Abstract
Methionine starvation can modulate gene methylation, cell cycle transition, and pathways related to survival following DNA damage. Methionine depletion by recombinant methioninase (rMETase) may have in vitro and in vivo efficacy against neuroblastoma (NB), especially when combined with chemotherapeutic drugs. rMETase from Pseudomonas Putida was produced in E. Coli and purified by ion-exchange chromatography. rMETase alone inhibited the proliferation of 15/15 NB cell lines in vitro. Among these 15 cell lines, only 66N demonstrated rMETase-induced apoptosis. rMETase alone suppressed LAN-1 and NMB-7 xenografts (p<0.01) and no toxicities were noted other than reversible weight loss. In vitro efficacy experiments combining rMETase and chemotherapeutic agents were carried out using SK-N-LD and SK-N-BE(1)N established at diagnosis, as well as LAN-1, SK-N-BE(2)C, and NMB-7 established at relapse. Microtubule depolymerization agents including vincristine, vinorelbine, vinblatine, and mebendazole showed synergism when tested in combination with rMETase in all 5 cell lines. Among DNA damaging agents, synergy with rMETase was observed only in cell lines established at diagnosis, and not at relapse. Cell cycle analysis showed that rMETase arrested G2 phase, and not M phase. In vivo efficacy experiments using LAN-1 and NMB-7 xenografts showed that rMETase rendered vincristine more effective than vincristine alone in tumor growth suppression (p<0.001). In conclusion, methionine depletion inhibited NB proliferation and arrested tumor cells at G2 phase. rMETase synergized with microtubule depolymerization agents. Moreover, synergism between rMETase and DNA damaging agents was dependent on whether cell lines were established at diagnosis or at relapse.
Keywords: recombinant methioninase, microtubule inhibitors, neuroblastoma therapy
INTRODUCTION
Methionine deprivation affects tumor cells with the propensity to divide and causes them to arrest predominantly in the G2 phase of the cell cycle and to eventually undergo apoptosis.1-5 Reduction of plasma methionine below 5 μmol/L arrests human xenograft growth in athymic mice.6 Methionine depletion can be achieved using parenteral L-methionine-deamino-mercaptomethane-lyase (methioninase, a methionine-cleaving enzyme derived from pseudomonas putida)7, 8 which is cloned and expressed in Escherichia coli.9, 10 A variety of tumors including Lewis lung carcinoma,11 several human colon cancer lines,12 and glioblastoma13, 14 have shown arrested growth by rMETase. In order to eradicate tumors, treatment has to be prolonged. However, prolonged deprivation of methionine is impractical because it may result in permanent liver damage15 and tumor will rapidly regrow after rMETase is withdrawn. Since methionine starvation can broadly affect protein synthesis, and modulate gene methylation, cell cycle transition and pathways related to survival following DNA damage in tumor,3, 6, 13, 14, 16, 17 but not in normal tissues, they may enhance the effects of chemotherapeutic drugs and decrease systemic toxicity. Indeed, there is growing evidence that certain classes of chemotherapeutic agents may be synergistically enhanced by methionine depletion; these include BCNU,3 doxorubicin,4 5-fluorouracil, mitomycin C,18 cisplatin,12 and nitrosoureas.13, 14
Neuroblastoma (NB) is an embryonic neoplasm of the sympathetic nervous system.19, 20 Metastatic NB is a highly proliferative cancer where gene methylation21, 22 and alkylator resistance23 are strongly correlated with poor prognosis. Drawing from the successful experience of bacteria-derived asparaginase in the current treatment of childhood acute lymphoblastic leukemia,24 we hypothesize that rMETase may have clinical utility against NB, which is methionine dependent. In this report, we describe the production and purification of pseudomonas putida rMETase, its in vitro and in vivo cytotoxicity against NB, alone and in combination with common chemotherapeutic agents, including microtubule depolymerization agents.
Materials and Methods
Plasmids
A pKK223-3 plasmid containing the gene for L-methioninase was kindly provided by Dr. Dennis Carson, University of Southern California (Los Angeles, CA) and Dr. Roger Harrison, University of Oklahoma (Norman, OK).10
Tumor Cell Lines
NB cell line LAN-1 was provided by Dr. Robert Seeger (Children's Hospital of Los Angeles; Los Angeles, CA), NB cell line NMB-7 by Dr. Liao of McMaster University (Hamilton, ON, Canada). SK-N-BE(1)N, SK-N-BE(2)C, SK-N-BE(2)N, SK-N-BE(2)S, LAI-5S and SH-EP-1, 55N and 66N were kindly provided by Dr. Robert Ross, Fordham University (New York, NY). NB cell lines SK-N-LD, SK-N-ED, SK-N-MM were established at Memorial Sloan-Kettering Cancer Center (MSKCC, New York, NY). IMR-32 and CHP-212 were purchased from American Type Culture Collection (Manassas, VA). Cells were cultured as previously described.25
Mouse Feed
The diet 518787 purchased from Dyets (Bethlehem, PA) was methionine and choline deficient and contained 1.7 g/kg DL-homocystine.6 In this report, it was referred to as Met(−)Hcyss(+)Chl(−).
Production of rMETase
rMETase fermentation was carried out in a 14 L Bioflow 3000 Fermentor (New Brunswick Scientific, Edison, NJ) using TB (Terrific Broth) medium containing 100 μg/ml ampicillin. Fermentation conditions and cell disruption were performed as previously described.26 rMETase was captured by QAE-Sepharose FF (Amersham Pharmacia Biotech, Piscataway, NJ) and then eluted with buffer A (20 mmol/L potassium phosphate buffer (pH 7.4), 20 μmol/L pyridoxal 5′-phosphate, 1 mmol/L EDTA (pH 8.0), 0.01% β-mercaptoethanol containing 0.3 mol/L NaCl. Pooled rMETase was heated at 60°C for 15 min. Endotoxin was removed by 1% triton X-114 phase separation,27 and followed by Detoxi-Gel™ Endotoxin Removing Gel (Pierce, Rockford, IL) to remove a small amount of endotoxin retained in the rMETase solution. The endotoxin level was determined by Limulus Amebocyte Lysate QCL-1000® (Cambrex Bio Science, Walkersville, MD). 2-ketobutyric acid (Sigma-Aldrich) was used to establish a standard curve for enzyme activity assay.28 One unit of rMETase was defined as the amount of enzyme that produced 1 μmol of α-ketobutyrate per minute at infinite concentration of L-methionine. Protein was quantified by BCA™ Protein Assay Kit (Pierce, Rockford, IL). A TSK-GEL G3000SWxl size exclusion column (30cm×7.8mm, 5 μ) (Tosoh Bioscience, Montgomeryville, PA) was used for HPLC analysis with 1×PBS as the mobile phase.
WST8 Assay for NB Cell Proliferation
WST-8 assay (Dojindo Molecular Technologies, MA) was validated by the manufacturer using MTT assay in NB cell line IMR-32 in the presence of DNA damaging agents and topoisomerase II inhibitor. Our laboratory also validated this assay using AlamarBlue and direct cell counting (RT-CES system, ACEA Biosciences, San Diego, CA) when LAN-1 and NMB-7 were treated with SN38.
In vitro proliferation inhibition of NB cell lines was carried out in 96-well plates (BD Biosciences, Bedford, MA). Twenty-four hours after the cells were seeded at a density of 4000-10000 cells/well, rMETase, chemotherapeutic drugs or their combinations were added to the wells. After 3-5 days of exposure, 1/10 volume WST8 was added and optical density (OD) was read at 450 nm. The proliferation rate of cells was calculated as follows: % Proliferation Rate = (OD450 of treated well − OD450 of medium only well)/(OD450 of non-treated well − OD450 of medium only well) × 100%. IC50 (half maximal inhibitory concentration) was calculated by SigmaPlot 8.0 (Systat Software, Inc., San Jose, CA) and combination index (CI) was calculated by CompuSyn (CompoSyn, Paramus, NJ).29
NB Cell Apoptosis and Cell Cycle Assay
The in vitro apoptosis and cell cycle experiments were carried out in 6-well plates (BD Biosciences). Twenty-four hours after the cells were seeded at a density of 2-5×105 cells/well, rMETase, vincristine or their combinations were added to the wells. After 12-48 hr of treatment, cells were stained with either 7-amino-actinomycin D (7AAD) and Annexin V or propidium iodide (PI) (BD PharMingen, San Diego, CA) for apoptosis assay or stained with PI and MPM-2/CY5 (Upstate Biotechnology, Charlottsville, VA) for cell cycle assay using flow cytometry with BD FACSCalibur System (BD Bioscience). For each sample, 10,000 events in gated area were collected. The data from flow cytometry were analyzed by Flowjo (Windows-version 5.7.2) software (Tree star, Ashland, OR).
Antitumor Effect with NB Xenografts
All animal experiments were carried out according to an Institutional Animal Care and Use Committee (IACUC)-approved protocol, and institutional guidelines for the proper and humane use of animals in research were followed. Athymic female nude mice were purchased from the National Cancer Institute. Mice with 5-10 mm established tumors were randomly separated into groups of 5-10 mice each. Based on pharmacokinetics study of rMETase in mice, after injection of 100Units rMETase, plasma rMETase half-life was 2.06 ± 0.10 hr. When mice were fed methionine-free diet, one injection of 100Unit rMETase could keep plasma methionine level nadir to 0.15 ± 0.06 μmol/L at 6 hours and recovered to 13.0% of pretreatment level at 10 hours. One injection of 100Unit rMETase could keep plasma methionine level below 5μM for ∼10 hours. That is why we chose twice daily dosing. When rMETase efficacy was tested against NB xenografts, 100 Units of rMETase was given iv bid ×4days/wk ×3weeks as a single agent. Mice were bled 4 hr after rMETase injection, which was carried out on the 4th day of the 1st and 3rd week of treatment. Methionine level was measured according to the method of Sun et al.30 In combination experiments, rMETase and vincristine were given separately or in combination at same time. 100 Units of rMETase was given iv bid ×2days/wk ×3weeks and vincristine were given iv qwk ×3weeks. In these experiments, Met(−)Hcyss(+)Chl(−) diet was given to the animals only during rMETase administration. The weight of mice was used as an index of toxicity. In addition, excessive weight loss may confound the conclusion on tumor shrinkage; thus, 15% maximum weight loss was used a guideline in designing methioninase dosing and timing of normal feeds. Tumor size and weight were measured twice per week and they were expressed as mean ± SEM (standard error of the mean) relative to tumor size or body weight values on day 0 (start of treatment). Mice were sacrificed when their tumor sizes exceeded 20 mm in diameter.
RESULTS
Fermentation, purification and characterization of rMETase
A 10 liter of fermentation culture produced ∼70,000 Units of rMETase in the supernatant from homogenized E coli cell extracts. The yield of rMETase was about 50%, with 30% loss during the column chromatography and 20% loss during the heat and endotoxin removal. rMETase was stable at 4°C for at least 2 months when stored in buffer A containing 0.3 mol/L NaCl (see Methods). After ultrafiltration, the final concentration of rMETase was about 800 Units/ml and endotoxin was less than 1 EU/ml. The enzyme activity ratio was 16.2 Units/mg.
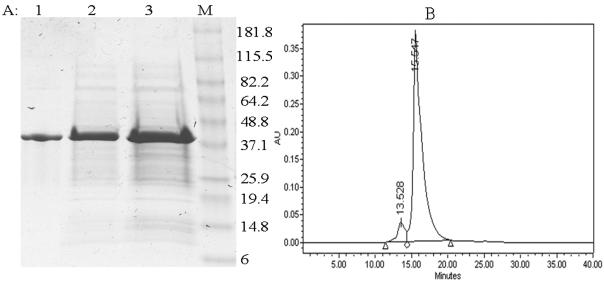
SDS-PAGE demonstrated that rMETase was almost pure after one step of ion-exchange chromatography (Fig. 1a). The major band indicated an apparent molecular weight of 43 KD. On size exclusion HPLC, the purified protein exhibited one major peak at a retention time of 15.547 min (Fig. 1b) with 92.5 % purity.
Figure 1. Molecular size and purity of rMETase.
A: SDS-PAGE. Lane 1: Purified rMETase; Lane 2: Supernatant from whole cell extract; Lane 3: Whole cell crude extract, Lane 4: Marker.
B: HPLC size exclusion profile of purified rMETase. The major peak at 15.547 min was rMETase.
Inhibition of in vitro proliferation of NB cells
The growth of 15 NB cell lines was inhibited by rMETase exposure in a dose-dependent manner. IC50 values were tabulated in Table I. All cell lines, except SK-N-BE(2)S which had a substrate adherent phenotype, were highly sensitive to rMETase. After 3 days of exposure, methionine levels on the 96-well plate were tested by HPLC. Only the wells with ≤0.175 Unit/ml rMETase had detectable methionine level (≥0.18 ± 0.01 μmol/L); the concentration of methionine in control medium containing 10% calf serum was 62.09 ± 0.94 μmol/L. Among these 15 cell lines, only 66N showed obvious and reproducible apoptosis induced by rMETase after 48 hours of exposure with a 22.4 % increase of Annexin V+/7AAD− population.
Table I.
Inhibition of NB cell proliferation by rMETase
| Cell Line | IC50 (Units/ml) |
|---|---|
| SK-N-BE(1)N | 0.04 ± 0.01 |
| SK-N-LD | 0.05 ± 0.01 |
| NMB-7 | 0.08 ± 0.01 |
| SH-EP-1 | 0.12 ± 0.05 |
| SK-N-BE(2)C | 0.13 ± 0.02 |
| LAN-1 | 0.18 ± 0.01 |
| IMR-32 | 0.20 ± 0.02 |
| CHP-212 | 0.20 ± 0.01 |
| 66N | 0.22 ± 0.03 |
| 55N | 0.27 ± 0.03 |
| SK-N-MM | 0.36 ± 0.03 |
| SK-N-ED | 0.39 ± 0.03 |
| LAI-5S | 0.48 ± 0.02 |
| SK-N-BE(2)N | 0.51 ± 0.11 |
| SK-N-BE(2)S | 5.12 ± 0.56 |
IC50 (half maximal inhibitory concentration) was expressed as mean ± SEM (standard error of the mean).
At high concentrations of rMETase, all NB cell lines will eventually die. After cells were treated with rMETase for 24 hours, it will take 3-4 days for the cell to recover by addition of 200uM methionine as demonstrated by flow cytometry analysis and cell number. These findings suggest that methionine depletion is the major mechanism of cell death.
Combining rMETase and chemotherapeutic drugs on neuroblastoma in vitro
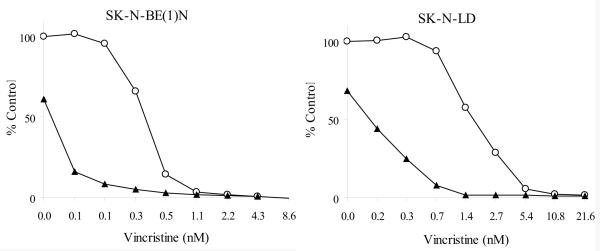
In vitro efficacy experiments were carried out using SK-N-LD and SK-N-BE(1)N established at diagnosis (p53 wild type), as well as LAN-1 (p53 deleted), SK-N-BE(2)C (p53 mutated), and NMB-7 (p53 wild type) all established at relapse. In these experiments, a fixed concentration of rMETase (approximately 20%-50% inhibition of cell growth) was used, while the concentrations of chemotherapeutic drugs were varied so as to kill 0 to 100% tumor cell in a serial dilution. Combination indice (CIs) calculated in non-constant ratio mode were shown in Table II and the effects of vincristine with or without rMETase on SK-N-BE(1)N and SK-N-LD proliferation were shown in Fig. 2. In all 5 NB cell lines tested, rMETase synergized with microtubule depolymerization agents (vincristine, vinorelbine, vinblatine, and mebendazole), but not microtubule stabilizing agents (paclitaxel, docetaxel, and fludelone). Gemcitabine also did not show synergy in these 5 cell lines. Cell lines established at diagnosis (SK-N-LD and SK-N-BE(1)N), but not those established at relapse (LAN-1, SK-N-BE(2)C, and NMB-7) showed synergism when rMETase combined with alkylators and topoisomerase inhibitors. In these 5 cell lines, their sensitivity to rMETase exposure was comparable to their sensitivity to chemotherapeutic drugs (data not shown).
Table II.
In vitro inhibition of proliferation of NB cell lines exposed to methionine depletion and chemotherapeutic drugs as determined by combination index (CI)*
| Chemotherapeutic agents |
Drug functions | SK-N-LD | SK-N- BE(1)N |
LAN-1 | SK-N- BE(2)C |
NMB-7 |
|---|---|---|---|---|---|---|
| Vincristine | microtubule depolymerization | 0.40 | 0.47 | 0.63 | 0.40 | 0.55 |
| Vinorelbine | microtubule depolymerization | 0.39 | 0.47 | 0.66 | 0.46 | 0.61 |
| Vinblatine | microtubule depolymerization | 0.40 | 0.50 | 0.65 | 0.63 | 0.61 |
| Mebendazole | microtubule depolymerization | 0.40 | 0.45 | 0.51 | 0.37 | 0.60 |
| Paclitaxel | microtubule stabilization | 0.88 | 1.14 | 1.91 | 1.25 | 1.38 |
| Docetaxel | microtubule stabilization | 0.72 | 0.81 | 1.10 | 1.92 | 0.99 |
| Iso-Fludelone | microtubule stabilization | 0.93 | 0.71 | 1.96 | 0.90 | 1.14 |
| Etoposide | topoisomerase II inhibitor | 0.48 | 0.58 | 0.98 | 12.78 | 1.57 |
| Topotecan | topoisomerase I inhibitor | 0.51 | 0.35 | 0.71 | 9.25 | 1.22 |
| SN38 | topoisomerase I inhibitor | 0.59 | 0.51 | 1.82 | 2.24 | 0.90 |
| Doxorubicin | DNA intercalation, topoisomerase II inhibitor |
0.59 | 0.59 | 0.64 | 11.99 | 0.81 |
| Cisplatin | DNA cross-linking | 0.67 | 0.65 | 0.71 | 1.79 | 1.37 |
| Thiotepa | alkylating agent | 0.60 | 0.60 | 1.00 | 2.11 | 1.34 |
| Gemcitabine | nucleoside analog | 0.85 | 1.11 | 1.08 | 76.99 | 1.12 |
CI was detailed in the Material and Methods section.
Synergism: CI ≤ 0.7 (in boldface)
Additive: 0.90 ≤ CI ≤ 1.10
Antagonism: CI > 1.10
Figure 2. Effect of vincristine with or without rMETase on NB cell proliferation of SK-N-BE(1)N and SK-N-LD.
Open circle: vincristine alone; Solid triangle: rMETase + vincristine combined treatment.
rMETase and vincristine on cell cycle arrest
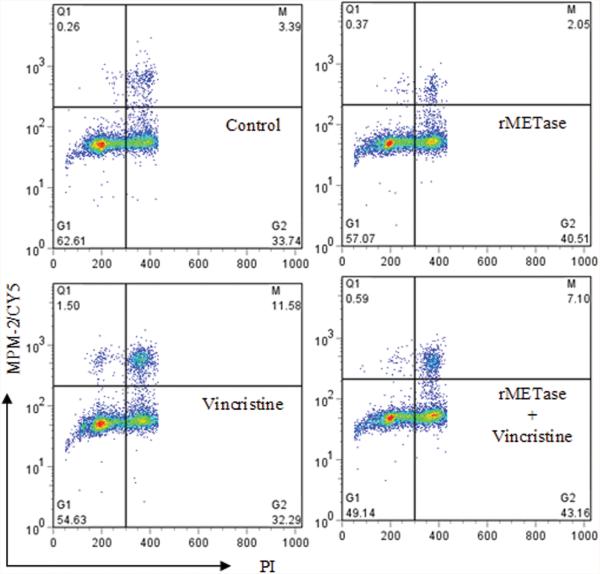
Based on the known function of chemotherapeutic drugs and the CI values (Table II), vincristine was chosen to study the mechanism involved when combined with rMETase. The effect of rMETase with or without vincristine on the cell cycle arrest was examined. After 18 hours of exposure to 0.2 Units/ml rMETase, SK-N-LD, SK-N-BE(1)N, LAN-1, SK-N-BE(2)C, and NMB-7 showed ∼10% increase of G2 population. However, unlike vincristine (2.5ng/ml), rMETase alone did not increase M phase arrest (Fig. 3). When rMETase and vincristine were combined, rMETase decreased the M phase arrest by vincristine, likely the result of a slower tumor cell cycle transition following G2 arrest by rMETase.
Figure 3. Effect of rMETase with or without vincristine on cell cycle arrest of SK-N-BE(2)C.
The flow cytometry results were presented in density plots, where the quadrants were set to include live cells. The lower left quadrant contained G1 phase population; the lower right quadrant contained G2 phase population; the upper right quadrant contained M phase population.
Efficacy of rMETase alone on NB Xenografts in athymic mice
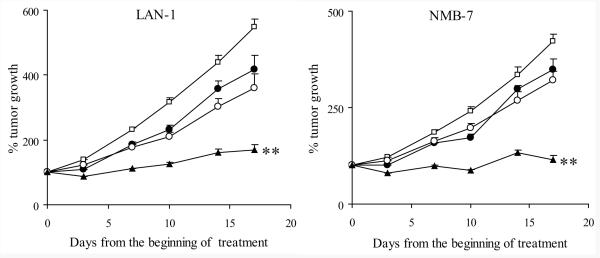
The schedule of rMETase administration was based on the toxicity of rMETase according to weight loss and pharmacokinetics of rMETase (see Methods). rMETase treatment was generally well tolerated. In xenografts, weight loss after methionine deprivation was the only dose limiting toxicity. When athymic mice planted with NB xenografts were treated with 100 Units rMETase iv bid ×4d/wk ×3wks, a ∼10% reversible weight loss was observed. Four hours after rMETase injection, plasma methionine concentration was 4.72 ± 0.47 μmol/L among rMETase-treated mice fed with normal chow, in contrast to 61.79 ± 2.14 μmol/L among the untreated mice (n=36). Mice fed with Met(−)Hcyss(+)Chl(−) diet had ∼20% reversible weight loss after injection of 100 Units rMETase iv bid ×4d/wk. Plasma methionine concentration was lowered to 0.40 ± 0.07 μmol/L among the rMETase-treated group, in contrast to 31.39 ± 3.78 μmol/L among the untreated mice (n=9). 100 Units rMETase iv bid ×4d/wk ×3wks demonstrated significant efficacy against LAN-1 and NMB-7 xenografts (p<0.01) (Fig. 4). Methionine levels 4 hours after rMETase injection were comparable among the 2 xenograft models (data not shown).
Figure 4. Efficacy of in vivo rMETase in NB xenografts.
Open square: Control (N=10 for LAN-1 and N=26 for NMB-7); Solid circle: rMETase treatment (N=10 for LAN-1 and N=34 for NMB-7); ** p<0.01 when AUC of rMETase group was compared to the control group.
Combining rMETase and vincristine in the suppression of NB xenografts
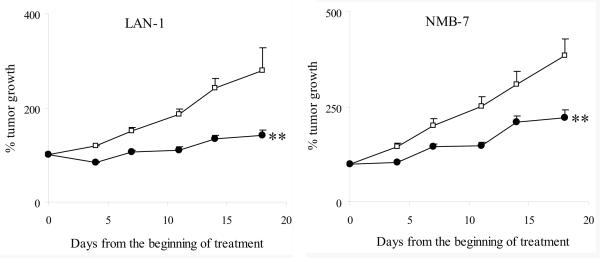
Based on our in vitro study and the CI values (Table II), vincristine was chosen as the best candidate for in vivo evaluation of treatment efficacy. Doxorubicin and thiotepa were also tested for comparison. When combined with chemotherapeutic agents, we chose doses of chemotherapeutic drugs with suboptimal anti-tumor effect so as to avoid a situation where the combined anti-tumor effect would exceed 100%. The following doses and regimens were chosen: Vincristine (0.8mg/kg) 31 and doxorubicin (6.5mg/kg) 32 iv qwk ×3wks, and thiotepa (3mg/kg) 33 iv ×2d/wk ×3wks were used. For these experiments, 100 Units/dose of rMETase was given iv bid ×2d/wk ×3wks in the presence of Met(−)Hcyss(+)Chl(−) diet. Administration of rMETase led to a ∼ 15% body weight loss for rMETase alone and the combination treatment groups; the animals regained their weight prior to the next cycle of treatment. The anti-tumor efficacy for single agents or in combination with rMETase was evaluated in 2 NB xenografts (Fig. 5). To evaluate in vivo efficacy of combination treatments by student's t-test, individual tumor growth rate versus time curve was transformed into area-under-the-curve (AUC) by SigmaPlot. AUCs showed that rMETase rendered vincristine more effective than vincristine alone in tumor growth suppression (p<0.001). In contrast, rMETase did not significantly enhance the efficacy of doxorubicin and thiotepa against tumor growth (data not shown).
Figure 5. Efficacy of in vivo combination treatment with rMETase and vincristine.
Open square: Control (N=9 for LAN-1 and NMB-7); Solid circle: rMETase alone (N=14 for LAN-1 and N=7 for NMB-7); Open circle: vincristine alone (N=8 for LAN-1 and N=7 for NMB-7); Solid triangle: rMETase + vincristine combined treatment (n=14 for LAN-1 and n=13 for NMB-7); ** p<0.001 when AUC of combination group was compared to other 3 groups, respectively.
DISCUSSION
The biochemical mechanism for methionine dependency has been studied extensively, but its effect on tumors derived from the sympathetic nervous system remains unclear. At high concentrations of rMETase, our studies showed that all NB cell lines will eventually die. At intermediate concentrations some will survive and some will linger on. Cells can be salvaged (death prevented) when methionine is added back to the medium, suggesting that methionine depletion is the major mechanism of cell death. We interpret these findings to mean that there is differential sensitivity of NB cell lines to methionine deprivation, as well as differences in their inherent resistance to apoptosis and cell death. We have looked for apoptosis at 24, 48 and 72 hours of rMETase treatment, and not beyond because massive cell loss usually has occurred by day 2. Observation at 48 hours after treatment with rMETase seemed to be most optimal time point for showing apoptosis. But we could document apoptosis in only 1 of 15 cell lines. We concluded that apoptosis is not the primary mechanism of cell death in the majority of NB cell lines tested here. This defect in apoptosis may relate to their caspase 3 and 8 deficiency,34 especially when MYCN was amplified, a genetic aberration typically found in these cell lines.
Methionine deprivation modulates the expression levels of many genes in tumor cells, 16, 17 some of which can provide rationales for combination approaches utilizing rMETase and chemotherapeutic drugs. We therefore undertook an analysis of standard chemotherapeutic agents against NB where the mechanisms of drug action were known. We tested for the presence of synergy between these chemotherapeutic agents and methionine deprivation using rMETase. We observed strong synergism between rMETase and vincristine, an M phase specific agent. We expanded these synergy studies using a panel of M phase specific agents (vincristine, vinorelbine, vinblastine, mebendazole, paclitaxel, docetaxel, and iso-fludelone) and found synergy only for microtubule depolymerization agents and not microtubule stabilizing agents. The mechanism of action likely involves both the methylation of tubulins and regulation of mitosis related gene by rMETase. S-adenosyl-L-methionine (SAM), produced from methionine and ATP, is the universal methyl donor in prokaryotes and eukaryotes and reduction of SAM compromises protein methylation. Methionine appears to be critical for the methylation of actins and tubulins in maintaining cellular stability35 and SAM is known to stabilize microtubule polymerization and reduce micronuclei formation.36 Thus, it is likely rMETase reduces methylation of tubulins, further destabilizing microtubules when tumor cells are subjected to depolymerization agents. On the other hand, rMETase can up-regulate (CDKN1A, and MDA1, etc) or down-regulate(CDKN2B, and Aurora Kinase B, etc).16, 17 The change in their genes expression levels is associated with the loss of mitosis in tumor cells.16 rMETase-induced genetic alteration shows the multifaceted effect of methionine deprivation,16, 17 which can explain the highly effective synergism between rMETase and a variety of diverse chemotherapies tested.
Downregulation of O6-methylguanine-DNA methyltransferase37 and reduction of SAM by methionine deprivation will decrease DNA methylation and affect DNA stability. Less stability of DNA increases the sensitivity of tumor cells to DNA damaging agents.38 When chemotherapeutic drugs were combined with methionine depletion, cell lines established at diagnosis had substantially smaller CIs and higher sensitivity than those established at relapse. It is highly likely that these resistant cell lines have already recruited pathways to repair DNA damage.39 One might expect the combination of rMETase and DNA damaging agents to be most useful if DNA repair pathways are inhibited. Poly(ADP-ribose) polymerase (PARP) inhibitor is cytotoxic for homologous-recombination (HR)-deficient cells by inhibiting base-excision repair.40 Our preliminary results suggested that PARP inhibitor synergized with rMETase in suppressing the proliferation of NB cell lines. A more detailed synergism analysis using cell lines established at diagnosis versus at relapse, with known DNA damage repair pathways will be critical for understanding the underlying mechanism of drug resistance.
For patients with high risk NB, acquired resistance to chemotherapy and the toxicity of chemotherapy in young children are clinical constraints. Late effects such as second cancer are clearly related to dose and intensity of chemoradiotherapy. Overcoming drug resistance or lowering drug doses to minimize toxicity can have a significant impact on patient survival. Given the toxicity profile and our results on preclinical efficacy of rMETase in human NB, methionine depletion especially when induced by rMETase may provide a useful adjunct to our current therapy of advanced stage disease. rMETase may be most effective when applied at initial diagnosis with diverse classes of chemotherapy. At relapse following exposure to chemotherapy, the synergy appeared to be restricted to microtubule depolymerization agents, a unique class of agents with primarily neurotoxicity, but minimal myelosuppression. The underlying mechanism of this cooperative effect is not currently understood. In contrast, neither microtubule stabilizing agents nor gemcitabine showed synergy despite their in vitro activity as single agents and their clinical efficacy in the treatment of NB.
In conclusion, methionine depletion inhibited NB proliferation and arrested tumor cells at G2 phase. However, apoptosis was not the primary mechanism of cell death in the majority of cell lines examined. As a single agent in vivo, methionine depletion inhibited NB xenografts growth. In combination therapy, methionine depletion synergized with microtubule depolymerization agents in vitro and rendered vincristine more effective than vincristine alone in tumor growth suppression in vivo. Synergism between methioninase and DNA damaging agents was dependent on whether cell lines were established at diagnosis or at relapse.
ACKNOWLEDGMENTS
We want to thank Dr. Irene Cheung (Memorial Sloan-Kettering Cancer Center, New York, NY) for critically reviewing the manuscript and Dr. Dennis Carson, University of Southern California (Los Angeles, CA) and Dr. Roger Harrison, University of Oklahoma (Norman, OK) for providing us with the pKK223-3 plasmid containing the gene for L-methioninase.
Grant sponsors: Supported in part by grants from the National Cancer Institute (CA61017, CA106450), Bethesda, MD; Hope Street Kids, Alexandria, VA; the Justin Zahn Fund, New York, NY; the Katie's Find A Cure Fund, New York, NY; and the Robert Steel Foundation, New York, NY.
Footnotes
Statements: In this report we showed that methioninase synergized with microtubule depolymerization agents in suppressing tumor cell growth. Moreover, synergism between methioninase and DNA damaging agents was dependent on whether cell lines were established at diagnosis or at relapse.
REFERENCES
- 1.Guo H, Lishko VK, Herrera H, Groce A, Kubota T, Hoffman RM. Therapeutic tumor-specific cell cycle block induced by methionine starvation in vivo. Cancer Res. 1993;53:5676–9. [PubMed] [Google Scholar]
- 2.Hoffman RM, Jacobsen SJ. Reversible growth arrest in simian virus 40-transformed human fibroblasts. Proc Natl Acad Sci U S A. 1980;77:7306–10. doi: 10.1073/pnas.77.12.7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kokkinakis DM, von Wronski MA, Vuong TH, Brent TP, Schold SC., Jr Regulation of O6-methylguanine-DNA methyltransferase by methionine in human tumour cells. Br J Cancer. 1997;75:779–88. doi: 10.1038/bjc.1997.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stern PH, Hoffman RM. Enhanced in vitro selective toxicity of chemotherapeutic agents for human cancer cells based on a metabolic defect. J Natl Cancer Inst. 1986;76:629–39. doi: 10.1093/jnci/76.4.629. [DOI] [PubMed] [Google Scholar]
- 5.Breillout F, Antoine E, Poupon MF. Methionine dependency of malignant tumors: a possible approach for therapy. J Natl Cancer Inst. 1990;82:1628–32. doi: 10.1093/jnci/82.20.1628. [DOI] [PubMed] [Google Scholar]
- 6.Kokkinakis DM, Schold SC, Jr., Hori H, Nobori T. Effect of long-term depletion of plasma methionine on the growth and survival of human brain tumor xenografts in athymic mice. Nutr Cancer. 1997;29:195–204. doi: 10.1080/01635589709514624. [DOI] [PubMed] [Google Scholar]
- 7.Lishko VK, Lishko OV, Hoffman RM. Depletion of serum methionine by methioninase in mice. Anticancer Res. 1993;13:1465–8. [PubMed] [Google Scholar]
- 8.Lishko VK, Lishko OV, Hoffman RM. The preparation of endotoxin-free L-methionine-alpha-deamino-gamma-mercaptomethane-lyase (L-methioninase) from Pseudomonas putida. Protein Expr Purif. 1993;4:529–33. doi: 10.1006/prep.1993.1069. [DOI] [PubMed] [Google Scholar]
- 9.Tan Y, Xu M, Tan X, Wang X, Saikawa Y, Nagahama T, Sun X, Lenz M, Hoffman RM. Overexpression and large-scale production of recombinant L-methionine-alpha-deamino-gamma-mercaptomethane-lyase for novel anticancer therapy. Protein Expr Purif. 1997;9:233–45. doi: 10.1006/prep.1996.0700. [DOI] [PubMed] [Google Scholar]
- 10.Hori H, Takabayashi K, Orvis L, Carson DA, Nobori T. Gene cloning and characterization of Pseudomonas putida L-methionine-alpha-deamino-gamma-mercaptomethane-lyase. Cancer Res. 1996;56:2116–22. [PubMed] [Google Scholar]
- 11.Yoshioka T, Wada T, Uchida N, Maki H, Yoshida H, Ide N, Kasai H, Hojo K, Shono K, Maekawa R, Yagi S, Hoffman RM, et al. Anticancer efficacy in vivo and in vitro, synergy with 5-fluorouracil, and safety of recombinant methioninase. Cancer Res. 1998;58:2583–7. [PubMed] [Google Scholar]
- 12.Tan Y, Sun X, Xu M, Tan X, Sasson A, Rashidi B, Han Q, Wang X, An Z, Sun FX, Hoffman RM. Efficacy of recombinant methioninase in combination with cisplatin on human colon tumors in nude mice. Clin Cancer Res. 1999;5:2157–63. [PubMed] [Google Scholar]
- 13.Kokkinakis DM, Hoffman RM, Frenkel EP, Wick JB, Han Q, Xu M, Tan Y, Schold SC. Synergy between methionine stress and chemotherapy in the treatment of brain tumor xenografts in athymic mice. Cancer Res. 2001;61:4017–23. [PubMed] [Google Scholar]
- 14.Kokkinakis DM, Wick JB, Zhou QX. Metabolic response of normal and malignant tissue to acute and chronic methionine stress in athymic mice bearing human glial tumor xenografts. Chem Res Toxicol. 2002;15:1472–9. doi: 10.1021/tx020033n. [DOI] [PubMed] [Google Scholar]
- 15.Kokkinakis DM. Methionine-stress: a pleiotropic approach in enhancing the efficacy of chemotherapy. Cancer letters. 2006;233:195–207. doi: 10.1016/j.canlet.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 16.Kokkinakis DM, Brickner AG, Kirkwood JM, Liu X, Goldwasser JE, Kastrama A, Sander C, Bocangel D, Chada S. Mitotic arrest, apoptosis, and sensitization to chemotherapy of melanomas by methionine deprivation stress. Mol Cancer Res. 2006;4:575–89. doi: 10.1158/1541-7786.MCR-05-0240. [DOI] [PubMed] [Google Scholar]
- 17.Kokkinakis DM, Liu X, Chada S, Ahmed MM, Shareef MM, Singha UK, Yang S, Luo J. Modulation of gene expression in human central nervous system tumors under methionine deprivation-induced stress. Cancer Res. 2004;64:7513–25. doi: 10.1158/0008-5472.CAN-04-0592. [DOI] [PubMed] [Google Scholar]
- 18.Goseki N, Yamazaki S, Shimojyu K, Kando F, Maruyama M, Endo M, Koike M, Takahashi H. Synergistic effect of methionine-depleting total parenteral nutrition with 5-fluorouracil on human gastric cancer: a randomized, prospective clinical trial. Jpn J Cancer Res. 1995;86:484–9. doi: 10.1111/j.1349-7006.1995.tb03082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kushner BH, Cheung NK. Neuroblastoma--from genetic profiles to clinical challenge. N Engl J Med. 2005;353:2215–7. doi: 10.1056/NEJMp058251. [DOI] [PubMed] [Google Scholar]
- 20.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–16. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 21.Alaminos M, Davalos V, Cheung NK, Gerald WL, Esteller M. Clustering of gene hypermethylation associated with clinical risk groups in neuroblastoma. J Natl Cancer Inst. 2004;96:1208–19. doi: 10.1093/jnci/djh224. [DOI] [PubMed] [Google Scholar]
- 22.Alaminos M, Davalos V, Ropero S, Setien F, Paz MF, Herranz M, Fraga MF, Mora J, Cheung NK, Gerald WL, Esteller M. EMP3, a myelin-related gene located in the critical 19q13.3 region, is epigenetically silenced and exhibits features of a candidate tumor suppressor in glioma and neuroblastoma. Cancer Res. 2005;65:2565–71. doi: 10.1158/0008-5472.CAN-04-4283. [DOI] [PubMed] [Google Scholar]
- 23.Wagner LM, McLendon RE, Yoon KJ, Weiss BD, Billups CA, Danks MK. Targeting Methylguanine-DNA Methyltransferase in the Treatment of Neuroblastoma. Clin Cancer Res. 2007;13:5418–25. doi: 10.1158/1078-0432.CCR-07-0418. [DOI] [PubMed] [Google Scholar]
- 24.Avramis VI, Tiwari PN. Asparaginase (native ASNase or pegylated ASNase) in the treatment of acute lymphoblastic leukemia. International journal of nanomedicine. 2006;1:241–54. [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung NK, Guo HF, Modak S, Cheung IY. Anti-idiotypic antibody facilitates scFv chimeric immune receptor gene transduction and clonal expansion of human lymphocytes for tumor therapy. Hybrid Hybridomics. 2003;22:209–18. doi: 10.1089/153685903322328938. [DOI] [PubMed] [Google Scholar]
- 26.Cheung N-KV, Modak S, Lin YK, Guo HF, Zanzonico P, Chung J, Zuo Y, Sanderson J, Wilbert S, Theodore LJ, Axworthy D, Larson SM. Single chain Fv-streptavidin substantially improved therapeutic index in multi-step targeting directed at disialoganglioside GD2. J Nucl Med. 2004 [PubMed] [Google Scholar]
- 27.Liu S, Tobias R, McClure S, Styba G, Shi Q, Jackowski G. Removal of endotoxin from recombinant protein preparations. Clin Biochem. 1997;30:455–63. doi: 10.1016/s0009-9120(97)00049-0. [DOI] [PubMed] [Google Scholar]
- 28.Takakura T, Mitsushima K, Yagi S, Inagaki K, Tanaka H, Esaki N, Soda K, Takimoto A. Assay method for antitumor L-methionine gamma-lyase: comprehensive kinetic analysis of the complex reaction with L-methionine. Anal Biochem. 2004;327:233–40. doi: 10.1016/j.ab.2004.01.024. [DOI] [PubMed] [Google Scholar]
- 29.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–81. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 30.Sun X, Tan Y, Yang Z, Li S, Hoffman RM. A rapid HPLC method for the measurement of ultra-low plasma methionine concentrations applicable to methionine depletion therapy. Anticancer Res. 2005;25:59–62. [PubMed] [Google Scholar]
- 31.Thompson J, George EO, Poquette CA, Cheshire PJ, Richmond LB, de Graaf SS, Ma M, Stewart CF, Houghton PJ. Synergy of topotecan in combination with vincristine for treatment of pediatric solid tumor xenografts. Clin Cancer Res. 1999;5:3617–31. [PubMed] [Google Scholar]
- 32.Harris SM, Scott JA, Brown JL, Charlton PA, Mistry P. Preclinical anti-tumor activity of XR5944 in combination with carboplatin or doxorubicin in non-small-cell lung carcinoma. Anti-cancer drugs. 2005;16:945–51. doi: 10.1097/01.cad.0000176499.17939.56. [DOI] [PubMed] [Google Scholar]
- 33.Fingert HJ, Pu AT, Chen ZY, Googe PB, Alley MC, Pardee AB. In vivo and in vitro enhanced antitumor effects by pentoxifylline in human cancer cells treated with thiotepa. Cancer Res. 1988;48:4375–81. [PubMed] [Google Scholar]
- 34.Iolascon A, Borriello A, Giordani L, Cucciolla V, Moretti A, Monno F, Criniti V, Marzullo A, Criscuolo M, Ragione FD. Caspase 3 and 8 deficiency in human neuroblastoma. Cancer Genet Cytogenet. 2003;146:41–7. doi: 10.1016/s0165-4608(03)00125-0. [DOI] [PubMed] [Google Scholar]
- 35.Moephuli SR, Klein NW, Baldwin MT, Krider HM. Effects of methionine on the cytoplasmic distribution of actin and tubulin during neural tube closure in rat embryos. Proc Natl Acad Sci U S A. 1997;94:543–8. doi: 10.1073/pnas.94.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramirez T, Garcia-Montalvo V, Wise C, Cea-Olivares R, Poirier LA, Herrera LA. S-adenosyl-L-methionine is able to reverse micronucleus formation induced by sodium arsenite and other cytoskeleton disrupting agents in cultured human cells. Mutation research. 2003;528:61–74. doi: 10.1016/s0027-5107(03)00099-x. [DOI] [PubMed] [Google Scholar]
- 37.Kokkinakis DM, Ahmed MM, Delgado R, Fruitwala MM, Mohiuddin M, Albores-Saavedra J. Role of O6-methylguanine-DNA methyltransferase in the resistance of pancreatic tumors to DNA alkylating agents. Cancer Res. 1997;57:5360–8. [PubMed] [Google Scholar]
- 38.Cellarier E, Durando X, Vasson MP, Farges MC, Demiden A, Maurizis JC, Madelmont JC, Chollet P. Methionine dependency and cancer treatment. Cancer treatment reviews. 2003;29:489–99. doi: 10.1016/s0305-7372(03)00118-x. [DOI] [PubMed] [Google Scholar]
- 39.Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, Villegas E, Jacquemont C, Farrugia DJ, Couch FJ, Urban N, Taniguchi T. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–20. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Livingston DM, Silver DP. Cancer: crossing over to drug resistance. Nature. 2008;451:1066–7. doi: 10.1038/4511066a. [DOI] [PubMed] [Google Scholar]