Abstract
Background
The genome sequence of Geobacter metallireducens is the second to be completed from the metal-respiring genus Geobacter, and is compared in this report to that of Geobacter sulfurreducens in order to understand their metabolic, physiological and regulatory similarities and differences.
Results
The experimentally observed greater metabolic versatility of G. metallireducens versus G. sulfurreducens is borne out by the presence of more numerous genes for metabolism of organic acids including acetate, propionate, and pyruvate. Although G. metallireducens lacks a dicarboxylic acid transporter, it has acquired a second putative succinate dehydrogenase/fumarate reductase complex, suggesting that respiration of fumarate was important until recently in its evolutionary history. Vestiges of the molybdate (ModE) regulon of G. sulfurreducens can be detected in G. metallireducens, which has lost the global regulatory protein ModE but retained some putative ModE-binding sites and multiplied certain genes of molybdenum cofactor biosynthesis. Several enzymes of amino acid metabolism are of different origin in the two species, but significant patterns of gene organization are conserved. Whereas most Geobacteraceae are predicted to obtain biosynthetic reducing equivalents from electron transfer pathways via a ferredoxin oxidoreductase, G. metallireducens can derive them from the oxidative pentose phosphate pathway. In addition to the evidence of greater metabolic versatility, the G. metallireducens genome is also remarkable for the abundance of multicopy nucleotide sequences found in intergenic regions and even within genes.
Conclusion
The genomic evidence suggests that metabolism, physiology and regulation of gene expression in G. metallireducens may be dramatically different from other Geobacteraceae.
Background
Geobacter metallireducens is a member of the Geobacteraceae, a family of Fe(III)-respiring Delta-proteobacteria that are of interest for their role in cycling of carbon and metals in aquatic sediments and subsurface environments as well as the bioremediation of organic- and metal-contaminated groundwater and the harvesting of electricity from complex organic matter [1,2]. G. metallireducens is of particular interest because it was the first microorganism found to be capable of a number of novel anaerobic processes including: (1) conservation of energy to support growth from the oxidation of organic compounds coupled to the reduction of Fe(III) or Mn(IV) [3,4]; (2) conversion of Fe(III) oxide to ultrafine-grained magnetite [3]; (3) anaerobic oxidation of an aromatic hydrocarbon [5,6]; (4) reduction of U(VI) [7]; (5) use of humic substances as an electron acceptor [8]; (6) chemotaxis toward metals [9]; (7) complete oxidation of organic compounds to carbon dioxide with an electrode serving as the sole electron acceptor ([10]; and (8) use of a poised electrode as a direct electron donor [11]. Although the complete genome sequence of the closely related Geobacter sulfurreducens is available [12] and can provide insights into some of the common metabolic features of Geobacter species, G. metallireducens and G. sulfurreducens are significantly different in many aspects of their physiology. G. sulfurreducens is known to use only four carbon sources: acetate, formate, lactate (poorly) and pyruvate (only with hydrogen as electron donor), whereas G. metallireducens uses acetate, benzaldehyde, benzoate, benzylalcohol, butanol, butyrate, p-cresol, ethanol, p-hydroxybenzaldehyde, p-hydroxybenzoate, p-hydroxybenzylalcohol, isobutyrate, isovalerate, phenol, propionate, propanol, pyruvate, toluene and valerate [2].
Therefore, in order to gain broader insight into the physiological diversity of Geobacter species, the genome of G. metallireducens was sequenced and compared to that of Geobacter sulfurreducens [12]. Both genome annotations were manually curated with the addition, removal and adjustment of hundreds of protein-coding genes and other features. Phylogenetic analyses were conducted to validate the findings, including homologs from the finished and unfinished genome sequences of more distantly related Geobacteraceae. This paper presents insights into the conserved and unique features of two Geobacter species, particularly the metabolic versatility of G. metallireducens and the numerous families of multicopy nucleotide sequences in its genome, which suggest that regulation of gene expression is very different in these two species.
Results and Discussion
Contents of the two genomes
The automated annotation of the G. metallireducens genome identified 3518 protein-coding genes on the chromosome of 3997420 bp and 13 genes on the plasmid (designated pMET1) of 13762 bp. Manual curation added 59 protein-coding genes plus 56 pseudogenes to the chromosome and 4 genes to the plasmid. Ten of the chromosomal genes were reannotated as pseudogenes and another 22 were removed from the annotation. In addition to the 58 RNA-coding genes in the automated annotation, manual curation identified 479 conserved nucleotide sequence features. Likewise, to the 3446 protein-coding genes in the automated annotation of the G. sulfurreducens genome [12], manual curation added 142 protein-coding genes and 19 pseudogenes. Five genes were reannotated as pseudogenes and 103 genes were removed from the annotation. In addition to the 55 RNA-coding genes in the automated annotation, manual curation identified 462 conserved nucleotide sequence features. Of the 3629 protein-coding genes and pseudogenes in G. metallireducens, 2403 (66.2%) had one or more full-length homologs in G. sulfurreducens.
The nucleotide composition of the 3563 intact protein-coding genes of G. metallireducens was determined in order to identify some of those that were very recently acquired. The average G+C content of the protein-coding genes was 59.5%, with a standard deviation of 5.9%. Only three genes had a G+C content more than two standard deviations above the mean (> 71.2%), but 146 genes had a G+C content more than two standard deviations below the mean (< 47.7%), most of which lack homologs in G. sulfurreducens and may be recent acquisitions (Additional file 1: Table S1). Clusters of such genes (shaded in Additional file 1: Table S1) were often interrupted or flanked by transposons with higher G+C content. The functions of most of these genes cannot be assigned at present, but 23 of them are predicted to act in cell wall biogenesis.
Plasmid pMET1 of G. metallireducens consists of a series of six predicted transcriptional units on one strand, tentatively attributed to the mobilization (Gmet_A3575-Gmet_A3574-Gmet_A3573-Gmet_A3572-Gmet_A3643), entry exclusion (Gmet_A3571), addiction (Gmet_A3570-Gmet_A3579-Gmet_A3642), partition (Gmet_A3568-Gmet_A3641), transposition (Gmet_A3567), and replication (Gmet_A3566-Gmet_A3565) functions of the plasmid, and one operon on the opposite strand, comprised of three genes of unknown function (Gmet_A3576-Gmet_A3577-Gmet_A3644). The predicted origin of replication, located 3' of the repA gene (Gmet_A3565), includes four pairs of iterons and a set of six hairpins, suggesting that pMET1 replicates by a rolling-circle mechanism, although it is significantly larger than most such plasmids [13]. Among the fifteen other nucleotide sequence features identified on the plasmid during manual curation was a palindromic putative autoregulatory site (TTTGTTATACACGTATAACAAA) located 5' of the addiction module. Other than the potential toxicity of the addiction module, the impact of pMET1 on the physiology of G. metallireducens is unknown.
Metabolism of acetate and other carbon sources
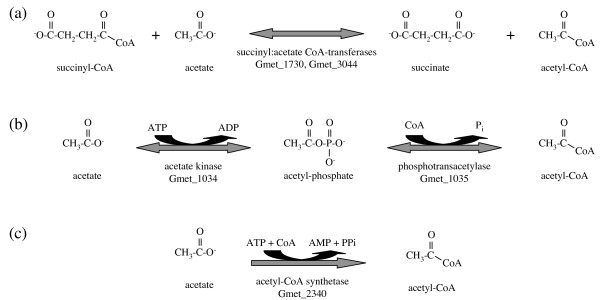
Acetate is expected to be the key electron donor supporting Fe(III) reduction in aquatic sediments and subsurface environments [14], and Geobacter species quickly become the predominant bacterial species when acetate is injected into subsurface environments to promote in situ bioremedation of uranium-contaminated groundwater [15,16]. Surprisingly, the initial activation of acetate by ligation with coenzyme A (CoA) in G. sulfurreducens occurs by two reversible pathways [17] (Figure 1), indicating that acetate may be inefficiently utilized at low concentrations. These two pathways are also present in G. metallireducens, along with a third, irreversible reaction that may permit efficient activation of acetate at low concentrations. The first pathway of acetate activation (Figure 1a) occurs through either of two succinyl:acetate CoA-transferases that can convert succinyl-CoA to succinate during oxidation of acetate by the tricarboxylic acid (TCA) cycle pathway, in the same capacity as succinyl-CoA synthetase but conserving energy in the form of acetyl-CoA rather than GTP or ATP [17]. Microarray data from both species suggest that expression of one succinyl:acetate CoA-transferase isoenzyme (Gmet_1730 = GSU0174) is constant and expression of the other (Gmet_3044 = GSU0490) is induced during acetate-fueled growth with electron acceptors other than soluble Fe(III), such as Fe(III) oxides, nitrate, or fumarate (D. Holmes, B. Postier, and R. Glaven, personal communications). The second pathway (Figure 1b) consists of two steps: acetate kinase (Gmet_1034 = GSU2707) converts acetate to acetyl-phosphate, which may be a global intracellular signal affecting various phosphorylation-dependent signalling systems, as in Escherichia coli [18]; and phosphotransacetylase (Gmet_1035 = GSU2706) converts acetyl-phosphate to acetyl-CoA [17]. G. metallireducens possesses orthologs of the enzymes of both pathways characterized in G. sulfurreducens [17], and also has an acetyl-CoA synthetase (Gmet_2340, 42% identical to the Bacillus subtilis enzyme [19]) for irreversible activation of acetate to acetyl-CoA at the expense of two ATP (Figure 1c). Thus, Geobacteraceae such as G. metallireducens may be better suited to metabolize acetate at the low concentrations naturally found in most soils and sediments.
Figure 1.
Pathways of acetate activation in G. metallireducens. (a) The succinyl:acetate CoA-transferase reaction. (b) The acetate kinase and phosphotransacetylase reactions. (c) The acetyl-CoA synthetase reaction.
Three enzymes distantly related to the succinyl:acetate CoA-transferases are encoded by Gmet_2054, Gmet_3294, and Gmet_3304, for which there are no counterparts in G. sulfurreducens. All three of these proteins closely match the characterized butyryl:4-hydroxybutyrate/vinylacetate CoA-transferases of Clostridium species [20]. However, their substrate specificities may be different because the G. metallireducens proteins and the Clostridium proteins cluster phylogenetically with different CoA-transferases of Geobacter strain FRC-32 and Geobacter bemidjiensis (data not shown). The presence of these CoA-transferases indicates that G. metallireducens has evolved energy-efficient activation steps for some unidentified organic acid substrates that G. sulfurreducens cannot utilize.
Numerous other enzymes of acyl-CoA metabolism are predicted from the genome of G. metalllireducens but not that of G. sulfurreducens (Additional file 2: Table S2), including six gene clusters, three of which have been linked to degradation of aromatic compounds that G. metallireducens can utilize [6,21-23] but G. sulfurreducens cannot [24]. All seven acyl-CoA synthetases of G. sulfurreducens have orthologs in G. metallireducens, but the latter also possesses acetyl-CoA synthetase, benzoate CoA-ligase (experimentally validated [23]), and seven other acyl-CoA synthetases of unknown substrate specificity. The G. metallireducens genome also includes eleven acyl-CoA dehydrogenases, three of which are specific for benzylsuccinyl-CoA (69% identical to the Thauera aromatica enzyme [25]), glutaryl-CoA (experimentally validated [26]) and isovaleryl-CoA (69% identical to the Solanum tuberosum mitochondrial enzyme [27]), whereas none can be identified in G. sulfurreducens. G. metallireducens also has nine pairs of electron transfer flavoprotein genes (seven of which are adjacent to genes encoding iron-sulfur cluster-binding proteins) that are hypothesized to connect acyl-CoA dehydrogenases to the respiratory chain, whereas G. sulfurreducens has only one. None of the seventeen enoyl-CoA hydratases of G. metallireducens is an ortholog of GSU1377, the sole enoyl-CoA hydratase of G. sulfurreducens. G. metallireducens also possesses eleven acyl-CoA thioesterases, of which G. sulfurreducens has orthologs of five plus the unique thioesterase GSU0196. Of the ten acyl-CoA thiolases of G. metallireducens, only Gmet_0144 has an ortholog (GSU3313) in G. sulfurreducens. BLAST searches and phylogenetic analyses demonstrated that several of these enzymes of acyl-CoA metabolism have close relatives in G. bemidjiensis, Geobacter FRC-32, Geobacter lovleyi and Geobacter uraniireducens, indicating that their absence from G. sulfurreducens is due to gene loss, and that this apparent metabolic versatility is largely the result of expansion of enzyme families within the genus Geobacter (data not shown). The ability of G. metallireducens and other Geobacteraceae to utilize carbon sources that G. sulfurreducens cannot utilize may be due to stepwise breakdown of multicarbon organic acids to simpler compounds by these enzymes.
Growth of G. metallireducens on butyrate may be attributed to reversible phosphorylation by either of two butyrate kinases (Gmet_2106 and Gmet_2128), followed by reversible CoA-ligation by phosphotransbutyrylase (Gmet_2098), a pathway not present in G. sulfurreducens, which cannot grow on butyrate [24]. These gene products are 42–50% identical to the enzymes characterized in Clostridium beijerinckii and Clostridium acetobutylicum [28,29].
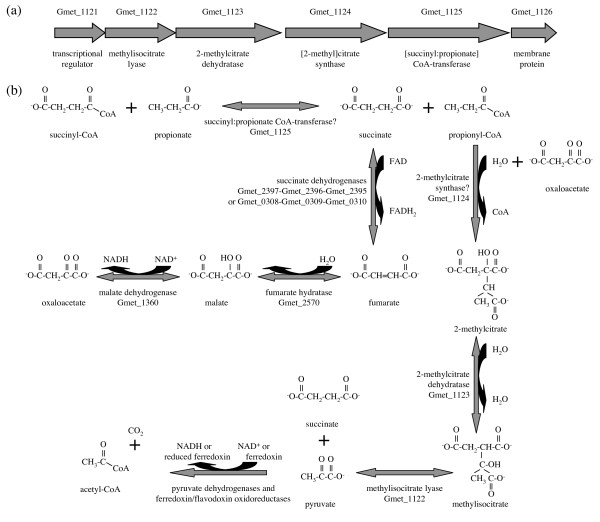
An enzyme very similar to succinyl:acetate CoA-transferase is encoded by Gmet_1125 within the same operon as methylisocitrate lyase (Gmet_1122), 2-methylcitrate dehydratase (Gmet_1123), and a citrate synthase-related protein hypothesized to be 2-methylcitrate synthase (Gmet_1124) [30] (Figure 2a), all of which are absent in G. sulfurreducens. This arrangement of genes, along with the ability of G. metallireducens to utilize propionate as an electron donor [31] whereas G. sulfurreducens cannot [24], suggests that the Gmet_1125 protein could be a succinyl:propionate CoA-transferase that, together with the other three products of the operon, would convert propionate (via propionyl-CoA) and oxaloacetate to pyruvate and succinate (Figure 2b). Upon oxidation of succinate to oxaloacetate through the TCA cycle and oxidative decarboxylation of pyruvate to acetyl-CoA, the pathway would be equivalent to the breakdown of propionate into six electrons, one molecule of carbon dioxide, and acetate, followed by the succinyl:acetate CoA-transferase reaction (Figure 2b). In a phylogenetic tree, the hypothetical succinyl:propionate CoA-transferase Gmet_1125 and gene Geob_0513 of Geobacter FRC-32, which is also capable of growth with propionate as the sole electron donor and carbon source (M. Aklujkar, unpublished), form a branch adjacent to succinyl:acetate CoA-transferases of the genus Geobacter (data not shown). In a similar manner, the hypothetical 2-methylcitrate synthase Gmet_1124 and gene Geob_0514 of Geobacter FRC-32 form a branch adjacent to citrate synthases of Geobacter species (data not shown), consistent with the notion that these two enzyme families could have recently evolved new members capable of converting propionate via propionyl-CoA to 2-methylcitrate.
Figure 2.
Growth of G. metallireducens on propionate. (a) The gene cluster predicted to encode enzymes of propionate metabolism. (b) The proposed pathway of propionate metabolism.
Gmet_0149 (GSU3448) is a homolog of acetate kinase that does not contribute sufficient acetate kinase activity to sustain growth of G. sulfurreducens [17] and has a closer BLAST hit to propionate kinase of E. coli (40% identical sequence) than to acetate kinase of E. coli. Although it does not cluster phylogenetically with either of the E. coli enzymes, its divergence from acetate kinase (Gmet_1034 = GSU2707) is older than the last common ancestor of the Geobacteraceae (data not shown). This conserved gene product remains to be characterized as a propionate kinase or something else.
The proposed pathway for growth of G. metallireducens on propionate (Figure 2) is contingent upon its experimentally established ability to grow on pyruvate [31]. G. sulfurreducens cannot utilize pyruvate as the carbon source unless hydrogen is provided as an electron donor [17]. Oxidation of acetyl-CoA derived from pyruvate in G. sulfurreducens may be prevented by a strict requirement for the succinyl:acetate CoA-transferase reaction (thermodynamically inhibited when acetyl-CoA exceeds acetate) to complete the TCA cycle in the absence of detectable activity of succinyl-CoA synthetase (GSU1058-GSU1059) [17]. With three sets of succinyl-CoA synthetase genes (Gmet_0729-Gmet_0730, Gmet_2068-Gmet_2069, and Gmet_2260-Gmet_2261), G. metallireducens may produce enough activity to complete the TCA cycle.
G. sulfurreducens and G. metallireducens may interconvert malate and pyruvate through a malate oxidoreductase fused to a phosphotransacetylase-like putative regulatory domain (maeB; Gmet_1637 = GSU1700), which is 51% identical to the NADP+-dependent malic enzyme of E. coli [32]. G. sulfurreducens has an additional malate oxidoreductase without this fusion (mleA; GSU2308) that is 53% identical to an NAD+-dependent malic enzyme of B. subtilis [33], but G. metallireducens does not.
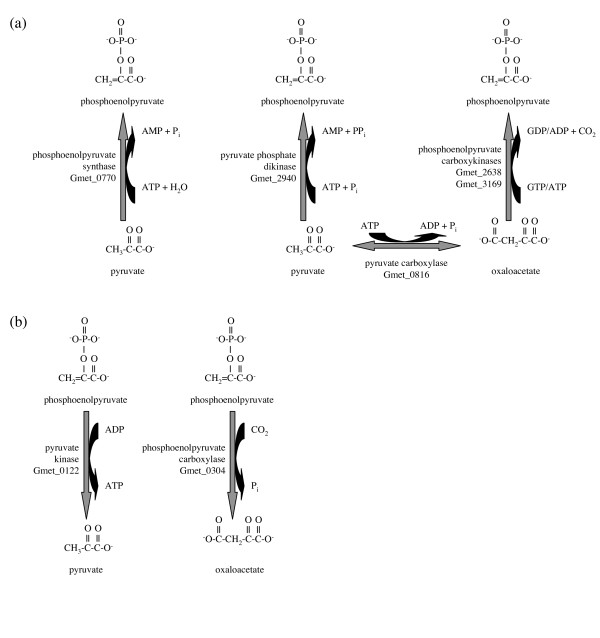
G. metallireducens possesses orthologous genes for all three pathways that activate pyruvate or oxaloacetate to phosphoenolpyruvate in G. sulfurreducens (Figure 3a): phosphoenolpyruvate synthase (Gmet_0770 = GSU0803), pyruvate phosphate dikinase (Gmet_2940 = GSU0580) and GTP-dependent phosphoenolpyruvate carboxykinase Gmet_2638 = GSU3385) [17]. It also encodes a homolog of the ATP-dependent phosphoenolpyruvate carboxykinase of E. coli (Gmet_3169, 48% identical) that has no homolog in G. sulfurreducens. In the catabolic direction, in addition to pyruvate kinase (Gmet_0122 = GSU3331) that converts phosphoenolpyruvate to pyruvate plus ATP, G. metallireducens has a homolog of E. coli phosphoenolpyruvate carboxylase (Gmet_0304, 30% identical, also found in Geobacter FRC-32) that may convert phosphoenolpyruvate to oxaloacetate irreversibly (Figure 3b) and contribute to the observed futile cycling of pyruvate/oxaloacetate/phosphoenolpyruvate [34] if not tightly regulated. Thus, control of the fate of pyruvate appears to be more complex in G. metallireducens than in G. sulfurreducens.
Figure 3.
Potential futile cycling of pyruvate/oxaloacetate and phosphoenolpyruvate in G. metallireducens. (a) Conversion of pyruvate to phosphoenolpyruvate. (b) Conversion of phosphoenolpyruvate to pyruvate or oxaloacetate.
Evidence of recent fumarate respiration in G. metallireducens
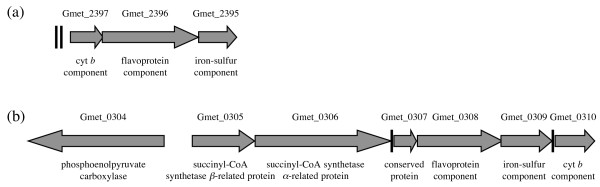
The succinate dehydrogenase complex of G. sulfurreducens also functions as a respiratory fumarate reductase, possibly in association with a co-transcribed b-type cytochrome [35]. G. metallireducens has homologous genes (Gmet_2397-Gmet_2395 = GSU1176-GSU1178), but is unable to grow with fumarate as the terminal electron acceptor unless transformed with a plasmid that expresses the dicarboxylic acid exchange transporter gene dcuB of G. sulfurreducens [35], which has homologues in Geobacter FRC-32, G. bemidjiensis, G. lovleyi, and G. uraniireducens. Surprisingly, G. metallireducens has acquired another putative succinate dehydrogenase or fumarate reductase complex (Gmet_0308-Gmet_0310), not found in other Geobacteraceae, by lateral gene transfer from a relative of the Chlorobiaceae (phylogenetic trees not shown), and evolved it into a gene cluster that includes enzymes of central metabolism acquired from other sources (Figure 4). Thus, G. metallireducens may have actually enhanced its ability to respire fumarate before recently losing the requisite transporter.
Figure 4.
Acquisition of a second fumarate reductase/succinate dehydrogenase by G. metallireducens. (a) The ancestral gene cluster. (b) The gene cluster acquired from a relative of the Chlorobiaceae, located near other acquired genes relevant to central metabolism: an uncharacterized enzyme related to succinyl-CoA synthetase and citrate synthase (Gmet_0305-Gmet_0306) and phosphoenolpyruvate carboxylase (Gmet_0304). Conserved nucleotide sequences (black stripes) were also identified in the two regions.
Nitrate respiration and loss of the modE regulon from G. metallireducens
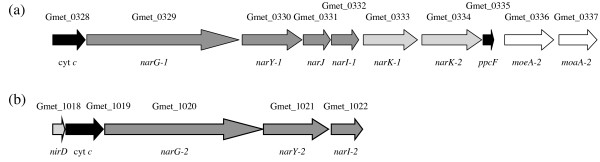
G. metallireducens is able to respire nitrate [4], whereas G. sulfurreducens cannot [24]. The nitrate reductase activity of G. metallireducens is attributed to the narGYJI genes (Figure 5a; Gmet_0329-Gmet_0332), which are adjacent to the narK-1 and narK-2 genes encoding a proton/nitrate symporter and a nitrate/nitrite antiporter (Gmet_0333 and Gmet_0334, respectively) predicted according to homology with the two halves of narK in Paracoccus pantotrophus [36]. A second narGYI cluster (Figure 5b; Gmet_1020 to Gmet_1022) is missing a noncatalytic subunit (narJ), and its expression has not been detected (B. Postier, personal communication). The first gene of both operons encodes a unique diheme c-type cytochrome (Gmet_0328 and Gmet_1019), suggesting that the nitrate reductase may be connected to other electron transfer components besides the menaquinol pool, perhaps operating in reverse as a nitrite oxidase. The product of the ppcF gene (Gmet_0335) in the intact nar operon, which is related to a periplasmic triheme c-type cytochrome involved in Fe(III) reduction in G. sulfurreducens [37], may permit electron transfer to the nitrate reductase from extracellular electron donors such as humic substances [38] or graphite electrodes [11]. The final two genes of the intact nar operon (Gmet_0336-Gmet_0337), encode the MoeA and MoaA enzymes implicated in biosynthesis of bis-(molybdopterin guanine dinucleotide)-molybdenum, an essential cofactor of the nitrate reductase.
Figure 5.
The respiratory nitrate reductase operons. (a) The major (expressed) operon also encodes the nitrate and nitrite transporters (narK-1, narK-2), two c-type cytochromes including ppcF, and two genes of molybdenum cofactor biosynthesis (moeA-2, moaA-2). (b) The minor operon (expression not detected) also encodes the Rieske iron-sulfur component of nitrite reductase (nirD) and a c-type cytochrome, but lacks a narJ gene.
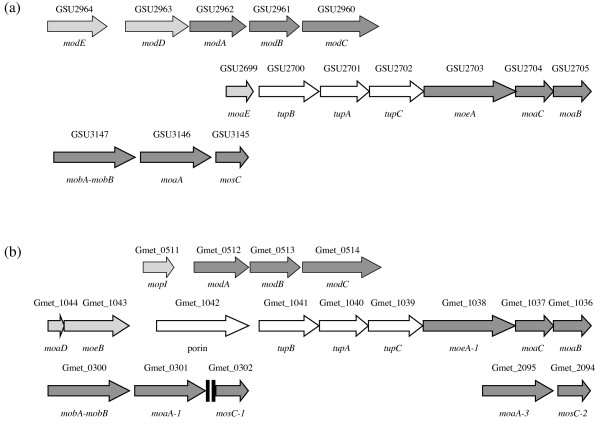
Phylogenetic analysis indicates that the moeA and moaA gene families have repeatedly expanded in various Geobacteraceae (data not shown). G. sulfurreducens has a single copy of each, but G. metallireducens has three closely related isoenzymes, of which moeA-1 (Gmet_1038 = GSU2703, 40% identical to the E. coli protein [39]) and moaA-1 (Gmet_0301 = GSU3146, 36% identical to the E. coli protein [40]) occupy a conserved location among other genes of molybdopterin biosynthesis (Table 1, Figure 6). A possible reason for the expansion in G. metallireducens and other Geobacteraceae is a need to upregulate molybdopterin biosynthesis for specific processes: moeA-2 and moaA-2 (Gmet_0336-Gmet_0337, 38% and 33% identity to the E. coli proteins) may support nitrate reduction; moaA-3 (Gmet_2095, 35% identity to E. coli) may function with nearby gene clusters for catabolism of benzoate [23] and p-cresol [22]; and moeA-3 (Gmet_1804, 37% identity to E. coli) may aid growth on benzoate, during which it is upregulated [21]. G. metallireducens differs from G. sulfurreducens in other aspects of molybdenum assimilation as well (Table 1): notably, G. sulfurreducens possesses a homolog of the moaE gene (GSU2699) encoding the large subunit of molybdopterin synthase, but lacks homologs of the small subunit gene moaD and the molybdopterin synthase sulfurylase gene moeB, whereas G. metallireducens lacks a moaE homolog but possesses homologs of moaD (Gmet_1043) and moeB (Gmet_1042). Comparison with the genomes of other Geobacteraceae suggests that these differences are due to loss of ancestral genes. How the nitrate reductase of G. metallireducens can function with the molybdopterin synthase complex being apparently incomplete is unknown.
Table 1.
Genes of molybdenum cofactor biosynthesis in G. sulfurreducens and G. metallireducens.
| Locus | Gene in G. sulfurreducens | Gene in G. metallireducens | Function |
|---|---|---|---|
| modE | GSU2964 | Gmet_05111 | regulation of molybdate-responsive genes |
| modD | GSU2963 | none | inner membrane protein, possible quinolinate phosphoribosyltransferase |
| modA | GSU2962 | Gmet_0512 | molybdate transport (periplasmic component) |
| modB | GSU2961 | Gmet_0513 | molybdate transport (membrane component) |
| modC | GSU2960 | Gmet_0514 | molybdate transport (ATP-binding component) |
| moaD | none | Gmet_1044 | dithiolene addition to molybdopterin (molybdopterin synthase small subunit) |
| moeB | none | Gmet_1043 | molybdopterin synthase sulfurylase |
| moaE | GSU2699 | none | dithiolene addition to molybdopterin (molybdopterin synthase large subunit) |
| moeA | GSU2703 | Gmet_1038; Gmet_0336; Gmet_1804 | molybdenum-sulfur ligation? |
| moaC | GSU2704 | Gmet_1037 | molybdopterin precursor Z synthesis |
| moaB | GSU2705 | Gmet_1036 | molybdopterin precursor Z synthesis |
| mobA | GSU3147 N-terminal domain | Gmet_0300 N-terminal domain | attachment of molybdopterin to guanosine |
| mobB | GSU3147 C-terminal domain | Gmet_0300 C-terminal domain | attachment of molybdopterin to guanosine |
| moaA | GSU3146 | Gmet_0301; Gmet_0337; Gmet_2095 | molybdopterin precursor Z synthesis |
| mosC | GSU3145 | Gmet_0302; Gmet_2094 | molybdenum sulfurase |
| pcmV | none | Gmet_2138 | possible 4-hydroxybenzoyl-CoA reductase molybdenum cofactor biosynthesis protein |
| pcmW | none | Gmet_2139 | possible 4-hydroxybenzoyl-CoA reductase molybdenum cofactor biosynthesis protein |
| pcmX | none | Gmet_2140 | uncharacterized protein related to MobA |
1Gmet_0511 is missing the N-terminal ModE domain but retains the C-terminal molybdopterin-binding MopI domains.
Figure 6.
G. sulfurreducens and G. metallireducens possess different genes for molybdenum cofactor biosynthesis. (a) G. sulfurreducens has the global regulator modE. (b) G. metallireducens has multiple copies of moeA, moaA, and mosC, and putative integration host factor binding sites (black stripes). Both genomes have conserved genes (dark grey) for molybdate transport (modABC) and molybdopterin biosynthesis (moeA, moaCB, mobA-mobB, mosC) alongside tup genes for tungstate transport (white), but neither genome has all the genes thought to be essential for bis-(molybdopterin guanine dinucleotide)-molybdenum biosynthesis (light grey). See also Table 1.
In G. sulfurreducens, putative binding sites for the molybdate-sensing ModE protein (GSU2964) have been identified by the ScanACE software [41,42] in several locations, and the existence of a ModE regulon has been predicted [43]. The genes in the predicted ModE regulon (Additional file 3: Table S3) include one of the two succinyl:acetate CoA-transferases, a glycine-specific tRNA (anticodon CCC, corresponding to 26% of glycine codons), several transport systems, and some nucleases. In G. metallireducens, there is no full-length modE gene, but a gene encoding the C-terminal molybdopterin-binding (MopI) domain of ModE (Gmet_0511) is present in the same location (Figure 6). Phylogenetic analysis shows that the Gmet_0511 gene product is the closest known relative of G. sulfurreducens ModE, and that it has evolved out of the Geobacteraceae/Chlorobiaceae cluster of full-length ModE proteins by loss of the N-terminal ModE-specific domain (data not shown). The ScanACE software detected only one of the ModE-binding sites of G. sulfurreducens at the corresponding location in the G. metallireducens genome, but some vestigial sites were apparent when other syntenous locations were visually inspected (Additional file 3: Table S3), indicating that the ModE regulon once existed in G. metallireducens, but recent loss of the ModE N-terminal domain is allowing the regulatory sites to disappear gradually over the course of genome sequence evolution due to the absence of selective pressure for these sites to remain conserved. Thus, genes that may be controlled globally by ModE in G. sulfurreducens and other Geobacteraceae to optimize molybdenum cofactor-dependent processes have recently acquired independence in G. metallireducens.
Amino acid biosynthesis and its regulation
The two genomes differ in several aspects of amino acid biosynthesis and its regulation. To make aspartate from oxaloacetate, a homolog of Bacillus circulans aspartate aminotransferase [44] is present in G. metallireducens (Gmet_2078; 65% identical), whereas a homolog of the Sinorhizobium meliloti enzyme [45] is found in G. sulfurreducens (GSU1242; 52% identical). Both species possess asparagine synthetase (Gmet_2172 = GSU1953 and Gmet_2024, 30% and 24% identical to asnB of B. subtilis [46]) and glutamine synthetase (Gmet_1352 = GSU1835, 61% identical to glnA of Fremyella diplosiphon [47]), as well as an aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase operon (Gmet_0076, Gmet_0075, Gmet_0073 = GSU3383, GSU3381, GSU3380, 36–53% identical to the homologous subunits in B. subtilis [48]) that includes glutamine synthetase adenylyltransferase (glnE; Gmet_0071 = GSU3378). The G. sulfurreducens glnE gene may be inactive due to a deletion of ~ 45 codons in the C-terminal domain.
For biosynthesis of lysine, threonine and methionine, G. metallireducens and other Geobacteraceae possess a linked pair of aspartate-4-semialdehyde dehydrogenase genes: Pseudomonas aeruginosa-type Gmet_0603 (69% identity) [49] and Mycobacterium bovis-type Gmet_0604 (47% identity) [50], but G. sulfurreducens has only the former (GSU2878). A haloacid dehalogenase family protein (Gmet_1630 = GSU1694) encoded between two genes of the threonine biosynthesis pathway could be the enzyme required to complete the pathway, a phosphoserine:homoserine phosphotransferase analogous to that of P. aeruginosa [51], and may overlap functionally with the unidentified phosphoserine phosphatase required to complete the biosynthetic pathway of serine.
Conserved nucleotide sequences (possible promoters and riboswitches) were identified on the 5' sides of several biosynthetic operons (Table 2). The lysine biosynthesis operon in G. sulfurreducens and other Geobacteraceae begins with a P. aeruginosa-type meso-diaminopimelate decarboxylase (GSU0158; 51% identity) [52], whereas G. metallireducens has two isoenzymes in other locations (Gmet_0219, 30% identical to the E. coli enzyme [53], with homologs in a few Geobacteraceae; Gmet_2019, 31% identical to the P. aeruginosa enzyme [52], unique to G. metallireducens). The recently identified L,L-diaminopimelate aminotransferase (dapL; Gmet_0213 = GSU0162) [54] is co-transcribed with the dapAB genes encoding the two preceding enzymes of lysine biosynthesis, but separated from them by a predicted short RNA element (Gmet_R1005 = GSU0160.1), also found in 23 other locations on the G. metallireducens chromosome (Additional file 4: Figure S1, Additional file 5: Table S4).
Table 2.
Conserved nucleotide sequences 5' of biosynthetic operons.
| Operon | Locus tag and sequence coordinates | |
|---|---|---|
| G. metallireducens | G. sulfurreducens | |
| aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase (gatCAB-mtnA-glnE-nth) | Gmet_P0076 93465..93502 |
GSU3383.1 3719308..3719345 |
| lysine (dapA) | Gmet_P0211 244588..244640 |
GSU0157.1 176066..176117 |
| aromatic amino acids (aroG-2) | Gmet_R0069 384337..384528 |
GSUR082 3450692..3450963 |
| cobalamin (cobUTSCB-thiC-2; cbiM-1-cbiQ-1-cbiO-1-cbiX-cobH-cbiD-cobLIM-cbiG-cobQ-cbiB-cobD) | Gmet_R0070 513498..513761 no match |
GSU3011.1 3302884..3303201 GSU3004.1 3296929..3297108 |
| methionine (metC-1-metC-2; metX)1 | Gmet_R0073 765279..765444 Gmet_R0129 3145553..3145656 |
GSUR063 1014004..1014271 GSU2461.2 2700118..2700220 |
| leucine (leuA)2 | Gmet_P1265 1425160..1425452 |
GSU1906.1 2085440..2085740 |
| leucine/isoleucine (leuCD) | Gmet_P1268 1428650..1428793 |
GSU1903.1 2082057..2082203 |
| coenzyme A (panBC) | Gmet_P1642 1843163..1843275 |
GSU1704.1 1868745..1868863 |
| pyrimidines (pyrRBC-carAB) | Gmet_P1768 1983157..1983191 |
GSU1269.1 1384886..1384920 |
| tryptophan (iorAB-paaK) | Gmet_P1827 2042198..2042288 |
GSU1739.1 1905464..1905561 |
| purines, pyrimidines (purMN, rimI-pyrKD) | Gmet_P1844 2056600..2056732 |
GSU1757.1 1920275..1920400 |
| guanine (guaBA) | Gmet_P2293 2600787..2600857 |
GSU2195.1 2408782..2408854 |
| serine (serA) | Gmet_P2378 2689446..2689518 |
GSU1197.1 1301091..1301163 |
| thiamin (thiE/D-thiC-1; thiS-1-thiG-tenI)3 | Gmet_R0131 3292750...3292897 Gmet_R0134 3319520..3319741 |
GSUR060 640780..640988 GSU0589.1 622533..622801 |
| arginine (argBDFG) | Gmet_P0203 3719308..3719345 |
GSU0149.1 167623..167663 |
1The sequence 5' of metC-1, metC-2, and metX is a SAM-responsive riboswitch.
2The sequence 5' of leuA is a T-box, an RNA structure that recognizes the aminoacylation state of tRNA.
3The sequence 5' of thiamin biosynthesis operons is a thiamin diphosphate-responsive riboswitch.
S-adenosylmethionine (SAM)-responsive riboswitches (Table 2) may regulate homoserine O-acetyltransferase (Gmet_2783 = GSU2462, 45% identical to the Leptospira meyeri enzyme [55]), the first dedicated enzyme of methionine biosynthesis, and also two linked cystathionine-γ-synthase/cystathionine-β-lyase genes (Gmet_0698 = GSU0944; Gmet_0699 = GSU0945, 49% and 51% identical to the Lactococcus lactis lyase [56,57]). Phylogenetic analysis could not distinguish the synthase from the lyase (data not shown), but their presence suggests that homocysteine can be made by transsulfuration of homoserine with cysteine, and not only by the putative O-acetylhomoserine sulfhydrylases (Gmet_0819 = GSU2425, Gmet_2390 = GSU1183 and Gmet_1566, 47%, 56% and 38% identical to the Emericella nidulans enzyme [58], respectively). In G. metallireducens, transsulfuration may also be controlled by a GC-rich element between Gmet_0698 and Gmet_0699, which contains four tandem repeats of the heptanucleotide GGGACCG and is found in 49 intergenic and intragenic locations in the genome (Additional file 6: Figure S2, Additional file 5: Table S4).
The leucine pathway-specific leuA gene (2-isopropylmalate synthase; Gmet_1265 = GSU1906, 49% identical to the E. coli enzyme [59]) may be controlled by feedback inhibition through a T-box [60] predicted to form an antiterminator structure in response to uncharged leucine-specific tRNA having the GAG anticodon (Gmet_R0037 = GSUR030) (Table 2), putatively the only tRNA capable of recognizing 55% of leucine codons in G. metallireducens and 48% in G. sulfurreducens (CTC and CTT).
There are three 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase isoenzymes to catalyze the first step of aromatic amino acid biosynthesis: one similar to aroF of E. coli (Gmet_2375 = GSU2291, 55% identity [61], but with a P148T substitution incompatible with feedback inhibition by tyrosine [62]) and two Thermotoga maritima-type enzymes (Gmet_0024 = GSU3333; Gmet_0346 = GSU3142, 51% and 46% identity [63], respectively). As one chorismate mutase is fused to prephenate dehydratase (pheA; Gmet_0862 = GSU2608, 41% identical to the Pseudomonas stutzeri fusion protein [64]), the other (Gmet_1955 = GSU1828, 30% identical to the chorismate mutase domain of the P. stutzeri fusion protein) may function predominantly in tyrosine biosynthesis, possibly regulated by the adjacent gene product (Gmet_1956 = GSU1829) that resembles the phenylalanine/tyrosine-responsive domain of T. maritima DAHP synthase [65]. Gmet_1956 orthologs phylogenetically cluster with the regulatory domains of Gmet_0024 orthologs (data not shown), suggesting that Gmet_0024 may be a tyrosine-inhibited DAHP synthase and Gmet_0346 may be inhibited by another end product such as phenylalanine. A predicted short RNA element (Gmet_R0069 = GSUR082, Table 2), found 5' of Gmet_0346 and its orthologs in several Geobacteraceae, may participate in regulation of this isoenzyme's expression.
In all non-Geobacteraceae that possess an indole-scavenging tryptophan synthase β2 protein, it is encoded apart from the trp operon containing the trpAB1 genes for the α (indole-producing) and β1 (indole-consuming) subunits of tryptophan synthase [66]. In G. metallireducens and G. sulfurreducens, however, the β2 gene trpB2 (Gmet_2493 = GSU2379, 60% identical to the T. maritima protein [67]) is the penultimate gene of the predicted trp operon and the trpB1 (Gmet_2482 = GSU2375, 66% identical to the Acinetobacter calcoaceticus protein [68]) and trpA (Gmet_2477 = GSU2371, 47% identical to the Azospirillum brasilense protein [69]) genes are separated from the 3' end of the operon and from each other by three or more intervening genes, most of which are not conserved between the two genomes (not shown). Next to the trpB2 gene of G. metallireducens is one of 24 pairs of a conserved nucleotide motif (Additional file 7: Figure S3, Additional file 5: Table S4) hypothesized to bind an unidentified global regulator protein. Other, evolutionarily related paired sites where another unidentified global regulator may bind (Additional file 8: Figure S4, Additional file 5: Table S4) are found in 21 locations. Between the proBA genes of G. metallireducens, encoding the first two enzymes of proline biosynthesis (Gmet_3198-Gmet_3199 = GSU3212-GSU3211, 41% and 45% identical to the E. coli enzymes [70]), is one of eight pairs of predicted binding sites for yet another unidentified global regulator (Additional file 9: Figure S5, Additional file 5: Table S4). In G. sulfurreducens, the space between proBA is occupied by a different conserved nucleotide sequence (not shown), found only in four other places in the same genome. Overall, a comparison of the two genomes offers insight into unique features of amino acid biosynthesis and its regulation that deserve further study.
Nucleotide metabolism
Differences in nucleotide metabolism were identified in the two genomes. G. metallireducens has acquired a possibly redundant large subunit of carbamoyl-phosphate synthetase (Gmet_0661, 50% identical to the P. aeruginosa protein [71]) in addition to the ancestral gene (Gmet_1774 = GSU1276, 65% identity to P. aeruginosa), Both genomes encode a second putative thymidylate kinase (Gmet_3250 = GSU3301) distantly related to all others, in addition to the one found in other Geobacteraceae (Gmet_2318 = GSU2229, 41% identical to the E. coli enzyme [72]). G. sulfurreducens has evidently lost the purT gene product of G. metallireducens and several other Geobacteraceae (Gmet_3193, 58% identical to the E. coli enzyme [73]), which incorporates formate directly into purine nucleotides instead of using the folate-dependent purN gene product (Gmet_1845 = GSU1759, 46% identical to the E. coli enzyme [74]).
Carbohydrate metabolism
Comparative genomics indicates that, similar to most Geobacter species, G. metallireducens possesses two glyceraldehyde-3-phosphate dehydrogenase isoenzymes: Gmet_1211 and Gmet_1946 (59% and 56% identical to gluconeogenic GapB and glycolytic GapA of Corynebacterium glutamicum [75], respectively), but G. sulfurreducens has an ortholog of only the latter (GSU1629). G. metallireducens also has a putative fructose 6-kinase (Gmet_2805, 39% identical to the E. coli enzyme [76]) that is not present in G. sulfurreducens. Remarkably, G. metallireducens possesses two isoenzymes each of UDP-glucose 4-epimerase (Gmet_1486; Gmet_2329 = GSU2240, 50% and 54% identical to the A. brasilense enzyme [77]), glutamine:fructose-6-phosphate aminotransferase (Gmet_1487; Gmet_0104 = GSU0270, 55% and 53% identical to the Thermus thermophilus enzyme [78]), GDP-mannose 4,6-dehydratase (Gmet_1488 = GSU0626; Gmet_1311, 61% and 72% identical to the E. coli enzyme [79]) and UDP-N-acetylglucosamine 2-epimerase (Gmet_1489 = GSU2243, 61% identical to the E. coli enzyme [80]; Gmet_1504, 39% identical to the Methanococcus maripaludis enzyme [81]). G. metallireducens has evolved a gene cluster of the four enzyme activities (Gmet_1486-Gmet_1489) from both ancestral gene duplication and lateral gene transfer (data not shown). The reason for this emphasis on interconversion of hexoses in G. metallireducens versus G. sulfurreducens is unknown.
Unlike the genomes of G. sulfurreducens and most other Geobacteraceae, which encode the enzymes of only the non-oxidative branch of the pentose phosphate pathway, the G. metallireducens genome includes a cluster of oxidative pentose phosphate pathway enzyme genes: 6-phosphogluconolactonase (Gmet_2618, 30% identical to the Pseudomonas putida enzyme [82]), glucose-6-phosphate dehydrogenase (Gmet_2619, 50% identical to the Nostoc punctiforme enzyme [83]), and 6-phosphogluconate dehydrogenase (Gmet_2620, 36% identical to YqeC of B. subtilis [84]), along with two ribose-5-phosphate isomerase isoenzymes (Gmet_2621 and Gmet_1604 = GSU1606, 39% and 44% identical to RpiB of E. coli [85]). Thus, G. metallireducens apparently generates biosynthetic reducing equivalents in the form of NADPH from carbohydrates. The NADPH supply of G. sulfurreducens, in contrast, may derive from the electron transfer chain via a ferredoxin:NADP+ reductase (GSU3058-GSU3057, each 52% identical to its Pyrococcus furiosus homolog [86]) that is found in other Geobacteraceae, but not in G. metallireducens.
Both G. sulfurreducens and G. metallireducens may protect themselves from desiccation by making trehalose from glucose storage polymers via maltooligose in three steps catalyzed by an alpha-amylase domain protein (Gmet_3469 = GSU2361), maltooligosyltrehalose synthase (Gmet_3468 = GSU2360, 35% identical to the Rhizobium leguminosarum enzyme [87]), and maltooligosyltrehalose trehalohydrolase (Gmet_3467 = GSU2358, 44% identical to the Arthrobacter strain Q36 enzyme [88]). G. sulfurreducens, P. propionicus and G. lovleyi may also make trehalose from glucose-6-phosphate by the sequential action of trehalose-6-phosphate synthase (GSU2337, containing a domain 37% identical to the Mycobacterium tuberculosis enzyme [89]) and trehalose-6-phosphatase (GSU2336, 29% identical to the E. coli enzyme [90]), which are missing in G. metallireducens. Thus, G. sulfurreducens is capable of achieving osmotolerance without consuming carbohydrate storage polymers, but G. metallireducens is not.
Biogenesis of c-type cytochromes and pili
The genome of G. metallireducens encodes 91 putative c-type cytochromes, of which 65 have homologs among the 103 c-type cytochromes of G. sulfurreducens. Of the c-type cytochrome genes implicated in Fe(III) and U(VI) reduction in G. sulfurreducens, those conserved in G. metallireducens are macA (Gmet_3091 = GSU0466) [91-93] and ppcA (Gmet_2902 = GSU0612) [37], whereas different c-type cytochrome sequences are found in syntenous locations where one would expect omcB and omcC (Gmet_0910 ≠ GSU2737; Gmet_0913 ≠ GSU2731) [94], and omcE (Gmet_2896 ≠ GSU0618) [95]. The G. metallireducens genome contains no genes homologous to omcS (GSU2504) and omcT (GSU2503) [95], and only a paralog (Gmet_0155 = GSU2743) of omcF (GSU2432) [96]. This lack of conservation is being investigated further (J. Butler, personal communication).
Notable differences between G. metallireducens and G. sulfurreducens are apparent in the biogenesis of c-type cytochromes, in biosynthesis of the heme group, and in reduction of disulfide bonds to allow covalent linkage to heme. In addition to the membrane-peripheral protoporphyrinogen IX oxidase of G. sulfurreducens and other Geobacteraceae, encoded by the hemY gene (Gmet_3551 = GSU0012, 38% identical to the Myxococcus xanthus enzyme [97]), G. metallireducens has a membrane-integral isoenzyme encoded by hemG (Gmet_2953, 43% identical to the E. coli enzyme [98]), with a homolog in Geobacter FRC-32. These two species also possess a putative disulfide bond reduction system not found in G. sulfurreducens and other Geobacteraceae, comprised of DsbA, DsbB, DsbE and DsbD homologs (Gmet_1380, Gmet_1381, Gmet_1383, Gmet_1384), encoded in a cluster alongside a two-component signalling system (Gmet_1378-Gmet_1379), an arylsulfotransferase (Gmet_1382), and a conserved protein of unknown function (Gmet_1385). Transcription of dsbA and dsbB is diminished during growth on benzoate [21], and phylogenetic analysis indicates that these DsbA and DsbB proteins belong to subfamilies distinct from those that have been characterized (R. Dutton, personal communication). Located apart from this cluster, DsbC/DsbG (Gmet_2250) of G. metallireducens has homologs in several Geobacteraceae, but not in G. sulfurreducens. However, CcdA/DsbD (Gmet_2451 = GSU1322) is present in both. Thus, the pathways of c-type cytochrome biogenesis may be significantly different in the two species and somehow linked to the degradation of aromatic compounds by G. metallireducens.
In both G. sulfurreducens and G. metallireducens, there are four c-type cytochrome biogenesis genes related to ResB of B. subtilis [99], each predicted to be co-transcribed with a gene encoding a ResC/HemX-like protein (hypothesized to be a heme transporter with eight predicted transmembrane segments) [100] and several multiheme c-type cytochrome genes (Additional file 10: Table S5). One more protein of the ResC/HemX-like family (Gmet_3232 = GSU3283) is encoded among enzymes of heme biosynthesis in both genomes. These gene arrangements suggest that each pair of c-type cytochrome biogenesis proteins may be dedicated to the efficient expression of the cytochromes encoded nearby. Two of the pairs are orthologously conserved (Gmet_2901-Gmet_2900 = GSU0613-GSU0614; Gmet_0592..Gmet_0594 = GSU2891-GSU2890); the other two pairs (Gmet_0572-Gmet_0573; Gmet_0578-Gmet_0579; GSU0704-GSU0705; GSU2881.1-GSU2880), which appear to derive from expansion of ancestral genes, may be relevant to the diversified c-type cytochrome repertoire of the two species. Interestingly, three of these gene pairs in G. metallireducens are arranged in proximity to each other in a cluster of ten operons with the same coding DNA strand (Gmet_0571 to Gmet_0601), suggesting that their expression may be co-ordinated by transcriptional readthrough (Additional file 10: Table S5). The purposes of various pairs of c-type cytochrome biogenesis proteins in Geobacteraceae remain to be determined.
The pili of G. sulfurreducens have been implicated in electron transfer [101,102] and biofilm formation [103]. Most genes attributed to pilus biogenesis in G. sulfurreducens have orthologs in G. metallireducens, suggesting that these roles of pili may be conserved. However, instead of the ancestral pilY1 gene found in G. sulfurreducens (GSU2038) and other Geobacteraceae, which may encode a pilus tip-associated adhesive protein [104], G. metallireducens possesses a phylogenetically distinct pilY1 gene in the same location (Gmet_0967; data not shown), surrounded by different genes of unknown function within a cluster of pilus biogenesis genes. Therefore, it remains possible that structural and functional differences between the pili of the two species will be identified in future.
Solute transport systems
Although the substrates of most solute transport systems of G. metallireducens and G. sulfurreducens are unknown, several features distinguish the two species (Additional file 11: Table S6). One of two predicted GTP-dependent Fe(II) transporters of the Geobacteraceae (feoB-1 Gmet_2444 = GSU1380), located next to the ferric uptake regulator gene (fur Gmet_2445 = GSU1379), is present in G. metallireducens; the other (feoB-2 GSU3268), with two feoA genes on its 5' side (GSU3268.1, GSU3270) potentially encoding an essential cytosolic component of the transport system [105], is not. Phylogenetic analysis showed that the FeoB-2 proteins of Geobacteraceae are closely related to the characterized Fe(II)-specific FeoB proteins of Porphyromonas gingivalis [106] and Campylobacter jejuni [107], whereas the FeoB-1 proteins of Geobacteraceae cluster apart from them (data not shown). FeoB-1 proteins are not closely related to the manganese-specific FeoB of P. gingivalis [106] either, and so their substrate specificity cannot be assigned at present.
In G. metallireducens, duplicate kup genes, predicted to encode low-affinity potassium/proton symporters, are found in one place (Gmet_0038 = GSU3342; Gmet_0039 = GSU2485, 29% and 31% identical to the E. coli protein [108]), apart from the kdpABCDE genes (Gmet_2433-Gmet_2437 = GSU2480-GSU2484, 38–49% identical to the homologs in E. coli [109,110]) encoding an osmosensitive potassium-translocating ATPase complex. In G. sulfurreducens, one of these kup genes (GSU2485) is located 3' of the kdp gene cluster, apparently under control of an osmosensitive riboswitch (GSU2484.1, sequence coordinates 2728254 to 2728393), and there is a third kup gene (GSU2350, 49% identity to E. coli) not found in other Geobacteraceae. G. sulfurreducens also has at least two potassium/proton antiporters (GSU1203, 34% identical to CvrA of Vibrio parahaemolyticus [111]; GSU2759, 31% identical to KefB of E. coli [112]) and a sodium/proton antiporter complex (mrpABCDEFG GSU2344-GSU2338, 29–48% identical to the homologs in B. subtilis [113]) that are not found in G. metallireducens. Three mechanosensitive ion channels are common to the two species (Gmet_1942 = GSU1633; Gmet_2581 = GSU2316; and Gmet_2522 = GSU2794); two more are unique to G. sulfurreducens (GSU1557; GSU1723). Thus, control of monovalent cation homeostasis appears to be more complex in G. sulfurreducens.
Several heavy metal efflux pumps are conserved between the two species, but their substrate specificity is uncertain. Transporters present in G. sulfurreducens but not G. metallireducens include that for uracil (GSU0932, 48% identical to the Bacillus caldolyticus protein [114]). Transporters present in G. metallireducens but not G. sulfurreducens include those for nitrate/nitrite (Gmet_0333-Gmet_0334) and chromate (Gmet_2732-Gmet_2731), which are each present as two paralogous genes rather than gene fusions such as their homologs that have been characterized in other bacteria [36,115].
Signalling, chemotaxis and global regulation
G. metallireducens possesses orthologs of the six sigma factors of RNA polymerase identified in G. sulfurreducens (Table 3), as well as a seventh factor (Gmet_2792) not found in other Geobacteraceae, related to the extracytoplasmic sigma-Z factor of B. subtilis [116]. Intriguingly, a particular anti-anti-sigma factor gene is frameshifted in both genomes: GSU1427 has frameshifts in the phosphatase domain, resulting in an in-frame protein, whereas the homologous Gmet_1229 is shifted out of frame in the kinase domain. These differences imply that global regulatory networks may be different in the two species.
Table 3.
Sigma factors of G. metallireducens and G. sulfurreducens.
| Locus Tag | Annotation | G. metallireducens gene | G. sulfurreducens gene |
|---|---|---|---|
| rpoH | RNA polymerase sigma-32 factor | Gmet_2854 | GSU0655 |
| rpoE | RNA polymerase sigma-24 factor, putative | Gmet_2612 | GSU0721 |
| rpoN | RNA polymerase sigma-54 factor | Gmet_1283 | GSU1887 |
| rpoD | RNA polymerase sigma-70 factor RpoD | Gmet_0395 | GSU3089 |
| rpoS | RNA polymerase sigma-38 factor, stationary phase | Gmet_1421 | GSU1525 |
| fliA | RNA polymerase sigma-28 factor for flagellar operon | Gmet_0429 | GSU3053 |
| none | RNA polymerase sigma-Z factor | Gmet_2792 | none |
The G. metallireducens genome encodes 83 putative sensor histidine kinases containing HATPase_c domains (Additional file 12: Table S7), of which 45 (54%) have orthologs among the 95 such proteins of G. sulfurreducens. There are 94 proteins with response receiver (REC) domains in G. metallireducens (Additional file 12: Table S7), out of which 66 (70%) have orthologs among the 110 such proteins of G. sulfurreducens. Twenty-seven of the REC domain-containing proteins and another 101 genes and four pseudogenes (Additional file 12: Table S7) were predicted to be transcriptional regulators in G. metallireducens. There are 20 putative diguanylate cyclases containing GGDEF domains, of which 16 (80%) have orthologs among the 29 putative diguanylate cyclases of G. sulfurreducens (Additional file 13: Table S8). Overall, the portion of the genome dedicated to signalling and transcriptional regulation in G. metallireducens is slightly less than in G. sulfurreducens, but still considerable and significantly different in content.
Several protein factors involved in chemotaxis-type signalling pathways are conserved between the two genomes: G. sulfurreducens and G. metallireducens each possess four or five CheA sensor kinases and ten CheY response receivers, almost all of which are orthologous pairs (Additional file 14: Table S9). In contrast, 17 of the 34 methyl-accepting chemotaxis proteins (MCPs) of G. sulfurreducens have no full-length matches in G. metallireducens (Additional file 14: Table S9). Due to apparent gene family expansion in G. sulfurreducens, its remaining 17 MCPs correspond to only 13 MCPs of G. metallireducens (Additional file 14: Table S9). The other five MCPs of G. metallireducens lack full-length matches in other Geobacteraceae (Additional file 14: Table S9). Whereas G. sulfurreducens may use its closely related MCPs to fine-tune its chemotactic responses, G. metallireducens may accomplish response modulation by having twice as many MCP methyltransferases (CheR) and methylesterases (CheB) as G. sulfurreducens (Additional file 14: Table S9).
Integration host factors (IHF) and histone-like (HU) DNA-binding proteins are global regulators of gene expression composed of two homologous proteins that bend DNA in specific locations [117]. IHF/HU binding sites are favoured by some mobile genetic elements for insertion. The genome of G. metallireducens encodes orthologs of the single HU protein, both IHF beta proteins, and one of two IHF alpha proteins of G. sulfurreducens (Table 4). Another HU gene and two additional IHF alpha genes are present in G. metallireducens but not G. sulfurreducens (Table 4). Three sets of putative global regulatory elements unique to the G. metallireducens genome (Additional files 7,8,9: Figures S3, S4 and S5, Additional file 5: Table S4) may be recognized by different combinations of IHF/HU proteins. A fourth set found in G. metallireducens (Additional file 15: Figure S6, Additional file 5: Table S4) is similar to multicopy sequences in many other genomes. Two transposons (ISGme8 and ISGme9) were found inserted near putative IHF/HU-binding sites of Class 1 (Additional file 5: Table S4). No such putative global regulatory sequence elements were identified in G. sulfurreducens. However, pirin, a Fe(II)-binding protein that associates with DNA in eukaryotic nuclei [118,119], is present in G. sulfurreducens as GSU0825, but in G. metallireducens only as a frameshifted fragment, Gmet_3471. These genetic differences indicate that the proteins that decorate and bend the chromosome are very different in the two species.
Table 4.
Integration host factor (IHF) and histone-like (HU) genes of G. metallireducens and G. sulfurreducens.
| Locus Tag | G. metallireducens gene | G. sulfurreducens gene |
|---|---|---|
| ihfA-1 | Gmet_1417 | GSU1521 |
| ihfA-2 | none | GSU2120 |
| ihfA-3 | Gmet_3057 | none |
| ihfA-4 | Gmet_3056* | none |
| ihfB-1 | Gmet_1833 | GSU1746 |
| ihfB-2 | Gmet_0868 | GSU2602 |
| hup-1 | Gmet_0355 | GSU3132 |
| hup-2 | Gmet_1608 | none |
*Gmet_3056 is frameshifted near the N-terminus, but may be expressed from an internal start codon.
The functions and associations of the various IHF alpha (ihfA), IHF beta (ihfB), and HU (hup) genes are yet unknown, as is their correspondence to any of the predicted regulatory sites illustrated in Figures S3, S4, S5, and S6.
Although no quorum sensing through N-acylhomoserine lactones (autoinducers) has ever been demonstrated for any Geobacteraceae, this kind of signalling may be possible for G. metallireducens because it possesses a LuxR family transcriptional regulator with an autoinducer-binding domain (Gmet_1513), and two divergently transcribed genes with weak sequence similarity to autoinducer synthetases (Gmet_2037 and Gmet_2038). Both Gmet_2037 and Gmet_2038 have atypically low G+C content (Additional file 1: Table S1) and may have been recently acquired by G. metallireducens. The presence of a conserved nucleotide sequence on the 5' side of Gmet_2037 and in 15 other locations on the chromosome (Additional file 16: Figure S7, Additional file 5: Table S4) suggests that Gmet_2037 may be an unusual autoinducer synthetase that is regulated by a riboswitch rather than an autoinducer-binding protein. This conserved sequence is also found on the 5' side of many genes (frequently c-type cytochromes) in the genomes of G. sulfurreducens, G. uraniireducens, and P. propionicus, and overlaps with predicted cyclic diguanylate-responsive riboswitches [120].
The genomes of G. metallireducens and G. sulfurreducens differ in several other aspects of regulation. Nine pairs of potential toxins and antitoxins were identified in the G. metallireducens genome (Additional file 17: Table S10), which may poison vital cellular processes in response to stimuli that interfere with their autoregulation. Only one of these was similar to one of the five potential toxin/antitoxin pairs of G. sulfurreducens. Both the CRISPR1 and CRISPR2 (clustered regularly interspaced short palindromic repeat) loci of G. sulfurreducens, thought to encode 181 short RNAs that may provide immunity against infection by unidentified phage and plasmids [121,122], have no parallel in G. metallireducens, which has CRISPR3 (also found in G. uraniireducens) instead, encoding only twelve putative short RNAs of more variable length and unknown target specificity (Additional file 18: Table S11). Another difference in RNA-level regulation is that a single-stranded RNA-specific nuclease of the barnase family (Gmet_2616) and its putative cognate inhibitor of the barstar family (Gmet_2617) are present in G. metallireducens but not G. sulfurreducens.
Several conserved nucleotide sequences were identified by comparison of intergenic regions between the G. sulfurreducens and G. metallireducens genomes, and those that are found in multiple copies (Additional file 19: Figure S8, Additional file 5: Table S4) may give rise to short RNAs with various regulatory or catalytic activities.
Conclusion
Inspection of the G. metallireducens genome indicates that this species has many metabolic capabilities not present in G. sulfurreducens, particularly with respect to the metabolism of organic acids. Many biosynthetic pathways and regulatory features are conserved, but several putative global regulator-binding sites are unique to G. metallireducens. The complement of signalling proteins is significantly different between the two genomes. Thus, the genome of G. metallireducens provides valuable information about conserved and variable aspects of metabolism, physiology and genetics of the Geobacteraceae.
Methods
Sequence analysis and annotation
The genome of G. metallireducens GS-15 [31] was sequenced by the Joint Genome Institute from cosmid and fosmid libraries. Two gene modeling programs – Critica (v1.05), and Glimmer (v2.13) – were run on both replicons [GenBank:NC007517, GenBank:NC007515], using default settings that permit overlapping genes and using ATG, GTG, and TTG as potential starts. The results were combined, and a BLASTP search of the translations vs. Genbank's non-redundant database (NR) was conducted. The alignment of the N-terminus of each gene model vs. the best NR match was used to pick a preferred gene model. If no BLAST match was returned, the longest model was retained. Gene models that overlapped by greater than 10% of their length were flagged for revision or deletion, giving preference to genes with a BLAST match. The revised gene/protein set was searched against the Swiss-Prot/TrEMBL, PRIAM, Pfam, TIGRFam, Interpro, KEGG, and COGs databases, in addition to BLASTP vs. NR. From these results, product assignments were made. Initial criteria for automated functional assignment set priority based on PRIAM, TIGRFam, Pfam, Interpro profiles, pairwise BLAST vs. Swiss-Prot/TrEMBL, KEGG, and COG groups. tRNAs were annotated using tRNAscan-SE (v1.23). rRNAs were annotated using a combination of BLASTN and an rRNA-specific database. The srpRNA was located using the SRPscan website. The rnpB and tmRNA were located using the Rfam database and Infernal. Riboswitches and other noncoding RNAs predicted in the G. sulfurreducens genome [GenBank:NC00293] were retrieved from the Rfam database [123] and used to annotate the corresponding sequences in G. metallireducens.
Operon organization was predicted using the commercial version of the FGENESB software (V. Solovyev and A. Salamov, unpublished; Softberry, Inc; 2003–2007), with sequence parameters estimated separately from the G. sulfurreducens and G. metallireducens genomes. Default parameters were used in operon prediction, including minimum ORF length of 100 bp.
Binding sites of the global regulator ModE (consensus ATCGCTATATANNNNNNTATATAACGAT) were predicted using ScanACE software [41,42] using the algorithm of Berg and von Hippel [124] and the footprinted matrix of E. coli ModE-regulated sites from the Regulon DB database v 4.0 [125]. Functional annotations of transport proteins were evaluated by referring to TCDB http://www.tcdb.org, and PORES http://garlic.mefos.hr/pores was used to annotate porins. Transposase families were assigned ISGme numbers for inclusion in the ISFinder database http://www-is.biotoul.fr.
Manual curation
The automated genome annotation of G. metallireducens was queried with the protein BLAST algorithm [126] using all predicted proteins in the automated annotation of the G. sulfurreducens genome [12] to identify conserved genes that aligned over their full lengths. The coordinates of numerous genes in both genomes were adjusted according to the criteria of full-length alignment, plausible ribosome-binding sites, and minimal overlap between genes on opposite DNA strands. The annotations of all other genes in G. metallireducens were checked by BLAST searches of NR. Discrepancies in functional annotation of conserved genes between the two genomes were also resolved by BLAST of NR and of the Swiss-Prot database. All hypothetical proteins were checked for similarity to previously identified domains, conservation among other Geobacteraceae, and absence from species other than Geobacteraceae. Genes that had no protein-level homologs in NR were checked (together with flanking intergenic sequences) by translated nucleotide BLAST in all six reading frames, and by nucleotide BLAST to ensure that conserved protein-coding or nucleotide features had not been missed. All intergenic regions of 120 bp or larger were also checked, which led to the annotation of numerous conserved nucleotide sequences numbered as follows: Gmet_R#### (for predicted RNAs and miscellaneous conserved sequences, a nonzero first digit indicating membership in a group of four or more sequences); Gmet_P#### (for conserved, putative regulatory sequences 5' of predicted operons, numbers corresponding to the first gene of the operon); Gmet_I[1-4]## [A, B] (for the four classes of putative global regulator binding sites, mostly found in pairs); Gmet_H4## (for putative global regulatory elements consisting of four tandem heptanucleotide repeats); and Gmet_C### (for the spacers of clustered regularly interspaced short palindromic repeats – CRISPR). Newly added features in the G. sulfurreducens genome were assigned unique numbers with decimal points (GSU####.#) in accordance with earlier corrections.
Phylogenetic analysis
Phylogenetic analysis of selected proteins was performed on alignments generated using T-COFFEE [127], manually corrected in Mesquite [128]. Phylogenetic trees were constructed by the neighbour-joining method using Phylip software [129], with 500 bootstrap replications.
Abbreviations
ATP: adenosine triphosphate; CoA: coenzyme A; CRISPR: clustered regularly interspaced short palindromic repeats; DAHP: 3-deoxy-D-arabino-heptulosonate-7-phosphate; DNA: deoxyribonucleic acid; GDP: guanosine diphosphate; GTP: guanosine triphosphate; HAD: haloacid dehalogenase; HU: histone-like DNA-binding proteins; IHF: integration host factors; MCP: methyl-accepting chemotaxis protein; NAD(H): nicotinamide adenine dinucleotide (reduced); NADP(H): nicotinamide adenine dinucleotide phosphate (reduced); RNA: ribonucleic acid; SAM: S-adenosylmethionine; TCA: tricarboxylic acid; tRNA: transfer RNA; UDP: uridine diphosphate
Authors' contributions
AL supervised the genome sequencing, GD performed genome sequence finishing, and ML supervised the automated annotation process. JK predicted ModE binding sites. MA performed manual curation of the genome annotations, sequence alignments and phylogenetic analyses, and wrote the manuscript. DL conceived of the study and offered guidance with the writing. All authors read, assisted with editing, and approved the final manuscript.
Supplementary Material
Table S1. Genes of G. metallireducens with atypical G+C content (more than two standard deviations from the mean). This table lists genes of G. metallireducens that have G+C content more than two standard deviations from the mean, and indicates by shading (alternated for contrast) those gene clusters that may be recent acquisitions.
Table S2. Enzymes of acyl-CoA metabolism in G. sulfurreducens and G. metallireducens. This table compares the genes predicted to function in acyl-CoA metabolism in G. sulfurreducens and G. metallireducens.
Table S3. Predicted binding sites of the global regulator ModE in the genome of G. sulfurreducens, which are mostly absent from the G. metallireducens genome. This table lists the predicted ModE-binding sites of G. sulfurreducens and compares them to the corresponding sequences in G. metallireducens.
Figure S1. A family of 24 predicted short RNA elements in the G. metallireducens genome. This is an alignment of 24 DNA sequences that were matched by nucleotide-level BLAST. Each RNA is found in an intergenic region, e.g. the 5' regions of genes affecting lysine/arginine metabolism, and contains a central palindromic structure GRCGTAGCGCTGCTACGCC. Similar sequences were found in the genomes of G. sulfurreducens, G. uraniireducens, and Desulfotalea psychrophila. The sequence strand and start and stop nucleotide positions are indicated.
Table S4. Genes found next to multicopy nucleotide sequences of unknown function in G. metallireducens. This table lists the genes adjacent to all of the multicopy nucleotide sequences identified in the G. metallireducens genome.
Figure S2. A family of 49 predicted regulatory RNA elements in G. metallireducens, containing four heptanucleotide repeats (consensus GGACCGG). This is an alignment of 49 DNA sequences that were matched by nucleotide-level BLAST. These elements are found within genes, sometimes more than once per gene, as well as between genes. The sequence strand and start and stop nucleotide positions are indicated.
Figure S3. Predicted global regulator binding sites (class 1). This is an alignment of 48 DNA sequences that were matched by nucleotide-level BLAST. Each site contains four tandem octanucleotide repeats (consensus GTTGCTYN), the outer two being poorly conserved. The distance between each pair of sites (on opposite strands) is variable. Each sequence begins at the right extremity of the top line (the 3' side of the "-" strand of the chromosome), loops on the left side (switching strands), and continues to the right extremity of the bottom line (the 3' side of the "+" strand of the chromosome); start and stop nucleotide positions are indicated. Insertion sequences of the ISGme8 or ISGme9 families may be found at a fixed distance from either or both sites of a pair; these occurrences are indicated on the appropriate lines.
Figure S4. Predicted global regulator binding sites (class 2). This is an alignment of 47 DNA sequences that were matched by nucleotide-level BLAST. Each of 21 paired sites, four sites that also belong to class 1, and one possibly vestigial unpaired site contains three tandem repeats (consensus TCTCCGTS[Y]). The distance between each pair of sites (on opposite strands) is variable. Each sequence begins at the right extremity of the top line (the 3' side of the "-" strand of the chromosome), loops on the left side (switching strands), and continues to the right extremity of the bottom line (the 3' side of the "+" strand of the chromosome); start and stop nucleotide positions are indicated.
Figure S5. Predicted global regulator binding sites (class 3). This is an alignment of 16 DNA sequences that were matched by nucleotide-level BLAST. Fifteen of the sites consist of five tandem heptanucleotide repeats (consensus MTYCTGA). Each sequence begins at the right extremity of the top line (the 3' side of the "-" strand of the chromosome), loops on the left side (switching strands), and continues to the right extremity of the bottom line (the 3' side of the "+" strand of the chromosome); start and stop nucleotide positions are indicated.
Table S5. Cytochrome c biogenesis gene clusters of G. sulfurreducens and G. metallireducens, and associated c-type cytochromes. This table compares the clusters of genes predicted to be involved in biogenesis of c-type cytochromes in G. sulfurreducens and G. metallireducens.
Table S6. Transport systems of G. sulfurreducens and G. metallireducens. This table compares the genes predicted to be involved in transport of solutes across the cell membrane and cell wall of G. sulfurreducens and G. metallireducens.
Table S7. Sensor histidine kinases (HATPase_c domain proteins), REC domain-containing proteins, and transcriptional regulators of G. metallireducens. This table compares the genes predicted to be involved in two-component signalling and transcriptional regulation in G. sulfurreducens and G. metallireducens.
Table S8. Diguanylate cyclases (GGDEF domain proteins) of G. sulfurreducens and G. metallireducens. This table compares the genes predicted to produce the intracellular messenger cyclic diguanylate in G. sulfurreducens and G. metallireducens.
Table S9. Chemotaxis-type signalling proteins of G. sulfurreducens and G. metallireducens. This table compares the genes predicted to participate in chemotaxis-type signalling in G. sulfurreducens and G. metallireducens.
Figure S6. Predicted global regulator binding sites (class 4). This is an alignment of 20 DNA sequences that were matched by nucleotide-level BLAST. Each site appears to be based on a pentanucleotide repeat (consensus CCYTC) that occurs four times on one strand and twice on the other. The sequence strand and start and stop nucleotide positions are indicated.
Figure S7. A predicted regulatory short RNA found in the 5' regions of c-type cytochromes and other proteins. This is an alignment of 16 DNA sequences that were matched by nucleotide-level BLAST. The location of Gmet_R3013 suggests that N-acylhomoserine lactone signalling may be under control of this RNA element. Similar sequences were found in the genomes of G. sulfurreducens, G. uraniireducens, and P. propionicus. The sequence strand and start and stop nucleotide positions are indicated.
Table S10. Toxin/antitoxin pairs of G. metallireducens and G. sulfurreducens. This table compares the genes predicted to encode toxin/antitoxin pairs in G. sulfurreducens and G. metallireducens.
Table S11. The CRISPR3 locus of G. metallireducens contains spacers of variable length. The thirteen clustered regularly interspaced short palindromic repeats (CRISPR) of G. metallireducens (consensus sequence GTAGCGCCCGCCTACATAGGCGGGCGAGGATTGAAAC) are far fewer than the thirty-eight of CRISPR1 and one hundred and forty-three of CRISPR2 in G. sulfurreducens.
Figure S8. Miscellaneous multicopy nucleotide sequences found in the G. metallireducens genome. These are alignments of 16 sets of miscellaneous DNA sequences in G. metallireducens that were matched by nucleotide-level BLAST. The sequence strand and start and stop nucleotide positions are indicated. (a) A palindromic sequence-containing family also found in the genomes of G. sulfurreducens, G. uraniireducens, and P. propionicus. (b) Sequences of this type were also found in the genomes of G. sulfurreducens and G. uraniireducens. (c) These sequences are unique to G. metallireducens. (d) The ends of these sequences form inverted repeats. Each sequence begins at the left extremity of the top line (the 5' side of the "+" strand of the chromosome), loops on the right side (switching strands), and continues to the left extremity of the bottom line (the 5' side of the "-" strand of the chromosome). A fragment related to Gmet_R6002 was found in the G. sulfurreducens genome. (e) These sequences are unique to G. metallireducens. (f) Sequences of this type were also found in the genomes of G. uraniireducens and G. bemidjiensis. (g) These sequences contain four octanucleotide repeats (consensus TWGTTGAY), two in tandem on each strand. (h) Sequences of this type were also found in the genome of G. sulfurreducens. (i) These sequences are unique to G. metallireducens. (j) These elements are located near each other. (k) These sequences are unique to G. metallireducens. (l-p) These elements are located near each other. Gmet_R0147 continues as Gmet_R0055, a tRNA-Asn gene (underlined).
Contributor Information
Muktak Aklujkar, Email: muktak@microbio.umass.edu.
Julia Krushkal, Email: jkrushka@utmem.edu.
Genevieve DiBartolo, Email: gen.dibartolo@gmail.com.
Alla Lapidus, Email: ALapidus@lbl.gov.
Miriam L Land, Email: landml@ornl.gov.
Derek R Lovley, Email: dlovley@microbio.umass.edu.
Acknowledgements
We thank Maddalena Coppi, Jessica Butler, Ned Young, Mounir Izallalen and Radhakrishnan Mahadevan for helpful discussions. We also thank Jose F. Barbe and Marko Puljic for technical assistance. This research was supported by the Office of Science (Biological and Environmental Research), U.S. Department of Energy (Grant No. DE-FC02-02ER63446).
References
- Lovley DR. Dissimilatory Fe(III) and Mn(IV) reduction. Microbiol Rev. 1991;55:259–287. doi: 10.1128/mr.55.2.259-287.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley DR, Holmes DE, Nevin KP. Dissimilatory Fe(III) and Mn(IV) reduction. Adv Microb Physiol. 2004;49:219–286. doi: 10.1016/S0065-2911(04)49005-5. [DOI] [PubMed] [Google Scholar]
- Lovley DR, Stolz JF, Nord GLJ, Phillips EJP. Anaerobic production of magnetite by a dissimilatory iron-reducing microorganism. Nature. 1987;330:252–254. doi: 10.1038/330252a0. [DOI] [Google Scholar]
- Lovley DR, Phillips EJ. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl Environ Microbiol. 1988;54:1472–1480. doi: 10.1128/aem.54.6.1472-1480.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley DR, Baedecker MJ, Lonergan DJ, Cozzarelli IM, Phillips EJP, Siegel DI. Oxidation of aromatic contaminants coupled to microbial iron reduction. Nature. 1989;339:297–299. doi: 10.1038/339297a0. [DOI] [Google Scholar]
- Lovley DR, Lonergan DJ. Anaerobic oxidation of toluene, phenol, and p-cresol by the dissimilatory iron-reducing organism, GS-15. Appl Environ Microbiol. 1990;56:1858–1864. doi: 10.1128/aem.56.6.1858-1864.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley DR, Phillips EJP, Gorby YA, Landa ER. Microbial reduction of uranium. Nature. 1991;350:413–416. doi: 10.1038/350413a0. [DOI] [Google Scholar]
- Lovley DR, Coates JD, Blunt-Harris EL, Phillips EJP, Woodward JC. Humic substances as electron acceptors for microbial respiration. Nature (Letters) 1996;382:445–447. doi: 10.1038/382445a0. [DOI] [Google Scholar]
- Childers SE, Ciufo S, Lovley DR. Geobacter metallireducens accesses insoluble Fe(III) oxide by chemotaxis. Nature. 2002;416:767–769. doi: 10.1038/416767a. [DOI] [PubMed] [Google Scholar]
- Bond DR, Holmes DE, Tender LM, Lovley DR. Electrode-reducing microorganisms that harvest energy from marine sediments. Science. 2002;295:483–485. doi: 10.1126/science.1066771. [DOI] [PubMed] [Google Scholar]
- Gregory KB, Bond DR, Lovley DR. Graphite electrodes as electron donors for anaerobic respiration. Environ Microbiol. 2004;6:596–604. doi: 10.1111/j.1462-2920.2004.00593.x. [DOI] [PubMed] [Google Scholar]
- Methé BA, Nelson KE, Eisen JA, Paulsen IT, Nelson W, Heidelberg JF, Wu D, Wu M, Ward N, Beanan MJ, Dodson RJ, Madupu R, Brinkac LM, Daugherty SC, DeBoy RT, Durkin AS, Gwinn M, Kolonay JF, Sullivan SA, Haft DH, Selengut J, Davidsen TM, Zafar N, White O, Tran B, Romero C, Forberger HA, Weidman J, Khouri H, Feldblyum TV, Utterback TR, Van Aken SE, Lovley DR, Fraser CM. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science. 2003;302:1967–1969. doi: 10.1126/science.1088727. [DOI] [PubMed] [Google Scholar]
- Khan SA. Plasmid rolling-circle replication: highlights of two decades of research. Plasmid. 2005;53:126–136. doi: 10.1016/j.plasmid.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Lovley DR, Chapelle FH. Deep subsurface microbial processes. Rev Geophys. 1995;33:365–381. doi: 10.1029/95RG01305. [DOI] [Google Scholar]
- Anderson RT, Vrionis HA, Ortiz-Bernad I, Resch CT, Long PE, Dayvault R, Karp K, Marutzky S, Metzler DR, Peacock A, White DC, Lowe M, Lovley DR. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl Environ Microbiol. 2003;69:5884–5891. doi: 10.1128/AEM.69.10.5884-5891.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes DE, O'Neil RA, Vrionis HA, N'Guessan LA, Ortiz-Bernad I, Larrahando MJ, Adams LA, Ward JA, Nicoll JS, Nevin KP, Chavan MA, Johnson JP, Long PE, Lovley DR. Subsurface clade of Geobacteraceae that predominates in a diversity of Fe(III)-reducing subsurface environments. ISME J. 2007;1:663–677. doi: 10.1038/ismej.2007.85. [DOI] [PubMed] [Google Scholar]
- Segura D, Mahadevan R, Juarez K, Lovley DR. Computational and experimental analysis of redundancy in the central metabolism of Geobacter sulfurreducens . PLoS Comput Biol. 2008;4:e36. doi: 10.1371/journal.pcbi.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ. The acetate switch. Microbiol Mol Biol Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy FJ, Waters DA, Takova TY, Henkin TM. Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis . Mol Microbiol. 1993;10:259–271. doi: 10.1111/j.1365-2958.1993.tb01952.x. [DOI] [PubMed] [Google Scholar]
- Gerhardt A, Cinkaya I, Linder D, Huisman G, Buckel W. Fermentation of 4-aminobutyrate by Clostridium aminobutyricum: cloning of two genes involved in the formation and dehydration of 4-hydroxybutyryl-CoA. Arch Microbiol. 2000;174:189–199. doi: 10.1007/s002030000195. [DOI] [PubMed] [Google Scholar]
- Butler JE, He Q, Nevin KP, He Z, Zhou J, Lovley DR. Genomic and microarray analysis of aromatics degradation in Geobacter metallireducens and comparison to a Geobacter isolate from a contaminated field site. BMC Genomics. 2007;8:180. doi: 10.1186/1471-2164-8-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters F, Heintz D, Johannes J, van Dorsselaer A, Boll M. Genes, enzymes, and regulation of para-cresol metabolism in Geobacter metallireducens . J Bacteriol. 2007;189:4729–4738. doi: 10.1128/JB.00260-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wischgoll S, Heintz D, Peters F, Erxleben A, Sarnighausen E, Reski R, van Dorsselaer A, Boll M. Gene clusters involved in anaerobic benzoate degradation of Geobacter metallireducens . Mol Microbiol. 2005;58:1238–1252. doi: 10.1111/j.1365-2958.2005.04909.x. [DOI] [PubMed] [Google Scholar]
- Caccavo F Jr, Lonergan DJ, Lovley DR, Davis M, Stolz JF, McInerney MJ. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl Environ Microbiol. 1994;60:3752–3759. doi: 10.1128/aem.60.10.3752-3759.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuthner B, Heider J. Anaerobic toluene catabolism of Thauera aromatica: the bbs operon codes for enzymes of beta oxidation of the intermediate benzylsuccinate. J Bacteriol. 2000;182:272–277. doi: 10.1128/JB.182.2.272-277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wischgoll S, Taubert M, Peters F, Jehmlich N, von Bergen M, Boll M. Decarboxylating and non-decarboxylating glutaryl-CoA dehydrogenases in the aromatic metabolism of obligately anaerobic bacteria. J Bacteriol. 2009. [DOI] [PMC free article] [PubMed]
- Faivre-Nitschke SE, Couee I, Vermel M, Grienenberger J-M, Gualberto JM. Purification, characterization and cloning of isovaleryl-CoA dehydrogenase from higher plant mitochondria. Eur J Biochem. 2001;268:1332–1339. doi: 10.1046/j.1432-1327.2001.01999.x. [DOI] [PubMed] [Google Scholar]
- Huang KX, Huang S, Rudolph FB, Bennett GN. Identification and characterization of a second butyrate kinase from Clostridium acetobutylicum ATCC 824. J Mol Microbiol Biotechnol. 2000;2:33–38. [PubMed] [Google Scholar]
- Oultram JD, Burr ID, Elmore MJ, Minton NP. Cloning and sequence analysis of the genes encoding phosphotransbutyrylase and butyrate kinase from Clostridium acetobutylicum NCIMB 8052. Gene. 1993;131:107–112. doi: 10.1016/0378-1119(93)90677-U. [DOI] [PubMed] [Google Scholar]
- Bond DR, Mester T, Nesbo CL, Izquierdo-Lopez AV, Collart FL, Lovley DR. Characterization of citrate synthase from Geobacter sulfurreducens and evidence for a family of citrate synthases similar to those of eukaryotes throughout the Geobacteraceae. Appl Environ Microbiol. 2005;71:3858–3865. doi: 10.1128/AEM.71.7.3858-3865.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley DR, Giovannoni SJ, White DC, Champine JE, Phillips EJ, Gorby YA, Goodwin S. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch Microbiol. 1993;159:336–344. doi: 10.1007/BF00290916. [DOI] [PubMed] [Google Scholar]
- Iwakura M, Tokushige M, Katsuki H. Studies on regulatory functions of malic enzymes. VII. Structural and functional characteristics of sulfhydryl groups in NADP-linked malic enzyme from Escherichia coli W. J Biochem. 1979;86:1239–1249. doi: 10.1093/oxfordjournals.jbchem.a132639. [DOI] [PubMed] [Google Scholar]
- Lerondel G, Doan T, Zamboni N, Sauer U, Aymerich S. YtsJ has the major physiological role of the four paralogous malic enzyme isoforms in Bacillus subtilis . J Bacteriol. 2006;188:4727–4736. doi: 10.1128/JB.00167-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YJ, Chakraborty R, Martin HG, Chu J, Hazen TC, Keasling JD. Flux analysis of central metabolic pathways in Geobacter metallireducens during reduction of soluble Fe(III)-nitrilotriacetic acid. Appl Environ Microbiol. 2007;73:3859–3864. doi: 10.1128/AEM.02986-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JE, Glaven RH, Esteve-Nunez A, Nunez C, Shelobolina ES, Bond DR, Lovley DR. Genetic characterization of a single bifunctional enzyme for fumarate reduction and succinate oxidation in Geobacter sulfurreducens and engineering of fumarate reduction in Geobacter metallireducens. J Bacteriol. 2006;188:450–455. doi: 10.1128/JB.188.2.450-455.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood NJ, Alizadeh T, Richardson DJ, Ferguson SJ, Moir JW. Two domains of a dual-function NarK protein are required for nitrate uptake, the first step of denitrification in Paracoccus pantotrophus . Mol Microbiol. 2002;44:157–170. doi: 10.1046/j.1365-2958.2002.02859.x. [DOI] [PubMed] [Google Scholar]
- Lloyd JR, Leang C, Hodges Myerson AL, Coppi MV, Ciufo S, Methé B, Sandler SJ, Lovley DR. Biochemical and genetic characterization of PpcA, a periplasmic c-type cytochrome in Geobacter sulfurreducens. Biochem J. 2003;369:153–161. doi: 10.1042/BJ20020597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley DR, Fraga JL, Coates JD, Blunt-Harris EL. Humics as an electron donor for anaerobic respiration. Environ Microbiol. 1999;1:89–98. doi: 10.1046/j.1462-2920.1999.00009.x. [DOI] [PubMed] [Google Scholar]
- Nohno T, Kasai Y, Saito T. Cloning and sequencing of the Escherichia coli chlEN operon involved in molybdopterin biosynthesis. J Bacteriol. 1988;170:4097–4102. doi: 10.1128/jb.170.9.4097-4102.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers SL, McNairn E, Blasco F, Giordano G, Boxer DH. Molecular genetic analysis of the moa operon of Escherichia coli K-12 required for molybdenum cofactor biosynthesis. Mol Microbiol. 1993;8:1071–1081. doi: 10.1111/j.1365-2958.1993.tb01652.x. [DOI] [PubMed] [Google Scholar]
- Robison K, McGuire AM, Church GM. A comprehensive library of DNA-binding site matrices for 55 proteins applied to the complete Escherichia coli K-12 genome. J Mol Biol. 1998;284:241–254. doi: 10.1006/jmbi.1998.2160. [DOI] [PubMed] [Google Scholar]
- Roth FP, Hughes JD, Estep PW, Church GM. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat Biotechnol. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- Yan B, Lovley DR, Krushkal J. Genome-wide similarity search for transcription factors and their binding sites in a metal-reducing prokaryote Geobacter sulfurreducens . Biosystems. 2007;90:421–441. doi: 10.1016/j.biosystems.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Battchikova N, Koivulehto M, Denesyuk A, Ptitsyn L, Boretsky Y, Hellman J, Korpela T. Aspartate aminotransferase from an alkalophilic Bacillus contains an additional 20-amino acid extension at its functionally important N-terminus. J Biochem. 1996;120:425–432. doi: 10.1093/oxfordjournals.jbchem.a021429. [DOI] [PubMed] [Google Scholar]
- Alfano JR, Kahn ML. Isolation and characterization of a gene coding for a novel aspartate aminotransferase from Rhizobium meliloti . J Bacteriol. 1993;175:4186–4196. doi: 10.1128/jb.175.13.4186-4196.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Fujita Y, Ehrlich SD. Three asparagine synthetase genes of Bacillus subtilis . J Bacteriol. 1999;181:6081–6091. doi: 10.1128/jb.181.19.6081-6091.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmorjani K, Liotenberg S, Houmard J, de Marsac NT. Molecular characterization of the gene encoding glutamine synthetase in the cyanobacterium Calothrix sp. PCC 7601. Biochem Biophys Res Commun. 1992;189:1296–1302. doi: 10.1016/0006-291X(92)90214-6. [DOI] [PubMed] [Google Scholar]
- Curnow AW, Hong K, Yuan R, Kim S, Martins O, Winkler W, Henkin TM, Soll D. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci USA. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TT, Williams S, Schweizer HP, Lam JS. Molecular genetic analysis of the region containing the essential Pseudomonas aeruginosa asd gene encoding aspartate-beta-semialdehyde dehydrogenase. Microbiology. 1997;143:899–907. doi: 10.1099/00221287-143-3-899. [DOI] [PubMed] [Google Scholar]
- Cirillo JD, Weisbrod TR, Pascopella L, Bloom BR, Jacobs WR Jr. Isolation and characterization of the aspartokinase and aspartate semialdehyde dehydrogenase operon from mycobacteria. Mol Microbiol. 1994;11:629–639. doi: 10.1111/j.1365-2958.1994.tb00342.x. [DOI] [PubMed] [Google Scholar]
- Singh SK, Yang K, Karthikeyan S, Huynh T, Zhang X, Phillips MA, Zhang H. The thrH gene product of Pseudomonas aeruginosa is a dual activity enzyme with a novel phosphoserine:homoserine phosphotransferase activity. J Biol Chem. 2004;279:13166–13173. doi: 10.1074/jbc.M311393200. [DOI] [PubMed] [Google Scholar]
- Martin C, Cami B, Yeh P, Stragier P, Parsot C, Patte JC. Pseudomonas aeruginosa diaminopimelate decarboxylase: evolutionary relationship with other amino acid decarboxylases. Mol Biol Evol. 1988;5:549–559. doi: 10.1093/oxfordjournals.molbev.a040515. [DOI] [PubMed] [Google Scholar]
- Stragier P, Danos O, Patte JC. Regulation of diaminopimelate decarboxylase synthesis in Escherichia coli. II. Nucleotide sequence of the lysA gene and its regulatory region. J Mol Biol. 1983;168:321–331. doi: 10.1016/S0022-2836(83)80021-7. [DOI] [PubMed] [Google Scholar]
- Hudson AO, Gilvarg C, Leustek T. Biochemical and phylogenetic characterization of a novel diaminopimelate biosynthesis pathway in prokaryotes identifies a diverged form of LL-diaminopimelate aminotransferase. J Bacteriol. 2008;190:3256–3263. doi: 10.1128/JB.01381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhy P, Martel A, Margarita D, Saint Girons I, Belfaiza J. Homoserine O-acetyltransferase, involved in the Leptospira meyeri methionine biosynthetic pathway, is not feedback inhibited. J Bacteriol. 1997;179:4396–4398. doi: 10.1128/jb.179.13.4396-4398.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobric N, Limsowtin GK, Hillier AJ, Dudman NP, Davidson BE. Identification and characterization of a cystathionine beta/gamma-lyase from Lactococcus lactis ssp. cremoris MG1363. FEMS Microbiol Lett. 2000;182:249–254. doi: 10.1111/j.1574-6968.2000.tb08903.x. [DOI] [PubMed] [Google Scholar]
- Fernandez M, van Doesburg W, Rutten GA, Marugg JD, Alting AC, van Kranenburg R, Kuipers OP. Molecular and functional analyses of the metC gene of Lactococcus lactis, encoding cystathionine beta-lyase. Appl Environ Microbiol. 2000;66:42–48. doi: 10.1128/AEM.66.1.42-48.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sie'nko M, Topczewski J, Paszewski A. Structure and regulation of cysD, the homocysteine synthase gene of Aspergillus nidulans. Curr Genet. 1998;33:136–144. doi: 10.1007/s002940050319. [DOI] [PubMed] [Google Scholar]
- Yura T, Mori H, Nagai H, Nagata T, Ishihama A, Fujita N, Isono K, Mizobuchi K, Nakata A. Systematic sequencing of the Escherichia coli genome: analysis of the 0–2.4 min region. Nucleic Acids Res. 1992;20:3305–3308. doi: 10.1093/nar/20.13.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis . Cell. 1993;74:475–482. doi: 10.1016/0092-8674(93)80049-K. [DOI] [PubMed] [Google Scholar]
- Shultz J, Hermodson MA, Garner CC, Herrmann KM. The nucleotide sequence of the aroF gene of Escherichia coli and the amino acid sequence of the encoded protein, the tyrosine-sensitive 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase. J Biol Chem. 1984;259:9655–9661. [PubMed] [Google Scholar]
- Weaver LM, Herrmann KM. Cloning of an aroF allele encoding a tyrosine-insensitive 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase. J Bacteriol. 1990;172:6581–6584. doi: 10.1128/jb.172.11.6581-6584.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Howe DL, Woodard RW. Thermotoga maritima 3-deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthase: the ancestral eubacterial DAHP synthase? J Biol Chem. 2003;278:27525–27531. doi: 10.1074/jbc.M304631200. [DOI] [PubMed] [Google Scholar]
- Fischer RS, Zhao G, Jensen RA. Cloning, sequencing, and expression of the P-protein gene (pheA) of Pseudomonas stutzeri in Escherichia coli: implications for evolutionary relationships in phenylalanine biosynthesis. J Gen Microbiol. 1991;137:1293–1301. doi: 10.1099/00221287-137-6-1293. [DOI] [PubMed] [Google Scholar]
- Shumilin IA, Bauerle R, Wu J, Woodard RW, Kretsinger RH. Crystal structure of the reaction complex of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase from Thermotoga maritima refines the catalytic mechanism and indicates a new mechanism of allosteric regulation. J Mol Biol. 2004;341:455–466. doi: 10.1016/j.jmb.2004.05.077. [DOI] [PubMed] [Google Scholar]
- Merkl R. Modelling the evolution of the archeal tryptophan synthase. BMC Evolutionary Biology. 2007;7:59. doi: 10.1186/1471-2148-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettwer S, Sterner R. A novel tryptophan synthase beta-subunit from the hyperthermophile Thermotoga maritima. Quaternary structure, steady-state kinetics, and putative physiological role. J Biol Chem. 2002;277:8194–8201. doi: 10.1074/jbc.M111541200. [DOI] [PubMed] [Google Scholar]
- Kishan V, Hillen W. Molecular cloning, nucleotide sequence, and promoter structure of the Acinetobacter calcoaceticus trpFB operon. J Bacteriol. 1990;172:6151–6155. doi: 10.1128/jb.172.10.6151-6155.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosselaere F, Lambrecht M, Vanderleyden J. Isolation and sequence analysis of the trpBA gene cluster, encoding tryptophan synthase, from Azospirillum brasilense. DNA Seq. 2000;11:287–293. doi: 10.3109/10425170009033245. [DOI] [PubMed] [Google Scholar]
- Deutch AH, Rushlow KE, Smith CJ. Analysis of the Escherichia coli proBA locus by DNA and protein sequencing. Nucleic Acids Res. 1984;12:6337–6355. doi: 10.1093/nar/12.15.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon DH, Lu CD, Walthall DA, Brown TM, Houghton JE, Abdelal AT. Structure and regulation of the carAB operon in Pseudomonas aeruginosa and Pseudomonas stutzeri: no untranslated region exists. J Bacteriol. 1994;176:2532–2542. doi: 10.1128/jb.176.9.2532-2542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynes JP, Tiraby M, Baron M, Drocourt D, Tiraby G. Escherichia coli thymidylate kinase: molecular cloning, nucleotide sequence, and genetic organization of the corresponding tmk locus. J Bacteriol. 1996;178:2804–2812. doi: 10.1128/jb.178.10.2804-2812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marolewski A, Smith JM, Benkovic SJ. Cloning and characterization of a new purine biosynthetic enzyme: a non-folate glycinamide ribonucleotide transformylase from E. coli . Biochemistry. 1994;33:2531–2537. doi: 10.1021/bi00175a023. [DOI] [PubMed] [Google Scholar]
- Smith JM, Daum HA 3rd. Identification and nucleotide sequence of a gene encoding 5'-phosphoribosylglycinamide transformylase in Escherichia coli K12. J Biol Chem. 1987;262:10565–10569. [PubMed] [Google Scholar]
- Omumasaba CA, Okai N, Inui M, Yukawa H. Corynebacterium glutamicum glyceraldehyde-3-phosphate dehydrogenase isoforms with opposite, ATP-dependent regulation. J Mol Microbiol Biotechnol. 2004;8:91–103. doi: 10.1159/000084564. [DOI] [PubMed] [Google Scholar]
- Sproul AA, Lambourne LT, Jean-Jacques DJ, Kornberg HL. Genetic control of manno(fructo)kinase activity in Escherichia coli . Proc Natl Acad Sci USA. 2001;98:15257–15259. doi: 10.1073/pnas.211569798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Troch P, Keijers V, Vanderleyden J. Sequence analysis of the Azospirillum brasilense exoB gene, encoding UDP-glucose 4'-epimerase. Gene. 1994;144:143–144. doi: 10.1016/0378-1119(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Fernandez-Herrero LA, Badet-Denisot MA, Badet B, Berenguer J. glmS of Thermus thermophilus HB8: an essential gene for cell-wall synthesis identified immediately upstream of the S-layer gene. Mol Microbiol. 1995;17:1–12. doi: 10.1111/j.1365-2958.1995.mmi_17010001.x. [DOI] [PubMed] [Google Scholar]
- Stevenson G, Andrianopoulos K, Hobbs M, Reeves PR. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Dieter U, Starman R, Barr K, Mayer H, Rick PD. Biosynthesis of enterobacterial common antigen in Escherichia coli. Biochemical characterization of Tn10 insertion mutants defective in enterobacterial common antigen synthesis. J Biol Chem. 1990;265:13490–13497. [PubMed] [Google Scholar]
- Namboori SC, Graham DE. Acetamido sugar biosynthesis in the Euryarchaea. J Bacteriol. 2008;190:2987–2996. doi: 10.1128/JB.01970-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruschka L, Adolf K, Burchhardt G, Dernedde J, Jurgensen J, Herrmann H. Analysis of the zwf-pgl-eda-operon in Pseudomonas putida strains H and KT2440. FEMS Microbiol Lett. 2002;215:89–95. doi: 10.1111/j.1574-6968.2002.tb11375.x. [DOI] [PubMed] [Google Scholar]
- Summers ML, Meeks JC, Chu S, Wolf RE Jr. Nucleotide sequence of an operon in Nostoc sp. strain ATCC 29133 encoding four genes of the oxidative pentose phosphate cycle. Plant Physiol. 1995;107:267–268. doi: 10.1104/pp.107.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni N, Fischer E, Laudert D, Aymerich S, Hohmann HP, Sauer U. The Bacillus subtilis yqjI gene encodes the NADP+-dependent 6-P-gluconate dehydrogenase in the pentose phosphate pathway. J Bacteriol. 2004;186:4528–4534. doi: 10.1128/JB.186.14.4528-4534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen KI, Hove-Jensen B. Ribose catabolism of Escherichia coli: characterization of the rpiB gene encoding ribose phosphate isomerase B and of the rpiR gene, which is involved in regulation of rpiB expression. J Bacteriol. 1996;178:1003–1011. doi: 10.1128/jb.178.4.1003-1011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Adams MW. Sulfide dehydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus: a new multifunctional enzyme involved in the reduction of elemental sulfur. J Bacteriol. 1994;176:6509–6517. doi: 10.1128/jb.176.21.6509-6517.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre HJ, Davies H, Hore TA, Miller SH, Dufour JP, Ronson CW. Trehalose biosynthesis in Rhizobium leguminosarum bv. trifolii and its role in desiccation tolerance. Appl Environ Microbiol. 2007;73:3984–3992. doi: 10.1128/AEM.00412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta K, Hattori K, Nakada T, Kubota M, Sugimoto T, Kurimoto M. Cloning and sequencing of trehalose biosynthesis genes from Arthrobacter sp. Q36. Biochim Biophys Acta. 1996;1289:10–13. doi: 10.1016/0304-4165(95)00139-5. [DOI] [PubMed] [Google Scholar]
- Pan YT, Carroll JD, Elbein AD. Trehalose-phosphate synthase of Mycobacterium tuberculosis. Cloning, expression and properties of the recombinant enzyme. Eur J Biochem. 2002;269:6091–6100. doi: 10.1046/j.1432-1033.2002.03327.x. [DOI] [PubMed] [Google Scholar]
- Kaasen I, McDougall J, Strom AR. Analysis of the otsBA operon for osmoregulatory trehalose synthesis in Escherichia coli and homology of the OtsA and OtsB proteins to the yeast trehalose-6-phosphate synthase/phosphatase complex. Gene. 1994;145:9–15. doi: 10.1016/0378-1119(94)90316-6. [DOI] [PubMed] [Google Scholar]
- Butler JE, Kaufmann F, Coppi MV, Nunez C, Lovley DR. MacA, a diheme c-type cytochrome involved in Fe(III) reduction by Geobacter sulfurreducens. J Bacteriol. 2004;186:4042–4045. doi: 10.1128/JB.186.12.4042-4045.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BC, Lovley DR. Investigation of direct vs. indirect involvement of the c-type cytochrome MacA in Fe(III) reduction by Geobacter sulfurreducens. FEMS Microbiol Lett. 2008;286:39–44. doi: 10.1111/j.1574-6968.2008.01252.x. [DOI] [PubMed] [Google Scholar]
- Shelobolina ES, Coppi MV, Korenevsky AA, DiDonato LN, Sullivan SA, Konishi H, Xu H, Leang C, Butler JE, Kim BC, Lovley DR. Importance of c -type cytochromes for U(VI) reduction by Geobacter sulfurreducens. BMC Microbiol. 2007;7:16. doi: 10.1186/1471-2180-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leang C, Coppi MV, Lovley DR. OmcB, a c -type polyheme cytochrome, involved in Fe(III) reduction in Geobacter sulfurreducens. J Bacteriol. 2003;185:2096–2103. doi: 10.1128/JB.185.7.2096-2103.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta T, Coppi MV, Childers SE, Lovley DR. Outer membrane c -type cytochromes required for Fe(III) and Mn(IV) oxide reduction in Geobacter sulfurreducens. Appl Environ Microbiol. 2005;71:8634–8641. doi: 10.1128/AEM.71.12.8634-8641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BC, Leang C, Ding YH, Glaven RH, Coppi MV, Lovley DR. OmcF, a putative c -type monoheme outer membrane cytochrome required for the expression of other outer membrane cytochromes in Geobacter sulfurreducens. J Bacteriol. 2005;187:4505–4513. doi: 10.1128/JB.187.13.4505-4513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey HA, Dailey TA. Protoporphyrinogen oxidase of Myxococcus xanthus. Expression, purification, and characterization of the cloned enzyme. J Biol Chem. 1996;271:8714–8718. doi: 10.1074/jbc.271.15.8714. [DOI] [PubMed] [Google Scholar]
- Sasarman A, Letowski J, Czaika G, Ramirez V, Nead MA, Jacobs JM, Morais R. Nucleotide sequence of the hemG gene involved in the protoporphyrinogen oxidase activity of Escherichia coli K12. Can J Microbiol. 1993;39:1155–1161. doi: 10.1139/m93-174. [DOI] [PubMed] [Google Scholar]
- Sun G, Sharkova E, Chesnut R, Birkey S, Duggan MF, Sorokin A, Pujic P, Ehrlich SD, Hulett FM. Regulators of aerobic and anaerobic respiration in Bacillus subtilis . J Bacteriol. 1996;178:1374–1385. doi: 10.1128/jb.178.5.1374-1385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Harvat EM, Stevens JM, Ferguson SJ, Saier MH Jr. Evolutionary origins of members of a superfamily of integral membrane cytochrome c biogenesis proteins. Biochim Biophys Acta. 2007;1768:2164–2181. doi: 10.1016/j.bbamem.2007.04.022. [DOI] [PubMed] [Google Scholar]
- Reguera G, McCarthy KD, Mehta T, Nicoll JS, Tuominen MT, Lovley DR. Extracellular electron transfer via microbial nanowires. Nature. 2005;435:1098–1101. doi: 10.1038/nature03661. [DOI] [PubMed] [Google Scholar]
- Reguera G, Nevin KP, Nicoll JS, Covalla SF, Woodard TL, Lovley DR. Biofilm and nanowire production leads to increased current in Geobacter sulfurreducens fuel cells. Appl Environ Microbiol. 2006;72:7345–7348. doi: 10.1128/AEM.01444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera G, Pollina RB, Nicoll JS, Lovley DR. Possible nonconductive role of Geobacter sulfurreducens pilus nanowires in biofilm formation. J Bacteriol. 2007;189:2125–2127. doi: 10.1128/JB.01284-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel T, Scheurerpflug I, Meyer TF. Neisseria PilC protein identified as type-4 pilus tip-located adhesin. Nature. 1995;373:357–359. doi: 10.1038/373357a0. [DOI] [PubMed] [Google Scholar]
- Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. Feo – transport of ferrous iron into bacteria. Biometals. 2006;19:143–157. doi: 10.1007/s10534-006-0003-2. [DOI] [PubMed] [Google Scholar]
- Dashper SG, Butler CA, Lissel JP, Paolini RA, Hoffmann B, Veith PD, O'Brien-Simpson NM, Snelgrove SL, Tsiros JT, Reynolds EC. A novel Porphyromonas gingivalis FeoB plays a role in manganese accumulation. J Biol Chem. 2005;280:28095–28102. doi: 10.1074/jbc.M503896200. [DOI] [PubMed] [Google Scholar]
- Naikare H, Palyada K, Panciera R, Marlow D, Stintzi A. Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect Immun. 2006;74:5433–5444. doi: 10.1128/IAI.00052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleyer M, Bakker EP. Nucleotide sequence and 3'-end deletion studies indicate that the K(+)-uptake protein kup from Escherichia coli is composed of a hydrophobic core linked to a large and partially essential hydrophilic C terminus. J Bacteriol. 1993;175:6925–6931. doi: 10.1128/jb.175.21.6925-6931.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse JE, Wieczorek L, Altendorf K, Reicin AS, Dorus E, Epstein W. Sequence homology between two membrane transport ATPases, the Kdp-ATPase of Escherichia coli and the Ca2+-ATPase of sarcoplasmic reticulum. Proc Natl Acad Sci USA. 1984;81:4746–4750. doi: 10.1073/pnas.81.15.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walderhaug MO, Polarek JW, Voelkner P, Daniel JM, Hesse JE, Altendorf K, Epstein W. KdpD and KdpE, proteins that control expression of the kdpABC operon, are members of the two-component sensor-effector class of regulators. J Bacteriol. 1992;174:2152–2159. doi: 10.1128/jb.174.7.2152-2159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radchenko MV, Waditee R, Oshimi S, Fukuhara M, Takabe T, Nakamura T. Cloning, functional expression and primary characterization of Vibrio parahaemolyticus K+/H+ antiporter genes in Escherichia coli. Mol Microbiol. 2006;59:651–663. doi: 10.1111/j.1365-2958.2005.04966.x. [DOI] [PubMed] [Google Scholar]
- Bakker EP, Booth IR, Dinnbier U, Epstein W, Gajewska A. Evidence for multiple K+ export systems in Escherichia coli . J Bacteriol. 1987;169:3743–3749. doi: 10.1128/jb.169.8.3743-3749.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Guffanti AA, Oudega B, Krulwich TA. mrp, a multigene, multifunctional locus in Bacillus subtilis with roles in resistance to cholate and to Na+ and in pH homeostasis. J Bacteriol. 1999;181:2394–2402. doi: 10.1128/jb.181.8.2394-2402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghim SY, Neuhard J. The pyrimidine biosynthesis operon of the thermophile Bacillus caldolyticus includes genes for uracil phosphoribosyltransferase and uracil permease. J Bacteriol. 1994;176:3698–3707. doi: 10.1128/jb.176.12.3698-3707.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes C, Ohtake H, Chu L, Misra TK, Silver S. Cloning, nucleotide sequence, and expression of the chromate resistance determinant of Pseudomonas aeruginosa plasmid pUM505. J Bacteriol. 1990;172:287–291. doi: 10.1128/jb.172.1.287-291.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin A, Bolotin A, Purnelle B, Hilbert H, Lauber J, Dusterhoft A, Ehrlich SD. Sequence of the Bacillus subtilis genome region in the vicinity of the lev operon reveals two new extracytoplasmic function RNA polymerase sigma factors SigV and SigZ. Microbiology. 1997;143(Pt 9):2939–2943. doi: 10.1099/00221287-143-9-2939. [DOI] [PubMed] [Google Scholar]
- Swinger KK, Rice PA. IHF and HU: flexible architects of bent DNA. Curr Opin Structl Biol. 2004;14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Pang H, Bartlam M, Zeng Q, Miyatake H, Hisano T, Miki K, Wong LL, Gao GF, Rao Z. Crystal structure of human pirin: an iron-binding nuclear protein and transcription cofactor. J Biol Chem. 2004;279:1491–1498. doi: 10.1074/jbc.M310022200. [DOI] [PubMed] [Google Scholar]
- Wendler WM, Kremmer E, Forster R, Winnacker EL. Identification of pirin, a novel highly conserved nuclear protein. J Biol Chem. 1997;272:8482–8489. doi: 10.1074/jbc.272.43.27091. [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Makarova K, Grishin N, Shabalina S, Wolf Y, Koonin E. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biology Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg OG, von Hippel PH. Selection of DNA binding sites by regulatory proteins. II. The binding specificity of cyclic AMP receptor protein to recognition sites. J Mol Biol. 1988;200:709–723. doi: 10.1016/0022-2836(88)90482-2. [DOI] [PubMed] [Google Scholar]
- Salgado H, Gama-Castro S, Martinez-Antonio A, Diaz-Peredo E, Sanchez-Solano F, Peralta-Gil M, Garcia-Alonso D, Jimenez-Jacinto V, Santos-Zavaleta A, Bonavides-Martinez C, Collado-Vides J. RegulonDB (version 4.0): transcriptional regulation, operon organization and growth conditions in Escherichia coli K-12. Nucleic Acids Res. 2004;32:D303–306. doi: 10.1093/nar/gkh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. Version 1.12. 2006.
- Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle. 2005.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Genes of G. metallireducens with atypical G+C content (more than two standard deviations from the mean). This table lists genes of G. metallireducens that have G+C content more than two standard deviations from the mean, and indicates by shading (alternated for contrast) those gene clusters that may be recent acquisitions.
Table S2. Enzymes of acyl-CoA metabolism in G. sulfurreducens and G. metallireducens. This table compares the genes predicted to function in acyl-CoA metabolism in G. sulfurreducens and G. metallireducens.
Table S3. Predicted binding sites of the global regulator ModE in the genome of G. sulfurreducens, which are mostly absent from the G. metallireducens genome. This table lists the predicted ModE-binding sites of G. sulfurreducens and compares them to the corresponding sequences in G. metallireducens.
Figure S1. A family of 24 predicted short RNA elements in the G. metallireducens genome. This is an alignment of 24 DNA sequences that were matched by nucleotide-level BLAST. Each RNA is found in an intergenic region, e.g. the 5' regions of genes affecting lysine/arginine metabolism, and contains a central palindromic structure GRCGTAGCGCTGCTACGCC. Similar sequences were found in the genomes of G. sulfurreducens, G. uraniireducens, and Desulfotalea psychrophila. The sequence strand and start and stop nucleotide positions are indicated.
Table S4. Genes found next to multicopy nucleotide sequences of unknown function in G. metallireducens. This table lists the genes adjacent to all of the multicopy nucleotide sequences identified in the G. metallireducens genome.
Figure S2. A family of 49 predicted regulatory RNA elements in G. metallireducens, containing four heptanucleotide repeats (consensus GGACCGG). This is an alignment of 49 DNA sequences that were matched by nucleotide-level BLAST. These elements are found within genes, sometimes more than once per gene, as well as between genes. The sequence strand and start and stop nucleotide positions are indicated.
Figure S3. Predicted global regulator binding sites (class 1). This is an alignment of 48 DNA sequences that were matched by nucleotide-level BLAST. Each site contains four tandem octanucleotide repeats (consensus GTTGCTYN), the outer two being poorly conserved. The distance between each pair of sites (on opposite strands) is variable. Each sequence begins at the right extremity of the top line (the 3' side of the "-" strand of the chromosome), loops on the left side (switching strands), and continues to the right extremity of the bottom line (the 3' side of the "+" strand of the chromosome); start and stop nucleotide positions are indicated. Insertion sequences of the ISGme8 or ISGme9 families may be found at a fixed distance from either or both sites of a pair; these occurrences are indicated on the appropriate lines.
Figure S4. Predicted global regulator binding sites (class 2). This is an alignment of 47 DNA sequences that were matched by nucleotide-level BLAST. Each of 21 paired sites, four sites that also belong to class 1, and one possibly vestigial unpaired site contains three tandem repeats (consensus TCTCCGTS[Y]). The distance between each pair of sites (on opposite strands) is variable. Each sequence begins at the right extremity of the top line (the 3' side of the "-" strand of the chromosome), loops on the left side (switching strands), and continues to the right extremity of the bottom line (the 3' side of the "+" strand of the chromosome); start and stop nucleotide positions are indicated.
Figure S5. Predicted global regulator binding sites (class 3). This is an alignment of 16 DNA sequences that were matched by nucleotide-level BLAST. Fifteen of the sites consist of five tandem heptanucleotide repeats (consensus MTYCTGA). Each sequence begins at the right extremity of the top line (the 3' side of the "-" strand of the chromosome), loops on the left side (switching strands), and continues to the right extremity of the bottom line (the 3' side of the "+" strand of the chromosome); start and stop nucleotide positions are indicated.
Table S5. Cytochrome c biogenesis gene clusters of G. sulfurreducens and G. metallireducens, and associated c-type cytochromes. This table compares the clusters of genes predicted to be involved in biogenesis of c-type cytochromes in G. sulfurreducens and G. metallireducens.
Table S6. Transport systems of G. sulfurreducens and G. metallireducens. This table compares the genes predicted to be involved in transport of solutes across the cell membrane and cell wall of G. sulfurreducens and G. metallireducens.
Table S7. Sensor histidine kinases (HATPase_c domain proteins), REC domain-containing proteins, and transcriptional regulators of G. metallireducens. This table compares the genes predicted to be involved in two-component signalling and transcriptional regulation in G. sulfurreducens and G. metallireducens.
Table S8. Diguanylate cyclases (GGDEF domain proteins) of G. sulfurreducens and G. metallireducens. This table compares the genes predicted to produce the intracellular messenger cyclic diguanylate in G. sulfurreducens and G. metallireducens.
Table S9. Chemotaxis-type signalling proteins of G. sulfurreducens and G. metallireducens. This table compares the genes predicted to participate in chemotaxis-type signalling in G. sulfurreducens and G. metallireducens.
Figure S6. Predicted global regulator binding sites (class 4). This is an alignment of 20 DNA sequences that were matched by nucleotide-level BLAST. Each site appears to be based on a pentanucleotide repeat (consensus CCYTC) that occurs four times on one strand and twice on the other. The sequence strand and start and stop nucleotide positions are indicated.
Figure S7. A predicted regulatory short RNA found in the 5' regions of c-type cytochromes and other proteins. This is an alignment of 16 DNA sequences that were matched by nucleotide-level BLAST. The location of Gmet_R3013 suggests that N-acylhomoserine lactone signalling may be under control of this RNA element. Similar sequences were found in the genomes of G. sulfurreducens, G. uraniireducens, and P. propionicus. The sequence strand and start and stop nucleotide positions are indicated.
Table S10. Toxin/antitoxin pairs of G. metallireducens and G. sulfurreducens. This table compares the genes predicted to encode toxin/antitoxin pairs in G. sulfurreducens and G. metallireducens.
Table S11. The CRISPR3 locus of G. metallireducens contains spacers of variable length. The thirteen clustered regularly interspaced short palindromic repeats (CRISPR) of G. metallireducens (consensus sequence GTAGCGCCCGCCTACATAGGCGGGCGAGGATTGAAAC) are far fewer than the thirty-eight of CRISPR1 and one hundred and forty-three of CRISPR2 in G. sulfurreducens.
Figure S8. Miscellaneous multicopy nucleotide sequences found in the G. metallireducens genome. These are alignments of 16 sets of miscellaneous DNA sequences in G. metallireducens that were matched by nucleotide-level BLAST. The sequence strand and start and stop nucleotide positions are indicated. (a) A palindromic sequence-containing family also found in the genomes of G. sulfurreducens, G. uraniireducens, and P. propionicus. (b) Sequences of this type were also found in the genomes of G. sulfurreducens and G. uraniireducens. (c) These sequences are unique to G. metallireducens. (d) The ends of these sequences form inverted repeats. Each sequence begins at the left extremity of the top line (the 5' side of the "+" strand of the chromosome), loops on the right side (switching strands), and continues to the left extremity of the bottom line (the 5' side of the "-" strand of the chromosome). A fragment related to Gmet_R6002 was found in the G. sulfurreducens genome. (e) These sequences are unique to G. metallireducens. (f) Sequences of this type were also found in the genomes of G. uraniireducens and G. bemidjiensis. (g) These sequences contain four octanucleotide repeats (consensus TWGTTGAY), two in tandem on each strand. (h) Sequences of this type were also found in the genome of G. sulfurreducens. (i) These sequences are unique to G. metallireducens. (j) These elements are located near each other. (k) These sequences are unique to G. metallireducens. (l-p) These elements are located near each other. Gmet_R0147 continues as Gmet_R0055, a tRNA-Asn gene (underlined).