Abstract
Summary
Eval is a flexible tool for analyzing the performance of gene annotation systems. It provides summaries and graphical distributions for many descriptive statistics about any set of annotations, regardless of their source. It also compares sets of predictions to standard annotations and to one another. Input is in the standard Gene Transfer Format (GTF). Eval can be run interactively or via the command line, in which case output options include easily parsable tab-delimited files.
Availability
To obtain the module package with documentation, go to http://genes.cse.wustl.edu/ and follow links for Resources, then Software. Please contact brent@cse.wustl.edu
Introduction
Automated gene annotation systems are typically based on large, complex probability models with thousands of parameters. Changing these parameters can change a system's performance as measured by the accuracy with which it reproduces the exons and gene structures in a standard annotation. While traditional sensitivity and specificity measures convey the accuracy of gene predictions [1,2], more information is often required for gaining insight into why a system is performing well or poorly. A deep analysis requires considering many features of a prediction set and its relation to the standard set, such as the distribution of number of exons per gene, the distribution of predicted exon lengths, and accuracy as a function of GC percentage. Such statistics can reveal which parameter sets are working well and which need tuning. We are not aware of any publicly available software systems that have this functionality. We therefore developed the Eval system to support detailed analysis and comparison of the large data sets generated by automated gene annotation systems [e.g., [3]].
Features
Statistics
Eval can generate a wide range of statistics showing the similarities and differences between a standard annotation set and a prediction set. It reports traditional performance measures, such as gene sensitivity and specificity, as well as measures focusing on specific features, including initial, internal, and terminal exons, and splice donor and acceptor sites (see Table 1 for a sampling of these statistics; for a complete list of all calculated statistics see online documentation). These specific measures can show why an annotation system is performing well or poorly on the traditional measures. They can also reveal specific weaknesses or strengths of the system – for example, that it is good at predicting the boundaries of genes but has problems with exon/intron structure because it does poorly on splice donor sites. Eval can also compute statistics on a single set of gene annotations (either predictions or standard annotations). These statistics reveal the average characteristics of the genes, such as their coding and genomic lengths, exon and intron lengths, number of exons, and so on. This is useful when tuning the parameters of annotation systems for optimal performance.
Table 1.
A sampling of the less common statistics calculated by Eval when comparing the output of TWINSCAN and GENSCAN on the "semi-artificial" gene set used in [1] to the gold standard annotation. Standard statistics such as gene and exon sensitivity and specificity are also calculated but are not shown.
| Feature | Statistic | TWINSCAN | GENSCAN |
| Transcripts | Exons Per Transcript | 6.46 | 5.93 |
| CDS Overlap Specificity | 96.55% | 70.59% | |
| CDS Overlap Sensitivity | 87.64% | 97.19% | |
| All Introns Matched Specificity | 26.90% | 8.60% | |
| All Introns Matched Sensitivity | 21.91% | 10.67% | |
| Start and Stop Codon Specificity | 44.14% | 17.65% | |
| Start and Stop Codon Sensitivity | 35.96% | 21.91% | |
| Initial Exons | Overlap Specificity | 70.16% | 35.47% |
| Overlap Sensitivity | 77.54% | 73.91% | |
| Terminal Exons | 5' Splice Specificity | 74.36% | 36.22% |
| 5' Splice Sensitivity | 74.64% | 71.01% | |
| Introns | 80% Overlap Specificity | 73.11% | 48.07% |
| 80% Overlap Sensitivity | 80.19% | 72.58% | |
| Nucleotides | Correct Specificity | 84.61% | 64.76% |
| Correct Sensitivity | 84.26% | 88.87% | |
| Splice Acceptors | Correct Specificity | 77.23% | 52.69% |
| Correct Sensitivity | 84.90% | 81.30% | |
| Splice Donors | Correct Specificity | 76.18% | 53.02% |
| Correct Sensitivity | 84.63% | 80.19% | |
| Start Codons | Correct Specificity | 61.97% | 34.90% |
| Correct Sensitivity | 49.44% | 37.64% | |
| Stop Codons | Correct Specificity | 82.22% | 47.95% |
| Correct Sensitivity | 62.36% | 58.99% |
Plots
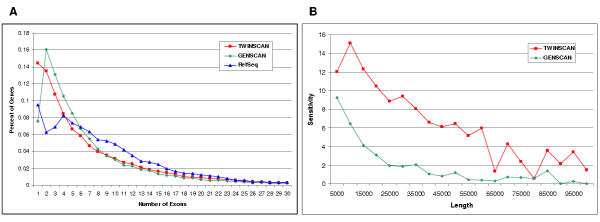
Eval can also produce two types of plots. One type is a histogram showing the distribution of a statistic. Histograms are useful for determining whether the annotation system is producing specific types of genes and exons in the expected proportions. For example, suppose that the average number of exons per gene in an automated annotation is slightly below that of a standard annotation. Comparing the two distributions can reveal whether that difference is due to an insufficient fraction of predictions with extremely large exon counts or an insufficient fraction with slightly above-average exon counts (Fig. 1a). The other type of plot categorizes exons or genes by their length or GC content and shows the statistic for each category. For example, plotting transcript sensitivity as a function of transcript length might reveal that an annotation system is performing poorly on long genes but well on short ones (Fig. 1b). Further analysis would be needed to determine whether this effect is due to intron length or exon count.
Figure 1.
Panel A. Distributions of exons-per-gene for TWINSCAN [4] and GENSCAN [5] gene predictions and RefSeq mRNA sequences aligned to the genome. The plot reveals that, although TWINSCAN predicts too few genes in the 5–20 exon range, it predicts the right proportion of genes with more than 25 exons. Panel B. Fraction of RefSeq genes that TWINSCAN and GENSCAN predict exactly right, as a function of the genomic length of the RefSeq, excluding UTRs. Both figures were made in Excel by importing Eval output as tab-separated files. Data in both panes was generated using the NCBI34 version of the human genome and TWINSCAN 1.2.
Multi-way comparisons (Venn diagrams)
Eval can also determine the similarities and differences among multiple annotation sets. For example, it can build clusters of genes or exons which share some property, such as being identical to each other or overlapping each other. Building clusters of identical genes from two gene predictors and a standard annotation can show how similar the predictors are in their correctly and incorrectly predicted genes. For example, it could reveal that the two programs predict the same or completely separate sets of correct and incorrect genes. If they predict correct gene sets with a small intersection and incorrect gene sets with a large intersection then they could be combined to create a system which has both a higher sensitivity and specificity than either one alone. Table 2 shows a different example - clustering of identical exons from the aligned human RefSeq mRNAs, TWINSCAN [3,4] predictions, and GENSCAN [5] predictions.
Table 2.
The results of building a Venn diagram based on exact exon matches among the aligned RefSeqs, TWINSCAN 1.2 predictions, and GENSCAN predictions, on the NCBI34 build of the human genome. All exons are first combined into clusters that have the same begin and end points. These clusters are then partitioned into the subset of exons annotated only by RefSeq (R), the subset annotated only by TWINSCAN (T), the subset annotated only by GENSCAN (G), the subset annotated by RefSeq and TWINSCAN but not GENSCAN (RT), etc. For each of these subsets, the table shows the number of clusters in the subset. It also shows the percentage all exons from each of the input sets that is included in that subset. The last column shows the fraction of all clusters included in that subset.
| Subset in partition | Cluster Count | % of RefSeq exons | % of Twinscan exons | % of Genscan exons | % of all clusters |
| R | 29,680 | 20.29% | 0.00% | 0.00% | 7.21% |
| T | 44,672 | 0.00% | 22.04% | 0.00% | 10.84% |
| G | 166,765 | 0.00% | 0.00% | 51.72% | 40.48% |
| RT | 15,141 | 10.55% | 7.47% | 0.00% | 3.68% |
| RG | 12,812 | 9.29% | 0.00% | 3.97% | 3.11% |
| TG | 57,795 | 0.00% | 28.52% | 17.92% | 14.03% |
| RTG | 85,069 | 59.88% | 41.97% | 26.38% | 20.65% |
Extraction of subsets
Eval can also extract subsets of genes that meet specific criteria for further analysis. Sets of genes that match another gene set by any of the following criteria can be selected: exact match, genomic overlap, CDS overlap, all introns match, one or more introns match, one or more exons match, start codon match, stop codon match, start and stop codon match. Boolean combinations of these criteria can also be specified. For example, the set of RefSeq genes that are predicted correctly by System1 but not by System2 can be extracted from annotations of the entire human genome with just a few commands. Once extracted, gene sets can be inspected individually using standard visualization tools.
Implementation
Eval is written in Perl and uses the Tk Perl module for displaying its graphical user interface. It is intended to run on Linux based systems, although it also runs under Windows. It requires the gnuplot utility to display the graphs it produces, but it can create the graphs as text files without this utility. The package comes with both command line and graphical interface. The command line interface provides quick access to the functions, while the graphical interface provides easier, more efficient access when running multiple analyses on the same data sets.
Annotations are submitted to Eval in GTF file format http://genes.cse.wustl.edu/GTF2.html, a community standard developed in the course of several collaborative genome annotations projects [6,7]. As such it can be run on the output of any annotation system. The Eval package contains a GTF validator which verifies correct GTF file format and identifies common syntactic and semantic errors in annotation files. It also contains Perl libraries for parsing, storing, accessing, and modifying GTF files and comparing sets of GTF files.
Although it is written in Perl, the Eval system runs relatively quickly. A standard Eval report comparing all TWINSCAN [3,4] genes predicted on the human genome to the aligned human RefSeqs processes ~40,000 transcripts and ~300,000 exons and completes in under five minutes on a machine with a 1.5 GHz Athlon processor and 2 GB of RAM.
Acknowledgments
Acknowledgments
This work was supported in part by grant DBI-0091270 from the National Science foundation to MRB and grant HG02278 from the National Institutes of Health to MRB.
Contributor Information
Evan Keibler, Email: evan@cse.wustl.edu.
Michael R Brent, Email: brent@cse.wustl.edu.
References
- Guigó R, Agarwal P, Abril JF, Burset M, Fickett JW. An assessment of gene prediction accuracy in large DNA sequences. Genome Res. 2000;10:1631–1642. doi: 10.1101/gr.122800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burset M, Guigo R. Evaluation of gene structure prediction programs. Genomics. 1996;34:353–367. doi: 10.1006/geno.1996.0298. [DOI] [PubMed] [Google Scholar]
- Flicek P, Keibler E, Hu Ping, Korf Ian, Brent Michael R. Leveraging the mouse genome for gene prediction in human: From whole-genome shotgun reads to a global synteny map. Genome Res. 2003;13:46–54. doi: 10.1101/gr.830003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I, Flicek P, Duan D, Brent MR. Integrating genomic homology into gene structure prediction. Bioinformatics. 2001;17 Suppl 1:S140–8. doi: 10.1093/bioinformatics/17.suppl_1.s140. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- Reese MG, Hartzell G, Harris NL, Ohler U, Abril JF, Lewis SE. Genome annotation assessment in Drosophila melanogaster. Genome Res. 2000;10:483–501. doi: 10.1101/gr.10.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]