Abstract
Central and peripheral injections of Ghrelin potently stimulates food intake via its receptor, GHSR1a expressed in the brain. In this study, we explored the role of GHSR1a in the paraventricular nucleus of the hypothalamus (PVN) by reducing their gene expression using the RNA interference (RNAi). pSUPER plasmids inserted with sh (short hairpin)-GHSR1a were injected into the PVN to reduce its expression. The transfected rats were monitored daily for their food intake and body weight throughout the experimental period lasting eight days. We found that knockdown of GHSR1a did not affect daily food intake but significantly reduced body weight and blood ghrelin levels. This suggests that the central ghrelin system could selectively regulate body weight without affecting energy intake.
Introduction
Ghrelin is predominantly produced in the stomach and in the gastrointestinal tract but small quantities have been found in the brain and many other tissues such as pituitary, pancreas, testes, etc [4, 6, 9, 11]. Ghrelin is a 28 amino acid peptide found in mainly two forms in blood plasma, one acylated at Ser3 with medium chain fatty acid, usually n-octanoic acid and the other, des-acyl ghrelin [5, 8]. Acylated ghrelin (AG) is the endogenous ligand for the growth hormone secretagogue receptor type 1a (GHSR1a) whereas des-acyl ghrelin (DAG) is not [17]. GHSR1a mediates AG stimulation of growth hormone (GH) release and food intake. GHSR1a is predominantly expressed in the pituitary, and at lower levels in the hypothalamic nuclei including the arcuate (ARC), paraventricular (PVN), ventromedial (VMH), and in some brainstem nuclei [10, 24].
Peripheral as well as central AG administration induces increases in food intake and body weight in rodents [21, 22]. AG’s orexigenic effect is completely abolished in NPY−/−, AgRP−/− mice demonstrating that NPY and AgRP are obligatory mediators for this effect [3]. Also, NPY and AgRP are found co-localized in the ARC, and ablation of the ARC by monosodium glutamate inhibited the orexigenic effect of intra-cerebroventricular (ICV) infused AG, but not the stimulation of GH from the pituitary, suggesting that mechanisms involved in the two effects are separate [18]. Antagonism of GHSR1a by ICV injection of GHRP-6 [D-Arg-1, D-Phe-5, Dtrp-7, 9 Leu-11], reduced food intake and body weight [1].
GHSR1a-KO mice showed a modest decrease in body weight when maintained on standard chow [17]. Yet in another study, GHSR−/− mice maintained on a High Fat Diet (HFD), had reduced food intake and body weight, which was attributed mainly to loss in fat mass compared to their wild types. The transgenic mice also showed a reduced respiratory quotient suggesting lipid mobilization [25].
In this study, we investigated the effect of knocking down ghsr1a gene in the PVN of rats using RNA interference (RNAi) technology. We transfected the PVN in vivo with plasmids to produce short hairpin RNA (shRNA) targeted to selectively knockdown ghsr1a mRNA and studied the effects of its knockdown on food intake and body weight in freely-fed male rats on standard chow diet. We hypothesized that reduction of ghsr1a expression in the PVN will reduce food intake and body weight.
Materials and Methods
Animals
Male Sprague-Dawley rats (SD) (Harlan, NC, USA) weighing 250–300 grams were housed in conventional hanging cages with a 12 hrs light/12 hrs dark photoperiod (lights on at 07:00) in a temperature-controlled room (21–22 °C). Rats had ad libitum access to standard chow and water throughout the experiment. Body weight and food intake, corrected for spillage, were monitored daily at 10:00 hrs from the time of arrival to the facility until the end of the experimental period except during the 2 days post surgery. All animal procedures were conducted in accordance with established guidelines of the University of Georgia Institutional Animal Care and Use Committee.
Construction of RNAi vectors
pSUPER (SUPpression of Endogenous RNA), carrying human H1 promoter, was purchased from OligoEngine (Seattle, WA, USA). To construct the sh-ghsr-pSUPER vector, a pair of 60-nt oligonucleotides, forward and reverse, each containing a unique 19-nt sequence derived from within the target mRNA transcripts of ghsr1a gene (Table 1), was annealed and ligated between the XhoI/BglII sites of the pSUPER. Within the 60-nt oligos, the 19-nt target sequence appears in both sense and anti-sense orientation, separated by a 9-nt spacer sequence.
Table 1.
Sequence of 60-nt oligonucleotides designed using the software provided online at the manufacturer website, http://www.oligoengine.com and cloned into the pSUPER
| Gene | Accession # | Sense strand | Anti-sense strand |
|---|---|---|---|
| GHSR1a | NM_032075 | GATCCCCcgactcactgcctgacgaa | TCGAGAAAAAcgactcactgcctgacgaa |
| TTCAAGAGAttcgtcaggcagtgagtcgTTTTTC | TCTCTTGAAttcgtcaggcagtgagtcgGGG |
Small letters indicate sequences of respective target genes
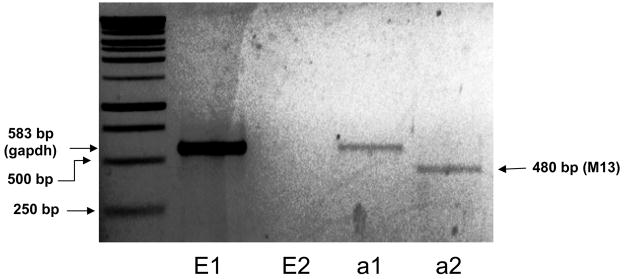
Briefly, annealed 60-nt oligos and pSUPER, which was double digested with BglII and XhoI (New England Biolabs, MA, USA), was gel purified and ligated using DNA ligase (Invitrogen, CA, USA) overnight. The plasmids were transformed into MAX Efficiency DH5α competent cells (Invitrogen, CA, USA) and were grown in LB-Agar plates supplemented with ampicillin (100μg/mL) (Invitrogen-Gibco, NY, USA). Several colonies were picked and PCR-screened using the M13 primers, where negative colony gave a 438 bp and positive around 480 bp products (fig 1). Positive colonies were further grown in LB media supplemented with ampicillin (100μg/mL) and the plasmids were extracted and purified using Qiagen plasmid Midi purification kit (Qiagen, CA, USA). Purified positive clones were sequenced and clones with perfect match to the inserted sequences were used in the study.
Figure 1.
1μg DNA containing sh-ghsr pSUPER (a) or just vehicle (E, not including pSUPER) was injected bilaterally at the PVN, suspended in 1μl of the NeuroFECT transfection reagent. After 8 days of transfection, RNA was isolated from the PVN and reverse transcribed. cDNAs were PCR-amplified using primers for M13. PVN samples injected with sh-ghsr pSUPER showed a PCR product at 480bp (lane a 2) whereas PVN samples injected with transfection reagent only (E 2) showed no PCR product. All samples were also PCR amplified with gapdh primers as positive controls (E 1 & a 1; 583bp).
Primer pair sequences for gapdh (forward:ctgagaatgggaagctggtcatca and reverse: tcatacttggcaggtttctccagga; and accession number is NM_017008)
In vivo brain transfection
Plasmids were resuspended in NeuroFECT transfection reagent (Genlantis, Gene Therapy Systems, Inc, San Diego, CA) at the ratio of 6:1 (NeuroFECT: DNA, wt/wt) and incubated at room temperature for 30 mins before injection into the brain of rats. In vivo transfection into rat brain with NeuroFECT was confirmed using sh-ghsr (fig 1). Briefly, a total volume of one μl each of the DNA: Transfection reagent complex was injected bilaterally into the PVN (coordinates; 1.9mm posterior from bregma, ±0.5mm lateral, and 8.5mm below the skull based on the rat brain atlas [15] over a period of 10 minutes in anesthetized rats that were fixed on a stereotaxic apparatus. Ten days post injection, brains were removed and PVN was bilaterally punched (puncher diameter: 1 millimeter) at 1.9 A/P bregma level and immediately frozen in liquid nitrogen. Total RNA was extracted using TRIzol reagent (Invitrogen, CA) and 1μg of total RNA from pooled bilateral PVN punches from each rat was reverse-transcribed using ImProm-II™ Reverse Transcription System (Promega, Madison, WI) followed by PCR reaction with GoTaq Green Master Mix (Promega, Madison, WI) using the primers for M13. A touchdown PCR was performed where the annealing temperature was raised by 8°C above the optimal for the first 25 cycles of PCR and for every subsequent 2 cycles, the annealing temperature was decreased by 1°C, thus minimizing non-specific PCR products.
In the study, SD rats were acclimatized for 1 week then divided into 2 groups, where each group consisted of 6 rats with similar body weights. The two different DNA: NeuroFECT complexes at similar ratios as mentioned above, including control-pSUPER: NeuroFECT and sh-ghsr-pSUPER: NeuroFECT were injected bilaterally into the PVN with the same procedure as mentioned above.
Radioimmunoassay of plasma ghrelin level
After decapitation of rats, trunk blood was collected and subsequently centrifuged at 1500× g for 20 min at 4°C. Blood plasma was transferred into new Eppendorf tubes containing EDTA/2Na (1.5mg/mL), aprotinin (500U/mL), and 1N HCl (26.5μL/mL) to prevent acylated ghrelin degradation and stored at −80°C until assay of total ghrelin by RIA kit from Linco Research (Millipore, MA, USA). Each plasma sample was assayed in triplicate and, only the samples that had the intra-assay coefficient of variation of below 10% were used in the statistical analysis.
Real-Time PCR
Total RNA was extracted from frozen PVN, interscapular brown adipose tissue (IBAT), epididymal white adipose tissue (EWAT), and sartorius muscle tissue (SMT) using the TRIzol reagent (Invitrogen, CA, USA). RNA was first quantified using the spectrophotometer (absorbance readings at 260) and subsequently screened for its integrity by running 2μg of each RNA sample on a 1.5% denaturing agarose gel. RNA samples that had intact ribosomal 28S and 18 S bands were treated with DNase using the DNA-free ™ kit (Ambion, TX, USA). Then, 1 μg of the RNA samples were primed with random hexamers and used to synthesize first strand cDNAs using the ImProm-II™ Reverse Transcription System (Promega, WI, USA) in a 20-μL reaction at 42° C for 45 min. At the end of the cDNA synthesis, the reverse transcription was inactivated by incubating at 70° C for 15 min.
Real time PCR was performed on MyiQ Single-color Real time PCR Detection system (BioRad, Hercules, CA). We measured the expression levels of ghsr1a in the PVN and ucp1, ucp2 and ucp3 in the IBAT, EWAT and SMT, respectively. 2 uL of the cDNA reaction was amplified in a 10 μL PCR reaction using the iQ™ SYBR® Green Supermix (BioRad, Hercules, CA). The primer pairs and the size of expected PCR product of the genes are shown in Table 2. Gapdh mRNA was amplified as a housekeeping gene. All the primer pairs were designed using the freely available online software, Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and were synthesized by Integrated DNA Technologies, Inc (IA, USA). PCR reactions were initiated with a 3 min incubation at 95° C to activate the hot-start enzyme, iTaq™ DNA polymerase. PCR fragments were amplified for 45 cycles of 95° C for 30 sec, 60° C for 30 sec, and 1 min at 72° C followed by a melting curve to confirm the specificity of the amplification products. Negative wells (without template) were also included with each primer set to ensure absence of primer-dimer formations. PCR efficiencies ranged between 95 to 103 %, thus we used the ΔΔCT approach, which measures the change in threshold values for the genes of interest in the samples relative to the housekeeping gene (gapdh).
Table 2.
Sequences of the primers used in the real time PCR analysis
| Gene | Forward | Reverse | Product size (bp) | Reference |
|---|---|---|---|---|
| ghsr1a | tgggtgtccagcgtcttcttcttt | Caaacaccaccacagcaagcatct | 166 | |
| gapdh | agacagccgcatcttcttgt | Cttgccgtgggtagagtcat | 207 | |
| Ucp1 | Tccctcaggattggcctctac | Gtcatcaagccagccgagat | [16] | |
| Ucp2 | gcattggcctctacgactctctgtaa | Atggtcttgtaggcttcgacagtg | 211 | |
| Ucp3 | Accctgttttggttgcaaag | Gagtccatcctgtccttcca | 176 |
Statistical analysis
Data are represented as mean± SE for each treatment group. The data from the food intake and body weight measurements were analyzed by a repeated measure one-factor ANOVA analysis followed by post hoc Fisher’s protected least significant difference (PLSD) test. Data from the RIA assay were analyzed by a one-way ANOVA analysis followed by post hoc Student-Newman-Keuls test. Data are expressed as least square means ± S.E.M., with consideration of significance at p<0.05. Statistical analyses were performed using either the SuperANOVA (Abacus Concepts, Inc., Berkeley, CA) and or Stat-View program (SAS Institute Inc., Cary, NC) where appropriate.
Results
Effect of sh-ghsr pSUPER on ghsr gene expression
We confirmed successful in vivo transfection of pSUPER vector using the NeuroFECT transfection reagent at the PVN by performing PCR with the M13 primers as described in the materials and methods section (Fig 5). Positive clones of sh-ghsr pSUPER along with empty vector (transfection reagent without vector) were transfected bilaterally into the PVN. Only sh-ghsr pSUPER transfected PVN samples (Fig 1, lane a2) show a PCR product at 480bp whereas the empty vector transfected PVN samples do not show a band at 480bp (Fig 1, lane E2). PCR was also performed with gapdh primers as positive control that shows a band at 583bp in both samples (Fig 1, lanes E1 & a1).
Figure 5.
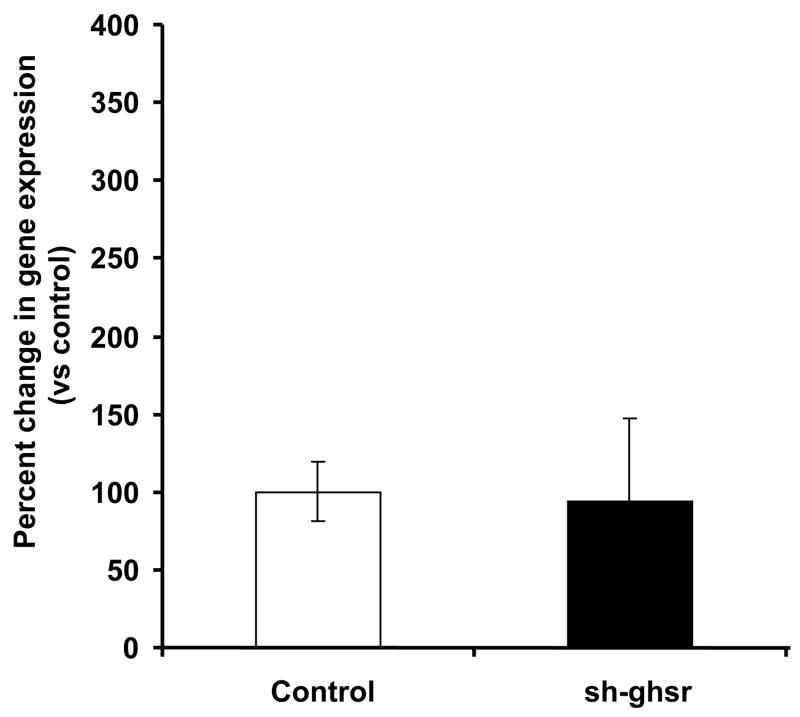
(a) ucp1 mRNA levels in the IBAT transfected with either control pSUPER (white bar) or sh-ghsr pSUPER (black bar) after 8 days of transfection. ucp1 expression is normalized with gapdh expression and represented as relative percent change compared to control. Data are represented as means ±SE. n = 4 per group.
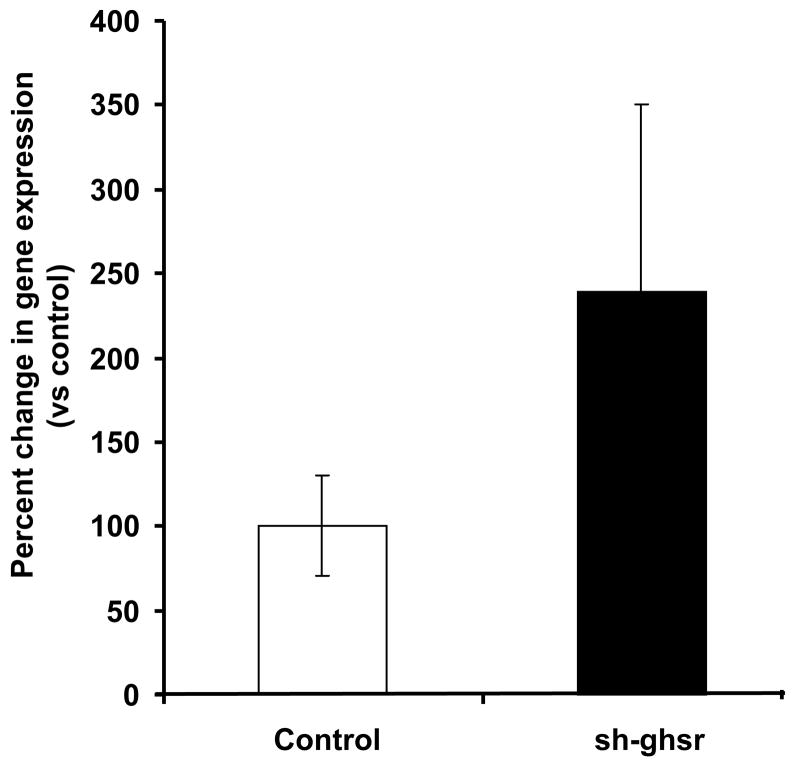
(b) ucp2 mRNA levels in the EWAT transfected with either control pSUPER (white bar) or sh-ghsr pSUPER (black bar) after 8 days of transfection. Ucp2 expression is normalized with gapdh expression and represented as relative percent change compared to control. Data are represented as means ±SE. n = 4 per group.
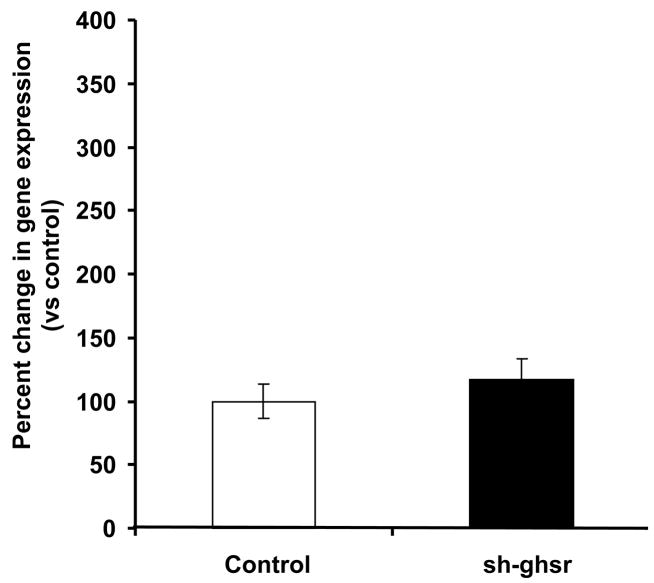
(c) ucp3 mRNA levels in the SMT transfected with either control pSUPER (white bar) or sh-ghsr pSUPER (black bar) after 8 days of transfection. Ucp3 expression is normalized with gapdh expression and represented as relative percent change compared to control. Data are represented as means ±SE. n = 4 per group.
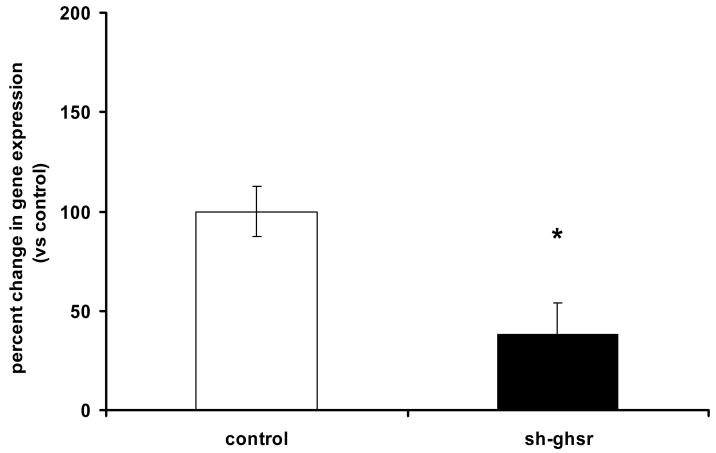
Transfection with sh-ghsr pSUPER significantly reduced ghsr mRNA expression by approximately 60% at the PVN after 8 days of transfection (Fig 2, p=0.04).
Figure 2.
(a) ghsr1a mRNA levels in the PVN transfected with control pSUPER (without insert; control group) and sh-ghsr pSUPER after 8 days of transfection. ghsr1a expression is normalized with gapdh expression and represented as relative percent change compared to control Data which are presented as means ±SE and Asterisks represent significant difference between bars (p<0.05). n = 4 per group.
Effect of sh-ghsr pSUPER on food intake and body weight
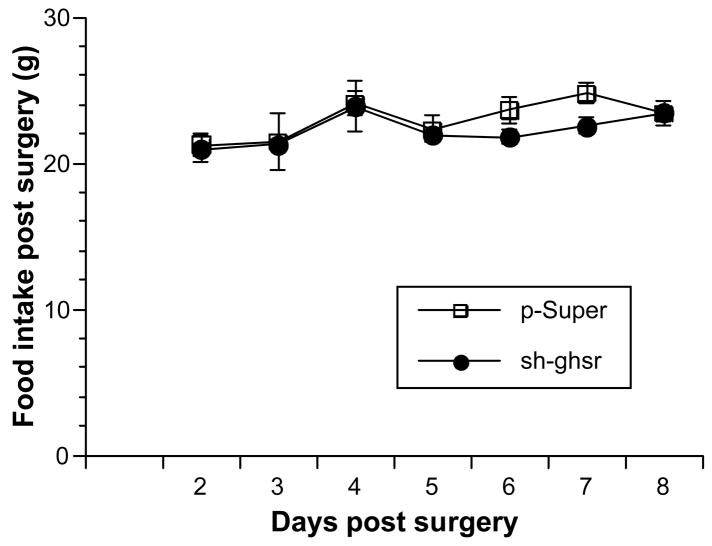
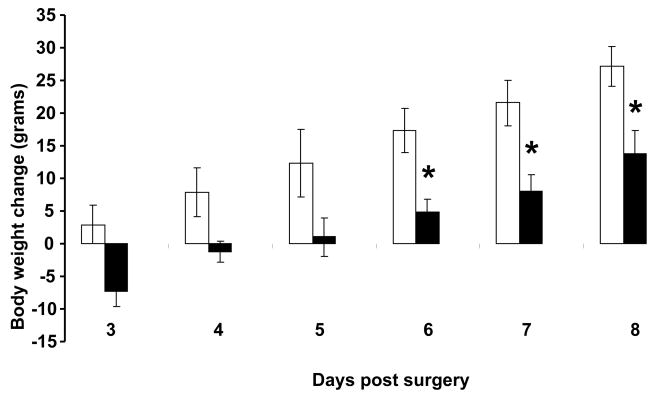
We found no change in 24h cumulative food intake between the sh-ghsr pSUPER vs. control (Fig 3a) group rats. However, we found a significant decrease in body weight gain in sh-ghsr pSUPER transfected rats compared to the control group (Fig 3b). Rats treated with sh-ghsr pSUPER showed reduced gain in body weight vs. control; by 75% reduction on day 6, which is 8 days post transfection (p=0.0276), by 70% on day 7 (p=. 0151) and by 50 % on day 8. Body weights of rats transfected with control pSUPER increased by 26.5g, whereas the sh-ghsr pSUPER rats gained approximately 14g over 8 days from the day of surgery. This is a remarkable difference of an average 12.5g reduction in body weight gain in sh-ghsr pSUPER group over an 8-day period when we consider that the 24h cumulative food intake remained unchanged in this group compared to controls.
Figure 3.
(a) Daily 24h food intake post surgery. Open squares represent control pSUPER and black circles represent sh-ghsr pSUPER transfected rats. Data are represented as means ±SE. n = 6 per group.
(b) Change in daily body weight in each individual rat from its original body weight at the time of surgery was calculated and the data presented in the figure is the means of the change in body weight in rats in a group from day 2 (24 h post surgery). White bars represent control pSUPER and gray bars represent sh-ghsr pSUPER transfected rats. Data are represented as means ±SE and Asterisks represent significant difference of experimental vs. control (p<0.05). n = 6 per group.
Effect of sh-ghsr pSUPER on blood ghrelin level
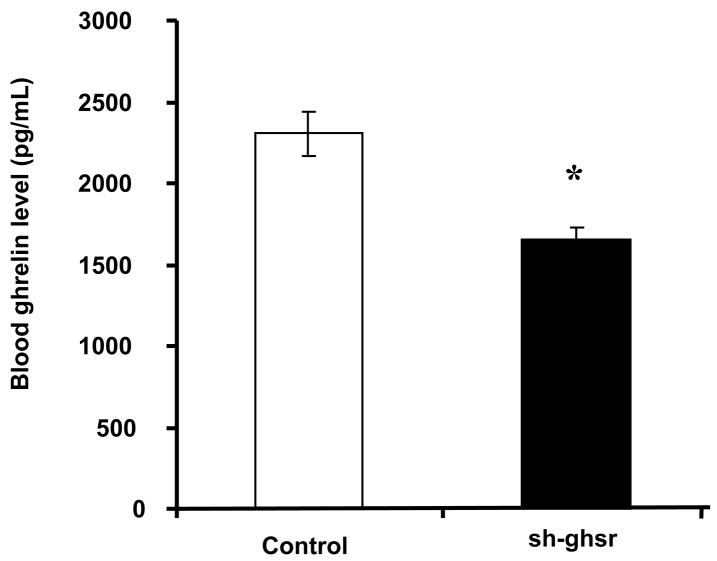
Blood total ghrelin is a good surrogate for measurement of acylated ghrelin, which binds to GHSR1a to stimulate food intake and growth hormone release. We found that blood ghrelin level was significantly decreased in the sh-ghsr pSUPER group (1658.57 ± 66.4(SE) pg/mL) compared to the control (2305.4 pg/mL ± 136.94(SE) pg/mL) (Fig 4, p=. 0003). Blood ghrelin level was reduced in the sh-ghsr pSUPER group by approximately 30% from the controls.
Figure 4.
Blood Total ghrelin levels measured in the trunk blood collected at the end of the 8 days of the experiment. White bars represent control pSUPER and black bars represent sh-ghsr pSUPER transfected rats. Data are represented as means ±SE and Asterisks represent significant difference of experimental vs control (p<0.05). n = 6 per group.
Effect of sh-ghsr pSUPER on UCPs in brown and white adipose tissues, and muscle tissue
We did not see differences in ucp1 expression levels in IBAT of the sh-ghsr pSUPER group compared to the controls (Fig 5a). We found a trend toward an increase in ucp2 expression in EWAT of the sh-ghsr pSUPER vs. the controls (Fig 5b) but did not reach significance. In the SMT, ucp3 expression levels also remained unchanged between the sh-ghsr pSUPER and the control groups (Fig 5c).
Discussion
We demonstrated that sh-ghsr pSUPER transfected in vivo into the PVN of rats potently and significantly reduced the ghsr mRNA expression by approximately 60% of that in the control as measured by real time PCR analysis. Plasmid encoded sh-RNAs have been demonstrated to induce a potent and specific RNAi in adult mice [13]. Moreover, pSUPER induced sh-RNAs have been demonstrated to reduce target mRNAs potently and specifically in rodents in vivo [2, 12].
Interestingly, knockdown of the ghsr at the PVN did not cause a decrease in food intake even though others have shown a reduction in food intake with central injections of ghsr antagonists [1]. Also GHSR-null mice showed no change in food intake when fed standard chow, but when fed a HFD, ate less food and accumulated less body weight and adiposity than control mice [25]. Also, only female mice when fed standard chow showed reduced body weight and adiposity suggesting a possible role of gender on the ghrelin system with respect to diet type and energy balance [25]. Nevertheless, we observed a significant decrease in body weight in our male rats when fed a standard chow suggesting that knockdown of ghsr in adult rats might play a role in body weight regulation. Furthermore, a similar but modest decrease in body weight was reported in novel GHSR-knockout mice when fed standard chow [17].
Interestingly, in the sh-ghsr pSUPER group we found that blood ghrelin level was significantly reduced compared to the control. PVN is one of the major sites for the autonomic nerves output, which connects the brain and the gut through its two branches including the vagus and sympathetic nervous system. Therefore, it is logical to expect that if such an interaction between central and peripheral ghrelin does exist, then autonomic nerves could be a likely mediator in this interaction. However, a blood borne pathway cannot be abandoned in this interaction. In a recent study, it was reported that vaccination against ghrelin potently and significantly reduced body weight gain without any affect on food intake, thus markedly reducing feed efficiency (i.e., body weight gain per unit energy intake) [26]. Taken together, we speculate that the decrease in blood ghrelin level observed in our sh-ghsr pSUPER group had some influence on their reduced body weight gain, however it remains to be determined if this is true or not.
Therefore, in order to understand the mechanism of the cause of decrease in body weight gain seen in the sh-ghsr pSUPER group, we looked into some of the genes that are involved in energy expenditure. We measured gene expression of ucp1 in brown adipose tissue because central administration of ghrelin has been shown to suppress sympathetic activity in brown adipose tissue and decrease ucp1 expression [23]. We also studied the expression of ucp2 in white adipose tissue because it has been linked to obesity and body weight regulation [14]. Finally, we also measured the expression of ucp3 in the soleus muscle because it is mainly found in skeletal muscle and suggested to be involved in energy expenditure [7]. Unexpectedly, we did not see any changes in ucp1, ucp2 or ucp3 expressions at their respective tissues compared to their control groups. However, there does appear to be a trend in increased expression of ucp2 in the EWAT with an approximately 2.5-fold increase in the sh-ghsr pSUPER group compared to the controls. But, this did not reach a significant level, likely due to variability within groups and also due to the small sample number.
Central ghrelin infusion decreased spontaneous locomotor activity in rats [19]. Therefore, reduction in central ghrelin receptor could cause an opposite effect, which is an increase in locomotor activity, thus increasing energy expenditure causing weight loss in our experimental rats. However, we did not measure the locomotor activity of our rats; therefore we do not know the affect of reduction of central ghrelin receptor on this behavior. Chronic central ghrelin infusion, independent of its effect on food intake, has been shown to markedly increase fat storage-promoting enzymes such as lipoprotein lipase, acetylCoA carboxylase α, fatty acid synthase and a correspondingly, decrease in enzymes responsible for fat oxidation such as carnitine palmitoyl transferase-1 α [20]. In line with these reports, we think an opposite phenomenon could be expected to occur in our sh-ghsr pSUPER group, where reduction in central ghrelin receptor caused a decrease in the fat storage-promoting enzymes but an increase in fat-oxidation enzymes. To test this, future studies will be required to focus on measuring the metabolic enzymes including those that have been mentioned above. In addition, use of indirect caloriemetry and dual-emission x-ray absorption (DEXA) may better reveal the mechanisms involved in this phenomenon. Lastly, it may also be the case that the loss in body weight gain we observed in our rats, may be due to the loss of body fluid. Since body fluid constitutes about ~60% of body weight in mammals, it is possible that the loss in body fluid could have accounted for the decreased body weight in these rats. However, in this study we did not measure either water intake or body fluid releases in the sh-ghsr pSUPER or control animals.
In conclusion, we found that knockdown of the central ghrelin receptor expression at the PVN of adult rats can affect its body weight without influencing food intake. This approach of RNA interference of ghsr1a expression in the brain of adult rats is advantageous over knockout models because it seems to reveal a new dimension of the central role of ghrelin, which would not have been possible to investigate in transgenic mice. More importantly, site-specific knockdown of ghsr1a at different nuclei in the hypothalamus has the potential to examine the role of the specific site on energy homeostasis induced by ghrelin.
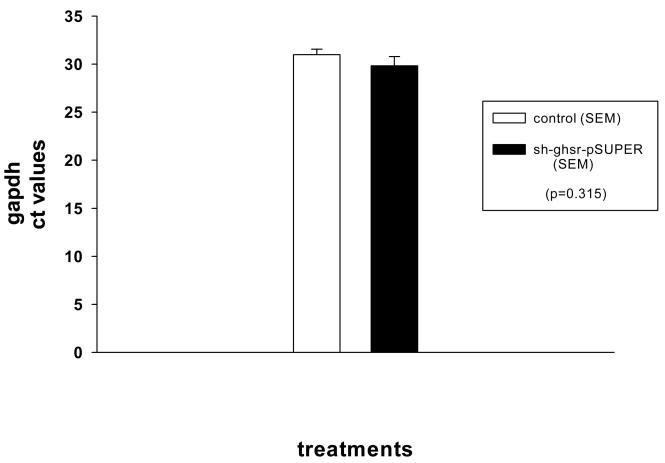
Figure A. Real time PCR gapdh CT values in PVN samples from control and sh-ghsr-pSUPER groups.
gapdh CT values in PVN samples run in duplicates from each group were statistically analyzed using the one-way ANOVA. Data is expressed as mean with ±SEM. White bar represents control group whereas black bar sh-ghsr-pSUPER group, p=0.315.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Asakawa A, Inui A, Kaga T, Katsuura G, Fujimiya M, Fujino MA, et al. Antagonism of ghrelin receptor reduces food intake and body weight gain in mice. Gut. 2003;52:947–52. doi: 10.1136/gut.52.7.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science (New York, NY. 2002;296:550–3. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 3.Chen HY, Trumbauer ME, Chen AS, Weingarth DT, Adams JR, Frazier EG, et al. Orexigenic action of peripheral ghrelin is mediated by neuropeptide Y and agouti-related protein. Endocrinology. 2004;145:2607–12. doi: 10.1210/en.2003-1596. [DOI] [PubMed] [Google Scholar]
- 4.Cowley MA, Smith RG, Diano S, Tschop M, Pronchuk N, Grove KL, et al. The distribution and mechanism of action of ghrelin in the CNS demonstrates a novel hypothalamic circuit regulating energy homeostasis. Neuron. 2003;37:649–61. doi: 10.1016/s0896-6273(03)00063-1. [DOI] [PubMed] [Google Scholar]
- 5.Date Y, Kojima M, Hosoda H, Sawaguchi A, Mondal MS, Suganuma T, et al. Ghrelin, a novel growth hormone-releasing acylated peptide, is synthesized in a distinct endocrine cell type in the gastrointestinal tracts of rats and humans. Endocrinology. 2000;141:4255–61. doi: 10.1210/endo.141.11.7757. [DOI] [PubMed] [Google Scholar]
- 6.Gnanapavan S, Kola B, Bustin SA, Morris DG, McGee P, Fairclough P, et al. The tissue distribution of the mRNA of ghrelin and subtypes of its receptor, GHS-R, in humans. J Clin Endocrinol Metab. 2002;87:2988. doi: 10.1210/jcem.87.6.8739. [DOI] [PubMed] [Google Scholar]
- 7.Hesselink MK, Mensink M, Schrauwen P. Human uncoupling protein-3 and obesity: an update. Obesity research. 2003;11:1429–43. doi: 10.1038/oby.2003.192. [DOI] [PubMed] [Google Scholar]
- 8.Hosoda H, Kojima M, Matsuo H, Kangawa K. Ghrelin and des-acyl ghrelin: two major forms of rat ghrelin peptide in gastrointestinal tissue. Biochem Biophys Res Commun. 2000;279:909–13. doi: 10.1006/bbrc.2000.4039. [DOI] [PubMed] [Google Scholar]
- 9.Hosoda H, Kojima M, Matsuo H, Kangawa K. Purification and characterization of rat des-Gln14-Ghrelin, a second endogenous ligand for the growth hormone secretagogue receptor. J Biol Chem. 2000;275:21995–2000. doi: 10.1074/jbc.M002784200. [DOI] [PubMed] [Google Scholar]
- 10.Howard AD, Feighner SD, Cully DF, Arena JP, Liberator PA, Rosenblum CI, et al. A receptor in pituitary and hypothalamus that functions in growth hormone release. Science (New York, NY. 1996;273:974–7. doi: 10.1126/science.273.5277.974. [DOI] [PubMed] [Google Scholar]
- 11.Kojima M, Hosoda H, Date Y, Nakazato M, Matsuo H, Kangawa K. Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature. 1999;402:656–60. doi: 10.1038/45230. [DOI] [PubMed] [Google Scholar]
- 12.Makimura H, Mizuno TM, Mastaitis JW, Agami R, Mobbs CV. Reducing hypothalamic AGRP by RNA interference increases metabolic rate and decreases body weight without influencing food intake. BMC neuroscience. 2002;3:18. doi: 10.1186/1471-2202-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. RNA interference in adult mice. Nature. 2002;418:38–9. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 14.Memon RA, Hotamisligil GS, Wiesbrock SM, Uysal KT, Faggioni R, Moser AH, et al. Upregulation of uncoupling protein 2 mRNA in genetic obesity: lack of an essential role for leptin, hyperphagia, increased tissue lipid content, and TNF-alpha. Biochimica et biophysica acta. 2000;1484:41–50. doi: 10.1016/s1388-1981(99)00195-x. [DOI] [PubMed] [Google Scholar]
- 15.Paxinos G, Watson C. The Rat Brain in stereotaxic coordinates. 4. Academic Press; 2003. [DOI] [PubMed] [Google Scholar]
- 16.Penn DM, Jordan LC, Kelso EW, Davenport JE, Harris RB. Effects of central or peripheral leptin administration on norepinephrine turnover in defined fat depots. American journal of physiology. 2006;291:R1613–21. doi: 10.1152/ajpregu.00368.2006. [DOI] [PubMed] [Google Scholar]
- 17.Sun Y, Wang P, Zheng H, Smith RG. Ghrelin stimulation of growth hormone release and appetite is mediated through the growth hormone secretagogue receptor. Proc Natl Acad Sci U S A. 2004;101:4679–84. doi: 10.1073/pnas.0305930101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamura H, Kamegai J, Shimizu T, Ishii S, Sugihara H, Oikawa S. Ghrelin stimulates GH but not food intake in arcuate nucleus ablated rats. Endocrinology. 2002;143:3268–75. doi: 10.1210/en.2002-220268. [DOI] [PubMed] [Google Scholar]
- 19.Tang-Christensen M, Vrang N, Ortmann S, Bidlingmaier M, Horvath TL, Tschop M. Central administration of ghrelin and agouti-related protein (83–132) increases food intake and decreases spontaneous locomotor activity in rats. Endocrinology. 2004;145:4645–52. doi: 10.1210/en.2004-0529. [DOI] [PubMed] [Google Scholar]
- 20.Theander-Carrillo C, Wiedmer P, Cettour-Rose P, Nogueiras R, Perez-Tilve D, Pfluger P, et al. Ghrelin action in the brain controls adipocyte metabolism. J Clin Invest. 2006;116:1983–93. doi: 10.1172/JCI25811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tschop M, Smiley DL, Heiman ML. Ghrelin induces adiposity in rodents. Nature. 2000;407:908–13. doi: 10.1038/35038090. [DOI] [PubMed] [Google Scholar]
- 22.Wren AM, Small CJ, Ward HL, Murphy KG, Dakin CL, Taheri S, et al. The novel hypothalamic peptide ghrelin stimulates food intake and growth hormone secretion. Endocrinology. 2000;141:4325–8. doi: 10.1210/endo.141.11.7873. [DOI] [PubMed] [Google Scholar]
- 23.Yasuda T, Masaki T, Kakuma T, Yoshimatsu H. Centrally administered ghrelin suppresses sympathetic nerve activity in brown adipose tissue of rats. Neurosci Lett. 2003;349:75–8. doi: 10.1016/s0304-3940(03)00789-4. [DOI] [PubMed] [Google Scholar]
- 24.Zigman JM, Jones JE, Lee CE, Saper CB, Elmquist JK. Expression of ghrelin receptor mRNA in the rat and the mouse brain. J Comp Neurol. 2006;494:528–48. doi: 10.1002/cne.20823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zigman JM, Nakano Y, Coppari R, Balthasar N, Marcus JN, Lee CE, et al. Mice lacking ghrelin receptors resist the development of diet-induced obesity. J Clin Invest. 2005;115:3564–72. doi: 10.1172/JCI26002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zorrilla EP, Iwasaki S, Moss JA, Chang J, Otsuji J, Inoue K, et al. Vaccination against weight gain. Proc Natl Acad Sci U S A. 2006;103:13226–31. doi: 10.1073/pnas.0605376103. [DOI] [PMC free article] [PubMed] [Google Scholar]