Abstract
Large-scale genetic interaction studies provide the basis for defining gene function and pathway architecture. Recent advances in the ability to generate double mutants en masse in S. cerevisiae have dramatically accelerated the acquisition of genetic interaction information and the biological inferences that follow. Here, we describe a method based on F-driven conjugation, which allows for high-throughput generation of double mutants in E. coli. This method, termed Genetic Interaction ANalysis Technology for E. coli (GIANT-coli), permits us to systematically generate and array double mutant cells on solid media, in high-density arrays. We show that colony size provides a robust and quantitative output of cellular fitness and that GIANT-coli can recapitulate known synthetic interactions and identify new negative (synthetic sickness/lethality) and positive (suppressive/epistatic) relationships. Finally, we describe a complementary strategy for suppressor mutant identification on a genome-wide level. Together, these methods permit rapid, large-scale genetic interaction studies in E. coli.
INTRODUCTION
Genetic interactions report on the extent to which the function of one gene depends on the presence of a second gene and have a long history of facilitating the identification and characterization of cellular pathways. In Saccharomyces cerevisiae, recent advances in the ability to systematically create double mutant strains have resulted in new technologies enabling genome-wide genetic interaction screens1. Both the plate based synthetic genetic array (SGA)2, 3 and diploid based synthetic lethality analysis on microarrays (dSLAM)4 approaches identify negative interactions, i.e. synthetic sick/lethal (SSL) pairs. E-MAP (Epistatic Mini Array Profiles)5, 6 extends this analysis by quantitatively assessing colony size so that positive interactions can be identified as well; in those cases the double mutant is healthier than would be expected based on the growth of the two single mutants. Together, these three approaches have led to a dramatic increase in the number of genetic interactions reported, and have provided functional insights into numerous cellular networks. The E-MAP technology has been extended to Schizosaccharomyces pombe7 and analogous approaches have been developed in C. elegans using RNAi technology8.
Genetic interaction screens are relatively rare in bacteria, and when employed, usually interrogate a limited query gene-set. Additionally, synthetic lethal double mutants are nonviable in the haploid state, meaning that their identification in prokaryotes is difficult using conventional genetic tools. Although conditional alleles are sometimes used, strategies must be developed on a case-by-case basis, preventing high-throughput approaches that facilitate the functional characterization of unknown genes. As a consequence reports of genetic interactions in E. coli are rare; we are aware of fewer than 200 reported synthetic lethal interactions, as compared to almost 20,000 in S. cerevisiae9, even though the two genomes are of roughly comparable size. Lack of genetic interaction data undoubtedly contributes to the lagging functional annotation of bacterial genomes. Even in E. coli and B. subtilis, arguably the best-studied prokaryotes, one third of the genes are of unknown function10, 11. The situation is worse for less studied bacteria and is exacerbated by the immense amount of information from genomic and metagenomic approaches. To-date, 626 bacterial genomes have been sequenced, and 961 are in progress (NCBI database). Next-generation sequencing technologies ensure that the pace of discovering new genes will accelerate. The development of high-throughput genetic interaction screens applicable to bacterial species will be invaluable in utilizing this information to understand gene function and pathway organization.
Here, we report a method, termed GIANT-Coli (Genetic Interaction ANalysis Technology for E. coli) that allows for high-throughput generation of double mutant strains for the first time in E. coli. This method can be used for large-scale quantitative analyses of genetic interactions and is compatible with both the 384 and 1536 high-density arrays. GIANT-Coli is based on the well-characterized Hfr conjugation gene transfer system and uses two comprehensive E. coli mutant libraries of ~4000 single-gene deletions. Additionally, we have developed facile methods to convert F− female (recipient) strains into Hfr males (donors) en masse and have devised a simple variant of our genetic interaction methodology that identifies single-gene knockouts able to suppress deletions that cause a conditional lethal phenotype. These methods will permit rapid exploration of the genetic interaction landscape in E. coli.
RESULTS
Rationale for Method of Gene Transfer
The major bottleneck in producing genetic interaction data is developing a robust method for mass generation of double gene knockouts. To accomplish that, we chose to transfer marked deletions from one E. coli strain to another using the efficient Hfr mating (conjugation) transfer system. Conjugation works well on solid agar surfaces, making it amenable to high throughput technology. The Hfr donor (male) has a chromosomally integrated conjugative F plasmid. Upon contact with a recipient cell lacking an F (F−; female), the Hfr donor is nicked at the origin of transfer (oriT) within the integrated F plasmid. Oriented transfer of a single strand of the circular E. coli chromosome proceeds from oriT. The transferred single-stranded DNA is replicated in the recipient and maintained only by integration via double crossover. As existing Hfr strains12 have multiple and often not completely defined mutations, we created an isogenic Hfr by transducing the previously described “pseudo Hfr”13 into our wildtype background. The “pseudo-Hfr” has the transfer region of F integrated at trp13 and since it transferred as efficiently as the classic high mating Hfr 3000 strain14 in our high-throughput mating methodology (Supplementary Fig. 1A, B), it was used in all subsequent experiments in this report.
Development of GIANT-coli
The high-throughput mating system has 3 steps (Fig. 1). In Step 1, the donor strain, a pseudo-Hfr containing a single gene deletion marked with the kanamycin resistance gene kan (Keio collection15), was mated on agar plates to ASKA “recipient strains”, a set of single-gene knockouts marked with the chloramphenicol resistance gene cat (N. Yamamoto et al. unpublished data) or vice versa. In our high-throughput format, recipient strains were robotically arrayed on agar plates in the desired format (384, 768 or 1536 colonies per plate), grown overnight and then transferred onto an agar plate previously inoculated with a lawn of an isogenic donor strain. These “mating plates” were incubated overnight to allow growth and mating of the parental strains. In Step 2, cells were transferred robotically from the mating plates onto plates containing kanamycin (“intermediate selection”). The rationale for this “intermediate selection” is explained in (A) below. In Step 3, cells from the intermediate selection plate were pinned onto a plate containing both antibiotics to select for double recombinants. The double recombinant colonies were imaged after growing for an experimentally determined time that ensures low background growth of synthetic lethal pairs and permits easy differentiation between sick and healthy mutants (see Supplementary Fig. 1E for detailed protocol). Note that these colonies are the visible manifestation of the cells arrayed on the plate and do not arise from a single cell.
Figure 1.
A flowchart depicting the different steps used in GIANT-coli. An Hfr donor (male) strain carrying a selectable marker (kan) replacing an ORF is mated on agar plates with arrayed F− recipients (females; 1536 per plate) carrying a different selectable marker (cat) replacing another ORF (Step1). Following mating, cells are subjected to an intermediate selection on the bacteriocidal antibiotic kanamycin (Step 2) and then to a final selection for double mutants using both antibiotics (Step 3). Images of two representative plates used for generating a mating plate are shown below the cartoon.
Three critical parameters were optimized so that this procedure performed robustly: 1) efficiency of mating, 2) efficient transfer of most of the chromosome and 3) low recovery of strains in which the locus examined has been duplicated, leading to recovery of “false positives”. First, for efficient and reproducible mating, the ratio between donor and recipient cells proved critical. We standardized growth phase, number of the donor cells, and time of growth of lawn prior to transferring arrayed recipients (Supplementary Fig. 1E). Second, we found that long matings on a solid surface partially obviate the problem of poor efficiency of transfer of markers far from oriT, as a marker 4.2 MB from trp::oriT (90% coverage of the chromosome) showed no substantial drop in the recovery of recombinants, as measured by colony size (data not shown). Third, we developed two strategies to minimize recovery of false positives, created by duplication of a region of the chromosome. We monitored the effectiveness of these strategies by their ability to prevent recipients with a single gene marked by both the donor and recipient antibiotics (e.g. A::kan and A::cat), which we refer to as “self-mating”.
(A) The intermediate selection step
Rapidly growing E. coli in LB initiates multiple (≤16) rounds of replication16, allowing for facile generation of strains with duplicated regions of the chromosome. Plating directly on the double antibiotic selects for and maintains such strains, thereby increasing the fraction of “self-mating” recombinants. As the “intermediate selection” plates have only the bacteriocidal antibiotic (kanamycin), duplicated regions that confer resistance to both antibiotics are not selected. Instead, they are eliminated by spontaneous resolution of this unstable state and by competition from growth of the kanamycin resistant (KanR) parent. Moreover, only one parent is transferred to the subsequent double antibiotic plate, eliminating new rounds of mating and further generation of rare duplications. This step also magnifies small differences in growth of the daughter double mutants, allowing for easier detection of genetic interactions. The intermediate selection virtually eliminates background from “self-mating” as well as growth observed with truly synthetic lethal pairs (Supplementary Fig. 1C, D).
(B) The minimal media protocol
As multiple genomes are the primary source of duplication events, when mating was performed on M9 glycerol medium where cells have ~1 genome per cell17, the background growth of self-mating and synthetic lethal pairs was substantially decreased (data not shown). The intermediate selection decreased this background even further.
Validation of GIANT-Coli
To assess our strategy for mapping genetic interactions in E. coli, we performed a 12 by 12 genetic cross, which provided 66 distinct, pair-wise double mutant strains as well as 12 self-matings. Our choice of genes (surA, ybaY, ycbS, ompC, yraI, cpxR, degP, pal, ompA, yfgL, yraP and basR) was based on: i) spatial compartmentalization to increase interaction probability; all gene-products studied here have roles associated with the envelope compartment, ii) gene pairs with known interactions as a positive control (surA-degP, surA-yfgL, degP-yfgL), iii) gene pairs close together in the chromosome to provide information about the linkage cut-off of our methodology (yraP-yraI, ompA-ycbS), iv) a marker far from oriT to evaluate whether chromosomal position affects recombination efficiency (ompC), and v) mutants with notable growth defects (surA, pal) to facilitate identification of double mutants exhibiting positive interactions.
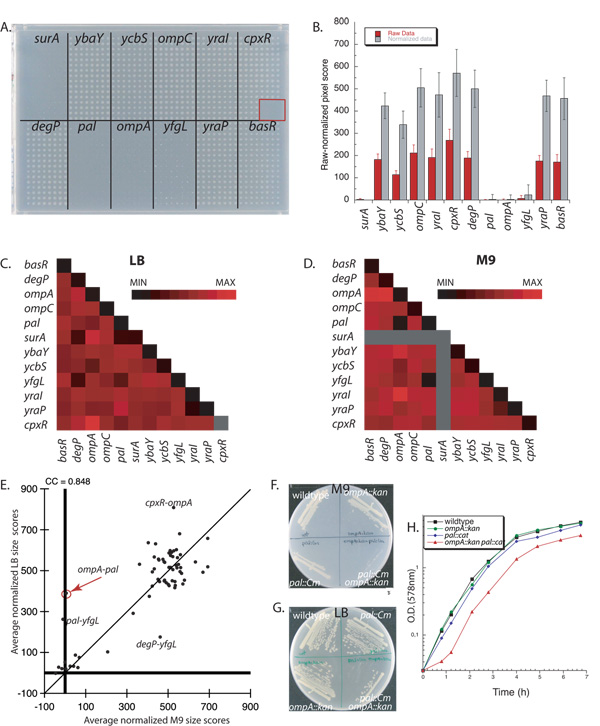
The small subset of genes allowed us to array each recipient multiple times on the same plate so that we could assess reproducibility, compare different plate formats (384 vs 1536) and media [rich (LB) versus minimal (M9-glycerol)] and evaluate growth differences between reciprocal mutant pairs. Representative plates are shown for crosses of pseudo-Hfr pal::kan with the 12 chloramphenicol resistant (CmR) recipients arrayed in 1536 (128 replicas of each mutant; Fig. 2A & Supplementary Fig. 2A) and 384 (32 replicas of each mutant; Supplementary Fig. 2C, E) formats. Note that growth in the self-mating pair (pal::kan × pal::cat) was negligible and comparable to that in the sterility control (red box in Fig. 2A), arguing that false positives were rare. Several new synthetic lethal interactions were apparent: pal-ompA and pal-yfgL on M9 plates (Fig. 2A & Supplementary Fig. 2C), and pal-surA on LB plates (Supplementary Fig. 2A, E). As the surA::cat clone used as recipient did not grow in minimal medium (Fig. 2A & Supplementary Fig. 2C), no inferences can be made about its interactions in M9 glycerol.
Figure 2.
A 12 × 12 genetic interaction matrix to validate GIANT-coli. A. A representative 1536-format, M9-glycerol plate showing the double mutants resulting from crossing pseudo-Hfr pal::kan with 12 CmR ASKA recipients arrayed in boxes of 16×8=128 replicas. The red box is a sterility control, since no recipients are arrayed in this spot. B. Quantification of (A). Error bars depict standard deviations (n > 240). C–D. Heat maps representing 12 × 12 crosses in LB (C) and M9-glycerol (D) based on the combined data from the 384 and 1536 plate formats and averaged results of reciprocal genetic interactions. The gray lines indicate that no results were extracted in M9-glycerol from surA::kan and surA::cat, as these clones grew very poorly in this medium. The color-coded bar ranges from a minimum size score (MIN) to a maximum (MAX) calculated for each dataset separately. E. Scatter plot of averaged normalized colony-size scores comparing growth in M9-glycerol versus LB for the 65 pairs (of 78 total) that grew in both media. Double mutants with substantially different growth in the two media are identified by name. The differential phenotype of the pal−ompA− double mutant is further analyzed in (F–H). F–H. The pal−ompA− double mutant was reconstructed by P1 transduction and the conditional interaction identified by GIANT-coli was recapitulated by lethality on M9-glycerol plates (F), smaller colony size in LB plates (G), and longer lag-phase and slightly slower growth rate in LB medium (H). Doubling times of wildtype, ompA::cat, pal::kan and ompA::cat pal::kan in (H) are approximately 28.5’, 30’, 31.5’ and 38’, respectively.
We quantified growth by obtaining an image of the plate and measuring pixel counts of the colony within a defined boundary. A key challenge is to distinguish differences that result from the growth properties of the double mutants from those arising from properties of the parental single mutants and from plate to plate variation. To do this, we used a dual normalization procedure to eliminate differences arising both from properties of the parental single mutants and from plate to plate variation. (Supplementary Fig. 3A & Supplementary Methods)18. In-plate (horizontal) normalization adjusts for growth differences between plates. Strain (vertical) normalization adjusts for growth and/or mating defects of the recipients. Comparison of raw and normalized data (Fig. 2B, Supplementary Fig. 2B, D & F) shows that strain normalization filters out the growth defect of pal mutants in M9 and the known deficiency of ompA mutants as mating recipients 19. The pal-ycbS and ompA-ycbS interactions scored as negative in the raw data were essentially neutral after normalization (Supplementary Fig. 3B). ~70% of the normalized scores exhibited acceptable standard deviations, i.e. within 25% of the plate mean (Fig. 2B, Supplementary Fig. 2B, D & F). The larger error bars apparent in a few cases resulted from either pinning problems (basR-pal pair in LB1536; Supplementary Fig. 2A, B) or from suppressors of synthetic sick and lethal interactions (ompA-pal pair in LB384 and LB1536; Supplementary Fig. 2A, B, E & F); such suppressors were never evident in more than 10% of the colonies pinned.
To visualize all genetic interaction data simultaneously we generated heat maps. The normalized colony-size scores that deviated the most from the median score in each 12×12 matrix (Fig. 2C, D and Supplementary Fig. 4A–D) were denoted either as black (slower growing, negative genetic interactions) or red (faster growing, positive genetic interactions). Heat maps shown in Fig. 2C and D had data from both 384 and 1536 plate formats and reciprocally constructed double mutants, as the data were concordant (Supplementary Fig. 4). We independently verified the 8 most reproducibly negative interactions, and 4 most reproducibly positive genetic interactions using established methodologies. These verification experiments confirmed all positive and negative interactions (Table 1, Supplementary Table 1, Fig. 2F–H & Supplementary Fig. 5) except for the two negative interactions that resulted from linkage effects: yraP-yraI separated by only 7.7 kilobases (synthetic lethal), and ompA-ycbS separated by 17.2 kilobases (synthetic sick). This verified an expected limitation of our methodology: recombination between closely linked markers is less frequent than that of markers that are far apart (see also linkage section below and Fig. 4C). Each verified interaction exhibited at least 20% impaired or enhanced growth for both reciprocally constructed double mutants. Two previously described negative interactions, surA-degP20 and degP-yfgL21, provided validation for the efficacy of the method. Inability to reproduce the synthetic lethal genetic interaction for surA-yfgL22 was not due to failure of our methodology as it was also not reproduced by standard P1 transduction (data not shown), and may be due to strain and/or allele differences.
Table 1.
Reproduction of synthetic interactions detected in the 12×12 genetic interaction experiment. New synthetic phenotypes were recapitulated by reconstructing the double mutants with P1 transduction and then examining either colony size on plates (A) or growth rate in liquid (B). Synthetic lethal interactions were validated by examining co-transduction of a linked marker (C). In this technique, the P1 donor has a selectable marker closely linked to the first gene deletion being tested, and the recipient is either the wildtype strain or has the second gene deletion. When two mutations are synthetically lethal, co-transduction of the linked markers from the donor strain is never observed when the recipient is deleted for the second gene because all events bringing in the first gene deletion are lethal (see also Supplementary Table 1). Nd = not-determined; lit = interactions reported in the literature. Negative and positive interactions are indicated respectively in bold and underlined fonts. yraP-yraI and ompA-ycbS were both recorded as synthetic sick-lethal in mating experiments, but this was due to linkage.
| Pairs | LB | M9 | ||
|---|---|---|---|---|
| interaction | verification | interaction | verification | |
| degP-surA | lethal | C, lit | nd | - |
| pal-surA | lethal | C | nd | - |
| pal-yfgL | lethal | C | lethal | C |
| pal-ompA | sick | A, B (30 & 37°C) | lethal | A, C |
| degP-yfgL | sick | A, B (mostly at 37°C), lit | - | - |
| degP-pal | slightly sick | A, B (both only at 37°C) | - | - |
| cpxR-pal | sick* | A, B | slightly sick | A |
| ompA-yraP | slightly sick | - | sick | A |
| pal-yraP | positive | A | - | A |
| ompA-degP | slightly positive | - | positive | A |
| ompA-surA | positive | A | nd | - |
| cpxR-ompA | Positive* | A | slightly positive | A |
only one way available; cpxR− is very poor recipient in LB and therefore that data were not evaluated
Figure 4.
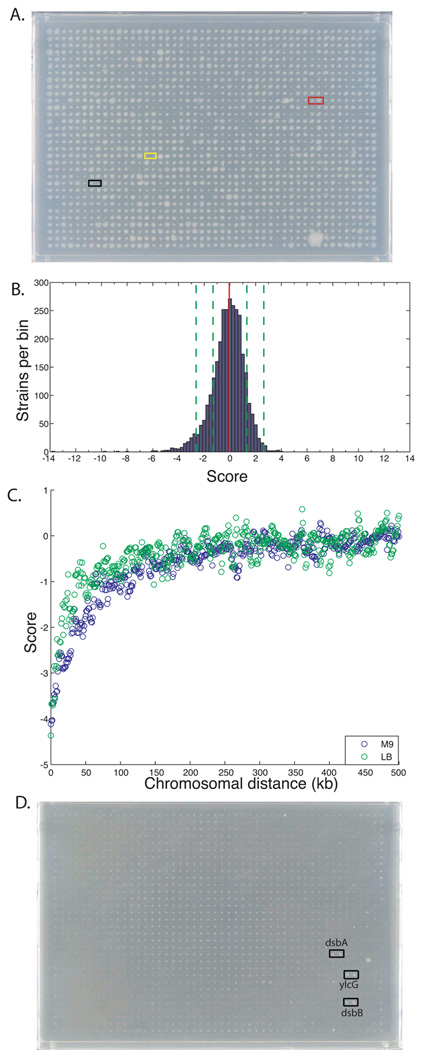
Genome-wide screens using GIANT-coli. A, B. Cross of pseudo-Hfr pal::cat ASKA mutant with the entire F− KanR Keio collection (3985 mutants) arrayed in 1536 format (1536 colonies per plate); each Keio mutant is present twice as adjacent duplicates (768 unique recipients per plate). A representative image of one M9 plate (out of six total) is shown in panel (A). Interactions identified in the 12 × 12 matrix recapitulated here are marked with differently colored boxes. Black: the self-mating pair, pal::cat pal::kan; yellow: the synthetic lethal pair, pal::cat ompA::kan; red: pal::cat degP::kan, a neutral interaction in minimal media. The distribution of all interaction scores of pal::cat with the Keio collection is shown as a histogram in (B). In the histogram the number of Keio mutants within a bin of 0.25 are plotted against the interaction scores; the mean of the distribution (red dotted line) and 1 and 2 standard deviations (green dotted lines) are shown. C. Linkage biases of Hfr mating in LB (green) and M9 (blue). Median interaction scores, extracted using a variation of the E-MAP analysis software18, are aggregated into 5 kb bins sliding with 1 kb steps and are plotted as a function of chromosomal distance in kbs between genes. The analysis is based on 14 genome-wide screens in M9 glycerol and 9 in LB (data not shown). D. Suppression analysis of yraP::cat lethality in 3% SDS. Pseudo-Hfr yraP::cat was mated with the Keio collection as in (A) above except that double mutants were selected on both antibiotics in the presence of 3% SDS. A representative image of one plate is shown; complete data are shown in Supplementary Table 2B.
There were several differences between results in M9 glycerol and LB (Fig. 2C, D), some of which have been validated (Fig. 2 E–H), arguing that screening double mutants in a variety of conditions will be worthwhile. The LB-specific effects appear to reflect growth rate rather than rich medium per se, as decreasing growth rate by lowering the temperature to 30°C partially restored growth of these double mutants in LB (see degP-pal in Supplementary Fig. 5), consistent with previous work indicating that slower growth partially compensates for outer membrane defects 21.
Using GIANT-coli at the genomic level
To extend GIANT-coli to larger gene-sets, we developed methods to rapidly convert single-gene deletion mutants into Hfr donor strains. We created a “double male” strain (Fig. 3A), which transfers from two origins: 1) an upstream oriT in a pseudogene linked to a tetracycline resistance marker (tetAR; see Supplementary Methods); and 2) from the pseudo-Hfr linked to an ampicillin or gentamicin resistance marker (bla and gen respectively). When transfer initiates from the upstream oriT, the downstream pseudo-Hfr and its oriT are transferred efficiently, as assessed by acquisition of the adjacent antibiotic resistance marker (bla or gen). We mated the double male with the entire Keio collection arrayed in 384 format and selected Hfr versions of the Keio collection on ampicillin-kanamycin or gentamicin-kanamycin plates. The same methodology was successfully used to generate Hfr derivatives of several CmR ASKA deletion mutants (data not shown).
Figure 3.
A toolkit that facilitates the use of GIANT-coli in genome-wide analyses. A. High-throughput conversion of an entire single-gene knockout F− library to an Hfr donor library. A double male strain is crossed with the Keio deletion library and a male (Hfr) Keio library is isolated by selecting on Amp-Kan or Gen-Kan plates (depending on whether the pseudo-Hfr locus is linked to gen or to bla). The entire process is carried out on agar plates. Transfer capabilities of a number of the newly generated pseudo-Hfr’s have been validated. B. Targeted integration of F-transfer functions at different chromosomal loci. Conditionally-replicating CIP vectors contain oriRγ (red circle), a ~40 kb BamH1 fragment of F (blue) including its 33 kb transfer region (dark blue) and oriT (blue circle), aadA, conferring streptomycin and spectinomycin resistance (violet) and ~300 bp of chromosomal homology (green). For pNTM3, the chromosomal region is from rhaM. CIPs are carried in a strain expressing the Π protein, which allows the plasmid to replicate from oriRγ. Upon mating to the F− Keio or ASKA deletion mutants, which lack Π, CIPs are unable to replicate; selection for streptomycin and/or spectinomycin resistance results in chromosomal integration dictated by the particular homology region present on the CIP.
An alternative approach employed a Chromosomal Integration Plasmid (CIP) to deliver the F-transfer region to various chromosomal locations. This plasmid is replicated from the Pi dependent R6K γ-ori and has the F transfer region, a ~300 bp of chromosomal homology, and a streptomycin-spectinomycin cassette for selection. CIPs are carried in a pir+ recA− host where they replicate as plasmids but do not integrate. Upon transfer to a pir−, recA+ F− strain, the plasmid integrates into the chromosome by homologous recombination (Fig. 3B & Supplementary Methods), simultaneously converting the F− strains into Hfr donors in a high-throughput manner (Takeuchi et al., unpublished data).
We tested whether we could recapitulate results from our 12 × 12 matrix when each strain is present only several times per plate by crossing Hfr::cat donors against the entire KanR Keio collection, both in rich and minimal media. To our satisfaction, important quality controls were met: i) the self-mating control was always one of the most negative interactions on the plate (e.g. see Fig. 4A); ii) we recapitulated almost all genetic interactions discovered in our 12 × 12 matrix (e.g., pal-ompA produced a synthetic lethal interaction and pal-degP a neutral interaction on minimal medium; Fig. 4A), and identified and verified many new ones (see Fig. 4B and Supplementary Table 2A for the results of the Pal screen on M9). We note that double mutants with a mucoid-slimy phenotype gave false readings in our screen (6 cases in the Pal screen on M9), since their colony size did not reflect their actual fitness. We are currently developing a program to identify such colonies, exclude them from our initial analysis and assign them a score reflective of extent of mucoidy.
Crossing several donors against the Keio collection gave sufficient data to make an initial assessment of the linkage cut-off of our methodology. To define linkage biases, we plotted interaction scores from 9×3985 (LB) and 14×3985 (M9) crosses as a function of the distance between genes in kilobases (kbs) (Fig. 4C). Gene-pairs separated by less than 60 kb in M9 and 30 kb in LB displayed negative interactions that result from a decrease in recombination efficiency. The lower cut-off in LB may reflect greater recombination resulting from the multiple genomes present under those conditions. As many functionally related genes are arranged in operons in bacteria, it is important to determine double mutant phenotypes of genes that are in close proximity to each on the chromosome. To accomplish this, we will compare the interaction scores of closely linked genes to a standard linkage curve generated from the hundred thousand gene-pairs tested (similar to Fig. 4C). This will allow us to distinguish whether the observed colony sizes differ significantly from the size predicted from the decreased recombination efficiency of closely linked genes.
Interestingly, the moderately separated ompA-ycbS pair (17.2 kb) gave more recombinants when the donor transferred the gene of the pair that is closer to oriT rather than the converse (i.e. more recombinants when the donor transferred ycbS than ompA; Supplementary Fig. 4A–D). At present, we lack sufficient data to determine whether this asymmetry is a general rule.
The methodology described in this report can be modified to identify suppressors of conditionally lethal mutants. This new application can be broadly used as it requires hand pinning the Keio collection on a few plates, does not rely on quantitative analysis of large datasets and can be completed in 3 days. Here, the conditional lethal mutant is used as a donor strain and mated to the entire library of deletion mutants with the general protocol described above, except that the final double antibiotic selection plates contain an inhibitory compound or are incubated in the condition that unmasks the lethality of the donor. As a proof of principle experiment, we mated the conditional lethal yraP::cat strain with the Keio collection, selecting double recombinants able to grow in 3% SDS, the condition causing yraP− lethality22 (Fig. 4D and Supplementary Table 2B). The same approach can be used to find suppressors of an essential gene. In this case the donor stain has an inactivated copy of the essential gene in the chromosome and a functional copy of this gene on a non-mobilizable plasmid that cannot be transferred to the recipient; double recombinants that grow identify suppressors that compensate for loss of the essential gene function.
DISCUSSION
The small dataset that we generated illustrates the great potential of this method to provide new information: quantitative genetic interaction analysis of only 66 reciprocal double mutants revealed 12 synthetic interactions (of which 10 are new), ranging from lethal to suppressive growth phenotypes. Genome-wide screens provided many additional candidates for genetic interactions. These results are in strong accord with the view that the paucity of documented genetic interactions in E. coli as compared to S. cerevisiae reflects differences in methodologies employed to search for such interactions, rather than fundamental differences in the genetic interaction networks of the these organisms.
Our initial findings only hint at the rich biology that remains to be uncovered by systematic exploration of synthetic relationships. Some of the interactions can be rationalized, such as the three positive interactions identified for the ompA mutant. SurA and to a lesser degree DegP are required for proper OmpA folding23, 24, while CpxR activates expression of DegP and DsbA25, 26, which are both required for proper OmpA folding24, 27. In all three backgrounds (cpxR−, surA− and degP−) removing OmpA could decrease envelope stress caused by accumulation of misfolded OmpA. Likewise, some suppressors of the yraP mutant are also suggestive of compensation. Skp, a periplasmic chaperone, and DsbA and DsbB, which orchestrate disulfide-bond formation in the periplasm, may suppress because misfolding of a downstream target of these chaperones buffers the envelope defect of yraP− cells, possibly by relieving an imbalance in membrane composition. As no known DsbA-DsbB and Skp substrates28, 29 were identified as suppressors, an essential outer membrane protein targeted by those chaperones (e.g. Imp or YaeT), not present in our query set, may be responsible for this phenotype.
In contrast, the importance of Pal was unanticipated because its precise function(s) is unknown. Pal is an outer-membrane (OM)-associated lipoprotein that tethers the OM to the peptidoglycan (PG)30, an interaction believed to stabilize the OM. Pal is also the OM anchor of the 5-protein Tol-Pal system that bridges the inner and outer membranes31 and is energized by proton motive force32. This protein complex is highly conserved among gram-negative bacteria and has recently been implicated in ensuring proper OM invagination during division33. Six newly identified synthetic interactions in our 12 × 12 matrix involve pal, several in a growth-conditional manner (pal-ompA; pal-yfgL; pal-surA; pal-degP; pal-cpxR; pal-yraP). Furthermore the fact that our genome-wide screen with the pal mutant identifies many additional genetic interactions including the entire biosynthetic pathways for the core oligosaccharide of LPS (lipopolysaccharide) and ADP-L-glycero-β-manno-heptose (another precursor of the LPS inner core), the biosynthetic pathway for the enterobacterial common antigen and several additional envelope proteins of known and unknown function (Supplementary Table 2A) suggests the possibility that the Tol-Pal system may be one of the central organizers of envelope functionality. The abundance of interactions identified for Pal illustrates the degree of information GIANT-coli can generate and gives a taste of its potential to provide insights into the function and integration of different cellular processes, when coherent data are collected and analyzed.
The addition of GIANT-coli to the genetic toolbox of E. coli ensures that systematic genetic interaction data can be rapidly accumulated and then interfaced with information from studies of individual pathways and regulatory systems as well as with large datasets including global phenotypic screens34, 35, protein-protein interaction data36, proteome chip data37, gene-expression profiling (Gene Expression Omnibus) and other forward genetics screens38, 39. We anticipate adapting this methodology to give additional readouts such as promoter activity (gfp or lacZ fusions), biofilm formation (Congo-red or calcofluor plates, crystal violet absorption), siderophore production (chrome azurol S plates), and growth inhibition (halo assays). Moreover, our goal is to extend GIANT-coli to other gram-negative and -positive bacteria. Hfr’s have been successfully used in Salmonella and Shigella; slight variations of GIANT-coli can be employed in cases where Hfr’s are not applicable, e.g. the marked gene deletion can be carried on a conjugative plasmid. We also want to develop a similar technology for naturally competent organisms (e.g. B. subtilis, S. pneumoniae, N. gonorrhoeae); preliminary experiments verify that DNA uptake can be used as a high-throughput approach to generate double mutants en masse in such organisms (M. Winkler, personal communication). Together, these methodologies should lead to rapid progress in discovery of gene function and network connections in the bacterial kingdom.
METHODS
A detailed description of strains used in this study and experimental procedures (growth conditions, array manipulations and data analysis procedures) can be found in Supplementary data online.
Supplementary Material
Supplementary Figure 1: Optimization of GIANT-coli
Supplementary Figure 2: GIANT-coli in different plate formats
Supplementary Figure 3: Data normalization
Supplementary Figure 4: Analysis of the 12 × 12 genetic interaction matrix in LB versus M9
Supplementary Figure 5: Verification of interactions identified in the 12 × 12 matrix
Supplementary Table 1: Reproduction of SSLs by co-transduction of a linked marker
Supplementary Table 2: Genetic interactions of pal in M9 and suppressors of the yraP− lethality in 3% SDS
Supplementary Table 3: Strains and primers used in this study
ACKNOWLEDGEMENTS
We wish to thank Sueyoung Lee, Angela Wong and Jeanyoung Lee for excellent technical assistance. We also thank C. J. Ingles, P.J. Kiley, C. Squires, S. A. Johnson, A. Hochschild & T. J. Silhavy for critically reading this manuscript and offering useful suggestions. This work was supported by Sandler Family Funding to C.A.G. and to N.J.K., NIH GM036278 to C.A.G., NIH GM62662 to B.L.W., CREST-JST, Grant-in-Aid for Scientific Research & Grant-in-Aid for Scientific Research on Priority Areas to H.M. A.T. is a recipient of an EMBO fellowship.
CONTRIBUTIONS
A.T., R.J.N., D.A.S., J.S.W., N.J.K. and C.A.G. designed research; A.T., R.J.N., D.A.S. and B.L. executed research; A.T., R.J.N., M.S., S.C. and H.B. analyzed data; N.Y., R.T., B.L.W. and H.M. contributed new reagents (single-gene knock out libraries & CIP plasmids); A.T., N.J.K. and C.A.G. wrote the paper; R.J.N., M.S., B.L.W. and J.S.W. edited the paper.
REFERENCES
- 1.Boone C, Bussey H, Andrews BJ. Exploring genetic interactions and networks with yeast. Nat Rev Genet. 2007;8:437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- 2.Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 3.Tong AH, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 4.Pan X, et al. A robust toolkit for functional profiling of the yeast genome. Mol Cell. 2004;16:487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- 5.Collins SR, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007 doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 6.Schuldiner M, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 7.Roguev A, Wiren M, Weissman JS, Krogan NJ. High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nat Methods. 2007;4:861–866. doi: 10.1038/nmeth1098. [DOI] [PubMed] [Google Scholar]
- 8.Byrne AB, et al. A global analysis of genetic interactions in Caenorhabditis elegans. J Biol. 2007;6:8. doi: 10.1186/jbiol58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breitkreutz BJ, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ara K, et al. Bacillus minimum genome factory - effective utilization of microbial genome information. Biotechnol Appl Biochem. 2006 doi: 10.1042/BA20060111. [DOI] [PubMed] [Google Scholar]
- 11.Ito M, Baba T, Mori H. Functional analysis of 1440 Escherichia coli genes using the combination of knock-out library and phenotype microarrays. Metab Eng. 2005;7:318–327. doi: 10.1016/j.ymben.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Low KB. chapter 127. In: Neidhardt FC, et al., editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM; 1996. [Google Scholar]
- 13.Francois V, Conter A, Louarn JM. Properties of new Escherichia coli Hfr strains constructed by integration of pSC101-derived conjugative plasmids. J Bacteriol. 1990;172:1436–1440. doi: 10.1128/jb.172.3.1436-1440.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bachmann BJ. chapter 133. In: Neidhardt FC, et al., editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM; 1996. [Google Scholar]
- 15.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen HJ, Youngren B, Hansen FG, Austin S. Dynamics of Escherichia coli chromosome segregation during multifork replication. J Bacteriol. 2007;189:8660–8666. doi: 10.1128/JB.01212-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nielsen HJ, Li Y, Youngren B, Hansen FG, Austin S. Progressive segregation of the Escherichia coli chromosome. Mol Microbiol. 2006;61:383–393. doi: 10.1111/j.1365-2958.2006.05245.x. [DOI] [PubMed] [Google Scholar]
- 18.Collins SR, Schuldiner M, Krogan NJ, Weissman JS. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoekstra WP, Havekes AM. On the role of the recipient cell during conjugation in Escherichia coli. Antonie Van Leeuwenhoek. 1979;45:13–18. doi: 10.1007/BF00400773. [DOI] [PubMed] [Google Scholar]
- 20.Rizzitello AE, Harper JR, Silhavy TJ. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J Bacteriol. 2001;183:6794–6800. doi: 10.1128/JB.183.23.6794-6800.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charlson ES, Werner JN, Misra R. Differential effects of yfgL mutation on Escherichia coli outer membrane proteins and lipopolysaccharide. J Bacteriol. 2006;188:7186–7194. doi: 10.1128/JB.00571-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onufryk C, Crouch ML, Fang FC, Gross CA. Characterization of six lipoproteins in the σE regulon. J Bacteriol. 2005;187:4552–4561. doi: 10.1128/JB.187.13.4552-4561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rouviere PE, Gross CA. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 1996;10:3170–3182. doi: 10.1101/gad.10.24.3170. [DOI] [PubMed] [Google Scholar]
- 24.Sklar JG, Wu T, Kahne D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007;21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Danese PN, Snyder WB, Cosma CL, Davis LJ, Silhavy TJ. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 1995;9:387–398. doi: 10.1101/gad.9.4.387. [DOI] [PubMed] [Google Scholar]
- 26.Pogliano J, Lynch AS, Belin D, Lin EC, Beckwith J. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 1997;11:1169–1182. doi: 10.1101/gad.11.9.1169. [DOI] [PubMed] [Google Scholar]
- 27.Bardwell JC, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 28.Leichert LI, Jakob U. Protein thiol modifications visualized in vivo. PLoS Biol. 2004;2:e333. doi: 10.1371/journal.pbio.0020333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qu J, Mayer C, Behrens S, Holst O, Kleinschmidt JH. The trimeric periplasmic chaperone Skp of Escherichia coli forms 1:1 complexes with outer membrane proteins via hydrophobic and electrostatic interactions. J Mol Biol. 2007;374:91–105. doi: 10.1016/j.jmb.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 30.Parsons LM, Lin F, Orban J. Peptidoglycan recognition by Pal, an outer membrane lipoprotein. Biochemistry. 2006;45:2122–2128. doi: 10.1021/bi052227i. [DOI] [PubMed] [Google Scholar]
- 31.Cascales E, Lloubes R. Deletion analyses of the peptidoglycan-associated lipoprotein Pal reveals three independent binding sequences including a TolA box. Mol Microbiol. 2004;51:873–885. doi: 10.1046/j.1365-2958.2003.03881.x. [DOI] [PubMed] [Google Scholar]
- 32.Cascales E, Lloubes R, Sturgis JN. The TolQ-TolR proteins energize TolA and share homologies with the flagellar motor proteins MotA-MotB. Mol Microbiol. 2001;42:795–807. doi: 10.1046/j.1365-2958.2001.02673.x. [DOI] [PubMed] [Google Scholar]
- 33.Gerding MA, Ogata Y, Pecora ND, Niki H, de Boer PA. The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol Microbiol. 2007;63:1008–1025. doi: 10.1111/j.1365-2958.2006.05571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inoue T, et al. Genome-wide screening of genes required for swarming motility in Escherichia coli K-12. J Bacteriol. 2007;189:950–957. doi: 10.1128/JB.01294-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bochner BR. New technologies to assess genotype-phenotype relationships. Nat Rev Genet. 2003;4:309–314. doi: 10.1038/nrg1046. [DOI] [PubMed] [Google Scholar]
- 36.Butland G, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 37.Chen CS, et al. A proteome chip approach reveals new DNA damage recognition activities in Escherichia coli. Nat Methods. 2008;5:69–74. doi: 10.1038/NMETH1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mazurkiewicz P, Tang CM, Boone C, Holden DW. Signature-tagged mutagenesis: barcoding mutants for genome-wide screens. Nat Rev Genet. 2006;7:929–939. doi: 10.1038/nrg1984. [DOI] [PubMed] [Google Scholar]
- 39.Girgis HS, Liu Y, Ryu WS, Tavazoie S. A comprehensive genetic characterization of bacterial motility. PLoS Genet. 2007;3:1644–1660. doi: 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Optimization of GIANT-coli
Supplementary Figure 2: GIANT-coli in different plate formats
Supplementary Figure 3: Data normalization
Supplementary Figure 4: Analysis of the 12 × 12 genetic interaction matrix in LB versus M9
Supplementary Figure 5: Verification of interactions identified in the 12 × 12 matrix
Supplementary Table 1: Reproduction of SSLs by co-transduction of a linked marker
Supplementary Table 2: Genetic interactions of pal in M9 and suppressors of the yraP− lethality in 3% SDS
Supplementary Table 3: Strains and primers used in this study