Abstract
Following an initial response to vancomycin therapy, a patient with meticillin-resistant Staphylococcus aureus (MRSA) bacteraemia developed endocarditis, failed a second course of vancomycin and then failed daptomycin therapy. An increase in the vancomycin minimum inhibitory concentrations of four consecutive MRSA blood isolates from 2 μg/mL to 8 μg/mL was shown by Etest. Population analysis of four successive blood culture isolates recovered over the 10-week period showed that the MRSA strain became progressively less susceptible to both vancomycin and daptomycin. Retrospectively, the macro Etest method using teicoplanin indicated a decrease in vancomycin susceptibility in the second blood isolate. The patient improved after treatment with various courses of trimethoprim/sulphamethoxazole, quinupristin/dalfopristin and linezolid. Early detection of vancomycin-heteroresistant S. aureus isolates, which appeared to have clinical significance in this case, continues to be a challenge for the clinical laboratory. Development of suitable practical methods for this should be given priority. Concurrent development of resistance to vancomycin and daptomycin, whilst rare, must be considered in a patient who is unresponsive to daptomycin following vancomycin therapy.
Keywords: Staphylococci, Vancomycin, Teicoplanin, Heteroresistance
1. Introduction
Vancomycin resistance in isolates of Staphylococcus aureus can be divided into three categories: high-level resistance [vancomycin-resistant S. aureus, with minimum inhibitory concentrations (MICs)≥16μg/mL], typically mediated by acquisition of the vanA resistance gene from enterococci; intermediate-level resistance [vancomycin-intermediate S. aureus (VISA)], defined by the Clinical and Laboratory Standards Institute (CLSI) as vancomycin MICs in the range of 4–8 μg/mL; and heterogeneous intermediate resistance (hVISA), where the broth microdilution reference MICs are ≤2 μg/mL but the isolates contain subpopulations of cells for which the vancomycin MICs are 4–8 μg/mL [1,2]. The hVISA phenotype is particularly difficult to detect in the laboratory as there are no standardised methods available [3,4]. Detection of VISA and hVISA isolates is important clinically since such isolates often do not respond to vancomycin treatment [1,5]. The likely mechanisms of non-vanA-mediated reduced susceptibility to vancomycin are multifactorial [6].
Several laboratory methods have been described to detect the presence of hVISA strains among clinical isolates of S. aureus, including: inoculating brain–heart infusion (BHI) agar plates containing various concentrations of vancomycin [7]; performing population analysis profiles (PAPs) using traditional plating techniques or spiral plating methods, often combined with area under the curve analyses (PAP-AUC) [8,9]; and modified Etest (AB BIODISK, Piscataway, NJ; now bioMérieux) assays (also called the macro Etest method), which use both vancomycin and teicoplanin strips and a high inoculum on BHI agar [10,11]. Of these methods, PAP-AUC is typically considered the gold standard for detecting hVISA, although this method is too time consuming and labour intensive for routine clinical laboratories to perform. The new glycopeptide resistance detection (GRD) (AB BIODISK, Solna, Sweden; now bioMérieux) Etest, which incorporates both vancomycin and teicoplanin gradients into a single strip, is designed to be used in conjunction with blood agar plates to simplify detection of hVISA strains. The accuracy of this test has been evaluated recently and holds promise for detecting hVISA strains [12].
Recently, several reports have linked reduced susceptibility to vancomycin in S. aureus to reduced susceptibility to daptomycin [13–16]. Here we present a case report in which a series of four meticillin-resistant S. aureus (MRSA) isolates, recovered from the blood of a patient with mitral valve endocarditis, show a gradual increase in vancomycin resistance, progressing from vancomycin susceptibility to heteroresistance to homogeneous intermediate-level resistance. The decrease in the susceptibility of the MRSA isolates to vancomycin was accompanied by a concomitant decrease in susceptibility to daptomycin, which was also associated with treatment failure. The purpose of this study was to characterise the four MRSA isolates recovered from the patient and to determine whether the reduced susceptibility to vancomycin and daptomycin could have been detected earlier in the course of the patient’s disease before the patient failed therapy.
2. Case report
A 60-year-old man presented to hospital with 3 days of fever, malaise and neck pain on 15 September 2004. His past medical history was significant for coronary artery disease, hypertension, congestive heart failure and type 2 diabetes mellitus. He had undergone coronary artery bypass grafting and a mitral valve annuloplasty in March 2003 and placement of an automatic implantable cardioverter–defibrillator (AICD) in July 2004. Two blood cultures from the day of admission grew MRSA on 16 September (isolate RWJ1). Vancomycin therapy (1 g every 12 h) was started on 16 September. Bacteraemia persisted until 19 September, but two blood cultures from 21 September were sterile. A transthoracic echocardiogram on 20 September did not show evidence of endocarditis. A transoesophageal echocardiogram (TEE) on 22 September showed a 1-cm mobile mass on the mitral valve with moderate mitral regurgitation but no evidence of vegetations on the defibrillator leads. A computed tomography scan of the neck did not show any evidence of metastatic infection.
The AICD was left in place and the patient was discharged home to complete a 6-week course of intravenous vancomycin for mitral valve endocarditis. Mild baseline renal insufficiency worsened during therapy and the vancomycin dose was adjusted accordingly. Vancomycin trough levels and renal function (as creatinine levels), respectively, were monitored as follows: 25 September, 6 vancomycin 15.9 μg/mL, creatinine 1.8 mg/dL; 11 October, 20.7 μg/mL and 2.0 mg/dL; 19 October, 18.9 μg/mL and 2.2 mg/dL; and 25 October, 15.0 μg/mL and 2.3 mg/dL. The course of vancomycin was completed on 28 October and the patient’s intravenous catheter was removed. Post-treatment blood surveillance cultures were performed on 2 November, although the patient reported feeling well and was afebrile.
The patient re-presented to the hospital on 5 November with an acute episode of shaking chills, fever, hypovolaemia and renal failure. Blood cultures from 2 November became positive on 5 November and a Gram stain of the blood revealed Gram-positive cocci in clusters. The organism was subsequently identified as MRSA (isolate RWJ2). The vancomycin MIC for the MRSA isolate from the 2 November blood culture was ≤2 μg/mL by automated susceptibility testing [MicroScan Dade (now Siemens Healthcare), West Sacramento, CA]. Vancomycin was initiated again upon re-admission; however, therapy was changed to intravenous daptomycin 6 mg/kg every 48 h on 9 November when concerns about the possibility of acute interstitial nephritis due to vancomycin led to initiation of haemodialysis. On 11 November a repeat TEE showed a 1.2 cm × 2.0 cm mitral valve vegetation, but there was no evidence of vegetations on the AICD wires. Owing to concern about an infected defibrillator, the defibrillator generator and leads were removed on 15 November and cultures from the generator pocket grew MRSA. Blood cultures continued to be positive for MRSA. A temporary central catheter was removed and an upper extremity ultrasound study was negative for deep venous thrombosis at the sites of the recent catheters and defibrillator leads; however, bacteraemia persisted. A third TEE on 23 November showed a decrease in the vegetation size to 1.5 cm × 0.8 cm. Rifampicin was added to daptomycin therapy on 28 November.
On 4 December daptomycin was discontinued and vancomycin therapy was restarted because the patient remained febrile and was not improving. No daptomycin susceptibility data were available for the MRSA isolates at this time. Rifampicin was discontinued because of the emergence of resistance. On 6 December the MRSA isolates from blood cultures on 4 December (isolate RWJ3) showed decreased susceptibility to vancomycin (MIC = 8 μg/mL) by Etest (AB BIODISK) and therapy was changed to linezolid and trimethoprim/sulphamethoxazole (SXT). The isolate recovered from a repeat blood culture on 6 December (isolate RWJ4) showed the same result. Blood cultures drawn on 19 December were negative; however, one of two blood cultures drawn on 23 December showed Gram-positive cocci in clusters on the Gram stain, but no growth was recovered on subculture.
Linezolid therapy was changed to quinupristin/dalfopristin (Q/D) on 3 January because the patient developed thrombocytopenia. SXT therapy was discontinued shortly thereafter because of a lack of any data for benefit of combination therapy with Q/D. The patient developed disabling myalgias and arthralgias while on Q/D and treatment was changed back to linezolid monotherapy. The patient remained on linezolid therapy until 3 February, which was 6 weeks after the last positive blood culture. Twelve months after completion of therapy the patient continues to do well with no evidence of recurrence of infection. The patient is currently undergoing peritoneal dialysis and has had placement of a new AICD.
3. Methods
3.1. Bacterial isolates
Staphylococcus aureus isolates RWJ1, RWJ2, RWJ3 and RWJ4, all from the same patient, were recovered from positive blood culture (BACTEC; BD Diagnostics, Sparks, MD) and identified as S. aureus by colony morphology, positive catalase and coagulase reactions, and inoculation of MicroScan Combo 20 panels (Siemens Healthcare). Antimicrobial susceptibility profiles were determined initially using MicroScan WalkAway Combo 20 panels. Vancomycin MICs were also determined using Etest strips (AB BIODISK) on Mueller–Hinton agar (BD Diagnostics) according to manufacturer’s instructions. Organisms were sent to the US Centers for Disease Control and Prevention (CDC) and the Project ICARE laboratory (Emory University, Atlanta, GA) for antimicrobial susceptibility testing using the CLSI broth microdilution reference method [2] with cation-adjusted Mueller–Hinton broth (BD Diagnostics) and population analysis studies. D-zone tests to detect inducible clindamycin resistance were performed as described by the CLSI [2]. The organisms were also inoculated onto BHI agar plates containing 6 μg/mL vancomycin. The following quality control (QC) organisms were used: S. aureus ATCC 25923; S. aureus ATCC 29213; and Enterococcus faecalis ATCC 29212.
3.2. Etest studies
The macro Etest method was performed as described by Hanaki et al. [11]. Organisms were suspended to a density of 2.0 McFarland standard, inoculated onto BHI agar (BD Diagnostics), and both vancomycin and teicoplanin Etest strips were placed on the plates. The plates were incubated at 35°C and read at 24 h and 48 h. Heteroresistance was defined as vancomycin and teicoplanin MICs ≥ 8 μg/mL or a teicoplanin MIC ≥12 μg/mL. GRD Etest strips (AB BIODISK) to detect vancomycin heteroresistance were used as described by the manufacturer.
3.3. Pulsed-field gel electrophoresis (PFGE) profiles
PFGE profiles were determined as described previously by McDougal et al. [17].
3.4. Population analysis profiles
PAPs were performed using BHI (BD Diagnostics) for vancomycin and daptomycin as described by Pfeltz et al. [18]. The concentration of antimicrobial agent in the agar plates ranged from 0.5 μg/mL to 128 μg/mL for vancomycin and from 0.5 μg/mL to 16 μg/mL for daptomycin. Experiments were performed in duplicate with similar results. QC organisms included S. aureus strains Mu50 and Mu3, which were a generous gift of Dr Keiichi Hiramatsu (Juntendo University, Tokyo, Japan).
4. Results
4.1. PFGE results and antimicrobial susceptibility profiles
All four MRSA isolates had indistinguishable SmaI PFGE profiles that were consistent with the USA100 strain type (data not shown).
Initial testing of the MRSA isolates by broth microdilution showed that all were susceptible to Q/D, linezolid and SXT. All organisms were resistant to erythromycin, gentamicin, oxacillin, penicillin and tetracycline. The antibiograms of the isolates for those antimicrobial agents with variable results (i.e. daptomycin, linezolid, rifampicin and vancomycin) determined by broth microdilution are shown in Table 1. Broth microdilution testing identified decreasing susceptibility in isolate RWJ4 (vancomycin MIC = 4 μg/mL). The vancomycin MICs determined by standard Etest method in the Robert Wood Johnson (RWJ) clinical laboratory and the Project ICARE laboratory are shown in Table 2. Decreasing susceptibility to vancomycin by Etest was recognised by the RWJ clinical laboratory in RWJ3 (vancomycin MIC = 4 μg/mL) and RWJ4 (8 μg/mL). The ICARE laboratory vancomycin MICs using the standard Etest method were lower (Table 2). Testing by the macro Etest method with vancomycin and teicoplanin indicated that RWJ2 was an hVISA strain since the teicoplanin MIC for this isolate was ≥12 μg/mL (Table 2). The GRD Etest method would not have designated RWJ2 as hVISA, although RWJ3 would have been classified as hVISA owing to its teicoplanin MIC. Although the vancomycin and daptomycin MICs increased, the isolates became more susceptible to linezolid during the same time frame (Table 1). The decrease in linezolid MICs was reproducible.
Table 1.
Antimicrobial susceptibility profiles of Staphylococcus aureus clinical isolates by broth microdilution
| Isolate no. | Date of isolation | MIC (μg/mL) |
|||
|---|---|---|---|---|---|
| Vancomycin | Daptomycin | Linezolid | Rifampicin | ||
| RWJ1 | 16 Sept. 2004 | 1.0 | ≤0.5 | 2.0 | ≤0.5 |
| RWJ2 | 5 Nov. 2004 | 2.0 | ≤0.5 | 4.0 | ≤0.5 |
| RWJ3 | 4 Dec. 2004 | 2.0 | 4.0 | 2.0 | >8.0 |
| RWJ4 | 6 Dec. 2004 | 4.0 | 4.0 | ≤1.0 | >8.0 |
MIC, minimum inhibitory concentration.
Table 2.
Susceptibility of Staphylococcus aureus clinical isolates to glycopeptides by Etest method
| Isolate | MIC (μg/mL) |
||||||
|---|---|---|---|---|---|---|---|
| GRD test on blood agar (VAN/TEIC) | RWJ on MHA, 0.5 McFarland (24 h) | ICARE on MHA, 0.5 McFarland (24 h) | ICARE on BHI agar, 2.0 McFarland (24 h) | ||||
| 24 h | 48 h | VAN | VAN | TEIC | VAN | TEIC | |
| RWJ1 | 1.0/3.0 | 1.0/3.0 | 2.0 | 2.0 | 1.5 | 4.0 | 8.0 |
| RWJ2 | 2.0/4.0 | 2.0/6.0 | 2.0 | 2.0 | 2.0 | 6.0 | 12 |
| RWJ3 | 2.0/6.0 | 2.0/8.0 | 4.0 | 2.0 | 4.0 | 12 | 12 |
| RWJ4 | 4.0/16 | 4.0/>32 | 8.0 | 3.0 | 16 | 12 | 32 |
MIC, minimum inhibitory concentration; GRD, glycopeptide resistance detection; VAN, vancomycin; TEIC, teicoplanin; RWJ, Robert Wood Johnson clinical laboratory; MHA, Mueller–Hinton agar; BHI, brain–heart infusion.
4.2. Population analysis profiles
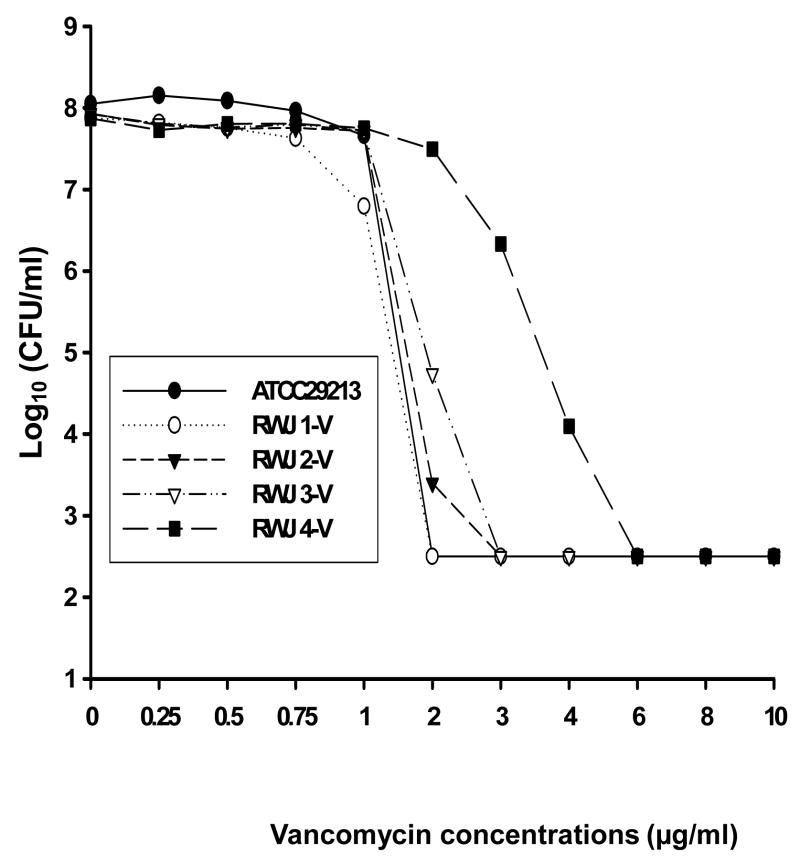
PAPs were determined for the four isolates using vancomycin (Fig. 1) and daptomycin (Fig. 2) to identify subpopulations of cells with reduced susceptibility to those antimicrobial agents. The population profiles showed a progressive increase in the vancomycin MICs for the four isolates (Fig. 1). The vancomycin MICs for RWJ1, RWJ2, RWJ3 and RWJ4 as determined by population analysis on BHI agar were 2, 3, 3 and 6 μg/mL, respectively.
Fig 1.
Population analysis of four Staphylococcus aureus isolates recovered from a patient with endocarditis. Analysis was performed using brain–heart infusion agar containing serial dilutions of vancomycin. CFU, colony-forming units.
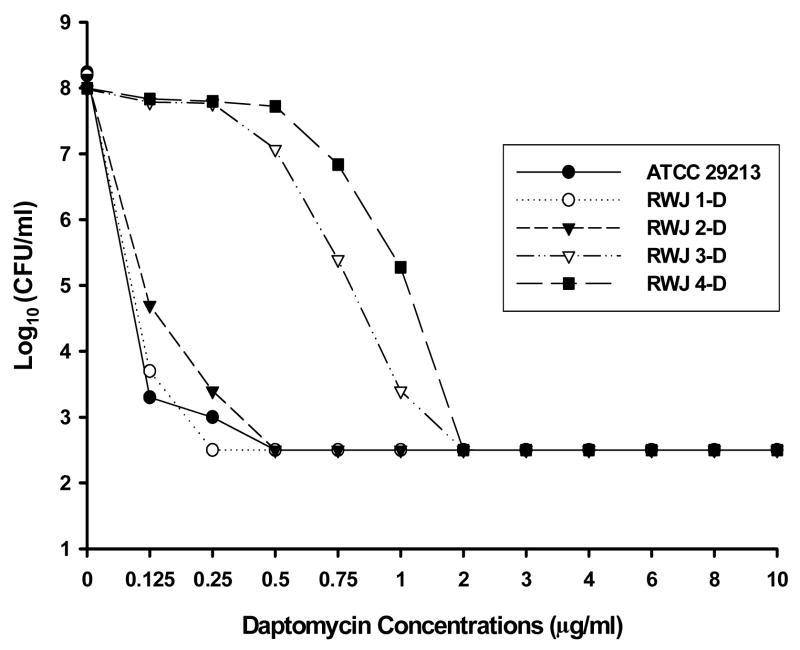
Fig 2.
Population analysis of four Staphylococcus aureus isolates recovered from a patient with endocarditis. Analysis was performed using brain–heart infusion agar containing serial dilutions of daptomycin. CFU, colony-forming units.
A similar increase in the proportion of cells of RWJ3 and RWJ4 that were capable of growing on 1μg/mL of daptomycin is shown in the population analysis in Fig. 2. The decreased susceptibility of RWJ2 to daptomycin in comparison with RWJ1 was more subtle and was reflected in an MIC increase from 0.25 μg/mL to 0.5 μg/mL. The daptomycin MICs for RWJ3 and RWJ4 were both 2 μg/mL, which would be considered non-susceptible by CLSI criteria [2].
5. Discussion
The clinical relevance of vancomycin heteroresistance has been debated in the literature [1,5,19]. Here we showed the failure of vancomycin to eradicate fully an MRSA strain even after an initial clinical response to therapy, likely due to both the presence of a foreign body (AICD) and the development of vancomycin heteroresistance. The heteroresistant phenotype changed to homogeneous intermediate-level resistance (i.e. it became a VISA isolate) on continued vancomycin therapy. Our findings of decreasing linezolid MICs in strains that sequentially demonstrated increasing MICs to vancomycin (Table 1) are similar to those recently reported by Watanabe et al. [20].
Our observations highlight several important issues regarding the detection of MRSA isolates with reduced susceptibility to vancomycin. First, both automated testing and standard broth microdilution susceptibility testing using Mueller–Hinton broth failed to indicate that the first two isolates had the potential for reduced susceptibility to vancomycin. Second, the standard Etest method demonstrated reduced susceptibility in the third isolate, whilst the macro Etest method would have classified the second isolate as an hVISA strain owing to the teicoplanin MIC of 12 μg/mL. Unfortunately, teicoplanin is rarely tested in the USA. The GRD Etest strip indicated heteroresistance starting with the third isolate, RWJ3; however, the GRD Etest was performed after multiple subcultures of the isolates and may have yielded higher MIC results with the second isolate had it been tested prior to being frozen [6].
It appears that reduced susceptibility to vancomycin and daptomycin developed in parallel in this strain, although we have no data regarding whether the mechanism of reduced susceptibility to the two antimicrobial agents is the same. Sakoulas et al. [14] reported a similar case of clinical daptomycin failure after treatment with vancomycin. Cui et al. [13] reported that the thickened cell wall of an hVISA strain was responsible for decreased susceptibility to daptomycin, and the same group documented that serial daptomycin selection generates daptomycin-non-susceptible strains in hVISA [21]. Both Jevitt et al. [22] and Patel et al. [23] noted associations between decreased susceptibility to vancomycin and decreased susceptibility to daptomycin. The three doubling-dilution increase in daptomycin MICs is similar to the results reported by Fowler et al. [24] in daptomycin clinical trials, where similar increases were noted among staphylococcal strains recovered from endocarditis patients showing microbiological failures.
In summary, detection of the hVISA phenotype remains a challenge for clinical laboratories. Using the macro Etest method with vancomycin and teicoplanin provided critical information about decreasing vancomycin resistance in this series of isolates, but the information was only available after the failure of vancomycin therapy. Development of suitable methods for the clinical laboratory to detect these strains should be given priority. Concurrent development of resistance to vancomycin and daptomycin, whilst rare, must be considered in a patient who is unresponsive to daptomycin following vancomycin therapy.
Acknowledgments
The authors thank Judy Rothberg for technical assistance.
Funding: Phase 5 of Project ICARE was supported in part by unrestricted research grants to the Rollins School of Public Health of Emory University by: Astra-Zeneca Pharmaceuticals, Wilmington, DE; Elan Pharmaceuticals, San Diego, CA; Johnson & Johnson Pharmaceutical Research & Development, LLC, Raritan, NJ; Pfizer Incorporated, New York, NY; and 3M Health Care Products, St Paul, MN.
Footnotes
The findings and conclusions in this publication are those of the authors and do not necessarily represent the views of the US Centers for Disease Control and Prevention (CDC).
Competing interests: None declared.
Ethical approval: Not required.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tenover FC, Moellering RC., Jr The rationale for revising the Clinical and Laboratory Standards Institute vancomycin minimal inhibitory concentration interpretive criteria for Staphylococcus aureus. Clin Infect Dis. 2007;44:1208–15. doi: 10.1086/513203. [DOI] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. Document M7-A7. 7. Wayne, PA: CLSI; 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard. [Google Scholar]
- 3.Wootton M, MacGowan AP, Walsh TR, Howe RA. A multicenter study evaluating the current strategies for isolating Staphylococcus aureus strains with reduced susceptibility to glycopeptides. J Clin Microbiol. 2007;45:329–32. doi: 10.1128/JCM.01508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howe RA, Wootton M, Walsh TR, Bennett PM, Macgowan AP. Heterogeneous resistance to vancomycin in Staphylococcus aureus. J Antimicrob Chemother. 2000;45:130–2. doi: 10.1093/jac/45.1.130. [DOI] [PubMed] [Google Scholar]
- 5.Howden BP, Ward PB, Charles PG, Korman TM, Fuller A, du Cros P, et al. Treatment outcomes for serious infections caused by methicillin-resistant Staphylococcus aureus with reduced vancomycin susceptibility. Clin Infect Dis. 2004;38:521–8. doi: 10.1086/381202. [DOI] [PubMed] [Google Scholar]
- 6.Pfeltz RF, Wilkinson BJ. The escalating challenge of vancomycin resistance in Staphylococcus aureus. Curr Drug Targets Infect Disord. 2004;4:273–94. doi: 10.2174/1568005043340470. [DOI] [PubMed] [Google Scholar]
- 7.Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S, et al. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet. 1997;350:1670–3. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 8.Wootton M. The need for accuracy in performing vancomycin intermediate resistant Staphylococcus aureus (VISA) and hetero-VISA detection methods. Int J Antimicrob Agents. 2006;28:586. doi: 10.1016/j.ijantimicag.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 9.Wootton M, Howe RA, Hillman R, Walsh TR, Bennett PM, MacGowan AP. A modified population analysis profile (PAP) method to detect hetero-resistance to vancomycin in Staphylococcus aureus in a UK hospital. J Antimicrob Chemother. 2001;47:399–403. doi: 10.1093/jac/47.4.399. [DOI] [PubMed] [Google Scholar]
- 10.Walsh TR, Bolmström A, Qwärnström A, Ho P, Wootton M, Howe RA, et al. Evaluation of current methods for detection of staphylococci with reduced susceptibility to glycopeptides. J Clin Microbiol. 2001;39:2439–44. doi: 10.1128/JCM.39.7.2439-2444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanaki H, Kuwahara-Arai K, Boyle-Vavra S, Daum RS, Labischinski H, Hiramatsu K. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J Antimicrob Chemother. 1998;42:199–209. doi: 10.1093/jac/42.2.199. [DOI] [PubMed] [Google Scholar]
- 12.Yusof A, Engelhardt A, Karlsson A, Bylund L, Vidh P, Mills K, et al. Evaluation of a new Etest vancomycin–teicoplanin strip for detection of glycopeptide-intermediate Staphylococcus aureus (GISA), in particular, heterogeneous GISA. J Clin Microbiol. 2008;46:3042–7. doi: 10.1128/JCM.00265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui L, Tominaga E, Neoh HM, Hiramatsu K. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:1079–82. doi: 10.1128/AAC.50.3.1079-1082.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakoulas G, Alder J, Thauvin-Eliopoulos C, Moellering RC, Jr, Eliopoulos GM. Induction of daptomycin heterogeneous susceptibility in Staphylococcus aureus by exposure to vancomycin. Antimicrob Agents Chemother. 2006;50:1581–5. doi: 10.1128/AAC.50.4.1581-1585.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman L, Alder JD, Silverman JA. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:2137–45. doi: 10.1128/AAC.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Julian K, Kosowska-Shick K, Whitener C, Roos M, Labischinski H, Rubio A, et al. Characterization of a daptomycin-nonsusceptible vancomycin-intermediate Staphylococcus aureus strain in a patient with endocarditis. Antimicrob Agents Chemother. 2007;51:3445–8. doi: 10.1128/AAC.00559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003;41:5113–20. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfeltz RF, Singh VK, Schmidt JL, Batten MA, Baranyk CS, Nadakavukaren MJ, et al. Characterization of passage-selected vancomycin-resistant Staphylococcus aureus strains of diverse parental backgrounds. Antimicrob Agents Chemother. 2000;44:294–303. doi: 10.1128/aac.44.2.294-303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein FW, Kitzis MD. Vancomycin-resistant Staphylococcus aureus: no apocalypse now. Clin Microbiol Infect. 2003;9:761–5. doi: 10.1046/j.1469-0691.2003.00734.x. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe Y, Neoh HM, Cui L, Hiramatsu K. Improved antimicrobial activity of linezolid against vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother. 2008;52:4207–8. doi: 10.1128/AAC.00676-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Camargo IL, Neoh HM, Cui L, Hiramatsu K. Serial daptomycin selection generates daptomycin-nonsusceptible Staphylococcus aureus strains with a heterogeneous vancomycin-intermediate phenotype. Antimicrob Agents Chemother. 2008;52:4289–99. doi: 10.1128/AAC.00417-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jevitt LA, Smith AJ, Williams PP, Raney PM, McGowan JE, Jr, Tenover FC. In vitro activities of daptomycin, linezolid, and quinupristin–dalfopristin against a challenge panel of staphylococci and enterococci, including vancomycin-intermediate S. aureus and vancomycin-resistant E. faecium. Microb Drug Resist. 2003;9:389–93. doi: 10.1089/107662903322762833. [DOI] [PubMed] [Google Scholar]
- 23.Patel JB, Jevitt LA, Hageman J, McDonald LC, Tenover FC. An association between reduced susceptibility to daptomycin and reduced susceptibility to vancomycin in Staphylococcus aureus. Clin Infect Dis. 2006;42:1652–3. doi: 10.1086/504084. [DOI] [PubMed] [Google Scholar]
- 24.Fowler VG, Jr, Boucher HW, Corey GR, Abrutyn E, Karchmer AW, Rupp ME, et al. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N Engl J Med. 2006;355:653–65. doi: 10.1056/NEJMoa053783. [DOI] [PubMed] [Google Scholar]