protein gel blots.
Protein Separation, Protein Gel Blots, and Protein Purification
All steps were performed at room temperature as described by Mongrand et al. (2004). Primary antibodies to REM were diluted 1/3000 (REMFL; 60 μg/mL), 1/2000 (REMpep; 18.3 μg/mL), 1/2000 (REMα130), and 1/20,000 (PMA), and anti-rabbit secondary antibodies coupled to horseradish peroxidase (Amersham Pharmacia) were added at 1/10,000 dilution. The antibodies raised against the ATPase PMA2 (Maudoux et al., 2000) may recognize PMA4 as well (Lefebvre et al., 2004). The 6-histidine-tagged and GST-tagged REMs (see Supplemental Table 1 online for cloning information) were expressed in Escherichia coli BL21 DE3 cells and purified using chelating sepharose, fast flow, and glutathione sepharose, respectively, according to the manufacturer's instructions (Amersham/Bioscience).
Sequence and Phylogenetic Analyses
Protein sequences were aligned using ClustalW with default parameters. Phylogenetic analysis was performed using the phylip package (http://evolution.genetics.washington.edu/phylip.html). Bootstrapping was performed on 2000 replicates with the seqboot program, 10 times jumbling on 250 data sets was done in the protpars program for protein sequence parsimony analysis, and a consensus tree was computed with the consense program. Parsimony analysis was conducted by a “subtree pruning and regrafting” search. A maximum likelihood approach (promlk program) based on the Jones-Taylor-Thornton model was used to infer true branch lengths, using the bootstrap data set and the parsimony consensus tree as input files.
Immunofluorescence Staining
Immunostaining procedures were performed on partially digested tomato roots as previously described (Hawes and Satiat-Jeunemaitre, 2001). A rabbit polyclonal antibody REMα130 was used at 1:600. Secondary antibodies were purchased from Sigma-Aldrich (fluorescein isothiocyanate–conjugated anti-rabbit used at 1:60). Slides were observed and images were collected using an upright laser scanning confocal microscope (TCS SP2; Leica Microsystems). The oil objectives used were ×40 numerical aperture 1.25 and ×63 numerical aperture 1.30, giving a resolution of ∼200 nm in the XY-plane and 400 nm along the z axis (pinhole 1 Airy unit).
Immunogold Labeling of Purified Plant Membranes
Tobacco leaf PM and DIM preparations were prefixed with 4% paraformaldehyde, blocked with 0.1% glycine and 5% BSA, and further incubated with antibodies to REMα130 (Reymond et al., 1996) at 1/800. Labeling was visualized with gold-conjugated GAR5 anti-rabbit secondary antibodies (Tebu-bio). Membranes were rinsed twice by centrifugation/resuspension with TBS/BSA between each step. Samples were either directly (in batch) dropped on parlodion-coated grids (homemade with 2% parlodion solution in isoamyle acetate) or embedded to be observed by transmission EM. In the latter case, samples were submitted to the progressive lowering temperature procedure. Briefly, samples were fixed 1 h at 4°C in 4% paraformaldehyde, 0.25 glutaraldehyde in 0.1 M phosphate buffer, pH 7.2, first rinsed in phosphate buffer with 0.5 M sucrose (three times, 5 min), second in 30% ethanol, and finally with 50% ethanol, prior to progressive dehydration by the progressive lowering temperature procedure. The temperature was decreased from 4 to −10°C. Samples were further dehydrated in 70% ethanol for 20 min and stained with 0.5% uranyl acetate in 70% ethanol for 1 h. The ethanol concentration was increased to 100% in two steps over 5 h and the samples progressively infiltrated with LR gold resin as follows: 20% in acetone for 1 h, 40% for 2 h; 60% 1 for 2 h, 80% 1 for 2 h, and 100% overnight.
EM, Chemical Fixation, and High-Pressure Freezing on Plant Tissue
Tomato shoot apices were treated with 3% glutaraldehyde and 1% OsO4. The samples were further embedded in Epon resin before immunogold labeling with antibodies to REMpp34. Tobacco tissues (root, leaves, and shoot) were high-pressure frozen with a Leica EMPACT system. Tissues were inserted in a flat copper carrier, fast frozen, and substituted in acetone containing 0.25% uranyle acetate for 36 h at −90°C. The temperature of the substitution medium was first raised to −60°C for 8 h, to −30°C for 13 h, and finally to −20°C. The substitution medium was then rinsed with acetone and replaced progressively with LR gold with 0.1% benzyl as follows: 20% in acetone for 1 h, 40% for 2 h; 60% 1 for 2 h, 80% 1 for 2 h, and 100% overnight.
Experiments with PVX
Mechanical virus inoculation, histochemical GUS staining, and quantification of PVX by ELISA were performed as described by Chapman et al. (1992), German-Retana et al. (2003), and Foxe and Prakash (1986), respectively.
The PVX derived from pTXS corresponds to the British isolate PVXUK3 (Kavanagh et al., 1992). Symptoms induced by this PVX isolate on tomato are very mild. GFP infection foci were observed using a Leica MZ16F binocular/DFC420 camera. ImageJ software (http://rsbweb.nih.gov/ij/) was used for area measurements. Protoplasts of transgenic tomato over- or underexpressing REM were prepared according to Kohm et al. (1993) and inoculated with polyethylene glycol with 4 μg of purified PVX RNA and cultured for 24 h.
Yeast Split-Ubiquitin Assay
Split ubiquitin assays were performed as described by Deslandes et al. (2003) except that yeasts were grown on SD-his/-trp + 90 mM fluoro-orotic acid to test the interaction.
GST Pull-Down Assay
GST-tagged REMFL was bound to sepharose beads according to the manufacturer's instructions. Four-week-old tobacco plants grown were used for Agrobacterium-mediated transient expression of TGBp1-3 and CP fused to GFP. Inoculated leaves were ground in liquid nitrogen. Plant proteins were extracted in the extraction buffer containing 50mM TrisHCl, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 1% TX100, 5 mM β-mercaptoethanol, 2.5 mM Na orthovanadate, 10 mM NaF, and protease inhibitor cocktail (Roche). Plant protein extracts were then filtered and cleared 5 min at 8000g. Beads (with or without GST.REMFL) were incubated overnight at 4C with 1 mL of protein extract and then rinsed three times with 2 mL of extraction buffer. Pull-down material was finally analyzed by protein gel blotting.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: StREM1.3, P93788; SlREM1.2, AAD28506; and EST sequence used for cloning of NtREM1.2, EB449751. Sequences used for phylogenetic analysis are as follows: NtREM1.2, EB451162; NtREM1.1, EB449751; SlREM1.1, AAD28507; StREM1.1, BI176676; StREM1.2, DN921711; and StREM6.1, CK269035.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Prototype Clade of REM in Solanaceae Species and Antibodies Developed for Its Study.
Supplemental Figure 2. Total Protein and Lipid Monitoring during Cyclodextrin Treatment.
Supplemental Figure 3. Negative Controls for in-Batch Immunogold Labeling of PM Vesicles.
Supplemental Figure 4. Localization of REM-GFP Constructs Transiently Expressed in Tobacco Epidermal Cells.
Supplemental Figure 5. Estimation of GFP-REM Spot Size by Image Analysis.
Supplemental Figure 6. Additional Pictures Showing Colocalization between YFP-REM and GFP-PDLP1.
Supplemental Figure 7. Immunofluorescence Labeling of REM in Tomato Dividing Root Cells Shows Accumulation of REM in the Phragmoplast at Late Telophase.
Supplemental Table 1. List of Constructs and Vectors Used in This Study.
Supplemental Table 2. List of Primers Used in This Work.
Supplemental Methods. Clustering Analysis on Electron Microscopy Images, and Calculation of the Percentage of REM Located in the Inner Leaflet.
Supplemental Data Set 1. Text File of the Alignment Used to Generate the Phylogenetic Tree Shown in Supplemental Figure 1A.
Supplementary Material
Acknowledgments
Laurent Deslandes and Benoit Lefebvre (Laboratoire des Interactions Plantes Micro-organismes, Toulouse) are thanked for kindly providing us tools for split ubiquitin and antibodies to PMA. Marc Boutry (Unité de Biochimie Physiologique, Louvain-la-Neuve-Belgium) is thanked for providing the BY-2 cell line expressing PMA4-GFP. We also thank Frédéric Gaymard (Biochimie et Physiologie Moléculaire des Plantes, Montpellier), Stéphane Blanc (Biologie et Génétique des Interactions Plante -Parasite, Montpellier), Benoit Lefebvre (Laboratoire des Interactions Plantes Micro-organismes, Toulouse), Véronique Germain and Nathalie Frangne (Physiologie et Biotechnologie Végétales, Villenave d'Ornon) for fruitful discussion and Mohamed-Amine Belka (Laboratoire de Biogenèse Membranaire, Bordeaux), Christophe Der (Plantes Microbes Environnement, Dijon), Vincent Lebret-Soler (Laboratoire Bordelais de Recherche en Informatique, Bordeaux), Chantal Faure, and Fater Youssef (Génomique, Diversité et Pouvoir Pathogène, Villenave d'Ornon) for technical assistance, with particular thanks to Martine Peypelut (Plateau Technique Imagerie/Cytologie, Villenave d'Ornon) for immunogold labelling experiments and Marie-Thérèse Crosnier (Laboratoire de Dynamique de la Compartimentation Cellulaire, Gif-sur-Yvette) for immunofluorescence staining. We greatly thank Marie-Thérèse Esquéré-Tugayé (Surfaces Cellulaires et Signalisation chez les Végétaux Toulouse), Jérome Joubes (Laboratoire de Biogenèse Membranaire, Bordeaux) and Isabelle Jupin (Laboratoire de Virologie Moléculaire, Paris) for critical readings of the manuscript. We thank the anonymous reviewers for their advice and comments, which greatly improved the work presented here. The immunofluorescence studies have been performed on the Imaging and Cell Biology Platform, Institut Fédératif de Recherche 87 “La Plante et son Environnement.” DNA sequencing was performed at the Genotyping and Sequencing facility of Bordeaux (grants CRA nos. 20030304002FA, 20040305003FA and FEDER no. 2003227). The Laboratoire de Biogenèse Membranaire acknowledges Altadis - Institut du Tabac, Advanced Technologies (Cambridge), Institut National de la Recherche Agronomique-Centre National de Ressources Génomiques Végétales (http://cnrgv.toulouse.inra.fr) and the European Sequencing of Tobacco Project (http://www.estobacco.info/) for the use of materials. Part of this work was funded by a grant from the Swiss National Science Foundation to PR (31-49450.96). We acknowledge the French Agence Nationale de la Recherche (contracts to SM Jeune Chercheur(e) 05 -45555 “Plant rafts” and to FSP Jeune Chercheur(e) 05-50611 “Vegeraft”).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Sébastien Mongrand (sebastien.mongrand@biomemb.u-bordeaux2.fr).
Online version contains Web-only data.
References
- Bariola, P.A., Retelska, D., Stasiak, A., Kammerer, R.A., Fleming, A., Hijri, M., Frank, S., and Farmer, E.E. (2004). Remorins form a novel family of coiled coil-forming oligomeric and filamentous proteins associated with apical, vascular and embryonic tissues in plants. Plant Mol. Biol. 55 579–594. [DOI] [PubMed] [Google Scholar]
- Batoko, H., Zheng, H.Q., Hawes, C., and Moore, I. (2000). A rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal golgi movement in plants. Plant Cell 12 2201–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Alfonso, Y., Cilia, M., San Roman, A., Thomas, C., Maule, A., Hearn, S., and Jackson, D. (2009). Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc. Natl. Acad. Sci. USA 106 3615–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat, R.A., Miklis, M., Schmelzer, E., Schulze-Lefert, P., and Panstruga, R. (2005). Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc. Natl. Acad. Sci. USA 102 3135–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat, R.A., and Panstruga, R. (2005). Lipid rafts in plants. Planta 223 5–19. [DOI] [PubMed] [Google Scholar]
- Borner, G.H., Sherrier, D.J., Weimar, T., Michaelson, L.V., Hawkins, N.D., Macaskill, A., Napier, J.A., Beale, M.H., Lilley, K.S., and Dupree, P. (2005). Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137 104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi, F., Frangne, N., Marc-Martin, S., Hawes, C., Neuhaus, J.M., and Paris, N. (2002). The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell 14 1077–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizard, J.P., Carapito, C., Delalande, F., Van Dorsselaer, A., and Brugidou, C. (2006). Proteome analysis of plant-virus interactome: Comprehensive data for virus multiplication inside their hosts. Mol. Cell. Proteomics 5 2279–2297. [DOI] [PubMed] [Google Scholar]
- Brown, D.A. (2006). Lipid rafts, detergent-resistant membranes, and raft targeting signals. Physiology (Bethesda) 21 430–439. [DOI] [PubMed] [Google Scholar]
- Chamberlain, L.H. (2004). Detergents as tools for the purification and classification of lipid rafts. FEBS Lett. 559 1–5. [DOI] [PubMed] [Google Scholar]
- Chapman, S., Kavanagh, T., and Baulcombe, D. (1992). Potato virus X as a vector for gene expression in plants. Plant J. 2 549–557. [DOI] [PubMed] [Google Scholar]
- Chazal, N., and Gerlier, D. (2003). Virus entry, assembly, budding, and membrane rafts. Microbiol. Mol. Biol. Rev. 67 226–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couchy, I., Bolte, S., Crosnier, M.T., Brown, S., and Satiat-Jeunemaitre, B. (2003). Identification and localization of a beta-COP-like protein involved in the morphodynamics of the plant Golgi apparatus. J. Exp. Bot. 54 2053–2063. [DOI] [PubMed] [Google Scholar]
- Deslandes, L., Olivier, J., Peeters, N., Feng, D.X., Khounlotham, M., Boucher, C., Somssich, I., Genin, S., and Marco, Y. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA 100 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillatti, J.J., Kiser, J., Rose, B., and Comai, L. (1987). Efficient transformation of tomato and the introduction and expression of a gene for herbicide tolerance. Plant Biol. 4 199–210. [Google Scholar]
- Foxe, M.J., and Prakash, J. (1986). Expression of resistance to potato virus X in isolated potato protoplasts. Arch. Virol. 88 167–174. [DOI] [PubMed] [Google Scholar]
- German-Retana, S., Redondo, E., Tavert-Roudet, G., Le Gall, O., and Candresse, T. (2003). Introduction of a NIa proteinase cleavage site between the reporter gene and HC-Pro only partially restores the biological properties of GUS- or GFP-tagged LMV. Virus Res. 98 151–162. [DOI] [PubMed] [Google Scholar]
- Grossmann, G., Opekarova, M., Novakova, L., Stolz, J., and Tanner, W. (2006). Lipid raft-based membrane compartmentation of a plant transport protein expressed in Saccharomyces cerevisiae. Eukaryot. Cell 5 945–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson, M.R., and Kohler, R.H. (2001). GFP imaging: Methodology and application to investigate cellular compartmentation in plants. J. Exp. Bot. 52 529–539. [PubMed] [Google Scholar]
- Hawes, C.R., and Satiat-Jeunemaitre, B. (2001). Trekking along the cytoskeleton. Plant Physiol. 125 119–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerklotz, H. (2002). Triton promotes domain formation in lipid raft mixtures. Biophys. J. 83 2693–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, A.R., Heppler, M.L., Ju, H.J., Krishnamurthy, K., Payton, M.E., and Verchot-Lubicz, J. (2004). Potato virus X TGBp1 induces plasmodesmata gating and moves between cells in several host species whereas CP moves only in N. benthamiana leaves. Virology 328 185–197. [DOI] [PubMed] [Google Scholar]
- Jacinto, T., Farmer, E.E., and Ryan, C.A. (1993). Purification of potato leaf plasma membrane protein pp34, a protein phosphorylated in response to oligogalacturonide signals for defense and development. Plant Physiol. 103 1393–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongebloed, U., Szederkenyi, J., Hartig, K., Schobert, C., and Komor, E. (2004). Sequence of morphological and physiological events during natural ageing and senescence of a castor bean leaf: Sieve tube occlusion and carbohydrate back-up precede chlorophyll degradation. Physiol. Plant. 120 338–346. [DOI] [PubMed] [Google Scholar]
- Joubes, J., Raffaele, S., Bourdenx, B., Garcia, C., Laroche-Traineau, J., Moreau, P., Domergue, F., and Lessire, R. (2008). The VLCFA elongase gene family in Arabidopsis thaliana: Phylogenetic analysis, 3D modelling and expression profiling. Plant Mol. Biol. 67 547–566. [DOI] [PubMed] [Google Scholar]
- Kavanagh, T., Goulden, M., Santa Cruz, S., Chapman, S., Barker, I., and Baulcombe, D. (1992). Molecular analysis of a resistance-breaking strain of potato virus X. Virology 189 609–617. [DOI] [PubMed] [Google Scholar]
- Kenworthy, A.K. (2008). Have we become overly reliant on lipid rafts? Talking point on the involvement of lipid rafts in T-cell activation. EMBO Rep. 9 531–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszniowska, S., Seiwert, B., and Schulze, W.X. (2009). Definition of Arabidopsis sterol-rich membrane microdomains by differential treatment with methyl-beta-cyclodextrin and quantitative proteomics. Mol. Cell. Proteomics 8 612–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohm, B.A., Goulden, M.G., Gilbert, J.E., Kavanagh, T.A., and Baulcombe, D.C. (1993). A potato virus X resistance gene mediates an induced, nonspecific resistance in protoplasts. Plant Cell 5 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy, K., Mitra, R., Payton, M.E., and Verchot-Lubicz, J. (2002). Cell-to-cell movement of the PVX 12K, 8K, or coat proteins may depend on the host, leaf developmental stage, and the PVX 25K protein. Virology 300 269–281. [DOI] [PubMed] [Google Scholar]
- Kusumi, A., and Suzuki, K. (2005). Toward understanding the dynamics of membrane-raft-based molecular interactions. Biochim. Biophys. Acta 1746 234–251. [DOI] [PubMed] [Google Scholar]
- Laloi, M., et al. (2007). Insights into the role of specific lipids in the formation and delivery of lipid microdomains to the plasma membrane of plant cells. Plant Physiol. 143 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhorst, M.F., Reuter, A., and Stuermer, C.A. (2005). Scaffolding microdomains and beyond: The function of reggie/flotillin proteins. Cell. Mol. Life Sci. 62 2228–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson, K. (1988). Lipid phase transitions in membranes involving intrinsic periodic curvature. Chem. Phys. Lipids 49 65–67. [DOI] [PubMed] [Google Scholar]
- Lefebvre, B., Batoko, H., Duby, G., and Boutry, M. (2004). Targeting of a Nicotiana plumbaginifolia H+-ATPase to the plasma membrane is not by default and requires cytosolic structural determinants. Plant Cell 16 1772–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre, B., Furt, F., Hartmann, M.A., Michaelson, L.V., Carde, J.P., Sargueil-Boiron, F., Rossignol, M., Napier, J.A., Cullimore, J., Bessoule, J.J., and Mongrand, S. (2007). Characterization of lipid rafts from Medicago truncatula root plasma membranes: A proteomic study reveals the presence of a raft-associated redox system. Plant Physiol. 144 402–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, A., Erlanger, M., Rosenthal, M., and Epel, B.L. (2007). A plasmodesmata-associated beta-1,3-glucanase in Arabidopsis. Plant J. 49 669–682. [DOI] [PubMed] [Google Scholar]
- Marguet, D., Lenne, P.F., Rigneault, H., and He, H.T. (2006). Dynamics in the plasma membrane: How to combine fluidity and order. EMBO J. 25 3446–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maudoux, O., Batoko, H., Oecking, C., Gevaert, K., Vandekerckhove, J., Boutry, M., and Morsomme, P. (2000). A plant plasma membrane H+-ATPase expressed in yeast is activated by phosphorylation at its penultimate residue and binding of 14-3-3 regulatory proteins in the absence of fusicoccin. J. Biol. Chem. 275 17762–17770. [DOI] [PubMed] [Google Scholar]
- Mongrand, S., Morel, J., Laroche, J., Claverol, S., Carde, J.P., Hartmann, M.A., Bonneu, M., Simon-Plas, F., Lessire, R., and Bessoule, J.J. (2004). Lipid rafts in higher plant cells: Purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 279 36277–36286. [DOI] [PubMed] [Google Scholar]
- Morel, J., Claverol, S., Mongrand, S., Furt, F., Fromentin, J., Bessoule, J.J., Blein, J.P., and Simon-Plas, F. (2006). Proteomics of plant detergent-resistant membranes. Mol. Cell. Proteomics 5 1396–1411. [DOI] [PubMed] [Google Scholar]
- Munro, S. (2003). Lipid rafts: Elusive or illusive? Cell 115 377–388. [DOI] [PubMed] [Google Scholar]
- Nagata, T., Nemoto, Y., and Hasezawa, S. (1992). Tobacco BY-2 cell line as the 'HeLa' cell in the cell biology of higher plants. Int. Rev. Cytol. 132 1–30. [Google Scholar]
- Nisole, S., Krust, B., and Hovanessian, A.G. (2002). Anchorage of HIV on permissive cells leads to coaggregation of viral particles with surface nucleolin at membrane raft microdomains. Exp. Cell Res. 276 155–173. [DOI] [PubMed] [Google Scholar]
- Oparka, K.J. (2004). Getting the message across: How do plant cells exchange macromolecular complexes? Trends Plant Sci. 9 33–41. [DOI] [PubMed] [Google Scholar]
- Parton, R.G., and Simons, K. (2007). The multiple faces of caveolae. Nat. Rev. Mol. Cell Biol. 8 185–194. [DOI] [PubMed] [Google Scholar]
- Pike, L.J. (2006). Rafts defined: A report on the Keystone Symposium on Lipid Rafts and Cell Function. J. Lipid Res. 47 1597–1598. [DOI] [PubMed] [Google Scholar]
- Prior, I.A., Muncke, C., Parton, R.G., and Hancock, J.F. (2003). Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 160 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele, S., Mongrand, S., Gamas, P., Niebel, A., and Ott, T. (2007). Genome-wide annotation of remorins, a plant-specific protein family: Evolutionary and functional perspectives. Plant Physiol. 145 593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran, L., and Simons, K. (2005). Lipid rafts and membrane dynamics. J. Cell Sci. 118 1099–1102. [DOI] [PubMed] [Google Scholar]
- Reymond, P., Kunz, B., Paul-Pletzer, K., Grimm, R., Eckerskorn, C., and Farmer, E.E. (1996). Cloning of a cDNA encoding a plasma membrane-associated, uronide binding phosphoprotein with physical properties similar to viral movement proteins. Plant Cell 8 2265–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche, Y., Gerbeau-Pissot, P., Buhot, B., Thomas, D., Bonneau, L., Gresti, J., Mongrand, S., Perrier-Cornet, J.M., and Simon-Plas, F. (2008). Depletion of phytosterols from the plant plasma membrane provides evidence for disruption of lipid rafts. FASEB J. 22 3980–3991. [DOI] [PubMed] [Google Scholar]
- Saez-Cirion, A., Nir, S., Lorizate, M., Agirre, A., Cruz, A., Perez-Gil, J., and Nieva, J.L. (2002). Sphingomyelin and cholesterol promote HIV-1 gp41 pretransmembrane sequence surface aggregation and membrane restructuring. J. Biol. Chem. 277 21776–21785. [DOI] [PubMed] [Google Scholar]
- Santa-Cruz, S., Chapman, S., Roberts, A.G., Roberts, I.M., Prior, D.A., and Oparka, K.J. (1996). Assembly and movement of a plant virus carrying a green fluorescent protein overcoat. Proc. Natl. Acad. Sci. USA 93 6286–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, R., London, E., and Brown, D. (1994). Interactions between saturated acyl chains confer detergent resistance on lipids and glycosylphosphatidylinositol (GPI)-anchored proteins: GPI-anchored proteins in liposomes and cells show similar behavior. Proc. Natl. Acad. Sci. USA 91 12130–12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano, R., and Portillo, F. (1990). Catalytic and regulatory sites of yeast plasma membrane H(+)-ATPase studied by directed mutagenesis. Biochim. Biophys. Acta 1018 195–199. [DOI] [PubMed] [Google Scholar]
- Shahollari, B., Varma, A., and Oelmuller, R. (2005). Expression of a receptor kinase in Arabidopsis roots is stimulated by the basidiomycete Piriformospora indica and the protein accumulates in Triton X-100 insoluble plasma membrane microdomains. J. Plant Physiol. 162 945–958. [DOI] [PubMed] [Google Scholar]
- Shaw, A.S. (2006). Lipid rafts: Now you see them, now you don't. Nat. Immunol. 7 1139–1142. [DOI] [PubMed] [Google Scholar]
- Simpson, C., Thomas, C., Findlay, K., Bayer, E., and Maule, A.J. (2009). An Arabidopsis GPI-anchor plasmodesmal neck protein with callose binding activity and potential to regulate cell-to-cell trafficking. Plant Cell 21 581–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C.L., Bayer, E.M., Ritzenthaler, C., Fernandez-Calvino, L., and Maule, A.J. (2008). Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol. 6 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot-Lubicz, J., Ye, C.M., and Bamunusinghe, D. (2007). Molecular biology of potexviruses: Recent advances. J. Gen. Virol. 88 1643–1655. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Lederer, C., and Baulcombe, D.C. (2000). A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103 157–167. [DOI] [PubMed] [Google Scholar]
- Widjaja, I., Naumann, K., Roth, U., Wolf, N., Mackey, D., Dangl, J.L., Scheel, D., and Lee, J. (2009). Combining subproteome enrichment and Rubisco depletion enables identification of low abundance proteins differentially regulated during plant defense. Proteomics 9 138–147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.