Summary
Many signal transduction networks control their output by switching regulatory elements on or off. To synchronize biological response with environmental stimulus, switching kinetics must be faster than changes in input. Two-component regulatory systems (used for signal transduction by bacteria, archaea, and eukaryotes) switch via phosphorylation or dephosphorylation of the receiver domain in response regulator proteins. Although receiver domains share conserved active site residues and similar three-dimensional structures, rates of self-catalyzed dephosphorylation span a ≥40,000-fold range in response regulators that control diverse biological processes. For example, autodephosphorylation of the chemotaxis response regulator CheY is 640-fold faster than Spo0F, which controls sporulation. Here we demonstrate that substitutions at two variable active site positions decreased CheY autodephosphorylation up to 40-fold and increased the Spo0F rate up to 110-fold. Particular amino acids had qualitatively similar effects in different response regulators. However, mutant proteins matched to other response regulators at the two key variable positions did not always exhibit similar autodephosphorylation kinetics. Therefore, unknown factors also influence absolute rates. Understanding the effects that particular active site amino acid compositions have on autodephosphorylation rate may allow manipulation of phosphoryl group stability for useful purposes, as well as prediction of signal transduction kinetics from amino acid sequence.
Keywords: CheY, dephosphorylation, response regulator, signal transduction, Spo0F, two-component regulatory systems
Introduction
Microorganisms from all three domains of life (Archaea, Bacteria, Eukarya), as well as some plant species, use two-component regulatory systems to accomplish signal transduction and control diverse processes such as behavior, development, pathogenesis, or physiology (Ashby, 2004; Catlett et al., 2003). In a typical two-component system [reviewed in (Stock et al., 2000)], a sensor kinase detects environmental input and represents this information by covalent attachment of a phosphoryl group to a cytoplasmic domain of the kinase. The phosphoryl group is then transferred to the receiver domain of a response regulator, where the presence or absence of a phosphoryl group is associated with a conformational change that modulates activity of a neighboring output domain. The receiver domain active site is composed of five conserved amino acids (Lee et al., 2001; Stock et al., 2000) (Fig. 1). In the Escherichia coli CheY receiver domain, Asp12, Asp13, and Asp57 bind a divalent cation that is required for phosphorylation and dephosphorylation (Lukat et al., 1990; Stock et al., 1993), Asp57 is the site of phosphorylation (Sanders et al., 1989), and Thr87 (Appleby and Bourret, 1998) and Lys109 (Lukat et al., 1991) participate in the phosphorylation-mediated conformational change by coordinating (along with the metal ion) the three phosphoryl group oxygens (Lee et al., 2001).
Fig. 1.
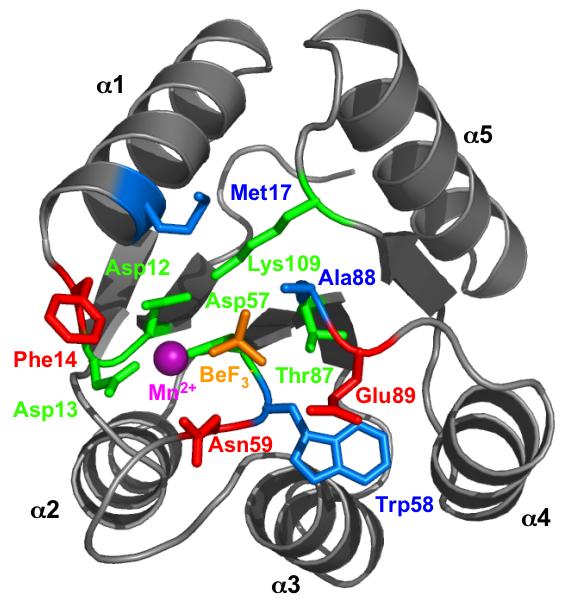
Receiver domain active site structure. E. coli CheY bound to the phosphoryl group analog BeF3- (orange) and Mn2+ (magenta) is shown (pdb 1FQW) (Lee et al., 2001). Conserved active site residues, green; variable active site residues tested experimentally, red; variable active site residues not tested in this work, blue. Figure created using PyMOL (DeLano, 2002).
Signal transduction networks, regardless of molecular mechanism, often share several functional features (Downward, 2001). One common component is a molecule that acts a binary switch. Switches are logically simple, but can be organized into circuits that yield complex outputs. In order to synchronize responses with stimuli, a switch element must be able to cycle between inactive and active states faster than the environment changes. Thus, the biochemical kinetics of information processing are related to the timescales of environmental stimulus and biological response. Response regulators are the switch elements of two-component regulatory systems. The fraction of the response regulator population that is phosphorylated at any given time is determined by the balance between the rates of phosphoryl group addition and removal. Response regulators obtain phosphoryl groups from sensor kinases as outlined above or by autophosphorylation (Lukat et al., 1992). Response regulators lose phosphoryl groups through self-catalyzed dephosphorylation (Hess et al., 1988) and, in some cases, reverse phosphotransfer (Georgellis et al., 1998; Sourjik and Schmitt, 1998) or via phosphatases (Perego, 2001; Pioszak and Ninfa, 2004; Szurmant et al., 2004; Zhao et al., 2002).
Successful signal transduction mechanisms are used repeatedly in nature for various purposes. For example, a single bacterial cell may have >100 different two-component regulatory systems simultaneously conducting distinct tasks in parallel (Goldman et al., 2006). In turn, biological processes regulated by analogous signaling mechanisms may operate on dramatically different timescales, with corresponding differences in the rates of signaling reactions. For example, autodephosphophorylation rate constants for different response regulators span a range of at least 40,000-fold (Table 1). Proteins involved in chemotaxis, which depends on split-second changes in behavior (Segall et al., 1982), predominate at the fast end of the scale, whereas response regulators that control transcription are found at the slow end. In some signaling pathways, phosphatases are responsible for the majority of response regulator dephosphorylation. Nevertheless, factors that influence the autocatalytic reaction are likely to be physiologically relevant because the phosphatases apparently stimulate the intrinsic response regulator autodephosphorylation reaction rather than utilize an entirely different mechanism (Pioszak and Ninfa, 2004; Zhao et al., 2002). Furthermore, the available data indicate that the magnitude of response regulator dephosphorylation rate stimulation by phosphatases [∼101-102-fold (Keener and Kustu, 1988; Silversmith et al., 2008; Zhu and Inouye, 2002)] may be modest relative to the overall 104-105-fold range of autodephosphorylation rates. Therefore, even in regulatory circuits where autodephosphorylation is not the primary route of phosphoryl group removal from response regulators, autodephosphorylation appears important to set a baseline rate near the physiological timescale of a particular system. How can different receiver domains, which share a common three-dimensional structure, conserved active site residues, and phosphoryl group reaction chemistry, catalyze autodephosphorylation at such dramatically different rates? In this study, we tested the hypothesis that variable active site residues, which differ between response regulators, contribute to the observed range of more than four orders of magnitude in autodephosphorylation rates.
Table 1.
Autodephosphorylation rate constants reported for various response regulators
| Response Regulator | Amino Acid At Positionb |
kdephosc (min-1) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Speciesa | ‘14’ | ‘17’ | ‘58’ | ‘59’ | ‘88’ | ‘89’ | Reference | ||
| R. sph | CheY6 | A | M | I | E | S | V | 20 | (Porter and Armitage, 2002) |
| E. coli | CheBd | S | M | L | E | S | L | 8 | (Stewart, 1993) |
| S. mel | CheY2 | Q | S | F | N | A | Q | 4 | (Sourjik and Schmitt, 1998) |
| S. mel | CheY1 | S | I | I | N | T | E | 4 | (Sourjik and Schmitt, 1998) |
| R. sph | CheY3 | S | V | Q | N | T | D | 3 | (Porter and Armitage, 2002) |
| R. sph | CheY5 | S | V | Q | N | T | E | 3 | (Porter and Armitage, 2002) |
| E. coli | CheY | F | M | W | N | A | E | 3 | (Silversmith et al., 2001) |
| R. sph | CheY2 | M | S | Y | N | G | T | 2 | (Porter and Armitage, 2002) |
| H. pyl | CheY1 | S | M | W | N | T | E | 2 | (Jimenez-Pearson et al., 2005) |
| R. sph | CheY4 | S | I | Q | N | T | E | 2 | (Porter and Armitage, 2002) |
| R. sph | CheB2 | S | V | L | E | S | L | 2 | (Porter and Armitage, 2002) |
| R. sph | CheY1 | S | I | I | N | T | E | 1 | (Porter and Armitage, 2002) |
| R. sph | CheB1 | S | A | L | E | S | H | 0.3 | (Porter and Armitage, 2002) |
| T. mar | CheY | A | M | I | T | A | M | 0.3 | (Swanson et al., 1996) |
| S. cer | SLN1 | N | N | V | Q | A | F | 0.05 | (Janiak-Spens et al., 1999) |
| S. cer | SSK1 | N | N | L | Q | A | S | 0.05 | (Janiak-Spens et al., 2000) |
| S. ent | NtrC | D | I | I | R | A | H | 0.04 | (Keener and Kustu, 1988) |
| E. coli | NtrCd | D | I | I | R | A | H | 0.03 | (Pioszak and Ninfa, 2004) |
| B. sub | Spo0A | N | L | I | I | A | F | 0.02 | (Perego, 2001) |
| H. pyl | CheY2 | S | D | I | E | S | R | 0.02 | (Jimenez-Pearson et al., 2005) |
| S. coel | AbsA2 | E | I | I | R | T | F | 0.01 | (Sheeler et al., 2005) |
| E. coli | PhoB | E | I | W | M | A | R | 0.009 | (Fisher et al., 1996) |
| R. cap | RegA | D | F | L | R | G | Y | 0.008 | (Bird et al., 1999) |
| B. sub | DegU | H | F | I | N | I | H | 0.008 | (Dahl et al., 1992) |
| S. mel | FixJ | E | V | L | R | G | H | 0.005 | (Weinstein et al., 1993) |
| E. coli | ArcA | E | T | I | N | G | R | 0.005 | (Georgellis et al., 1998) |
| E. coli | NarLd | H | L | L | N | V | S | 0.004 | (Cavicchioli et al., 1995) |
| B. sub | Spo0F | Q | I | M | K | A | Y | 0.004 | (Zapf et al., 1998) |
| R. sph | PrrA | D | F | L | R | G | Y | 0.002 | (Comolli et al., 2002) |
| E. coli | OmpR | D | L | L | M | A | K | 0.002 | (Ames et al., 1999) |
| E. fae | VanR | E | I | I | M | G | K | 0.0008 | (Wright et al., 1993) |
| T. mar | DrrA | D | I | I | M | A | K | 0.0005 | (Goudreau et al., 1998) |
B. sub, Bacillus subtilis; E. coli, Escherichia coli; E. fae, Enterococcus faecium; H. pyl, Helicobacter pylori; R. cap, Rhodobacter capsulatus; R. sph, Rhodobacter sphaeroides; S. cer, Saccharomyces cerevisiae; S. coel, Streptomyces coelicolor; S. ent, Salmonella enterica; S. mel, Sinorhizobium meliloti; T. mar, Thermotoga maritima
E. coli CheY numbering. Residues are color coded by putative association with autodephosphorylation rates: green = “fast”, red = “slow”, black = no association. The designation of a specific amino acid as “fast” or “slow” was determined independently for each position as follows. First, a tentative cut-off between “fast” and “slow” (indicated by dashed line) was established based on a natural break in the Table entries. All chemotaxis proteins except one are above this cut-off and all non-chemotaxis proteins are below this point. Furthermore, the largest gap in kdephos values (as determined by the ratio of successive kdephos entries) occurs here (0.3/0.05 = 6). Second, amino acids were grouped according to sidechain chemical similarity: acidic (D,E); amide (N,Q); basic (H,K,R); hydroxyl (S,T); small (A,G); aromatic (F,W,Y); and large hydrophobic (I,L,M,V). Third, for each position, if an amino acid group was represented more than once and >75% of the members of that group were on the same side of the cut-off, then the group was designated “fast” or “slow” accordingly. Fourth, the potential impact of changing the cut-off was examined. For five positions, adjustment of the cut-off to slightly faster or slower values did not increase the number of amino acids that could be given a classification. However, for position ‘59’, the large number of amide residues could not be classified based on the original criteria, even though most (9/14 = 64%) were above the initial cut-off. Inclusion of the two entries immediately below the initial cut-off raises the percentage (11/14 = 79%) above the 75% threshold for designation, so an exception was made and N or Q residues at position ‘59’ were assigned to the “fast” class. Note that most amino acids at positions ‘17’ and ‘58’ could not be designated as either “fast” or “slow”, which suggests these positions have little influence on autodephosphorylation rate.
Rate constants were rounded to one significant figure.
Rates measured at 37 °C were converted to an approximate rate at 25 °C by dividing by 5, based on rate differences between 25 and 37 °C observed for DrrA and acetyl phosphate (Goudreau et al., 1998). All other rates were measured between 20-30 °C.
Results
Variable active site residues may influence autodephosphorylation rate
Active site residues that may affect phosphoryl group stability were identified through a combination of receiver domain structural information and published autodephosphorylation rate data. Residues close to the phosphoryl group could impact autodephosphorylation rate by influencing the energy of the transition or ground states or altering the approach of the attacking water molecule. Using the CheY·BeF3- crystal structure (Lee et al., 2001) as a model, six candidate variable surface residues [positions 14, 17, 58, 59, 88 and 89] were identified within 10 Å of the phosphoryl group analog BeF3- (Fig. 1). Amongst 32 response regulators with reported autodephosphorylation rates, we then looked for a possible correlation between rate and amino acid identity at each of the six variable active site positions (Table 1). (Note that E. coli CheY numbering is used to indicate corresponding positions for each response regulator). Positions ‘17’ and ‘58’ were assumed not to contribute significantly to the observed differences in rate because at each of these positions similar residues were found in response regulators throughout the entire range of observed rates. For example, at both positions ‘17’ and ‘58’, Ile was found in response regulators with fast, intermediate, or slow autodephosphorylation rates. In contrast, positions ‘14’, ‘59’, ‘88’, and ‘89’ showed an apparent nonrandom distribution of residues between response regulators with fast autodephosphorylation rates compared to those with slow rates. For example, the amino acids at position ‘89’ were often negatively charged in response regulators with faster rates and positively charged in those with slower rates (Table 1).
Single substitutions at positions ‘59’ or ‘89’ generally had modest effects on autodephosphorylation rate
The literature data are consistent with the hypothesis that variable active site residues ‘14’, ‘59’, ‘88’, and ‘89’ affect response regulator autodephosphorylation rate. To test if the observed relationships were actually causal, we made amino acid substitutions at positions ‘14’, ‘59’, and ‘89’ in two different response regulators and then measured the loss of 32P-labelled phosphoryl groups from the mutant proteins as a function of time. E. coli CheY and Bacillus subtilis Spo0F were chosen for analysis because (i) the wildtype autodephosphorylation rates are very different (Fig. 2, column 1), (ii) both response regulators consist of a receiver domain alone, with no output domain, and (iii) the respective CheA and KinA kinases do not possess phosphatase activity (Hess et al., 1988; Wang et al., 2001a). Each of the listed characteristics simplifies interpretation of results. We did not investigate position ‘88’ in this study, because position ‘88’ is an Ala in both CheY and Spo0F and therefore is unlikely to contribute to the 640-fold difference in autodephosphorylation rates observed between these two response regulators. Omitting position ‘88’ also reduced the combinatorial complexity of mutants to be analyzed. To determine if the autodephosphorylation rate of one response regulator can easily be converted to that of another simply by matching a few key amino acids, most substitutions were chosen to reflect amino acids present at positions ‘14’, ‘59’, and ‘89’ in response regulators representing a range of autodephosphorylation rates. Nineteen CheY and 12 Spo0F mutant proteins were assayed in addition to the wildtype versions.
Fig. 2.
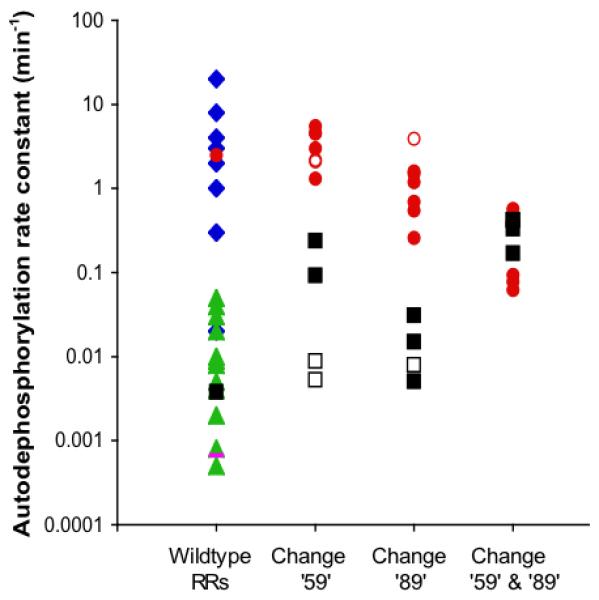
Response regulator autodephosphorylation rate constants. Note logarithmic scale. Column 1 shows wildtype values from Table 1: chemotaxis proteins (CheB or CheY) from various species, blue diamonds; E. coli CheY, red circle; transcriptional regulators, green triangles; B. subtilis Spo0F, black square. The rate constant for spontaneous dephosphorylation of the model acyl phosphate compound acetyl phosphate (Goudreau et al., 1998) is shown as a magenta bar for comparison. Each solid circle or square in columns 2-4 respectively represents a mutant CheY (red circle) or Spo0F (black square) protein from this study with an amino acid substitution at position ‘59’ (column 2), ‘89’ (column 3) or both (column 4). Open circles or squares represent previously published mutant proteins cited in the text.
Ala or Arg substitutions at position 59 previously were found to have no discernable impact on CheY autodephosphorylation rate (Silversmith et al., 2001). Similarly, five additional CheY single substitutions at position 59 had only modest (≤ two-fold) effects on rate (Table 2; Fig. 2, column 2). The negatively charged Asp substitution is noteworthy. The fastest reported response regulator autodephosphorylation rate constant that we are aware of is 20 min-1 in Rhodobacter sphaeroides CheY6, which contains a negatively charged Glu at position ‘59’ (Porter and Armitage, 2002). However, faster rates are conceivable. The haloacid dehalogenase (HAD) superfamily of enzymes shares some structural features with receiver domains, including an analogous set of conserved active site residues and, in many cases, an aspartyl-phosphate intermediate (Burroughs et al., 2006; Wang et al., 2001b). Furthermore, HAD enzymes usually contain an Asp at position ‘59’. In the HAD enzyme phosphoserine phosphatase, crystallographic evidence suggests that Asp‘59’ acts as a general base to activate the attacking water molecule, with the consequence that aspartyl-phosphate hydrolysis occurs at a rate of 840 min-1 (Cho et al., 2001; Wang et al., 2002). Nucleophilic attack by a water molecule is also the likely mechanism of autodephosphorylation in response regulators (Stock et al., 1993; Wolanin et al., 2003). We therefore tested the effect of replacing Asn59 with Asp on the autophosphatase activity of CheY, but this substitution resulted in only a two-fold rate increase (Table 2).
Table 2.
Autodephosphorylation rates of CheY mutants
| Substitution Position(s) | Amino Acid At Position |
Fold Change From Wildtype | |||
|---|---|---|---|---|---|
| 14 | 59 | 89 | kdephos (min-1)a | ||
| Wildtype | F | N | E | 2.5 ± 0.3 | NA |
| 59 | F | D | E | 5.5 ± 0.4 | +2.2 |
| F | M | E | 4.6 ± 0.4 | +1.8 | |
| F | E | E | 4.5 ± 0.2 | +1.8 | |
| F | K | E | 3.0 ± 0.1 | +1.2 | |
| F | L | E | 1.3 ± 0.1 | -1.9 | |
| 89 | F | N | L | 1.6 ± 0.1 | -1.6 |
| F | N | K | 1.2 ± 0.3 | -2.1 | |
| F | N | R | 0.69 ± 0.04 | -3.6 | |
| F | N | H | 0.55 ± 0.04 | -4.5 | |
| F | N | Y | 0.26 ± 0.01 | -9.6 | |
| 59/89 | F | D | L | 0.57 ± 0.08 | -4.4 |
| F | E | H | 0.50 ± 0.03 | -5.0 | |
| F | M | R | 0.094 ± 0.012 | -27 | |
| F | M | K | 0.078 ± 0.004 | -32 | |
| F | K | Y | 0.062 ± 0.006 | -40 | |
| 14/89 | H | N | H | 0.47 ± 0.00 | -5.3 |
| 14/59/89 | E | M | R | 0.11 ± 0.00 | -23 |
| Q | K | Y | 0.085 ± 0.009 | -29 | |
| D | M | K | 0.080 ± 0.005 | -31 | |
Mean ± standard deviation. Wildtype CheY was measured 8 times and each CheY mutant was measured 2-4 times.
In contrast to CheY, two substitutions in Spo0F at position ‘59’ (‘59’KE or ‘59’KN) resulted in substantial changes in rate, 62 and 24 times faster than wildtype, respectively (Table 3). Zapf et al. (Zapf et al., 1998) previously reported a similar result for Spo0F ‘59’KN. A potential explanation could be that the positively charged Lys might stabilize the phosphoryl group against removal. However, neither replacement of Spo0F Lys‘59’ with Ala or Met (Zapf et al., 1998) nor introduction of Lys59 into CheY (Table 2) affected phosphoryl group stability.
Table 3.
Autodephosphorylation rates of Spo0F mutants
| Substitution Position(s) | Amino Acid At Position |
Fold Change From Wildtype | |||
|---|---|---|---|---|---|
| ‘14’ | ‘59’ | ‘89’ | kdephos (min-1)a | ||
| Wildtype | Q | K | Y | 0.0039b ± 0.0011 | NA |
| ‘59’ | Q | E | Y | 0.24 ± 0.01 | +62 |
| Q | N | Y | 0.093b ± 0.005 | +24 | |
| ‘89’ | Q | K | E | 0.031 ± 0.001 | +7.9 |
| Q | K | L | 0.015 ± 0.002 | +3.8 | |
| Q | K | H | 0.0051± 0.0001 | +1.3 | |
| ‘59’/‘89’ | Q | E | L | 0.42 ± 0.04 | +110 |
| Q | N | E | 0.42 ± 0.05 | +110 | |
| Q | N | L | 0.33 ± 0.02 | +85 | |
| Q | N | H | 0.17 ± 0.01 | +44 | |
| ‘14’/‘59’/‘89’ | S | E | L | 0.49 ± 0.10 | +130 |
| F | N | E | 0.29 ± 0.03 | +74 | |
| H | N | H | 0.13 ± 0.00 | +33 | |
Mean ± standard deviation. Wildtype Spo0F was measured 4 times and each mutant Spo0F was measured 2-3 times.
Rates reported by Zapf et al. (Zapf et al., 1998) were 0.0039 and 0.087 min-1 for wildtype and ‘59’KN substitution, respectively.
The single substitutions made at position ‘89’ had moderate (≤ 10-fold) impact on the autodephosphorylation rates of CheY and Spo0F (Tables 2 and 3; Fig. 2, column 3). Ala or Gln substitutions at position 89 of CheY (Silversmith et al., 2003) and an Ala substitution at position ‘89’ of Spo0F (Tzeng and Hoch, 1997) also were previously found to have little effect.
Double substitutions at positions ‘59’ and ‘89’ significantly changed autodephosphorylation rates
Simultaneous substitutions at both positions 59 and 89 in CheY had a statistically significant (P < 0.05) greater effect on autodephosphorylation rates than changing a single position alone. A paired t test of the rates exhibited by CheY mutants with double vs. the constituent single substitutions gave P values of 0.0002 and 0.049 for positions 59 and 89, respectively. Three combinations of residues at positions 59 and 89 each changed the rate more than 25-fold (Table 2). The CheY 59NK-89EY mutant protein (a mimic of Spo0F) exhibited the largest change, a 40-fold decrease in rate from wildtype. Spo0F double substitutions at positions ‘59’ and ‘89’ also showed significant differences in rates compared to the corresponding single substitutions alone (P values of 0.03 and 0.01 for positions ‘59’ and ‘89’, respectively, paired t test). The Spo0F mutant proteins ‘59’KN-‘89’YE (a mimic of CheY) and ‘59’KE-‘89’YL (a mimic of CheB) both had rates 110 times faster than that of wildtype Spo0F (Table 3). Thus, simply by incorporating the residues at ‘59’ and ‘89’ of CheY into Spo0F or vice versa, the autodephosphorylation rates of the mutant proteins approached the reaction rates of the mimicked proteins. Wildtype CheY autodephosphorylates 640 times faster than wildtype Spo0F, whereas the CheY 59NK-89EY (Spo0F mimic) rate was only 16 times faster than Spo0F (Tables 2 and 3). Similarly, wildtype Spo0F autodephosphorylates 250 to 1,000 times slower than wildtype CheY proteins from various species that contain Asn at position ‘59’ and Glu at position ‘89’ (Table 1), but the Spo0F ‘59’KN-‘89’YE (CheY mimic) rate was only two to 10 times slower than that of the CheY proteins.
CheY positions 59 and 89 influence each other, whereas Spo0F positions ‘59’ and ‘89’ do not
To further understand the relationship between autodephosphorylation rate and the combinations of residues at positions ‘59’ and‘89’, we mathematically analyzed the contributions of individual positions to the reaction rate using double mutant cycles. If substitutions are made at two independent positions that do not interact either directly or indirectly, then the sum of the changes in activation energies of each single substitution (ΔΔG‡1+ΔΔG‡2) will equal the change in activation energy of the double substitution (ΔΔG‡1+2) (Mildvan et al., 1992; Mildvan, 2004; Wells, 1990). Significant deviation from the sum (non-additive effect) implies that one residue influences the contribution of the other residue to the reaction. Because ΔG‡ is directly proportional to the logarithm of the rate constant, the logarithm of the ratios of mutant and wildtype kdephos values can be used as a measure of ΔΔG‡. All four sets of Spo0F mutants showed modest differences between the actual (double mutant) and expected (sum of single mutants) values, suggesting little or no interactive effects, whereas four out of five sets of CheY mutants exhibited larger differences indicative of mutual influences between residues 59 and 89 (Table 4).
Table 4.
Additive or non-additive effects of positions ‘59’ and ‘89’ on autodephosphorylation rate
| Response Regulator | Substitutions | Expected Valuea | Actual Valueb | Absolute Value of Difference |
|---|---|---|---|---|
| CheY | 59D-89L | 0.15 | -0.64 | 0.79 |
| 59E-89H | -0.40 | -0.70 | 0.30 | |
| 59M-89R | -0.29 | -1.42 | 1.13 | |
| 59M-89K | -0.05 | -1.51 | 1.46 | |
| 59K-89Y | -0.90 | -1.61 | 0.71 | |
| Spo0F | ‘59’E-‘89’L | 2.37 | 2.03 | 0.34 |
| ‘59’N-‘89’E | 2.28 | 2.03 | 0.25 | |
| ‘59’N-‘89’H | 1.49 | 1.64 | 0.15 | |
| ‘59’N-‘89’L | 1.96 | 1.93 | 0.03 |
Position ‘14’ had little effect on autodephosphorylation rate
In the initial review of published data (Table 1), the amino acids at position ‘14’ appeared to correlate with autodephosphorylation rate. However, substitutions at position ‘14’ in CheY or Spo0F had less than a two-fold effect on autodephosphorylation rate when compared to CheY or Spo0F proteins with matched residues at positions ‘59’ and ‘89’ (Tables 2 and 3, Table S1).
Discussion
Variable active site positions ‘59’ and ‘89’ strongly influence autodephosphorylation rate
During signal transduction, response regulators generally switch from the active to inactive state via dephosphorylation. It is critical that the kinetics of this conversion match the timescale of the biological process involved. In spite of extensive structural and functional similarities, response regulators exhibit a range of more than 104-fold in autodephosphorylation rates (Table 1). We found that two variable residues, located in close spatial proximity to the phosphoryl group (Fig. 1), contribute substantially to this reaction rate. Twenty different pairs of amino acids at positions ‘59’ and ‘89’ tested in the CheY and/or Spo0F response regulators resulted in sets of proteins spanning ∼100-fold ranges of autodephosphorylation rates for each response regulator (Tables 2 and 3). Although the 20 different pairs examined here comprise only 5% of the 400 mathematically possible combinations, amino acids do not occur at equal frequencies at positions ‘59’ and ‘89’ in response regulators. The tested combinations actually occur in 24% of the entries in a database of 6,310 non-redundant response regulators (Pers. Comm., Kristin Wuichet & Igor Zhulin), and therefore reflect a significant fraction of the ‘59’/‘89’ pairs found in nature. Substitutions tested in CheY, which has a relatively fast autodephosphorylation rate, resulted in modest rate increases or larger rate decreases. Many tested substitutions in Spo0F, which has a relatively slow autodephosphorylation rate, resulted in considerable rate enhancements. No substitutions that retard Spo0F autodephosphorylation were observed among the limited test set, although response regulators with slower autodephosphorylation rates do exist (Table 1).
Location of β4/α4 loop explains the non-interactive tendencies of positions ‘59’ and ‘89’ observed in Spo0F mutants
Comparison of autodephosphorylation rates in proteins bearing single or double amino acid substitutions suggest that CheY positions 59 and 89 influence one another whereas Spo0F positions ‘59’ and ‘89’ do not (Table 4). The interactive effects (or lack thereof) between positions ‘59’ and ‘89’ on autodephosphorylation rates may arise from differences in the distances between the ‘59’ and ‘89’ sidechains in CheY and Spo0F. Structures of CheY and several other response regulators in the phosphorylated (or BeF3- bound) versus unphosphorylated state show a substantial (>4 Å) movement of the β4/α4 loop, which contains position ‘89’, toward the active site. The shift is accompanied by a change in the ‘89’ side chain from an outward to an inward orientation, thereby considerably decreasing the distance between residues ‘59’ and ‘89’ (Bachhawat et al., 2005; Birck et al., 1999; Hastings et al., 2003; Lee et al., 2001). Although the sidechains at positions 59 and 89 are ∼5 Å apart in wildtype CheY·BeF3- (Lee et al., 2001), in CheY 59NR·BeF3- a salt bridge exists between Arg59 and Glu89 (Silversmith et al., 2003), which suggests that, at least for CheY derivatives, the amino acids present at positions 59 and 89 determines whether an interaction occurs. Similarly, in the PhoB·BeF3- (Bachhawat et al., 2005), ArcA·BeF3- (Toro-Roman et al., 2005), Spo0A·PO32- (Lewis et al., 1999), DctD·BeF3- (Park et al., 2002), and FixJ·PO32- (Birck et al., 1999) structures, the sidechains of positions ‘59’ and ‘89’ are in close proximity (<4 Å) (Fig. 3), suggesting that interactions between these two positions commonly occur in phosphorylated receiver domains. In contrast, the β4/α4 loop is in a different position in Spo0F·BeF3- structures (Gardino et al., 2003; Varughese et al., 2006), thereby maintaining the outward orientation of residue ‘89’, which in turn results in a separation of >8Å between positions ‘59’ and ‘89’ (Fig. 3). The ability of position ‘89’ to influence Spo0F autodephosphorylation rate in spite of its distance from the phosphoryl group suggests an indirect interaction. One possibility is that position ‘89’ may alter the mobility of the β4/α4 loop in Spo0F (Feher and Cavanagh, 1999) and thereby affect the location of other sidechains such as Glu‘91’, which projects into the same space as position ‘89’ of CheY. The potential role of position ‘91’ in Spo0F autodephosphorylation has not yet been investigated.
Fig. 3.
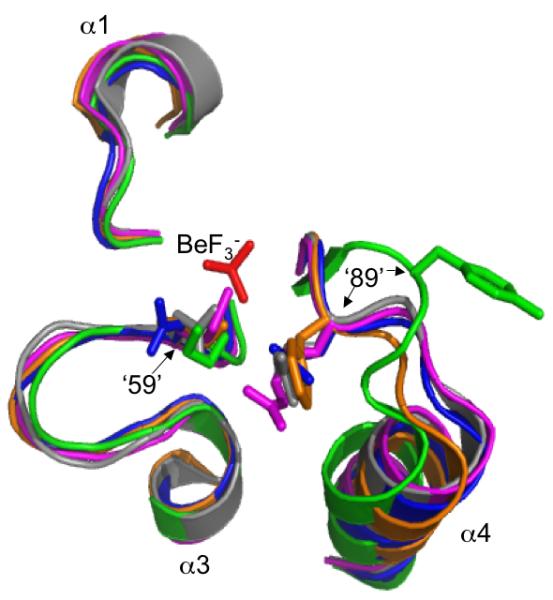
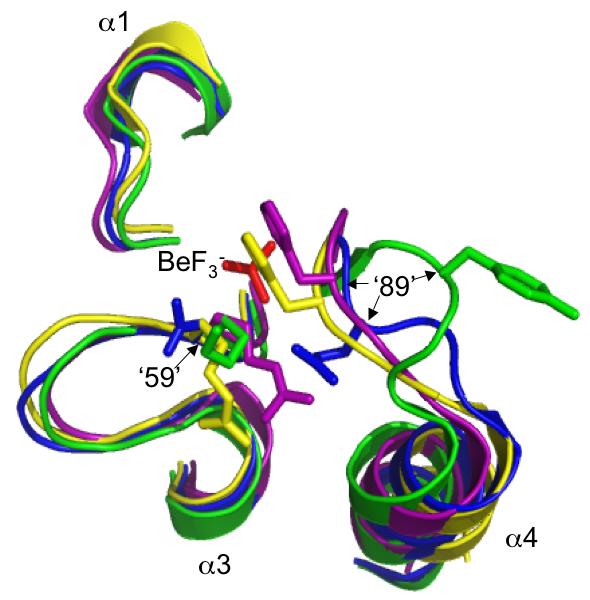
Orientations of residues at positions ‘59’ and ‘89’ in various activated receiver domain structures. Sections of superimposed backbones are displayed for (A) CheY·BeF3- (blue), Spo0F·BeF3- (green), PhoB·BeF3- (magenta), ArcA·BeF3- (grey), and Spo0A·PO32- (orange) and (B) CheY·BeF3- (blue), Spo0F·BeF3- (green), DctD·BeF3- (yellow), FixJ·PO32- (purple). For clarity, only the β1-α1, β3-α3, and β4-α4 loops surrounding the BeF3-/PO32- group (red) are shown. Sidechains are shown for position ‘59’, located at the C-terminal end of β3, and position ‘89’, on the β4-α4 loop. Note that the distance between positions ‘59’ and ‘89’ in Spo0F is greater than the ∼4 Å distance observed in other receiver domains (distances not shown). The specific software, method of superimposition, literature references, and pdb files used to create this figure are described in Experimental procedures.
Position ‘14’ does not appear to influence autodephosphorylation rate
The apparent correlation between the amino acids at position ‘14’ and autodephosphorylation rate in Table 1 made position ‘14’ a plausible candidate as an additional factor that influences response regulator phosphoryl group stability. However, the amino acid substitutions tested at position ‘14’ had little actual effect on rate (Tables 2 and 3, Table S1). This apparent discrepancy may arise from the high specificity that response regulators show toward their paired sensor kinases, with little cross talk from other kinases (Skerker et al., 2005; Yamamoto et al., 2005). There is structural evidence suggesting that position ‘14’ is important for response regulator interaction with paired kinases. Although high-resolution structural details of the interaction between response regulators and histidine phosphorylation sites are not available for sensor kinases, presumably similar interactions with histidine phosphotransferase proteins have been described. For example, Gln‘14’ of Spo0F (Varughese et al., 2006) forms a hydrogen bond with Spo0B, a dimeric phosphotransferase whose structure resembles that of histidine kinases. Furthermore, Asn‘14’ of SLN1 (Xu et al., 2003) forms a hydrogen bond with the monomeric phosphotransferase YPD1. In all three types of proteins (dimeric sensor kinases, dimeric histidine phosphotransferases, and monomeric histidine phosphotransferases), the histidine phosphorylation site is located on a four-helix bundle. If the hydrogen bond interactions observed between response regulators and phosphotransferase proteins also occur between response regulators and sensor kinases, then it may account for the prevalence of Asn, Asp, Gln, Glu, and His residues at position ‘14’ in transcriptional response regulators (Table 1). On the other hand, chemotaxis sensor kinases display a distinct organization from transcriptional sensor kinases (Dutta et al., 1999; Grebe and Stock, 1999). Therefore, the different sets of amino acids observed at position ‘14’ in chemotaxis response regulators, which cluster with the fast autodephosphorylation rates, versus transcriptional response regulators, which cluster with the slow autodephosphorylation rates, may simply reflect differences in interactions with their paired sensor kinases.
Exploiting the rules of response regulator autodephosphorylation
Construction of constitutively active or dominant negative small GTPases, based on knowledge of key positions applicable to most family members, has found widespread utility in dissecting signaling networks (Campbell et al., 2005). Similarly, if there are simple rules for how variable residues control autodephosphorylation rate, then this information could be used to manipulate the phosphoryl group stability of any response regulator via site-directed mutagenesis, which might be a useful tool for the in vivo investigation of two-component signaling systems. Although the absolute magnitude of autodephosphorylation rate associated with a particular substitution depends on the context of other active site residues and the response regulator backbone, the relative impact of a specific amino acid was fairly consistent. Therefore, the experimental data reported here could be used to make preliminary predictions about the consequences of certain amino acid substitutions. Comparing the rates of mutants with ‘89’ substitutions in the context of a fixed amino acid at ‘59’ (Table S2) yields a consensus order for autodephosphorylation rate from fastest to slowest of Glu > Leu,Lys > Arg,His > Tyr and is consistent for both CheY and Spo0F even though residue 89 occupies different spatial positions in the crystal structures of these two response regulators (Fig. 3). The order of residues at position ‘59’ is less evident, because all CheY single substitutions tested at this position had little effect on rate. However, by comparing the consequences of residues at ‘59’ while keeping a fixed residue at ‘89’ (Table S3), a rank of Glu > Asn > Met, Lys was revealed. Finally, there are six examples of the same ‘59’/‘89’ combinations in both CheY and Spo0F. Five of the six exhibit the same rank order in autodephosphorylation rate: ‘59’N-‘89’E, ‘59’N-‘89’L > ‘59’N-‘89’H, 59‘N’-‘89’Y > ‘59’K-‘89’Y (Table S4).
The utility of manipulating response regulator autodephosphorylation rates for in vivo studies will depend on whether alteration of positions ‘59’ and ‘89’ affects other functions of the particular response regulator that has been modified, including interaction with other proteins in the signaling network. Changing positions ‘59’ or ‘89’ appears to affect response regulator function in some cases and not in others: First, with regard to sensor kinases, positions ‘59’ and ‘89’ are hypothesized to be among the response regulator residues important for specific recognition of a paired sensor kinase (Hoch and Varughese, 2001; Li et al., 2003). We observed little or no impairment of phosphotransfer from CheA-P or KinA-P to mutant CheY or Spo0F proteins in vitro (see Experimental procedures), but our assay method was relatively insensitive to phosphotransfer kinetics. Second, with regard to output proteins, many CheY mutants with single substitutions at positions 59 or 89 can be phosphorylated and interact productively with the FliM flagellar switch protein in vivo, as inferred from ability to support clockwise flagellar rotation (Silversmith et al., 2001; Silversmith et al., 2003) (data not shown). However, the five CheY mutants in this study with substitutions at both positions 59 and 89 showed predominantly counterclockwise flagellar rotation (data not shown), consistent with reduced phosphorylation and/or function. Several Spo0F mutants with substitutions at positions ‘59’ or ‘89’ efficiently exchange phosphoryl groups with the downstream protein Spo0B in vitro (Tzeng and Hoch, 1997; Zapf et al., 1998). However, Ala substitutions at position ‘59’ or ‘89’ of Spo0F prevent sporulation in vivo, a defect that has been attributed to reduced phosphorylation by KinA (Tzeng and Hoch, 1997; Zapf et al., 1998). Third, with regard to phosphatases, changing CheY Glu89 prevents CheZ-stimulated dephosphorylation, but not binding to CheZ (Silversmith et al., 2001; Silversmith et al., 2003). In Spo0F, Ala substitutions at positions ‘59’ or ‘89’ reduce RapB-mediated stimulation of dephosphorylation five to 10-fold compared to wildtype (Tzeng et al., 1998). If positions ‘59’ and ‘89’ are important for phosphatase-stimulated dephosphorylation of other response regulators, then alteration of these residues could be a useful strategy to increase response regulator phosphorylation in vivo for those systems that include phosphatases. In summary, our study suggests candidates for engineering altered autodephosphorylation rates, but their practical utility will have to be evaluated on a case-by-case basis.
The findings of this study have further potential application. There has been substantial interest recently in engineering bacterial regulatory networks with particular properties, essentially creating programmable cellular behavior in which a specific input results in a desired output (Anderson et al., 2006; Kaern et al., 2003; Kobayashi et al., 2004; Voigt, 2006). A biosensor circuit can be envisioned in which an unstable output is mediated by response regulator activation. The availability of interchangeable receiver domain modules with a range of different characteristic phosphoryl group half-lives would facilitate construction of circuits with outputs of different duration following a given stimulus. In order for autodephosphorylation to control signal decay, the circuit would not contain a separate phosphatase. Our studies suggest mutants can be found that will interact sufficiently well with a sensor kinase and output protein to create a functioning circuit.
Finally, it is simple with current technology to identify genes encoding response regulators. However, it is often difficult or impossible to ascertain through bioinformatic means the specific output that is controlled by a particular response regulator. Complete elucidation of the features that control response regulator autodephosphorylation rate may make it possible to infer the timescale (seconds, minutes, or hours) of signal transduction (and hence the timescale of the controlled biological process) from response regulator amino acid sequence alone. In turn, such a predictive capability would permit exploration of the sensory capacity of an organism in relation to its ecological niche or physiological characteristics (e.g. does a slow-growing species process information at a slow rate?).
Factors other than positions ‘59’ and ‘89’ must contribute to phosphoryl group stability
Comparisons of autodephosphorylation rates between CheY or Spo0F mutant proteins created in this study and response regulators with matching amino acids at positions ‘59’ and ‘89’ showed some cases of remarkably good agreement (e.g. less than two-fold rate difference between E. coli CheY 59NE-89EH and R. sphaeroides CheB1) (Table S5). At the other extreme, there was a 160-fold difference between the autodephosphorylation rates of T. maritima DrrA and the matched E. coli CheY 59NM-89EK mutant. Such discrepancies suggest that the amino acid combinations at positions ‘59’ and ‘89’ do not solely account for the intrinsic dephosphorylation rate of response regulators. The data in Table 1 suggest the potential contributions of position ‘88’ are another obvious area for future investigation. However, many of the rate discrepancies noted in Table S5 exist between response regulators that are also matched at position ‘88’. Therefore, other as yet unidentified factors that influence autodephosphorylation rate must exist.
Some differences in rate may plausibly be attributed to differences in the positioning of residues ‘59’ and ‘89’ with respect to the active site. One example is His‘89’, which has been proposed to affect phosphoryl group stability by sterically inhibiting access of the nucleophilic water molecule in FixJ (Birck et al., 1999) (Fig. 3B), or possibly serving as a base to activate the water molecule in NtrC (Hastings et al., 2003). Similarly, the different spatial orientations of position ‘89’ observed in CheY·BeF3- (Lee et al., 2001) and Spo0F·BeF3- (Gardino et al., 2003; Varughese et al., 2006) (Fig. 3) may explain the range in rates (three-fold to 22-fold) observed for the nine pairs of CheY and Spo0F proteins with matched amino acids at positions ‘59’ and ‘89’ (Table S5). In contrast, no such obvious differences in structure are observed between CheY·BeF3- (Lee et al., 2001), ArcA·BeF3- (Toro-Roman et al., 2005), and PhoB·BeF3- (Bachhawat et al., 2005) (Fig. 3A), yet CheY mutants autodephosphorylate over 100 times and over 10 times faster than the respective ArcA and PhoB proteins they were designed to mimic (Table S5). However, subtle differences between response regulator structures that alter the precise geometry of active site side chains or backbone amides could conceivably affect the energetics of interactions with the phosphoryl group in the transition state and hence translate into a large effect on reaction rates. The eventual ability to predict absolute dephosphorylation rate, rather than relative rate, would enhance the potential applications described above.
Experimental procedures
Bacterial strains, plasmids and mutant construction
Construction of cheY mutations encoding 59ND, 59NE, 59NK, 89EH, and 89EK substitutions have been described previously (Silversmith et al., 2001; Silversmith et al., 2003). The cheY89EL mutation was created as described for cheY89EH but was not previously reported. New mutants were created by Stratagene® Quick-Change site directed mutagenesis kit in a pRBB40 derivative, pRS3, containing wildtype cheY and cheZ (Boesch et al., 2000).
Spo0F mutants were created as described above for CheY. The template, supplied by J. Cavanagh, was a pET28a(+) (Novagen®) vector containing spo0F cloned into the NdeI and BamHI restriction sites to create an N-terminal His6 tag with a thrombin cleavage site.
M. Perego supplied a plasmid containing full length B. subtilis kinA cloned from a NcoI-XhoI PCR fragment into pET28a(+), with two codons added to the XhoI site to create an in-frame C-terminal His6 tag.
Protein purification
CheY wildtype and mutant proteins were purified using a published method (Hess et al., 1991), starting from E. coli ΔcheY strain KO641recA containing the appropriate plasmid. CheA was purified from strain KO685/pDV4 as described (Hess et al., 1991) with the addition of an ion exchange step. After Affi-Gel Blue (Bio-Rad®) affinity chromatography, fractions containing CheA were applied to a HiTrap Q Fast Flow column (GE Healthcare®) equilibrated with TEDG-10 buffer [50 mM Tris-HCl pH 7.5, 0.5 mM EDTA, 2 mM DTT and 10% (v/v) glycerol] and eluted with a gradient of 0-500 mM NaCl in TEDG-10. Appropriate fractions were collected and then applied to a Superose 12 (GE Healthcare ®) FPLC gel filtration column. Protein concentrations were calculated using an empirically determined extinction coefficient of 0.727 (mg/ml)-1 cm-1 for CheY (Silversmith et al., 2001) or 0.223 (mg/ml)-1 cm-1 for CheA [estimated with ProtParam software (Gasteiger et al., 2005)].
His6 tag versions of KinA, wildtype Spo0F, and Spo0F mutants were expressed and purified from E. coli strain BL21(DE3). One liter of LB [1% (w/v) tryptone, 0.5% (w/v) yeast extract, 1% (w/v) NaCl] containing 30 μg/ml kanamycin was inoculated with 5 ml of overnight culture and grown to an optical density at 600 nm of 0.75. The culture was induced with 1 mM IPTG and grown for 4 hours at 37 °C. Cells were collected by centrifugation and resuspended in 25 ml of Lysis buffer (50 mM NaH2PO4 pH 8.0, 500 mM NaCl, 5 mM imidazole, 5 mM β-mercaptoethanol). The cells were disrupted by sonication and cellular debris was removed by centrifugation. The supernatant was then subjected to nickel column chromatography (Ni-NTA Agarose, Qiagen®) in accordance with manufacturer’s instructions. Protein was eluted in a stepwise fashion with 50, 100 and 150 mM imidizole, and the appropriate fractions were combined and concentrated. For KinA, the combined fractions were dialyzed into 50 mM Tris-HCl pH 8.0, 20 mM KCl and 1 mM dithiothreitol and then were chromatographed on a Superose 12 (GE Healthcare ®) size exclusion column in the same buffer. The final KinA concentration was determined based on an estimated extinction coefficient at 280 nm of 0.662 (mg/ml)-1 cm-1. After nickel affinity purification, the His6 tag was removed from all Spo0F proteins by the addition of 0.01 U thrombin (Novagen®) per 10 μg protein and allowed to incubate at 25 °C for 18 hours. This leaves three extra residues (Gly-Ser-His) on the N-terminus of Spo0F. The cleavage reaction was stopped by the addition of Sigma® protease inhibitor cocktail at 40 μl cocktail per 1 ml protein solution. Spo0F was separated from thrombin and the His6 tag peptide on a Superose 12 (GE Healthcare®) size exclusion column in MES buffer (50 mM MES pH 6.6, 100 mM NaCl, 0.5 mM EDTA). The final Spo0F concentration was determined using an estimated extinction coefficient at 280 nm of 0.360 (mg/ml)-1 cm-1.
Dephosphorylation assays
[32P]CheA-P and [32P]KinA-P were prepared as previously described for CheA-P (Silversmith et al., 1997). Briefly, 170 μg of kinase (CheA or KinA) was incubated for one hour with ∼600 μCi [γ-32P]ATP (GE Healthcare ®), 35 mM KCl, 3.5 mM MgCl2, and 35 mM Tris (pH 8.0). Residual ATP was separated from the [32P]kinase by ammonium sulfate precipitation, followed by gel filtration.
Reactions to measure CheY autodephosphorylation kinetics were described previously (Silversmith et al., 1997). Briefly, a molar excess of CheY (400 pmol) was added to approximately 28 pmol [32P]CheA-P in reaction buffer (100 mM Tris-HCl pH 7.5, 10 mM MgCl2) in a total of 80 μl. For wildtype and all CheY mutant proteins, phosphotransfer was completed within 10 seconds of mixing with [32P]CheA-P. A small amount (∼4%) of the initial [32P]CheA-P remained throughout the time course and never transferred to CheY, as previously observed (Stewart, 1997). Because there was no ATP present to regenerate CheA-P and all 32P transferred to CheY at once, the subsequent decline in [32P]CheY-P directly reflects the CheY autodephosphorylation rate. Aliquots (10 μl) of the reaction were removed at six different time points and quenched with 10 μl of 2x SDS sample buffer to stop the reaction. Samples were electrophoresed on 18% Tris-glycine polyacrylamide gels and dried. Relative CheY-P concentrations for each time point were measured by phosphorimaging, converted to a percentage of the initial concentration, and graphed against time. All CheY mutant proteins displayed autodephosphorylation kinetics that fit well to a single exponential decay, as calculated with Excel software to determine the rate constant.
Spo0F autodephosphorylation assays were conducted similarly to CheY assays except [32P]KinA-P was used rather than [32P]CheA-P, four nmol of Spo0F were used in an attempt to ensure complete and rapid phosphotransfer from KinA to Spo0F, and the reaction buffer contained 20 mM MgCl2 (Grimshaw et al., 1998). An average of ∼75% of radiolabel from [32P]KinA-P transferred to Spo0F within 10 seconds of mixing for wildtype and all but three mutant Spo0F proteins. The residual [32P]KinA-P decayed at an average rate of 0.0251 ± 0.0119 min-1, whereas an independent experiment without Spo0F displayed a loss of [32P]KinA-P at an experimentally indistinguishable rate of 0.0195 min-1. Because decay of the residual [32P]KinA-P was not affected by Spo0F, phosphotransfer from [32P]KinA-P to Spo0F was at or near completion by the first time point, and ∼25% of [32P]KinA-P was incapable of transferring phosphoryl groups even with a 140 fold molar excess of Spo0F. Three Spo0F mutant proteins (‘59’KE-‘89’YL, ‘59’KN-‘89’YE, ‘59’KN-‘89’YL) exhibited somewhat lower initial phosphotransfer (68%, 68% and 61%, respectively). A minor correction was applied to calculation of autodephosphorylation rates for these three mutants, based on the slightly less than expected phosphotransfer.
Wildtype Spo0F demonstrated a slow autodephosphorylation rate (t1/2 ∼ 3 hours). It was necessary to correct for spontaneous dephosphorylation of denatured Spo0F in sample buffer, because samples from early time points remained in sample buffer longer than late time points prior to electrophoresis, and therefore lost more Spo0F-P due to spontaneous dephosphorylation. To determine the dephosphorylation rate of denatured Spo0F, 3.5 pmol [32P]KinA-P was added at various times to 50 pmol wildtype Spo0F in reaction buffer in a total volume of 10 μl. After a five minute incubation (to allow complete phosphotransfer), each reaction was quenched with an equal volume of SDS sample buffer and then allowed to incubate at 25°C for 0, 4, 8, 12, 20, 24 or 30 hours. Initiation of the reactions was arranged so that all incubations finished at the same time, at which point the samples were electrophoresed simultaneously on a SDS gel. Analysis of the amount of radioactivity in Spo0F was carried out as described above and gave the dephosphorylation rate constant for denatured Spo0F-P as 0.0014 min-1. The autodephosphorylation rate constant for Spo0F was then determined by adding the experimental rate constant from the autodephosphorylation reactions to the rate constant obtained for denatured Spo0F-P in sample buffer. This correction was made for wildtype Spo0F and Spo0F ‘89’YH. For Spo0F mutants with half-lives of less than one hour the correction was negligible and therefore not applied.
All except one of the Spo0F mutant proteins displayed autodephosphorylation kinetics that fit well to a single exponential decay. The exception was Spo0F ‘59’KN-‘89’YH, which demonstrated biphasic dephosphorylation kinetics. The rate constant for loss of the first 90% of this Spo0F-P mutant was 0.17 min-1, and is the value reported for these experiments. Loss of the remaining 10% occurred at a rate constant of 0.0075 min-1. The basis for the biphasic decay is unknown.
Structure superimpositions
The Cα carbon atoms of PhoB·BeF3- (pdb 1ZES) (Bachhawat et al., 2005), ArcA·BeF3- (pdb 1XHF) (Toro-Roman et al., 2005), Spo0A·PO32- (pdb 1QMP) (Lewis et al., 1999), DctD·BeF3- (pdb 1L5Y) (Park et al., 2002), FixJ·PO32- (pdb 1D5W) (Birck et al., 1999), and Spo0F·BeF3- (pdb 2FTK) (Varughese et al., 2006) were superimposed to the Cα atoms of CheY·BeF3- (pdb 1FQW) (Lee et al., 2001) using the PyMOL® (DeLano, 2002) align function, with RMS values for each structure aligned to CheY of 0.913, 1.004, 1.326, 1.134, 1.591, and 1.477 Å, respectively.
Acknowledgements
We thank Ruth Silversmith for expert advice, John Cavanagh for the spo0F clone, Marta Perego for the kinA clone, Carolyn Chu for construction of the cheY89EL mutation, Kristin Wuichet and Igor Zhulin for access to their database of response regulator amino acid sequences, Brenda Temple for help with PyMOL superimpositions, and Channing Der, Ashalla Freeman, and Yael Pazy for useful discussions. This work was supported by National Institutes of Health grant GM050860 (to R.B.B.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
References
- Ames SK, Frankema N, Kenney LJ. C-terminal DNA binding stimulates N-terminal phosphorylation of the outer membrane protein regulator OmpR from Escherichia coli. Proc Natl Acad Sci U S A. 1999;96:11792–11797. doi: 10.1073/pnas.96.21.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol. 2006;355:619–627. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- Appleby JL, Bourret RB. Proposed signal transduction role for conserved CheY residue Thr87, a member of the response regulator active-site quintet. J Bacteriol. 1998;180:3563–3569. doi: 10.1128/jb.180.14.3563-3569.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashby MK. Survey of the number of two-component response regulator genes in the complete and annotated genome sequences of prokaryotes. FEMS Microbiol Lett. 2004;231:277–281. doi: 10.1016/S0378-1097(04)00004-7. [DOI] [PubMed] [Google Scholar]
- Bachhawat P, Swapna GV, Montelione GT, Stock AM. Mechanism of activation for transcription factor PhoB suggested by different modes of dimerization in the inactive and active states. Structure. 2005;13:1353–1363. doi: 10.1016/j.str.2005.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birck C, Mourey L, Gouet P, Fabry B, Schumacher J, Rousseau P, Kahn D, Samama JP. Conformational changes induced by phosphorylation of the FixJ receiver domain. Structure. 1999;7:1505–1515. doi: 10.1016/s0969-2126(00)88341-0. [DOI] [PubMed] [Google Scholar]
- Bird TH, Du S, Bauer CE. Autophosphorylation, phosphotransfer, and DNA-binding properties of the RegB/RegA two-component regulatory system in Rhodobacter capsulatus. J Biol Chem. 1999;274:16343–16348. doi: 10.1074/jbc.274.23.16343. [DOI] [PubMed] [Google Scholar]
- Boesch KC, Silversmith RE, Bourret RB. Isolation and characterization of nonchemotactic CheZ mutants of Escherichia coli. J Bacteriol. 2000;182:3544–3552. doi: 10.1128/jb.182.12.3544-3552.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L. Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J Mol Biol. 2006;361:1003–1034. doi: 10.1016/j.jmb.2006.06.049. [DOI] [PubMed] [Google Scholar]
- Campbell PM, Singh A, Williams FJ, Frantz K, Ulfu AS, Kelley GG, Der CJ. Genetic and pharmacologic dissection of ras effector utilization in oncogenesis. Methods Enzymol. 2005;407:195–217. doi: 10.1016/S0076-6879(05)07017-5. [DOI] [PubMed] [Google Scholar]
- Catlett NL, Yoder OC, Turgeon BG. Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot Cell. 2003;2:1151–1161. doi: 10.1128/EC.2.6.1151-1161.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavicchioli R, Schroder I, Constanti M, Gunsalus RP. The NarX and NarQ sensor-transmitter proteins of Escherichia coli each require two conserved histidines for nitrate-dependent signal transduction to NarL. J Bacteriol. 1995;177:2416–2424. doi: 10.1128/jb.177.9.2416-2424.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Wang W, Kim R, Yokota H, Damo S, Kim SH, Wemmer D, Kustu S, Yan D. BeF3- acts as a phosphate analog in proteins phosphorylated on aspartate: structure of a BeF3- complex with phosphoserine phosphatase. Proc Natl Acad Sci U S A. 2001;98:8525–8530. doi: 10.1073/pnas.131213698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comolli JC, Carl AJ, Hall C, Donohue T. Transcriptional activation of the Rhodobacter sphaeroides cytochrome c2 gene P2 promoter by the response regulator PrrA. J Bacteriol. 2002;184:390–399. doi: 10.1128/JB.184.2.390-399.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl MK, Msadek T, Kunst F, Rapoport G. The phosphorylation state of the DegU response regulator acts as a molecular switch allowing either degradative enzyme synthesis or expression of genetic competence in Bacillus subtilis. J Biol Chem. 1992;267:14509–14514. [PubMed] [Google Scholar]
- DeLano WL. The PyMOL molecular graphics system. 2002 [WWW document]. www.pymol.org.
- Downward J. The ins and outs of signalling. Nature. 2001;411:759–762. doi: 10.1038/35081138. [DOI] [PubMed] [Google Scholar]
- Dutta R, Qin L, Inouye M. Histidine kinases: diversity of domain organization. Mol Microbiol. 1999;34:633–640. doi: 10.1046/j.1365-2958.1999.01646.x. [DOI] [PubMed] [Google Scholar]
- Feher VA, Cavanagh J. Millisecond-timescale motions contribute to the function of the bacterial response regulator protein Spo0F. Nature. 1999;400:289–293. doi: 10.1038/22357. [DOI] [PubMed] [Google Scholar]
- Fisher SL, Kim SK, Wanner BL, Walsh CT. Kinetic comparison of the specificity of the vancomycin resistance VanS for two response regulators, VanR and PhoB. Biochemistry. 1996;35:4732–4740. doi: 10.1021/bi9525435. [DOI] [PubMed] [Google Scholar]
- Gardino AK, Volkman BF, Cho HS, Lee SY, Wemmer DE, Kern D. The NMR solution structure of BeF3--activated Spo0F reveals the conformational switch in a phosphorelay system. J Mol Biol. 2003;331:245–254. doi: 10.1016/s0022-2836(03)00733-2. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The Proteomics Protocols Handbook. Humana Press; Totowa, NJ: 2005. pp. 571–607. [Google Scholar]
- Georgellis D, Kwon O, De Wulf P, Lin EC. Signal decay through a reverse phosphorelay in the Arc two-component signal transduction system. J Biol Chem. 1998;273:32864–32869. doi: 10.1074/jbc.273.49.32864. [DOI] [PubMed] [Google Scholar]
- Goldman BS, Nierman WC, Kaiser D, Slater SC, Durkin AS, Eisen JA, Ronning CM, Barbazuk WB, Blanchard M, Field C, Halling C, Hinkle G, Iartchuk O, Kim HS, Mackenzie C, Madupu R, Miller N, Shvartsbeyn A, Sullivan SA, Vaudin M, Wiegand R, Kaplan HB. Evolution of sensory complexity recorded in a myxobacterial genome. Proc Natl Acad Sci U S A. 2006;103:15200–15205. doi: 10.1073/pnas.0607335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudreau PN, Lee PJ, Stock AM. Stabilization of the phospho-aspartyl residue in a two-component signal transduction system in Thermotoga maritima. Biochemistry. 1998;37:14575–14584. doi: 10.1021/bi980869i. [DOI] [PubMed] [Google Scholar]
- Grebe TW, Stock JB. The histidine protein kinase superfamily. Adv Microb Physiol. 1999;41:139–227. doi: 10.1016/s0065-2911(08)60167-8. [DOI] [PubMed] [Google Scholar]
- Grimshaw CE, Huang S, Hanstein CG, Strauch MA, Burbulys D, Wang L, Hoch JA, Whiteley JM. Synergistic kinetic interactions between components of the phosphorelay controlling sporulation in Bacillus subtilis. Biochemistry. 1998;37:1365–1375. doi: 10.1021/bi971917m. [DOI] [PubMed] [Google Scholar]
- Hastings CA, Lee SY, Cho HS, Yan D, Kustu S, Wemmer DE. High-resolution solution structure of the beryllofluoride-activated NtrC receiver domain. Biochemistry. 2003;42:9081–9090. doi: 10.1021/bi0273866. [DOI] [PubMed] [Google Scholar]
- Hess JF, Bourret RB, Oosawa K, Matsumura P, Simon MI. Protein phosphorylation and bacterial chemotaxis. Cold Spring Harb Symp Quant Biol. 1988;53:41–48. doi: 10.1101/sqb.1988.053.01.008. [DOI] [PubMed] [Google Scholar]
- Hess JF, Bourret RB, Simon MI. Phosphorylation assays for proteins of the two-component regulatory system controlling chemotaxis in Escherichia coli. Methods Enzymol. 1991;200:188–204. doi: 10.1016/0076-6879(91)00139-n. [DOI] [PubMed] [Google Scholar]
- Hoch JA, Varughese KI. Keeping signals straight in phosphorelay signal transduction. J Bacteriol. 2001;183:4941–4949. doi: 10.1128/JB.183.17.4941-4949.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiak-Spens F, Sparling JM, Gurfinkel M, West AH. Differential stabilities of phosphorylated response regulator domains reflect functional roles of the yeast osmoregulatory SLN1 and SSK1 proteins. J Bacteriol. 1999;181:411–417. doi: 10.1128/jb.181.2.411-417.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiak-Spens F, Sparling DP, West AH. Novel role for an HPt domain in stabilizing the phosphorylated state of a response regulator domain. J Bacteriol. 2000;182:6673–6678. doi: 10.1128/jb.182.23.6673-6678.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Pearson MA, Delany I, Scarlato V, Beier D. Phosphate flow in the chemotactic response system of Helicobacter pylori. Microbiology. 2005;151:3299–3311. doi: 10.1099/mic.0.28217-0. [DOI] [PubMed] [Google Scholar]
- Kaern M, Blake WJ, Collins JJ. The engineering of gene regulatory networks. Annu Rev Biomed Eng. 2003;5:179–206. doi: 10.1146/annurev.bioeng.5.040202.121553. [DOI] [PubMed] [Google Scholar]
- Keener J, Kustu S. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc Natl Acad Sci U S A. 1988;85:4976–4980. doi: 10.1073/pnas.85.14.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ. Programmable cells: interfacing natural and engineered gene networks. Proc Natl Acad Sci U S A. 2004;101:8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Cho HS, Pelton JG, Yan D, Berry EA, Wemmer DE. Crystal structure of activated CheY. Comparison with other activated receiver domains. J Biol Chem. 2001;276:16425–16431. doi: 10.1074/jbc.M101002200. [DOI] [PubMed] [Google Scholar]
- Lewis RJ, Brannigan JA, Muchova K, Barak I, Wilkinson AJ. Phosphorylated aspartate in the structure of a response regulator protein. J Mol Biol. 1999;294:9–15. doi: 10.1006/jmbi.1999.3261. [DOI] [PubMed] [Google Scholar]
- Li L, Shakhnovich EI, Mirny LA. Amino acids determining enzyme-substrate specificity in prokaryotic and eukaryotic protein kinases. Proc Natl Acad Sci U S A. 2003;100:4463–4468. doi: 10.1073/pnas.0737647100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukat GS, Stock AM, Stock JB. Divalent metal ion binding to the CheY protein and its significance to phosphotransfer in bacterial chemotaxis. Biochemistry. 1990;29:5436–5442. doi: 10.1021/bi00475a004. [DOI] [PubMed] [Google Scholar]
- Lukat GS, Lee BH, Mottonen JM, Stock AM, Stock JB. Roles of the highly conserved aspartate and lysine residues in the response regulator of bacterial chemotaxis. J Biol Chem. 1991;266:8348–8354. [PubMed] [Google Scholar]
- Lukat GS, McCleary WR, Stock AM, Stock JB. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci U S A. 1992;89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildvan AS, Weber DJ, Kuliopulos A. Quantitative interpretations of double mutations of enzymes. Arch Biochem Biophys. 1992;294:327–340. doi: 10.1016/0003-9861(92)90692-p. [DOI] [PubMed] [Google Scholar]
- Mildvan AS. Inverse thinking about double mutants of enzymes. Biochemistry. 2004;43:14517–14520. doi: 10.1021/bi048052e. [DOI] [PubMed] [Google Scholar]
- Park S, Meyer M, Jones AD, Yennawar HP, Yennawar NH, Nixon BT. Two-component signaling in the AAA + ATPase DctD: binding Mg2+ and BeF3- selects between alternate dimeric states of the receiver domain. Faseb J. 2002;16:1964–1966. doi: 10.1096/fj.02-0395fje. [DOI] [PubMed] [Google Scholar]
- Perego M. A new family of aspartyl phosphate phosphatases targeting the sporulation transcription factor Spo0A of Bacillus subtilis. Mol Microbiol. 2001;42:133–143. doi: 10.1046/j.1365-2958.2001.02611.x. [DOI] [PubMed] [Google Scholar]
- Pioszak AA, Ninfa AJ. Mutations altering the N-terminal receiver domain of NRI (NtrC) that prevent dephosphorylation by the NRII-PII complex in Escherichia coli. J Bacteriol. 2004;186:5730–5740. doi: 10.1128/JB.186.17.5730-5740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter SL, Armitage JP. Phosphotransfer in Rhodobacter sphaeroides chemotaxis. J Mol Biol. 2002;324:35–45. doi: 10.1016/s0022-2836(02)01031-8. [DOI] [PubMed] [Google Scholar]
- Sanders DA, Gillece-Castro BL, Stock AM, Burlingame AL, Koshland DE., Jr. Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J. Biol. Chem. 1989;264:21770–21778. [PubMed] [Google Scholar]
- Segall JE, Manson MD, Berg HC. Signal processing times in bacterial chemotaxis. Nature. 1982;296:855–857. doi: 10.1038/296855a0. [DOI] [PubMed] [Google Scholar]
- Sheeler NL, MacMillan SV, Nodwell JR. Biochemical activities of the absA two-component system of Streptomyces coelicolor. J Bacteriol. 2005;187:687–696. doi: 10.1128/JB.187.2.687-696.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silversmith RE, Appleby JL, Bourret RB. Catalytic mechanism of phosphorylation and dephosphorylation of CheY: kinetic characterization of imidazole phosphates as phosphodonors and the role of acid catalysis. Biochemistry. 1997;36:14965–14974. doi: 10.1021/bi9715573. [DOI] [PubMed] [Google Scholar]
- Silversmith RE, Smith JG, Guanga GP, Les JT, Bourret RB. Alteration of a nonconserved active site residue in the chemotaxis response regulator CheY affects phosphorylation and interaction with CheZ. J Biol Chem. 2001;276:18478–18484. doi: 10.1074/jbc.M011418200. [DOI] [PubMed] [Google Scholar]
- Silversmith RE, Guanga GP, Betts L, Chu C, Zhao R, Bourret RB. CheZ-mediated dephosphorylation of the Escherichia coli chemotaxis response regulator CheY: role for CheY glutamate 89. J Bacteriol. 2003;185:1495–1502. doi: 10.1128/JB.185.5.1495-1502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silversmith RE, Levin MD, Schilling E, Bourret RB. Kinetic characterization of catalysis by the chemotaxis phosphatase CheZ: Modulation of activity by the phosphorylated CheY substrate. J Biol Chem. 2008;283:756–765. doi: 10.1074/jbc.M704400200. [DOI] [PubMed] [Google Scholar]
- Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Schmitt R. Phosphotransfer between CheA, CheY1, and CheY2 in the chemotaxis signal transduction chain of Rhizobium meliloti. Biochemistry. 1998;37:2327–2335. doi: 10.1021/bi972330a. [DOI] [PubMed] [Google Scholar]
- Stewart RC. Activating and inhibitory mutations in the regulatory domain of CheB, the methylesterase in bacterial chemotaxis. J Biol Chem. 1993;268:1921–1930. [PubMed] [Google Scholar]
- Stewart RC. Kinetic characterization of phosphotransfer between CheA and CheY in the bacterial chemotaxis signal transduction pathway. Biochemistry. 1997;36:2030–2040. doi: 10.1021/bi962261k. [DOI] [PubMed] [Google Scholar]
- Stock AM, Martinez-Hackert E, Rasmussen BF, West AH, Stock JB, Ringe D, Petsko GA. Structure of the Mg2+-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry. 1993;32:13375–13380. doi: 10.1021/bi00212a001. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Swanson RV, Sanna MG, Simon MI. Thermostable chemotaxis proteins from the hyperthermophilic bacterium Thermotoga maritima. J Bacteriol. 1996;178:484–489. doi: 10.1128/jb.178.2.484-489.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H, Muff TJ, Ordal GW. Bacillus subtilis CheC and FliY are members of a novel class of CheY-P-hydrolyzing proteins in the chemotactic signal transduction cascade. J Biol Chem. 2004;279:21787–21792. doi: 10.1074/jbc.M311497200. [DOI] [PubMed] [Google Scholar]
- Toro-Roman A, Mack TR, Stock AM. Structural analysis and solution studies of the activated regulatory domain of the response regulator ArcA: a symmetric dimer mediated by the alpha4-beta5-alpha5 face. J Mol Biol. 2005;349:11–26. doi: 10.1016/j.jmb.2005.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng YL, Hoch JA. Molecular recognition in signal transduction: the interaction surfaces of the Spo0F response regulator with its cognate phosphorelay proteins revealed by alanine scanning mutagenesis. J Mol Biol. 1997;272:200–212. doi: 10.1006/jmbi.1997.1226. [DOI] [PubMed] [Google Scholar]
- Tzeng YL, Feher VA, Cavanagh J, Perego M, Hoch JA. Characterization of interactions between a two-component response regulator, Spo0F, and its phosphatase, RapB. Biochemistry. 1998;37:16538–16545. doi: 10.1021/bi981340o. [DOI] [PubMed] [Google Scholar]
- Varughese KI, Tsigelny I, Zhao H. The crystal structure of beryllofluoride Spo0F in complex with the phosphotransferase Spo0B represents a phosphotransfer pretransition state. J Bacteriol. 2006;188:4970–4977. doi: 10.1128/JB.00160-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt CA. Genetic parts to program bacteria. Curr Opin Biotechnol. 2006;17:548–557. doi: 10.1016/j.copbio.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Wang L, Fabret C, Kanamaru K, Stephenson K, Dartois V, Perego M, Hoch JA. Dissection of the functional and structural domains of phosphorelay histidine kinase A of Bacillus subtilis. J Bacteriol. 2001a;183:2795–2802. doi: 10.1128/JB.183.9.2795-2802.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Kim R, Jancarik J, Yokota H, Kim SH. Crystal structure of phosphoserine phosphatase from Methanococcus jannaschii, a hyperthermophile, at 1.8 Å resolution. Structure. 2001b;9:65–71. doi: 10.1016/s0969-2126(00)00558-x. [DOI] [PubMed] [Google Scholar]
- Wang W, Cho HS, Kim R, Jancarik J, Yokota H, Nguyen HH, Grigoriev IV, Wemmer DE, Kim SH. Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic “snapshots” of intermediate states. J Mol Biol. 2002;319:421–431. doi: 10.1016/S0022-2836(02)00324-8. [DOI] [PubMed] [Google Scholar]
- Weinstein M, Lois AF, Ditta GS, Helinski DR. Mutants of the two-component regulatory protein FixJ of Rhizobium meliloti that have increased activity at the nifA promoter. Gene. 1993;134:145–152. doi: 10.1016/0378-1119(93)90088-k. [DOI] [PubMed] [Google Scholar]
- Wells JA. Additivity of mutational effects in proteins. Biochemistry. 1990;29:8509–8517. doi: 10.1021/bi00489a001. [DOI] [PubMed] [Google Scholar]
- Wolanin PM, Webre DJ, Stock JB. Mechanism of phosphatase activity in the chemotaxis response regulator CheY. Biochemistry. 2003;42:14075–14082. doi: 10.1021/bi034883t. [DOI] [PubMed] [Google Scholar]
- Wright GD, Holman TR, Walsh CT. Purification and characterization of VanR and the cytosolic domain of VanS: a two-component regulatory system required for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry. 1993;32:5057–5063. doi: 10.1021/bi00070a013. [DOI] [PubMed] [Google Scholar]
- Xu Q, Porter SW, West AH. The yeast YPD1/SLN1 complex: insights into molecular recognition in two-component signaling systems. Structure. 2003;11:1569–1581. doi: 10.1016/j.str.2003.10.016. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Hirao K, Oshima T, Aiba H, Utsumi R, Ishihama A. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J Biol Chem. 2005;280:1448–1456. doi: 10.1074/jbc.M410104200. [DOI] [PubMed] [Google Scholar]
- Zapf J, Madhusudan M, Grimshaw CE, Hoch JA, Varughese KI, Whiteley JM. A source of response regulator autophosphatase activity: the critical role of a residue adjacent to the Spo0F autophosphorylation active site. Biochemistry. 1998;37:7725–7732. doi: 10.1021/bi9729615. [DOI] [PubMed] [Google Scholar]
- Zhao R, Collins EJ, Bourret RB, Silversmith RE. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat Struct Biol. 2002;9:570–575. doi: 10.1038/nsb816. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Inouye M. The role of the G2 box, a conserved motif in the histidine kinase superfamily, in modulating the function of EnvZ. Mol Microbiol. 2002;45:653–663. doi: 10.1046/j.1365-2958.2002.03061.x. [DOI] [PubMed] [Google Scholar]


