Abstract
Two critical properties of stem cells are self-renewal and multipotency. The maintenance of their “stemness” state and commitment to differentiation are therefore tightly controlled by intricate molecular networks. Epigenetic mechanisms, including DNA methylation, chromatin remodeling and the noncoding RNA-mediated process, have profound regulatory roles in mammalian gene expression. Recent studies have shown that epigenetic regulators are key players in stem cell biology and their dysfunction can result in human diseases such as cancer and neurodevelopmental disorders. Here, we review the recent evidences that advance our knowledge in epigenetic regulations of mammalian stem cells, with focus on embryonic stem cells and neural stem cells.
Introduction
Stem cells refer to the cells that have two fundamental properties that define their “stemness” characteristics: self-renewal and multipotency. During development, stem cells and resulting progenitor cells are responsible for generating all the tissues and cells of an organism. In adult, stem cells exist in many tissues throughout life and may play critical roles in tissue regeneration and repair. Mammalian embryonic stem cells (ESCs) derived from the inner mass of the embryonic blastocyst are pluripotent, which means that they have the ability to generate all types of cells in an adult animal. Tissue-specific stem cells have limited ability for differentiation and generally are committed to create the mature cells in the tissues where they reside. For example, neural stem cells (NSCs) could give rise to major cell types in the central nervous system (CNS), including neurons, astrocytes, and oligodendrocytes, whereas hematopoietic stem cells (HSCs) have the potential to produce all lineages of blood cells, such as Erythrocytes, B-lymphocytes, T-lymphocytes.
The development of a mammalian organism begins from the zygote (fertilized egg). The zygotye is totipotent, which means that it has the potential to develop into a complete organism, as well as extra embryonic tissues. The first visible cell differentiation in a developing zygote is the formation of the blastocyst that consists of trophoblasts, inner cell mass, and blastocyst cavity [1]. ESCs are experimentally derived from inner cell mass of blastocysts by following specific in vitro culture conditions. The closest resemblance, but not identical counter part, of ESCs in vivo is epiblasts that appear at E6-6.5 (embryonic day) in mice. Under an optimal in vitro maintenance condition, ESCs are pluripotent with the ability to generate stem cells and subsequent differentiated cells of all three germ layers, ectoderm, mesoderm, and endoderm both in culture dishes and upon transplantation into developing embryos [1]. Unlike the zygote, ESCs can not differentiate into extra-embryonic tissues such as yolk sac [2]. ESCs are ideal stem cells for both research and cell-based therapies both because that they can be cultured almost indefinitely without altering their stem cell properties and because that they have the potential to regenerate all types of cells and organs in an adult animal or a human being. However, despite great public interest and significant scientific advances in understanding ESCs, many critical questions remain. How do ESCs maintain their pluripotency? During ESC differentiation, how are those genes essential for pluripotency silenced, while other genes specific for differentiated cells activated? How do some of the genes essential for pluripotency become silenced during fetal development but are expressed again in the next generation germ cells? These are some of the most critical questions that need to be answered in ESC field.
Neurogenesis is defined as the process of generating new neurons from NSCs, which consists of the proliferation and fate determination of NSCs, migration and survival of young neurons, and maturation and integration of newly matured neurons [3,4]. To date, it is well accepted that adult mammalian brains have two regions with persistent neurogenic capabilities: one is the subventricular zone (SVZ) in the lateral ventricle and another one is the subgranular zone (SGZ) in the dentate gyrus of the hippocampus. The presence of neurogenesis in these two regions have been demonstrated in many species including primates [5,6] and humans [7,8]. Due to a lack of NSC-specific marker, the identity of true NSCs in vivo is still unclear. Experimental evidence suggests that adult NSCs may have originated from neuroepithelial cells or radial glia during initial neurogenic phase of embryos. These cells evolve into a subset of astrocytes that preserve their stem cell properties in the neurogenic niche of adult brains [9]. Recent advances have revealed that many mechanisms are involved in the regulation of neurogenesis: physiological activities including running, learning and memory, and enriched environment [10]; hormones [11]; growth factors such as Fgf and Vegf [12,13]; transcription factors such as Tlx [14], Bmi-1 [15], and Sox2 [16–18]; and diseases including epilepsy [19] and stroke [20,21].
One fundamental question in understanding neurogenesis is why the adult brain has such limited neurogenic potential compared to that in the embryonic brain. The reason might be that adult neurogenesis is mechanistically different from embryonic neurogenesis. For adult neurogenesis, multipotent NSCs are in intimate contact with the surrounding glia and the fate of NSCs is affected by their microenvironment (so-called “stem niche”) [22–25]. For example, mice lacking sonic hedgehog [26], Tlx [14], Bmi-1 [15], and Mbd1 [27] have profound deficits in postnatal neurogenesis but not in embryonic neural development. In addition, adult NSCs also process different intrinsic properties as compared to their embryonic counterparts. For example, isolated embryonic NSCs can generate much more diverse types of neurons than adult NSCs upon transplantation into embryonic brains [28]. This is likely due to the differences in the genetic and epigenetic programs between adult and embryonic NSCs. Consistent with this, we found that Mbd1−/− adult NSCs expressed abnormally higher levels of basic fibroblast growth factor (Fgf-2) and exhibited increased aneuploidy while NSCs isolated from Mbd1−/− embryonic day 14 (E14) or neonate (P1) brains did not [27]. Understanding the molecular mechanism of adult neurogenesis and adult NSCs is a critical step towards their therapeutic applications.
Although significant advances have been achieved concerning the biology of stem cells, how stem cells maintain their “stemness” remains to be answered. Recently, epigenetic regulations of development and cell fate determination have come to the center stage of stem cell biology and we will review how epigenetic mechanisms could determine ‘stem cells signature’ and play crucial roles in regulating gene expression and stem cell functions.
DNA Methylation and Epigenetic Reprogramming
The concept of epigenetics has been evolving since the first day it was proposed. Today, Epigenetic modifications refer to meiotically and mitotically heritable changes in gene expression that are not coded in the DNA sequence itself [29]. In mammals, epigenetic processes mainly include DNA methylation [30], histone modification [31], and noncoding RNA-mediated processes (Table 1)[32].
Table 1.
Brief Summary of Epigenetic Modifications and their Function in Gene Expression
| Epigenetic modification | Position or target | Function in gene expression |
|---|---|---|
| DNA methylation | CpG islands | Repression |
| Histone modification | H3(K4,K36,K79) | Activation |
| Methylation | H3(K9,K27), H4(K20) | Repression |
| H3(R17,R23), H4(R3) | Activation | |
| Acetylation | H3(K9,K14,K18,K56), H4(K5,K8,K13,K16) | Activation |
| microRNA | mRNA | Repression |
DNA methylation is a covalent modification of cytosine at position C5 in CpG dinucleotides. In mammals, over 70% of CpG dinucleotides are methylated and nearly all DNA methylation occurs on CpG dinulceotides that are underrepresented in the genome with the exception of CpG islands (CpG clusters). Unmethylated CpG islands are usually found in the promoters and the first exons [33]. DNA methylation is catalyzed by several DNA methyltransferase (DNMTs). The de novo establishment of DNA methylation relies on DNMT3a and 3b, whereas the maintenance of DNA methylation depends on DNMT1 that specifically recognizes semi-methylated DNA and methylates the remaining strand [34,35]. DNMT2 does not have methyltransferase activity and its function is not clear [37]. DNMT3L (DNMT3-like) lacks enzymatic activity, but could act as a cofactor for DNMT3a and 3b and modulate their enzymatic activity. In addition to its methyltransferase activity, DNMT1 can also form a complex with HDACs and act as transcriptional repression factor, suggesting that DNMTs may have more complex functions than catalyzing genomic DNA methylation [35,36]. Compared to DNA methylation, our knowledge of DNA demethylation is much more limited. It has been reported that MBD2 has DNA demethylase activity [38], however, it remains controversial whether DNA demethylation is reversible and dynamically regulated.
Mammalian DNA methylation has been implicated in a diverse range of cellular functions, including tissue-specific gene expression, cell differentiation, cell fate determination, genomic imprinting, and X chromosome inactivation [39]. Dnmt3a−/− mice displayed normal developmental phenotype at birth, but died at about 4 weeks after birth. Dnmt3b−/− mice exhibited developmental defects including growth impairment and rostral neural tube defects with variable severity at later stages [40]. In human, DNMT3b mutation results in immunodeficiency, centromere instability, and facial abnormality (ICF) syndrome with severe mental retardation [41,42]. DNA methylation represses gene transcription either through directly blocking the access of transcription factors to their binding sites or through indirectly recruiting methyl-CpG binding proteins (MBDs). MBDs family includes at least MBD1, MBD2, MBD3, MBD4, MECP2, and Keiso [43]. Mbd1−/− mice have reduced neurogenesis and learning deficits [27] while Mbd2−/− mice has a reduced intestinal tumorigenesis, defect in cytokine production, and impaired maternal behavior [44,45]. Mbd3−/− mice are embryonic lethal [46], whereas mutation of Mbd4, a DNA repair protein, leads to increased tumor incidence on susceptible genetic background [47]. Mutations in MECP2 leads to neurodevelopmental deficits in both humans (Rett Syndrome) and in rodents [48,49].
DNA methylation during embryonic reprogramming and in ESCs
DNA methylation patterns are dynamically regulated both during normal development and in diseases and properly established and maintained DNA methylation is critical for embryonic development [50]. Upon fertilization, the paternal genome is rapidly and actively demethylated whereas the maternal genome is slowly and passively demethylated. Then both genomes reach their peak methylation levels at the balstocyst stage, with the genome of the inner cell mass reaches the level of DNA methylation similar to somatic cells of the adult while that of trophoblasts has relatively lower methylation level [51]. DNA methylation levels and patterns are abnormal in many cloned animals especially in their trophoblasts, which could be a major reason for the low efficiency in animal cloning [52]. The second phase of DNA methylation change occurs in the primordial germ cells (PGCs) [53]. PGCs are generated from epiblasts and first arise in the posterior primitive streak on E7.5 in mice. Early PGCs have the same epigenetic marks as those of other epiblasts, including random X chromosome inactivation, imprinted gene expression and DNA methylation. On E8.5, PGCs migrate towards the genital ridge and arrive there by E11.5. During the migration, PGCs erase genome-wide DNA methylation and histone H3 lysine 9 di-methylation (H3K9me2), and instead acquire high levels of tri-methylation of H3K27 (H3K27me3), and re-obtain the totipotency [53].
The biphasic methylation is an essential component of epigenetic reprogramming and a critical step of embryonic development. ESCs express a cohort of genes specific to their pluripotent property such as Octamer binding transcription factor 4 (OCT4), SOX2, and NANOG. These genes are master regulators that control the expression of distinct yet overlapping downstream genes critical for stem cell properties and cell lineage determination [54]. Recent exciting discoveries showed that exogenous expression of OCT4, SOX2, c-MYC, and KLF4 could induce pluripotency in fibroblasts derived from either embryonic or adult mice or humans [55–58]. These induced pluripotent stem cells (iPS) exhibit many in vitro properties as ESCs but with differential in vivo pluripotency depending on their epigenetic programs. The iPS cell lines have high level of DNA methylation pattern at Oct 4 promoter similar to that of fibroblasts exhibit low germ line transmission efficiency, whereas the iPSs that exhibit low OCT4 promoter methylation similar to that of ESCs have high germ line incorporation efficiency [57, 59]. Although it is not clear how the introduction of these genes could result in epigenetic changes during the induction of iPS and it is very likely that the mechanisms underlying the epigenetic changes are different between ESCs and iPS, these studies suggest that epigenetic mechanism is a critical component in defining the “stemness” of ESCs.
DNA methylation in NSCs
During the embryonic stage of brain development, neurogenesis and gliogenesis appear in a temporally defined sequence [28]. Neurons appear first (around E10 in mice) and astrocytes are generated afterwards (around E15 in mice). Recent studies have revealed some potential mechanisms underlying this developmental control. It is well established that gp130-STAT3 signaling pathway promotes embryonic NSCs (neuroepithelial cells) to differentiate into astrocytes [60,61]. Interestingly, STAT3 binding site in the GFAP promoter is methylated at E11.5 in neuroepithelial cells, which blocks the binding of STAT3 to the GFAP promoter hence astrocytic differentiation. The same GFAP promoter becomes hypomethylated at E14.5, which allows the binding of STAT3 and the expression of astrocytic genes at later developmental stage [61–63]. The GFAP promoter and Exon 1 are also highly methylated in neurons which inhibit the expression of astrocytic gene in neurons [64]. Dnmt1 deficiency in NPCs of developing CNS (E9-10) induces DNA hypomethylation in over 90% of the CNS cells. Due to hypomethylation and over-activation of the gliogenic pathway JAK-STAT pathway, glial cell generation is significantly increased [65].
How DNA methylation regulates adult neurogenesis is less understood. It is possible that differential epigenetic mechanisms are modulating adult NSCs compared to their embryonic counter parts. The essential roles of DNA methylation in adult neurogenesis was first revealed in Mbd1 knock out mice. Mbd1−/− mice did not have detectable deficits during early development, but adult Mbd1−/− NSCs exhibit reduced neuronal differentiation and increased genomic instability [27]. Furthermore, Mbd1−/− mice exhibit decreased neurogenesis and impaired spatial learning [27]. Recently, we found that Mbd1 regulates neurogenesis and neuronal differentiation through regulating the expression of a stem cell mitogen, basic fibroblast growth factor (Fgf-2). In the absence of Mbd1, the promoter of Fgf-2 in NSCs was hypomethylated and the level of stem cells-expressed Fgf-2 was increased (Li et al., unpublished data). Overexpressing Fgf-2 promoted proliferation but inhibited the differentiation of adult NSCs (Li et al., unpublished data). During cell differentiation, MECP2 expression is increased and its expression levels correlate with neuronal maturation [66]. Although the generation of new neurons is not affected by Mecp2 mutation, the maturation of young neurons is severely affected [66–68]. DNMT3a protein is strongly expressed in neural precursor cells, post-mitotic CNS neurons, and oligodendrocytes, but only weakly expressed or completely absent in astrocytes [69]. The expression pattern of Dnmt3a and the fact that mice lacking Dnmt3a die within a few weeks of birth suggest a critical role of Dnmt3a in neurogenesis and CNS development [70]. These studies demonstrate that DNA methylation is necessary for the normal embryonic neurodevelopment.
More than 90% of methylated Cytosines in the human genome occur in retrotransposon, and DNA methylation represses the transcription of transposable elements. Long interspersed nuclear element-1 (LINE-1 or L1) elements, which comprised about 20% of mammalian genomes, are the major class of non-long terminal repeated (LTR) retrotransposons. It has been found that MECP2 silences L1 transcription and L1 retrotransposition [71]. One recent study found that during the differentiation from neural precursor cells to neurons L1 retrotransposition events alters the expression of neuronal genes, and influences neuronal cell fate [72]. DNA methylation could modulate NSC differentiation through its regulation of L1 retrotransposition.
Histone Modifications and the Chromatin State of Stem Cells
In eukaryotic cell, 146 bp DNA wraps histone octamer to form the nucleosome, the basic unit of chromatin. Chromatin has two different states: Euchromatin and Heterochromatin. Euchromatin is enriched with actively transcribed genes and is less condensed. Compared with Euchromatin, heterochromatin contains largely repetitive sequences and few transcribed genes, and is highly condensed. Heterchromatin is largely localized at centromere and telomere [73] and can also influence expression of euchromatin genes in a region-specific and sequence-independent manners. Heterochromatin structure and maintenance are essential for cellular genomic stability and important cellular processes such as accurate chromosome segregation during mitosis, X-chromosome inactivation, and correct repression and expression of imprinted genes, etc [74].
Compared with DNA methylation, the post-translational modifications of histone display high levels of diversity and complexity. The core histones H2A, H2B, H3, and H4 are subject to dozens of different modifications, including acetylation, methylation, and phosphorylation, etc [75]. Among these modifications, lysine (K) acetylation and methylation are the best-understood histone modifications. Lysine acetylation is generally involved in gene activation, whereas the effects of lysine methylation depend on the lysine residues. For example, methylation of H3K4 [76], H3K36 [77], or H3K79 [78] correlates with the active gene transcription, but methylation at H3K9, H3K27, or H4K20 usually links to gene repression. In addition, mono-, di-, and trimethylation at the same Lysine residues lead to different levels of gene activation or repression and are involved in distinct cellular pathways. The term “histone code” highlights the importance of histone modification in gene expression regulation.
Critical roles of histone modification enzymes
The initial histone modification studies were focused largely on histone acetylation that is catalyzed by two opposing enzymes, histone acetyltransferease (HAT) and histone deacetylase (HDAC). To date, at least 6 HATs and HAT complexes and 18 HDACs have been identified in mammals [79,80]. Many active transcription factors either recruit HATs or utilize their own internal HAT domains (e.g., CREB binding protein) to catalyze H3 and H4 acetylation and lead to accessible chromatin structure and gene activation. On the other hand, HDAC is frequently involved in gene repression. For example, Neuron-specific genes share the conserved 21–23-base pair DNA response element, RE-1 (repressor element 1). Neuronal restricted silencing factor (NRSF) binds to RE-1 and forms a repressing complex which mediates the repression through recruiting HDAC1/2 and Sin3A [81–84]. REST is expressed in ESCs and the differentiation of ESCs into neural progenitors and neurons requires the degradation of REST [81]. Treatment of adult hippocampal neural progenitors by Valproic acid (VPA), an HDACs inhibitor, lead to increased neuronal differentiation, but decreased astrocyte and oligodendrocyte differentiation [85]. More recently, it was found that VPA treatment leads to decreased seizure-induced neurogenesis through regulating REST and its target genes [86]. In the developing brain, VPA administration also induces the significant hypomyelination and delays the differentiation of oligodendrocytes through inhibiting the activity of HDACs [87]. Additionally, the development of corpus callosum is companied with the histone deacetylation [87].
Similarly, histone methylation and demethylation enzymes are also involved in cellular functions. One of the best studied histone methylation is the methylation of H3K9 by SUV39h, SetDB (mouse ESET), and G9a. These enzymes are recruited to methylated DNA, likely by MBDs, and catalyze methylation of the histones, followed by the binding of HP1 to form heterochromatin structure [88,89]. In ESCs, proper H3K9 methylation is critical for regulated expression of imprinted genes [90]. Recently, the discovery of histone demethylases has greatly advanced our understanding in chromatin-mediated gene regulation [91]. For example, Lysine-specific demethylase 1 (LSD1) stimulates androgen-receptor-dependent transcription; however, knockdown of LSD1 protein levels abrogates androgen-induced transcriptional activation and cell proliferation through relieving repressive histone marks by demethylation of H3K9 [92]. Chromatin remodeling conducted by chromatin remodeling enzymes is likely an integral part of classical transcription activation and repression.
PcG proteins and bivalent chromatin state
Polycomb Group (PcG) proteins are originally identified in the fruit fly as repressors of Hox genes. PcG proteins form three complexes: Polycomb repressive complex 2 (PRC2), PRC1, and PhoRC [93]. PRC2 is involved in the initiation of silencing and contains histone deacetylase and histone H3K27 methyltransferase activities, and PRC1 is implicated in maintaining gene repression [94]. One of the most exciting recent discoveries is the involvement of PcG proteins in stem cell function by forming and maintaining the bivalent chromatin state of stem cells [95,96].
It has long been proposed that histone modification patterns could define the state of the genome during development and cellular transformation [50]. The discovery of “bivalent” state in ESCs has revolutionized our understanding in the regulation of gene expression. In ESCs, while ESC-specific genes are marked by histones related to gene activation, those genes related to downstream differentiation are occupied by both active (methylated-H3K4) and repressive (trimethylated-H3K27) chromatin markers [97,98]. This so called “bivalent chromatin state” allows the cell/tissue-specific genes to be ‘primed’ for expression but ‘held in check’ by opposing chromatin modifications [95,96]. A more recent study using genome wide chromatin analyses of ESCs, NSCs, and embryonic fibroblast indicate that such bivalent state is not unique to ESCs [90]. In NSCs, which are more restricted stem cells compared to ESCs, bivalent chromatin state still exists but on different sets of genes. In NSCs, those ESC-specific genes are occupied by methylated-H3K9 that is permanent repressive marks, but the genes important for downstream neuronal and glial differentiation are marked by bivalent chromatin marks. Therefore, bivalent chromatin state is likely a common mechanism in many types of stem and progenitor cells for maintaining their differentiation potential and PcG proteins are critical factors in this regulatory mechanism. In fact, the disruption of some members of PRC1 or PRC2, such as Suz12, has been shown to result in embryonic lethality [99]. In ESCs, PcGs repress many developmental regulators, such as components of transforming growth factor-β, bone morphogenic protein, and Wnt signaling pathways [100]. Bmi-1, a component of PRC1, is expressed in the germinal zone of adult brains, as well as in neural progenitors, but not in differentiated cells, and is required for the self-renewal of subpopulations NSCs through repressing the expression of p16Ink4a-Rb and p19Arf −p53 pathways [15,101]. Bmi-1 mutant mice exhibit progressive postnatal growth retardation and neurological defects [102], which is possibly related to the decreased proliferation of progenitor cells in the newborn cortex and the newborn and adult SVZ, [15], and increased astrocytes production in vivo and in vitro [103].
Noncoding RNAs and Translational Regulation
Transcription factors are essential players in regulating stem cells functions [104,105]. Recently, post-transcriptional gene regulation is emerging as another essential regulator of stem cell development [106]. It is likely that the coordinate regulation of gene clusters relies on transcriptional regulation for both initial expression and post-transcriptional control for refinement. In 1960s, messenger RNAs (mRNAs) were demonstrated to carry the genetic information, while ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs) that did not code protein but facilitated the protein synthesis were termed as non-coding RNAs. Twenty years later, small nuclear RNAs (snRNAs) were isolated and found to be involved in pre-mRNA splicing. Today, non-coding RNA world has changed dramatically and some new members, such as microRNAs (miRNAs), small nucleolar RNAs (snoRNAs), short interfering RNAs (siRNAs), repeat-associated small interfering (rasi) RNA, and PIWI interacting RNA (piRNA) etc have been discovered. The biological functions of most of these newly discovered noncoding RNAs are just revealed.
The biogenesis and functions of miRNAs are the best studied among these newly discovered noncoding RNAs. The gene encoding miRNA is transcribed into primary miRNA by RNA polymerase II, and primary miRNA is then processed by RNase III Drosha into 70–100 nucleotide precursor miRNA. Precursor miRNA is transported into cytoplasm by exportin-5 and further processed by RNase III Dicer to form mature miRNA [107]. Mature miRNAs are single-stranded and composed of 17–25 nucleotides. Nearly 500 known human miRNA sequences have been identified. miRNAs incorporate into a miRNA-induced silencing complexex (miRISCs), and directs miRISC to the target mRNAs based on sequence homology, and leads to either degradation of mRNA or the translational suppression of target genes by pairing with sequences in the 3′ untranslated region of target mRNA [107–109].
miRNAs are especially attractive candidates for regulating stem cell functions. Because miRNA can repress genes expression based on imperfect match, they can simultaneously regulate many targets, allowing for coordinated control of many genes during stem cell differentiation. Dicer mutant mouse embryos die at as early as E7.5 [110]. Dicerdeficient ESCs fail to differentiate both in vitro and in vivo and exhibit defects in both DNA methylation and histone modification patterns [111]. Argonaute proteins (AGO1-4) are key components of RISC complexes for miRNA pathway and have been shown to be required for maintaining germline stem cells in Drosophila, C. elegans, and mice [112]. The loss of Eif2c2, one of the Argonaute proteins, leads to neural tube closure defect, demonstrating the critical function of miRNA pathways in mammalian CNS development [113].
miRNAs are involved in multiple cellular activities, including developmental timing, cell death, cell proliferation, haematopoiesis, apoptosis, neural patterning and cell specification [108,114]. Distinct miRNAs are specifically expressed in ESCs, neural progenitor cells and then neurons indicating a role for miRNAs in the nervous system development and stem cell biology [115]. The function of individual miRNA in mammalian neurodevelopment has also been explored. miR-124a and miR-9 are found to affect ESC-derived neurogenesis [116]. As a proposed mechanism of such action, miR-124a could down-regulate phosphatase small C-terminal domain phosphatase 1 that inhibits the neurogenesis [117]. miR-134 is specifically expressed in the brain and its expression increases during the development. Studies also revealed that miRNAs, such as miR-134, regulate the synapse development, including synaptic protein synthesis [118,119]. miRNAs are especially abundant in the adult brain, suggesting a key role for them in neuronal function and plasticity [106]. Components of the miRNA biogenesis machinery are expressed in brain regions that have ongoing neurogenesis and expression of several miRNAs coincide with the onset of neurogenesis and/or gliogenesis [120–122]. Furthermore, It has been shown that REST regulates the transcription of miRNAs, such as brain-specific miR-124a [123]. During the neuronal differentiation process, the level of REST decreases while the expression of miR-124a, suggesting that REST mediated expression regulation of miRNA could play important roles in NSC differentiation.
Crosstalks among Epigenetic Mechanisms
The crosstalk among epigenetic pathways was initially established in invertebrate animals and plants, and later confirmed in mammals. In Arabidopsis thaliana, the mutation of KRYPTONITE, a gene coding H3K9 methyltransferase, leads to the loss of Cytosine DNA methylation, which resembles the phenotype produced by the mutation of DNA methyltransferase CMT3 [124]. Heterochromation is repressive for gene transcription and is characterized by DNA methylation and enriched H3K9 trimethylation and hetrochromation protein 1 (HP1). The formation and maintenance of heterochromatin require the coordination of DNA methylation, H3K9 methylation, and RNAi machinery [125]. The first evidence for the crosstalk between DNA methylation and histone modification in mammals is provided by studying Suv39h mutant mice. In Suv39h1 mutant ESCs, Dnmt3b failed to localize to pericentric heterochromatin regions which lead to decreased DNA methylation [126]. Although one study reported a contradictory result [127], MECP2 has been shown to recruit the Sin3-HDAC complex to methylated DNA and this effect could be reversed HDAC inhibitor, trichostatin A [128,129].
Functional interaction between small noncoding RNAs and other epigenetic mechanisms in gene expression regulation have recently been demonstrated in plants and invertebrates. Double strand RNA can induce both DNA and histone methylation in plants and yeast [130]. Small RNAs can, therefore, lead to mitotically heritable transcriptional silencing by the formation of heterochromatin in yeast [131–133]. However, the crosstalk between small noncoding RNA pathway and other epigenetic mechanisms in mammals have only been shown in a few examples. Clustered miRNAs have been found to be localized in imprinted regions of human and mouse genome, suggesting potential roles of DNA methylation in regulating the expression of these miRNAs [134]. Saito et al found that treatment by DNA methylation inhibitor 5-aza-2′-deoxycytidine and histone deacetylase (HDAC) inhibitor 4-phenylbutyric acid lead to increased expression of some miRNAs in human cancer cells [135]. Mecp2 deficient neurons have altered expression levels of a subset of miRNAs [115]. On the other hand, DNMT3a, 3b and DNMT1 have been proposed as potential regulatory targets for miRNAs [136]. We have found that Mecp2 mutation leads to altered expression of a cohort of miRNAs in adult NSCs (Szulwach and Li et al, unpublished observation). These data, along with the heterochromatin deficits in Dicer mutant mice [111, 137], suggest that functional interaction between small RNA and epigenetic modulations is likely a critical mechanism regulating mammalian gene expression.
Concluding Remarks and Perspectives
Recent progress in understanding the establishment and lineage differentiation of stem cell has revealed important role of epigenetic regulations in these processes. It is evident that epigenetic mechanisms are essential for maintaining stem cell signature. While DNA methylation, histone modification and non-coding RNAs may regulate different molecular aspects of stem cells, these epigenetic mechanisms interact with each other and constitute a network that is critical for the function of stem cells and mammalian development (Fig. 1). Further studies should lead to better understanding of how these epigenetic mechanisms and genetic information coordinately regulate the gene activation and repression at both global and individual gene levels. Newly developed technology with high throughput and high sensitivity capabilities will significantly assist in this endeavor. Better understandings in stem cells and brain development will ultimately help to elucidate the underlying causes and possible treatment of many neurodevelopmental disorders.
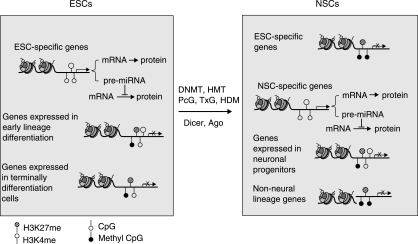
FIG. 1.
Schematic summary of epigenetic mechanisms that define the signature of stem cells and progenitor cells. Left panel: In ESCs, ESC-specific genes have lower DNA methylation levels and are occupied by active chromatin markers such as H3K4me that correlate with their active transcriptional state. Both ESC-specific mRNAs and miRNAs are transcribed to maintain the stemness of the ECSs. The genes that are expressed in terminally differentiated cells are methylated and occupied by repressive chromatin marker (H3K27me, hatched circle). However, those genes that are expressed in early differentiating progenitors (NSCs as example) are associated with both active (H3K4me, open circle) and repressive (H3K27me, hatched circle) chromatin markers that define a bivalent histone state, which could be a signature of cells with differentiation potential. Right panel: In NSCs, those genes that are specific for either ESCs or non-neural lineages are occupied by repressive chromatin marker (H3K27me) and are not expressed, while those genes that are specific for NSCs are associated with active chromatin markers (H3K4me) and are expressed. Those genes that will be expressed in more differentiating neural cells (neurons as example) are associated with both active (H3K4me, open circle) and repressive (H3K27me, hatched circle) chromatin markers that define a bivalent histone state. Therefore the bivalent state is a dynamically changing process that is specifically associated with the stage of the cellular differentiation. Many epigenetic factors are critical in defining and regulating the epigentic state of the above. These factors include but not limited to DNMTs (DNA methyltransferases), HMT (histone methyltransferase), PcG (polycomb protein, with H3K27 methyltransferase activity), TxG (trithorax protein, with H4K4 methyltransferase activity), and HDM (histone demethylase).
References
- 1.Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 2.Turksen K. Troy TC. Human embryonic stem cells: isolation, maintenance, and differentiation. Methods Mol Biol. 2006;331:1–12. doi: 10.1385/1-59745-046-4:1. [DOI] [PubMed] [Google Scholar]
- 3.Gage FH. Mammalian neural stem cells. Science. 2000;287:1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- 4.Ming GL. Song H. Adult neurogenesis in the mammalian central nervous system. Annu Rev Neurosci. 2005;28:223–250. doi: 10.1146/annurev.neuro.28.051804.101459. [DOI] [PubMed] [Google Scholar]
- 5.Kornack DR. Rakic P. The generation, migration, and differentiation of olfactory neurons in the adult primate brain. Proc Natl Acad Sci USA. 2001;98:4752–4757. doi: 10.1073/pnas.081074998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pencea V. Bingaman KD. Freedman LJ. Luskin MB. Neurogenesis in the subventricular zone and rostral migratory stream of the neonatal and adult primate forebrain. Exp Neurol. 2001;172:1–16. doi: 10.1006/exnr.2001.7768. [DOI] [PubMed] [Google Scholar]
- 7.Curtis MA. Kam M. Nannmark U. Anderson MF. Axell MZ. Wikkelso C. Holtås S. van Roon-Mom WM. Björk-Eriksson T. Nordborg C. Frisén J. Dragunow M. Faull RL. Erikssonl PS. Human neuroblasts migrate to the olfactory bulb via a lateral ventricular extension. Science. 2007;315:1243–1249. doi: 10.1126/science.1136281. [DOI] [PubMed] [Google Scholar]
- 8.Eriksson PS. Perfilieva E. Björk-Eriksson T. Alborn AM. Nordborg C. Peterson DA. Gage FH. Neurogenesis in the adult human hippocampus. Nat Med. 1998;4:1313–1317. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- 9.Alvarez-Buylla A. Garcia-Verdugo JM. Tramontin AD. A unified hypothesis on the lineage of neural stem cells. Nat Rev Neurosci. 2001;2:287–293. doi: 10.1038/35067582. [DOI] [PubMed] [Google Scholar]
- 10.Kempermann G. Kuhn HG. Gage FH. More hippocampal neurons in adult mice living in an enriched environment. Nature. 1997;386:493–495. doi: 10.1038/386493a0. [DOI] [PubMed] [Google Scholar]
- 11.Cameron HA. Tanapat P. Gould E. Adrenal steroids and N-methyl-D-aspartate receptor activation regulate neurogenesis in the dentate gyrus of adult rats through a common pathway. Neuroscience. 1998;82:349–354. doi: 10.1016/s0306-4522(97)00303-5. [DOI] [PubMed] [Google Scholar]
- 12.Reynolds BA. Tetzlaff W. Weiss S. A multipotent EGF-responsive striatal embryonic progenitor cell produces neurons and astrocytes. J Neurosci. 1992;12:4565–4574. doi: 10.1523/JNEUROSCI.12-11-04565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vescovi AL. Reynolds BA. Fraser DD. Weiss S. bFGF regulates the proliferative fate of unipotent (neuronal) and bipotent (neuronal/astroglial) EGF-generated CNS progenitor cells. Neuron. 1993;11:951–966. doi: 10.1016/0896-6273(93)90124-a. [DOI] [PubMed] [Google Scholar]
- 14.Shi Y. Chichung Lie D. Taupin P. Nakashima K. Ray J. Yu RT. Gage FH. Evans RM. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature. 2004;427:78–83. doi: 10.1038/nature02211. [DOI] [PubMed] [Google Scholar]
- 15.Molofsky AV. Pardal R. Iwashita T. Park IK. Clarke MF. Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pevny L. Placzek M. SOX genes and neural progenitor identity. Curr Opin Neurobiol. 2005;15:7–13. doi: 10.1016/j.conb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 17.Graham V. Khudyakov J. Ellis P. Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 18.Ferri AL. Cavallaro M. Braida D. Di Cristofano A. Canta A. Vezzani A. Ottolenghi S. Pandolfi PP. Sala M. DeBiasi S. Nicolis SK. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131:3805–3819. doi: 10.1242/dev.01204. [DOI] [PubMed] [Google Scholar]
- 19.Parent JM. Yu TW. Leibowitz RT. Geschwind DH. Sloviter RS. Lowenstein DH. Dentate granule cell neurogenesis is increased by seizures and contributes to aberrant network reorganization in the adult rat hippocampus. J Neurosci. 1997;17:3727–3738. doi: 10.1523/JNEUROSCI.17-10-03727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin K. Wang X. Xie L. Mao XO. Zhu W. Wang Y. Shen J. Mao Y. Banwait S. Greenberg DA. Evidence for stroke-induced neurogenesis in the human brain. Proc Natl Acad Sci USA. 2006;103:13198–13202. doi: 10.1073/pnas.0603512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Y. Jin K. Childs JT. Xie L. Mao XO. Greenberg DA. Neuronal nitric oxide synthase and ischemia-induced neurogenesis. J Cereb Blood Flow Metab. 2005;25:485–492. doi: 10.1038/sj.jcbfm.9600049. [DOI] [PubMed] [Google Scholar]
- 22.Weiss S. Dunne C. Hewson J. Wohl C. Wheatley M. Peterson AC. Reynolds BA. Multipotent CNS stem cells are present in the adult mammalian spinal cord and ventricular neuroaxis. J Neurosci. 1996;16:7599–7609. doi: 10.1523/JNEUROSCI.16-23-07599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmer TD. Markakis EA. Willhoite AR. Safar F. Gage FH. Fibroblast growth factor-2 activates a latent neurogenic program in neural stem cells from diverse regions of the adult CNS. J Neurosci. 1999;19:8487–8797. doi: 10.1523/JNEUROSCI.19-19-08487.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shihabuddin LS. Horner PJ. Ray J. Gage FH. Adult spinal cord stem cells generate neurons after transplantation in the adult dentate gyrus. J Neurosci. 2000;20:8727–8735. doi: 10.1523/JNEUROSCI.20-23-08727.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lie DC. Dziewczapolski G. Willhoite AR. Kaspar BK. Shults CW. Gage FH. The adult substantia nigra contains progenitor cells with neurogenic potential. J Neurosci. 2002;22:6639–6649. doi: 10.1523/JNEUROSCI.22-15-06639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahn S. Joyner AL. In vivo analysis of quiescent adult neural stem cells responding to Sonic hedgehog. Nature. 2005;437:894–897. doi: 10.1038/nature03994. [DOI] [PubMed] [Google Scholar]
- 27.Zhao X. Ueba T. Christie BR. Barkho B. McConnell MJ. Nakashima K. Lein ES. Eadie BD. Willhoite AR. Muotri AR. Summers RG. Chun J. Lee KF. Gage FH. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc Natl Acad Sci USA. 2003;100:6777–6782. doi: 10.1073/pnas.1131928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Temple S. The development of neural stem cells. Nature. 2001;414:112–117. doi: 10.1038/35102174. [DOI] [PubMed] [Google Scholar]
- 29.Levenson JM. Sweatt JD. Epigenetic mechanisms in memory formation. Nat Rev Neurosci. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- 30.Reik W. Dean W. Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 31.Jenuwein T. Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 32.Bernstein E. Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- 33.Jones PA. Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 34.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 35.Jaenisch R. Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl.):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 36.Robertson KD. Ait-Si-Ali S. Yokochi T. Wade PA. Jones PL. Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 37.Okano M. Xie S. Li E. Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res. 1998;26:2536–2540. doi: 10.1093/nar/26.11.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhattacharya SK. Ramchandani S. Cervoni N. Szyf M. A mammalian protein with specific demethylase activity form CpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 39.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 40.Okano M. Bell DW. Haber DA. Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 41.Ueda Y. Okano M. Williams C. Chen T. Georgopoulos K. Li E. Roles for Dnmt3b in mammalian development: a mouse model for the ICF syndrome. Development. 2006;133:1183–1192. doi: 10.1242/dev.02293. [DOI] [PubMed] [Google Scholar]
- 42.Hansen RS. Wijmenga C. Luo P. Stanek AM. Canfield TK. Weemaes CM. Gartler SM. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci USA. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klose RJ. Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 44.Hutchins AS. Mullen AC. Lee HW. Sykes KJ. High FA. Hendrich BD. Bird AP. Reiner SL. Gene silencing quantitatively controls the function of a developmental trans-activator. Mol Cell. 2002;10:81–91. doi: 10.1016/s1097-2765(02)00564-6. [DOI] [PubMed] [Google Scholar]
- 45.Sansom OJ. Berger J. Bishop SM. Hendrich B. Bird A. Clarke AR. Deficiency of Mbd2 suppresses intestinal tumorigenesis. Nat Genet. 2003;34:145–147. doi: 10.1038/ng1155. [DOI] [PubMed] [Google Scholar]
- 46.Hendrich B. Guy J. Ramsahoye B. Wilson VA. Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–723. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Millar CB. Guy J. Sansom OJ. Selfridge J. MacDougall E. Hendrich B. Keightley PD. Bishop SM. Clarke AR. Bird A. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science. 2002;297:403–405. doi: 10.1126/science.1073354. [DOI] [PubMed] [Google Scholar]
- 48.Amir RE. Van den Veyver IB. Wan M. Tran CQ. Francke U. Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 49.Moretti P. Zoghbi HY. MeCP2 dysfunction in Rett syndrome and related disorders. Curr Opin Genet Dev. 2006;16:276–281. doi: 10.1016/j.gde.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 50.Spivakov M. Fisher AG. Epigenetic signatures of stem-cell identity. Nat Rev Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- 51.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 52.Hochedlinger K. Jaenisch R. Nuclear reprogramming and pluripotency. Nature. 2006;441:1061–1067. doi: 10.1038/nature04955. [DOI] [PubMed] [Google Scholar]
- 53.Morgan HD. Santos F. Green K. Dean W. Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(Spec No 1):R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 54.Boiani M. Scholer HR. Regulatory networks in embryo-derived pluripotent stemcells. Nat Rev Mol Cell Biol. 2005;6:872–884. doi: 10.1038/nrm1744. [DOI] [PubMed] [Google Scholar]
- 55.Takahashi K. Tanabe K. Ohnuki M. Narita M. Ichisaka T. Tomoda K. Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 56.Takahashi K. Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 57.Wernig M. Meissner A. Foreman R. Brambrink T. Ku M. Hochedlinger K. Bernstein BE. Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 58.Yu J. Vodyanik MA. Smuga-Otto K. Antosiewicz-Bourget J. Frane JL. Tian S. Nie J. Jonsdottir GA. Ruotti V. Stewart R. Slukvin II. Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 59.Okita K. Ichisaka T. Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 60.Fukuda S. Abematsu M. Mori H. Yanagisawa M. Kagawa T. Nakashima K. Yoshimura A. Taga T. Potentiation of astrogliogenesis by STAT3-mediated activation of bone morphogenetic protein-Smad signaling in neural stem cells. Mol Cell Biol. 2007;27:4931–4937. doi: 10.1128/MCB.02435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takizawa T. Nakashima K. Namihira M. Ochiai W. Uemura A. Yanagisawa M. Fujita N. Nakao M. Taga T. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 2001;1:749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- 62.Shimozaki K. Namihira M. Nakashima K. Taga T. Stage- and site-specific DNA demethylation during neural cell development from embryonic stem cells. J Neurochem. 2005;93:432–439. doi: 10.1111/j.1471-4159.2005.03031.x. [DOI] [PubMed] [Google Scholar]
- 63.Namihira M. Nakashima K. Taga T. Developmental stage dependent regulation of DNA methylation and chromatin modification in a immature astrocyte specific gene promoter. FEBS Lett. 2004;572:184–188. doi: 10.1016/j.febslet.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 64.Setoguchi H. Namihira M. Kohyama J. Asano H. Sanosaka T. Nakashima K. Methyl-CpG binding proteins are involved in restricting differentiation plasticity in neurons. J Neurosci Res. 2006;84:969–979. doi: 10.1002/jnr.21001. [DOI] [PubMed] [Google Scholar]
- 65.Fan G. Martinowich K. Chin MH. He F. Fouse SD. Hutnick L. Hattori D. Ge W. Shen Y. Wu H. ten Hoeve J. Shuai K. Sun YE. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development. 2005;132:3345–3356. doi: 10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- 66.Thatcher KN. Lasalle JM. Dynamic changes in Histone H3 lysine 9 acetylation localization patterns during neuronal maturation require MeCP2. Epigenetics. 2006;1:24–31. doi: 10.4161/epi.1.1.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kishi N. Macklis JD. MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Mol Cell Neurosci. 2004;27:306–321. doi: 10.1016/j.mcn.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 68.Smrt RD. Eaves-Egenes J. Barkho BZ. Santistevan NJ. Zhao C. Aimone JB. Gage FH. Zhao X. Mecp2 deficiency leads to delayed maturation and altered gene expression in hippocampal neurons. Neurobiol Dis. 2007;27:77–89. doi: 10.1016/j.nbd.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feng J. Chang H. Li E. Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- 70.Li E. Bestor TH. Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 71.Yu F. Zingler N. Schumanna G. Strätling WH. Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription. Nucleic Acids Res. 2001;29:4493–4501. doi: 10.1093/nar/29.21.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Muotri AR. Chu VT. Marchetto MC. Deng W. Moran JV. Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 73.Grewal SI. Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grewal SI. Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 75.Lachner M. O'Sullivan RJ. Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci. 2003;116:2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 76.Santos-Rosa H. Schneider R. Bannister AJ. Sherriff J. Bernstein BE. Emre NC. Schreiber SL. Mellor J. Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 77.Krogan NJ. Kim M. Tong A. Golshani A. Cagney G. Canadien V. Richards DP. Beattie BK. Emili A. Boone C. Shilatifard A. Buratowski S. Greenblatt J. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schubeler D. MacAlpine DM. Scalzo D. Wirbelauer C. Kooperberg C. van Leeuwen F. Gottschling DE. O'Neill LP. Turner BM. Delrow J. Bell SP. Groudine M. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee KK. Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 80.Xu WS. Parmigiani RB. Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 81.Ballas N. Grunseich C. Lu DD. Speh JC. Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 82.Lunyak VV. Burgess R. Prefontaine GG. Nelson C. Sze SH. Chenoweth J. Schwartz P. Pevzner PA. Glass C. Mandel G. Rosenfeld MG. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- 83.Lunyak VV. Rosenfeld MG. No rest for REST: REST/NRSF regulation of neurogenesis. Cell. 2005;121:499–501. doi: 10.1016/j.cell.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 84.Rice JC. Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- 85.Hsieh J. Nakashima K. Kuwabara T. Mejia E. Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci USA. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jessberger S. Nakashima K. Clemenson GD., Jr. Mejia E. Mathews E. Ure K. Ogawa S. Sinton CM. Gage FH. Hsieh J. Epigenetic modulation of seizure-induced neurogenesis and cognitive decline. J Neurosci. 2007;27:5967–5975. doi: 10.1523/JNEUROSCI.0110-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shen S. Li J. Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol. 2005;169:577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Agarwal N. Hardt T. Brero A. Nowak D. Rothbauer U. Becker A. Leonhardt H. Cardoso MC. MeCP2 interacts with HP1 and modulates its heterochromatin association during myogenic differentiation. Nucleic Acids Res. 2007;35:5402–5408. doi: 10.1093/nar/gkm599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fujita N. Watanabe S. Ichimura T. Tsuruzoe S. Shinkai Y. Tachibana M. Chiba T. Nakao M. Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J Biol Chem. 2003;278:24132–24138. doi: 10.1074/jbc.M302283200. [DOI] [PubMed] [Google Scholar]
- 90.Mikkelsen TS. Ku M. Jaffe DB. Issac B. Lieberman E. Giannoukos G. Alvarez P. Brockman W. Kim TK. Koche RP. Lee W. Mendenhall E. O'Donovan A. Presser A. Russ C. Xie X. Meissner A. Wernig M. Jaenisch R. Nusbaum C. Lander ES. Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Klose RJ. Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 92.Metzger E. Wissmann M. Yin N. Müller JM. Schneider R. Peters AH. Günther T. Buettner R. Schüle R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 93.Schuettengruber B. Chourrout D. Vervoort M. Leblanc B. Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 94.Valk-Lingbeek ME. Bruggeman SW. van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 95.Boyer LA. Plath K. Zeitlinger J. Brambrink T. Medeiros LA. Lee TI. Levine SS. Wernig M. Tajonar A. Ray MK. Bell GW. Otte AP. Vidal M. Gifford DK. Young RA. Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 96.Lee TI. Jenner RG. Boyer LA. Guenther MG. Levine SS. Kumar RM. Chevalier B. Johnstone SE. Cole MF. Isono K. Koseki H. Fuchikami T. Abe K. Murray HL. Zucker JP. Yuan B. Bell GW. Herbolsheimer E. Hannett NM. Sun K. Odom DT. Otte AP. Volkert TL. Bartel DP. Melton DA. Gifford DK. Jaenisch R. Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Azuara V. Perry P. Sauer S. Spivakov M. Jørgensen HF. John RM. Gouti M. Casanova M. Warnes G. Merkenschlager M. Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 98.Bernstein BE. Mikkelsen TS. Xie X. Kamal M. Huebert DJ. Cuff J. Fry B. Meissner A. Wernig M. Plath K. Jaenisch R. Wagschal A. Feil R. Schreiber SL. Lander ES. A bivalent chromatin structure marks key developmental genes inembryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 99.Pasini D. Bracken AP. Jensen MR. Lazzerini Denchi E. Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sparmann A. van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 101.Molofsky AV. He S. Bydon M. Morrison SJ. Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.van der Lugt NM. Domen J. Linders K. van Roon M. Robanus-Maandag E. te Riele H. van der Valk M. Deschamps J. Sofroniew M. van Lohuizen M. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 protooncogene. Genes Dev. 1994;8:757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 103.Zencak D. Lingbeek M. Kostic C. Tekaya M. Tanger E. Hornfeld D. Jaquet M. Munier FL. Schorderet DF. van Lohuizen M. Arsenijevic Y. Bmi1 loss produces an increase in astroglial cells and a decrease in neural stem cell population and proliferation. J Neurosci. 2005;25:5774–5783. doi: 10.1523/JNEUROSCI.3452-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Howe ML. Mehmud ZF. Saha S. Buratovich M. Stutius EA. Schmidt HD. Lenon AL. Reddicks C. Ivanov GS. Przyborski SA. Ozer JS. Transcription Factor IIA tau is associated with undifferentiated cells and its gene expression is repressed in primary neurons at the chromatin level in vivo. Stem Cells Dev. 2006;15:175–190. doi: 10.1089/scd.2006.15.175. [DOI] [PubMed] [Google Scholar]
- 105.Weissman IL. Anderson DJ. Gage F. Stem and progenitor cells: origins,phenotypes, lineage commitments, and transdifferentiations. Annu Rev Cell Dev Biol. 2001;17:387–403. doi: 10.1146/annurev.cellbio.17.1.387. [DOI] [PubMed] [Google Scholar]
- 106.Cheng LC. Tavazoie M. Doetsch F. Stem cells: from epigenetics to microRNAs. Neuron. 2005;46:363–367. doi: 10.1016/j.neuron.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 107.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 108.Kosik KS. The neuronal microRNA system. Nat Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 109.Eulalio A. Huntzinger E. Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 110.Bernstein E. Kim SY. Carmell MA. Murchison EP. Alcorn H. Li MZ. Mills AA. Elledge SJ. Anderson KV. Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 111.Kanellopoulou C. Muljo SA. Kung AL. Ganesan S. Drapkin R. Jenuwein T. Livingston DM. Rajewsky K. Dicerdeficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Carmell MA. Xuan Z. Zhang MQ. Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- 113.Liu J. Carmell MA. Rivas FV. Marsden CG. Thomson JM. Song JJ. Hammond SM. Joshua-Tor L. Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 114.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 115.Wu H. Xu J. Pang ZP. Ge W. Kim KJ. Blanchi B. Chen C. Südhof TC. Sun YE. Integrative genomic and functional analyses reveal neuronal subtype differentiation bias in human embryonic stem cell lines. Proc Natl Acad Sci USA. 2007;104:13821–13826. doi: 10.1073/pnas.0706199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Krichevsky AM. Sonntag KC. Isacson O. Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Visvanathan J. Lee S. Lee B. Lee JW. Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ashraf SI. McLoon AL. Sclarsic SM. Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 119.Schratt GM. Tuebing F. Nigh EA. Kane CG. Sabatini ME. Kiebler M. Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 120.Smirnova L. Gräfe A. Seiler A. Schumacher S. Nitsch R. Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- 121.Giraldez AJ. Cinalli RM. Glasner ME. Enright AJ. Thomson JM. Baskerville S. Hammond SM. Bartel DP. Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 122.Miska EA. Alvarez-Saavedra E. Townsend M. Yoshii A. Sestan N. Rakic P. Constantine-Paton M. Horvitz HR. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Conaco C. Otto S. Han JJ. Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci USA. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jackson JP. Lindroth AM. Cao X. Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 125.Matzke MA. Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 126.Lehnertz B. Ueda Y. Derijck AA. Braunschweig U. Perez-Burgos L. Kubicek S. Chen T. Li E. Jenuwein T. Peters AH. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–1200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 127.Hu K. Nan X. Bird A. Wang W. Testing for association between MeCP2 and the brahma-associated SWI/SNF chromatin-remodeling complex. Nat Genet. 2006;38:962–964. doi: 10.1038/ng0906-962. author reply 964–967. [DOI] [PubMed] [Google Scholar]
- 128.Nan X. Ng HH. Johnson CA. Laherty CD. Turner BM. Eisenman RN. Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 129.Jones PL. Veenstra GJ. Wade PA. Vermaak D. Kass SU. Landsberger N. Strouboulis J. Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 130.Morris KV. Chan SW. Jacobsen SE. Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 131.Hall IM. Noma K. Grewal SI. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci USA. 2003;100:193–198. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hall IM. Shankaranarayana GD. Noma K. Ayoub N. Cohen A. Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- 133.Volpe TA. Kidner C. Hall IM. Teng G. Grewal SI. Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 134.Seitz H. Royo H. Bortolin ML. Lin SP. Ferguson-Smith AC. Cavaillé J. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 2004;14:1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Saito Y. Liang G. Egger G. Friedman JM. Chuang JC. Coetzee GA. Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatinmodifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 136.Rajewsky N. MicroRNA target predictions in animals. Nat Genet. 2006;38(Suppl.):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 137.Fukagawa T. Nogami M. Yoshikawa M. Ikeno M. Okazaki T. Takami Y. Nakayama T. Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]