Abstract
Microfluidic technologies’ ability to miniaturize assays and increase experimental throughput have generated significant interest in the drug discovery and development domain. These characteristics make microfluidic systems a potentially valuable tool for many drug discovery and development applications. Here, we review the recent advances of microfluidic devices for drug discovery and development and highlight their applications in different stages of the process, including target selection, lead identification, preclinical tests, clinical trials, chemical synthesis, formulations studies, and product management.
Introduction
The process of drug discovery and development continues to pose a significant challenge because the biological hypothesis must ultimately be tested in humans[1]. In 2006, only 18 new molecular entities and 4 new biologic license applications were approved by the US Food and Drug Administration (FDA)[2,3]. Recent developments, such as combinatorial chemistry, have greatly enhanced our ability to generate drug candidates. Furthermore, the sequencing of the human genome has opened new doors to understanding the nature of biological interactions in disease. Consequently, new drug candidates from the top 10 pharmaceutical companies entering clinical trials increased by 52% from 1998–2002 to 2003–2005[2]. Accordingly, investigational new drug (IND) applications to begin clinical testing increased by 45% from 2003 to 2005[2]. Considering the time for drug candidates to develop successfully into new drugs takes more than 10 years, the number of new drugs are expected to increase in the next few years. Therefore, there seems to be an emerging opportunity to discover and develop new drugs that may be aided by the development of novel tools.
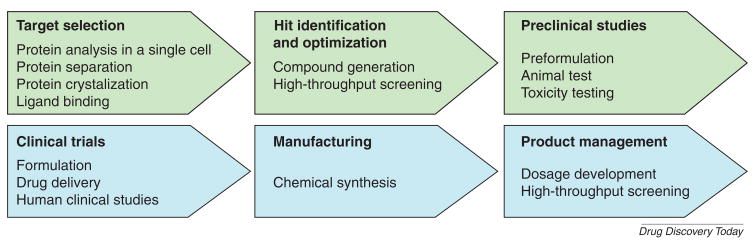
In general, the process of generating new therapeutics consists of two stages, drug discovery and drug development (Fig. 1). The discovery stage includes target selection, lead identification, and preclinical studies, while the development stage includes clinical trials, manufacturing, and product lifecycle management. The first step in drug discovery is to identify and select a ‘target’, such as a gene or protein, which can potentially be affected by a drug molecule[4]. Once the target is established, a promising molecule, or a ‘lead’ is identified that can interact with the target. The lead is optimized by screening many similar compounds and assessed based on its strategic portfolio fit, preliminary safety, technical issues and commercial opportunity. Moreover, the drug candidate will be tested for safety and efficacy in animal studies. After starting with thousands of compounds, only a few drug candidates continue into the development stage for clinical trials and manufacturing.
Figure 1. Microfluidic applications in drug discovery and development.
Drug discovery involves target selection, lead identification, optimization and preclinical studies. Drug development includes clinical trials, manufacturing, and product management process.
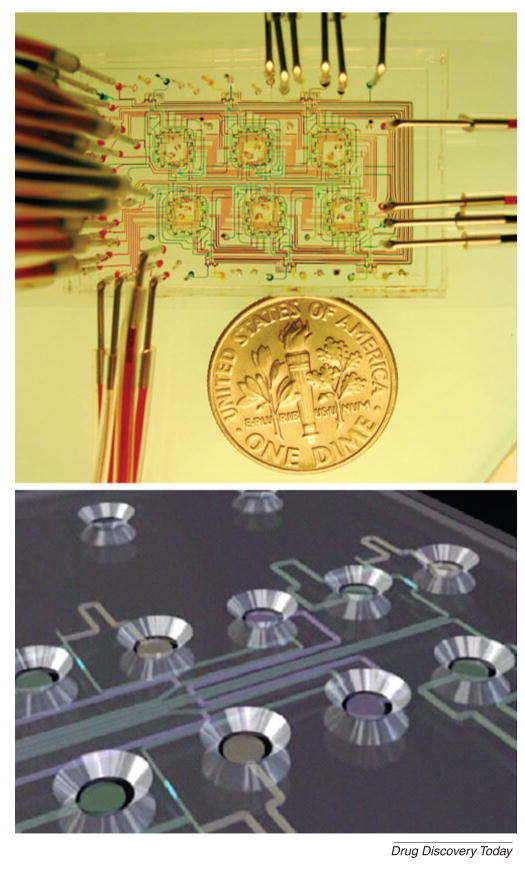
Much of the drug discovery process has been standardized using macroscopic systems that aim to provide the required throughput by incorporating automatic analysis and robotics. For the last 15 years, high-throughput screening (HTS) methods have been the gold standard. However, these methods have limitations, such as the necessary processing time and the need for expensive equipment and processes. To overcome these limitations, microtechnologies, such as microfluidics, are potentially useful. Microfluidic devices can be used to manipulate fluids within channels that are of the order of tens to hundreds of micrometers[5]. Multiple components, such as pumps, valves, mixers and heaters can be incorporated within fluidic systems to enable the easy manipulation of fluids. Thus, microfluidic channels can be used to perform experiments with higher throughput than is conventionally-achievable, while using minimal reagents and achieving fast reaction times (Fig. 2)[5,6]. Therefore, miniaturized microfluidic devices are emerging as useful tools for studying target selection, lead identification and optimization and preclinical test and dosage development[7,8]. In this paper, we review the recent developments and applications of microfluidic devices for drug discovery and development. We initially discuss the applications of microfluidic systems for the drug discovery process by identifying emerging areas in target selection and validation, lead identification and preclinical studies and subsequently discuss the use of these technologies for the drug development process.
Figure 2. An integrated microfluidic system.
(A) A microfluidic chemostat device containing high-density on-chip valves used to study microbial growth. (Reprinted by permission from Macmillan Publishers Ltd: [Nature] (Whitesides, 442: 368–373) copyright 2006)[5]. (B) Schematic of Caliper LabChip device in which microchannels connect with individual circular microwells to screen drug candidates (Reprinted by permission from Macmillan Publishers Ltd: [Nature] (Smith et al., 446: 219–222) copyright 2007)[6].
Target selection and validation
A drug target is usually a molecular structure, chemically definable as a molecular mass that can interact with therapeutic agents [9]. The types of drug targets that can interact with small-molecule therapeutic agents include proteins, polysaccharides, lipids and nucleic acids. Of these candidate targets, polysaccharides, lipids, and nucleic acids are investigated less frequently than proteins, because of a lack of understanding of the involvement of these molecules in disease and a lack of small-molecule therapeutic agents [10,11].
Both computational and experimental methods can be developed to identify druggable targets on the basis of ligand binding [12,13]. The computational approaches use information from the crystal structure of the target binding site, while experimental approaches use HTS applications of target candidates against a diverse collection of compounds. For example, a computational model of the binding sites of two proteins (homoserine dehydrogenase (HSD) and hematopoietic prostaglandin D synthase (H-PGDS) showed that HSD would be a difficult target, whereas H-PGDS would be druggable [13]. To verify this result, two HTS experiments were conducted, using a collection of 11,000 drug-like chemicals against the two target candidates. The results showed that 11 chemicals (about 0.1%) targeted H-PGDS while no compounds were effective against HSD, which indicated H-PGDS was a druggable target[13]. Therefore, protein structural and affinity studies can be important for target selection and validation. Here, we discuss the application of microfluidics as a useful tool in enabling these studies.
Protein analysis in a single cell
To select specific targets, the biological signal transduction pathways and protein-protein interactions within cells must be understood. The quantification of single-cell contents requires the detection of minute quantities of proteins and related molecules. To address this issue, microfluidic devices can be used [14]. These devices are integrated to manipulate, lyse, label, separate and quantify the protein contents of single cells using single-molecule fluorescence counting. For example, a microfluidic device was used to measure the number of epitope-tagged human β2-adrenergic receptors (β2ARs), an important pharmacologic target in a number of airway and cardiovascular diseases [14]. In addition, microfluidic devices integrated with an electrophoretic separation process have also been developed for the analysis of amino acids from the lysed contents of a single cell [15,16]. The study of cells in microfluidics devices has become an active area of research and promises to enable new and improved methods for basic biomedical study[17].
Protein separation and crystallization
Protein separation and crystallization is often required for the characterization of the structure of drug targets. Miniaturized microfluidic systems can be used for studying protein separation and crystallization due to the ability to integrate multiple functions, their higher speed, efficiency, automation, and portability[18]. Two-dimensional polyacrylamide gel electrophoresis (2-D PAGE) has been used as a conventional method of protein separation [19,20]. However, this method has a number of limitations, such as low throughput and sensitivity[21]. As a result, the technique requires relatively large amounts of samples, which at this stage in the development process, may often be in short supply. To overcome these limitations, miniaturized integrated microfluidic systems have been used. Two microfluidic-based molecule separation approaches are capillary electrophoresis and size-based separation. For example, peptide mixtures were separated in a microfluidic device by using micellar electrokinetic chromatography (MEKC) and capillary zone electrophoresis [20]. Additionally, a microfluidic system incorporating isoelectric focusing and capillary gel electrophoresis has been reported [22]. Solutions in this device were controlled by multiple on-chip valves that prevented intermixing between the two separation buffers. On-chip valves isolated isoelectrically-focused proteins, which could then be moved to a capillary channel and separated in as little as 20 minutes. Another microfluidic approach for protein separation was through the use of self-assembled colloidal sieves. A microfluidic colloidal self-assembly device has been developed to create ordered three-dimensional (3D) fluidic sieves in a microfluidic device to separate DNA and proteins[19].
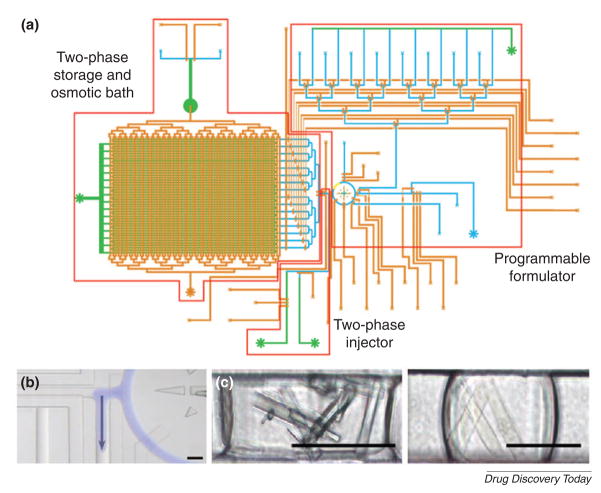
Protein crystallization is often the rate-limiting step in determining the structure of macromolecules, due to the inherent flexibility of macromolecules, and the relatively small hydrophilic loops that provide fewer potential crystal contacts [23–25]. To address this challenge, several microfluidic devices have been used. For example, a microfluidic system has been developed, in which droplets containing protein, precipitants and additives were generated inside immiscible fluids[26]. In this two-phase system, aqueous droplets were sheared in a flowing stream of oil. Inside these aqueous droplets, molecules (i.e. thaumatin) could be induced to crystallize. This was confirmed by the analysis of X-ray diffraction patterns of thaumatin protein crystal in the aqueous droplets[27]. In addition, a microfluidic platform integrated with 480 on-chip valves and 144 parallel reactions were developed for protein crystallization[28]. Pneumatically-actuated valves were used to isolate compounds and enable diffusive mixing. Proteins were crystallized using the free interface diffusion. Similarly, a microfluidic screening platform incorporated with 16 addressable channels, on-chip valves, and peristaltic pumps was developed for inducing protein crystallization (Fig. 3)[29]. Protein crystals of glucose isomerase and catalase were created in such a microfluidic channel.
Figure 3. Protein crystallization in a microfluidic screening platform.
(A) Schematic of microfluidic screening platform with many addressable channels. (B) An aqueous droplet was introduced (blue) into an immiscible carrier fluid using a peristaltic pump. (C) Protein crystals were formed in a microfluidic channel. Crystals were generated from glucose isomerase (left) and catalase (right). Scale bar is 100 μm. (Reprinted with permission from Lau et al. Copyright 2006 American Chemical Society)[29].
A droplet-based microfluidic approach has been also reported. In this case, many distinct reagents were sequentially introduced as ~ 140 nl plugs into a microfluidic device[30]. From 10 μl of a protein solution, approximately 1,300 crystallization trials were tested within 20 minutes. This method was compatible with growth, manipulation, and extraction of high-quality crystals of membrane proteins[30]. The knowledge of the phase behavior of a protein can help to create a rational screen that increases the success rate of crystallizing proteins. This strategy was applied, with a 75% success rate, to the crystallization of 12 diverse proteins, most of which had failed to be crystallized with traditional techniques[31].
Ligand binding
Much of drug discovery is based on the selective binding of low molecular weight molecules to bioactive macromolecules[32]. Typically, ligand-binding studies are performed by exposing the target to various concentrations of a compound. The extent of compound binding to a target is quantified as a function of the half-maximal inhibitory concentration (IC50) or the equilibrium dissociation constant (Kd) for the drug–target binary complex.
Microfluidic devices can be used for ligand binding studies to minimize interaction times, improve sensitivity, and to increase throughput. For example, assessing the degree of molecular interactions in a high-throughput format is difficult, especially for transient and low-affinity interactions. To demonstrate the application of microfluidic systems in addressing these challenges, a high-throughput microfluidic platform has been used to characterize DNA binding energy using four eukaryotic transcription factors. This system was used to test the binding of individual transcription factors to DNA and to predict their in vivo function [33]. A nano-replica molding process has also been used to produce polymer microfluidic channels containing label-free photonic crystal biosensors in a multiwell plate platform[34]. The system can measure the kinetic binding interaction of protein A with IgG molecules of high, medium and low affinity. This approach offered a method for minimizing the volume of reagent required to functionalize the biosensor surface, while retaining compatibility with the microplate assay format that is most commonly used in biological research [34].
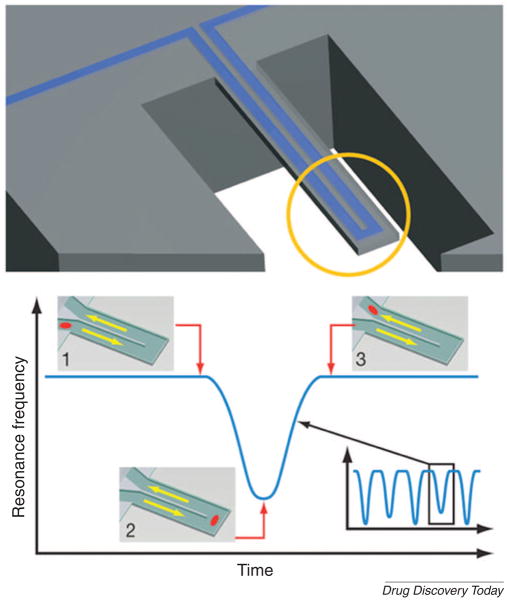
The quantification of specific ligand binding interactions depends on precise measurement of binding energy enthalpically [35] or by the use of NMR spectra [36], such data is often difficult to obtain. To address this problem, magnetic nanosensors and microfluidic cantilevers may be applicable. Magnetic nanosensors have been used to detect molecular interactions in the reversible self-assembly of disperse magnetic particles [37]. These magnetic nanosensors can be used in microfluidic systems, as affinity ligands for HTS applications. Alternatively, a microfluidic cantilever chip has made it possible to weigh and analyze biomolecules, single cells and single nanoparticles in fluid (Fig. 4)[38]. In solutions, viscosity complicates the measurement of the weight of small objects and this device eliminated viscous damping by placing the solution inside a hollow resonator that was surrounded by a vacuum. The suspended microchannel resonators can weigh single nanoparticles in water with sub-femtogram resolution. Although still in the early stages of development, this method provides a promising approach for the precise quantification of binding interactions.
Figure 4. A microfluidic cantilever for studying biomolecules, single cells, and nanoparticles.
(A) Schematic design of a microfluidic cantilever that can be used to determine mass changes. (B) Particles flow through without binding to the surface. Mass changes of particles were quantified by measuring resonance frequency. (Reprinted by permission from Macmillan Publishers Ltd: [Nature] (Burg et al., 446: 1066–1069) copyright 2007)[38].
Hit identification and optimization
The identification of small-molecule hits and optimization into leads are key processes in drug discovery [39]. The potential pool size of drug candidates is extremely large and has been estimated to be in the order 1063[40]. Generating and optimizing drug candidates using macroscale methods is time-consuming and labor intensive. To address these challenges, microscale technologies have been developed. Microscale technologies may be useful, due to their short reaction times and smaller reagent volumes. Moreover, these systems can be used to synthesize chemical libraries and for high-throughput screening to increase the likelihood of discovering new drugs.
Compound generation
To generate lead compounds, microreactors can be used to perform a large number of chemical syntheses [41–43]. For example, a miniaturized-synthesis and total analysis system that incorporated a chemical microprocessor with time-of-flight mass spectrometry (TOF-MS) has been used for the generation of compound libraries [41]. This system enabled real-time processing of the many reactions because it integrated continuous-flow synthesis and on-line analysis within a microfabricated structure. In addition, a glass microreactor has been used to prepare rapidly (within 20minutes), peptide derivatives from α–amino acids using solution phase chemistry [42]. Moreover, chemical libraries have also been synthesized, using an automated single-channel glass microreactor that integrated pumping, dilution, detection and analytical components [43].
The ability to generate synthetic genes has had a significant impact on generating lead compounds. In this case, a microfluidic device is an enabling tool for synthetic biology applications such as DNA synthesis [44–46]. For example, a microfluidic picoarray device has been developed for the synthesis and purification of oligonucleotides[45]. This device was composed of fluidic channels and 3,698 individual pico-reaction chambers for parallel synthesis of oligonucleotides. This programmable microfluidic device enabled ultra-fast generation of a large number of oligonucleotides. In addition, mRNA isolation and cDNA synthesis in microfluidic devices has been reported [46]. A microfluidic device fabricated by multi-layer soft lithography contained lysis, mRNA isolation/cDNA synthesis/purification and product collection modules. Purified mRNA and paramagnetic beads derivatized with oligo(dT)25 sequences were loaded into the chamber [46]. The bead-mRNA complexes were sent to output channels for real-time quantitative polymerase chain reaction (RT-qPCR) analysis. Solid-phase cDNA synthesis was implemented with purified mRNA of fibroblast cells by using the beads. This device can be useful for highly parallel single-cell gene expression analysis. Therefore, automated microfluidic microreactors can be used to synthesize combinatorial libraries in a sequential manner, as well as analyze multiple analogue reactions [43].
Microfluidic systems can be also used to understand and assess natural drug candidates. Natural products are an important source of new chemical entities with a world market of about US $ 60 billion and an annual growth rate of between 5% and 15%[47]. To identify the physiological action of individual natural compounds from a complex array of constituent molecules in the source material, microfluidic devices can be used. For example, multielectrode microchips have been developed for the screening of herbal medicines [48]. This study demonstrated the sensitivity, selectivity and robustness of microchips for high content screening of complex mixtures of neuroactive substances. Such chips have been used for the multiparametric assessment of herbal extracts from various plants. Also, carbon nanotubes have been used as sensitive amperometric detectors for the separation and detection of honokiol and magnolol in herbal medicines. These detectors offered lower operating potentials and enhanced signals to surface fouling [49]. In addition, a microfluidic reactor for the multi-step synthesis of natural products has been developed [50]. The silica-immobilization of enzymes generated stable heterogenous catalysts for the combinatorial synthesis of 2-aminophenoxazin-3-one.
High-throughput screening (HTS)
HTS is one of the most important methods for identifying ‘hit’ compounds [51]. Over the past two decades, HTS has been established as a major tool for pharmaceutical companies to screen the properties of new chemical entities [52]. Traditionally, HTS systems are performed by using multiple-well plates (e.g. 96, 384, and 1,536 well-plates), however, miniaturization of the multiple-well plates has been limited due to difficulties of dispensing nanoliter volumes of liquid into the wells. In addition, multiple-well systems still require relatively large volumes of high-cost compounds. To overcome these limitations, microfluidic devices can be useful. There are a number of microfluidic technologies that can be used to enable HTS studies. These include multiplexed systems, microwell arrays, plug-based methods and gradient-generating devices.
Multiplexed systems
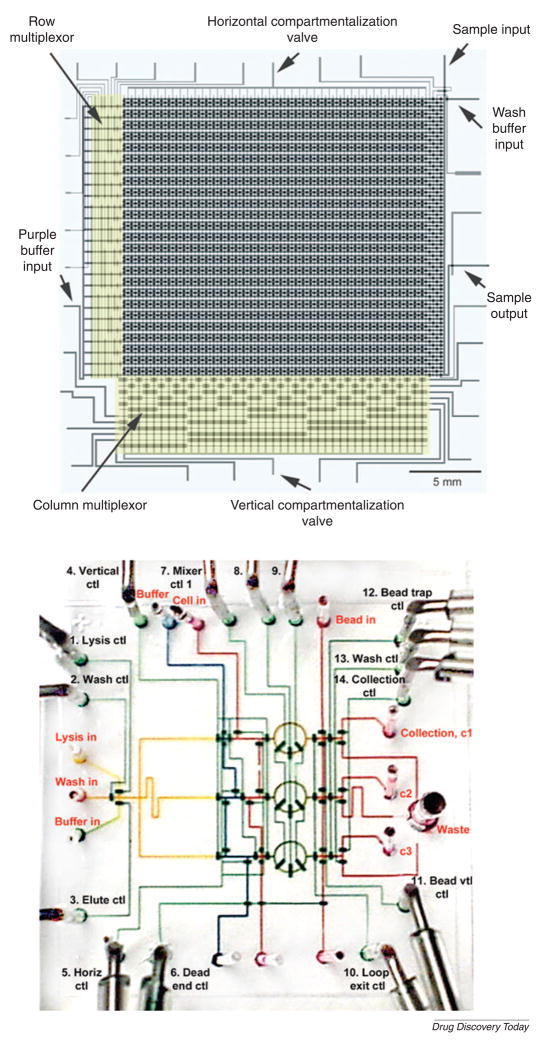
High-throughput microfluidic multiplexed systems fabricated by multi-layer soft lithography are comprised of thousands of on-chip valves and hundreds of individual single chambers (Fig. 5A)[53,54]. In these systems, valves and actuators can be used to control independently the delivery of the fluids to each chamber. The fluidic multiplexor, a combinatorial array of binary valves, precisely manipulated the complex combination of fluids. Using this microfluidic device, GFP-expressing Escherichia coli cells were screened. Similarly, an integrated microfluidic device for mRNA and DNA purification has been developed by using on-chip valves (Fig. 5B)[55]. A key component of this device is a fluidic rotary mixer that can be used to mix different solutions and perform multiple parallel processes of DNA recovery. Using such a chip, all processes, including cell isolation, DNA and mRNA purification were performed on bacterial cells.
Figure 5. Microfluidic large-scale integration.
(A) A high-density microfluidic device integrated with microfluidic multiplexors. Valves are formed at the intersection of control channels with fluidic channels. (Reprinted with permission from Thorsen et al. Copyright 2002 Science)[53]. (B) DNA purification chip. This device is used for multiple parallel processes of DNA recovery from living bacterial cells. (Reprinted by permission from Macmillan Publishers Ltd: [Nature Biotechnology] (Hong et al., 22: 435–439) copyright 2004)[55].
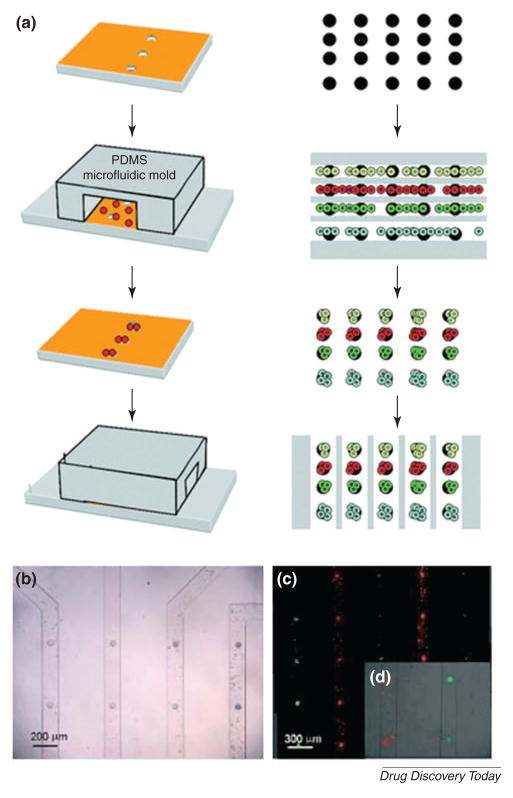
Multiplexed analysis has also been used for the simultaneous quantification of different targets within a single sample. Conventional methods for multiplexed analysis are expensive and the encoding/decoding of the samples is complicated. To address these limitations, a microfluidic flow-based lithographic technique has been developed for the high-throughput analysis of multifunctional encoded particles (Fig. 6)[56]. In this approach, two poly(ethylene glycol) (PEG) pre-polymer solutions flowed side-by-side in a microchannel and then cross-linked by UV light filtered through a photomask bearing the combination codes of 0 and 1 so that the pattern of the photomask was cast onto the PEG pre-polymer solutions. The resulting microstructures were comprised of two parts, one of which bore the barcode, while the other bore a probe for the target molecules. Upon incubation of the particles with samples, different targets could bind to their respective probes linked to the unique identifier barcode. These suspensions were passed through a detector, which could read the barcode and calculate the target concentration. Using this process, DNA oligomers of 500 attomoles (10−18 moles) were successfully detected.
Figure 6. Multifunctional encoded particles for analyzing high-throughput biomolecules.
(A) Schematic diagram of particle synthesis through photo-polymerization across laminar streams. Fluorescence images of single-probe (B) and probe-gradient (C). Scale bar is 100 μm. (Reprinted with permission from Pregibon et al. Copyright 2007 Science)[56].
Microwell arrays and plug-based methods
To enable the testing of cells and particles within microfluidic channels, microstructures can be used. These structures generate regions of low shear stress within the channels that enable the docking of various entities. Thus, the ability to pattern cells and particles in the channels can be used to perform HTS experiments[57]. For example, single cells were anchored and analyzed within poly(dimethylsiloxane) (PDMS)-based microwells[58] to enable clonal tracking of neuronal progenitor cells in microfabricated wells to analyze cell proliferation and differentiation [59]. In another instance, the response of multiple cell types (e.g. hepatocytes, fibroblasts, and embryonic stem cells) to different compounds was analyzed by seeding cells within microwells that were integrated within an array of reversibly sealed microfluidic channels (Fig. 7)[60]. The ability to position many cell types on a single chip could be useful for studying the effects of a series of compounds on different cell types. In addition, other types of microstructures that can minimize shear stress have been used to isolate single cells. A high-density microfluidic device has been developed in which single cells were isolated within cup-shaped trapping sites for analyzing enzyme kinetics[61].
Figure 7. Cell docking within microchannels containing microwells.
(A) Schematic drawing of reversibly sealed microfluidic device that can be aligned on an array of microwells. (B) Embryonic stem cells could be docked within the microchannels. (C) fluorescent image of cells labeled with membrane dyes CFSE (green) and SYTO (red). (D) A cell-free solution is flowed through the channels to remove non-adhered cells. (Khademhosseini et al. Lab Chip, 5, 1380–1386) -Reproduced by permission of The Royal Society of Chemistry[60].
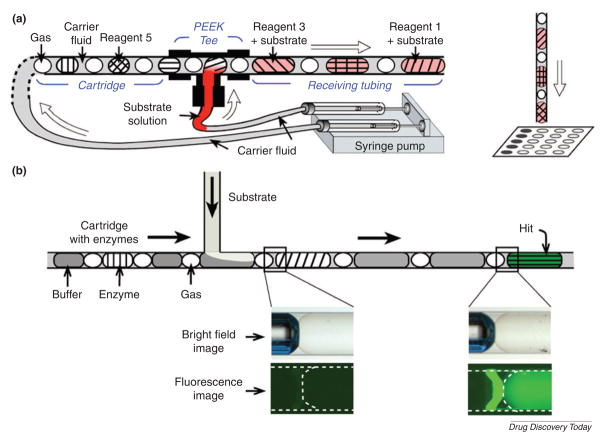
Plug-based microfluidic cartridges have been also used for HTS applications (Fig. 8)[62]. This system has many advantages over conventional multi-well plate methods, such as handling picoliter or nanoliter volumes, incubating reagents without evaporation and minimizing the exposure of reagents to the atmosphere. ‘Plugs’ are droplets surrounded by carrier fluid [63]. With such plugs, a single input of 320 nl can be split into 16 output plugs with the volume of 20 nl each. The low cost and simplicity of the plug method can be useful for enzymatic assays and protein crystallization. Therefore, both microwell and plug-based approaches have different advantages but the same potential to improve the conventional multi-well plate method.
Figure 8. Microfluidic cartridges with nanoliter plugs of reagents.
(A) The experimental setup using preloaded cartridges of nanoliter plugs for screening reagents. After incubation, the plugs are deposited onto a plate for analyzing and screening reagents. (B) Schematic drawing of a fluorescence assay using microfluidic cartridges with plugs with enzymes. (Reprinted with permission from Chen et al. Copyright 2006 Elsevier)[62].
Concentration gradients
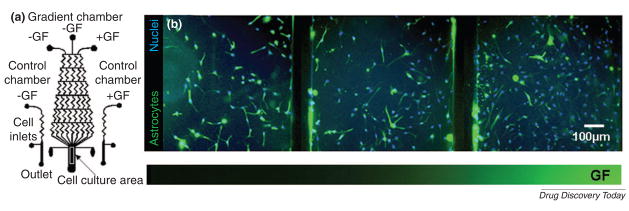
Concentration gradients also play an important role in drug screening and cell-based studies. A gradient-generating microfluidic device can generate multiple doses simultaneously and control stem cell behavior. By using gradients, cells can be stimulated with controlled temporal and spatial resolution to study the effects of drug concentration on chemotaxis[64]. The microfluidic device for generating pharmacological gradient profiles is important for lead optimization[65]. This gradient device was integrated with scanning-probe patch-clamp systems and was used to analyze the pharmacological screening of voltage-gated human ether-à-go-go related gene (hERG) K+ channels and ligand-gated γ-aminobutyric acid (GABA) receptors in response to a concentration gradient of Ba2+ and GABA. Besides pharmacological applications, gradients can be useful for cell-based studies such as human neural stem cell (NSC) differentiation (Fig. 9)[66]. Growth and differentiation of human NSCs exposed to different gradient profiles of growth factor mixtures were successfully controlled and analyzed in the microfluidic device. Therefore, these high-throughput microfluidic approaches can be useful for studying lead identification and drug candidate optimization.
Figure 9. A gradient-generating microfluidic device for analysis of stem cells.
(A) Schematic of the microfluidic device integrated with control and gradient chambers. (B) Differentiation of human NSCs exposed to gradients of growth factor mixtures in the microfluidic device. Fluorescence images show human NSCs stained with an astrocytes marker (green) and nuclear stain (blue). [Chung et al. Lab Chip, 5, 401–406] - Reproduced by permission of The Royal Society of Chemistry[66].
Preclinical studies
In the preclinical stage, the drug candidate’s toxicological and pharmacological properties are evaluated through in vitro and in vivo tests [67,68].
In vitro tests
Cell-based studies using microfluidic devices can be useful for assessing the interaction of the lead with normal or diseased cells. In toxicology, a major challenge is to recreate the cell–cell interactions that are present in living organisms, as well as the complex pharmacological and pharmacokinetic interactions between various organs[17]. For example, the toxic effect of drugs in one tissue often depends on the metabolic activity of another tissue. To mimic these interactions, a microfluidic system containing a network of interconnected chambers can be used [60,69,70]. In this approach, each compartment can be engineered to represent a specific organ such as liver, lung, and fat cells to mimic the physiologically-relevant features (i.e. circulation and interchange of metabolites) in the body.
Cell-based microfluidic devices can be also used to test the synergistic effect of combinatorial drugs [71]. Combinatorial drugs offer new hope for a number of diseases, but the range of possible combinations is too large to be investigated in expensive clinical studies. Using a microfluidic device, the screening process becomes cheaper and easier than in vivo methods. With different cell types, the effect of various drug combinations can be investigated.
In vivo tests
Although cell-based in vitro tests can provide useful preliminary data, animal tests are still required to test the pharmacological properties of drug candidates. For in vivo tests, the drug content within animal tissues needs to be determined. In general, three steps are required: sampling, processing, and analysis.
Obtaining blood samples from animals may be challenging, especially for small animals such as mice and rats. Blood loss must be minimized so that the physiological state of the animal is not disturbed. To obtain small blood samples, automated systems, based on a microfluidic platform that can be connected to the animal blood vessels through a catheter, have been developed[72,73].
Microfluidic systems can also be used to extract drug compounds for analysis of animal tissues and organs such as liver and brain[74–76]. For example, the separation of an antivenom antibody, which precipitates from a suspension containing contaminating soluble proteins, was achieved by a microfiltration device [77]. The overall yield of this approach was 10% better than that obtained from centrifugation because of reduced material losses from the integration of operations including separation, buffer exchange and final recovery. Furthermore, losses resulted from separation and extraction with organic solvent in methods, such as liquid-liquid extraction, can be minimized by the use of microfluidic devices. For example, a microfluidic device based on capillary forces and selective wetting surfaces achieved effective liquid-liquid phase separation [76].
Similar to sample processing, microfluidic methods can be applied to analyze biological samples. High performance liquid chromatography (LC) coupled with mass spectrometry (MS) is a powerful tool to determine drug concentrations in animal and human tissues[78]. Advanced microfluidics can improve the performance of LC/MS/MS systems[79]. The direct coupling of LC columns to low-flow electrospray ionization (ESI) mass spectrometer interfaces enabled a significant reduction in analysis time and volume, while maintaining the signal-to-noise ratio [79]. The nanospray method can be used to estimate drug metabolites in drug discovery when reference standards are not available. For a series of drug molecules (e.g. codeine, dextromethorphan, tolbutamide, phenobarbital, cocaine, morphine, and their respective metabolites), a nanospray method exhibited a distinct trend toward equimolar response[78]. In addition, microfluidics has been used for multiplexed MS analysis. Systems for high-throughput microfluidics-MS coupling via matrix assisted laser desorption/ionization (MALDI) has been developed during the last ten years[80]. These approaches have greatly enhanced the ability to analyze samples and have great potential in improving many aspects of the drug discovery process.
Clinical trials
A drug candidate must go through extensive studies in humans to demonstrate its safety and effectiveness. Physicians carry out clinical trials by working closely with patients and the sponsor company. During the trial, pharmacokinetic (PK) and pharmacodynamic (PD) profiles of the drug candidate are established in humans. PK characterizes the adsorption, distribution, metabolism, and elimination properties of a drug candidate, while PD defines the physiological and biological response to the administered drug candidate[81,82]. The basic parameter for PK is the drug concentration in blood while PD parameters vary with different types of disease. For example, the levels of glucose in circulating blood and dopamine release in brain, both of which are PD parameters, can be monitored for the treatment of diabetes [83] and neurodegenerative diseases [84] respectively. Blood sampling and processing is required to measure both PK and PD parameters. Here we review the applications of microfluidic systems in formulation study and human blood sampling.
Formulation study
Drug delivery and formulation studies are of great importance in the development of a pharmaceutical reagent [85]. Small molecules are generally formulated as oral preparations, however they may be formulated in various other ways such as creams for topical and transdermal delivery, polymeric systems (for various administration routes), and preparations for injection (for administering via the intravenous (i.v.), intramuscular (i.m.), intraductal (i.d.) and other routes). In addition to these technologies, microfabrication approaches can also be used to fabricate controlled-release drug delivery systems.
Silicon-based microchips have been fabricated for releasing single or multiple chemicals on demand using electrical stimuli[86]. These engineered microchips can be used to facilitate drug release and maintain circulating drug concentration at a level that sustains a constant biological effect. Specifically, drugs are maintained in small reservoirs, from which the drugs can be released based on electrical stimulation. In addition, degradable polymers have also been used to initiate the release process based on the degradation of polymeric reservoir covers. Furthermore, silicone rubber devices with microchannels have been used for medicine-delivering implants[87,88]. In these microfluidic devices, the flow of liquids was controlled to release the drug in a desired manner. Microfluidic systems can also be used to deliver drugs in a minimally-invasive manner by transdermal means. In these approaches, microneedle arrays, which penetrate into skin without pain, can be used to deliver the drugs through the skin[89].
Novel microfluidic methods were also reported for the preparation of lipid vesicles and polymeric nano- and microparticles for drug delivery. Liposomes, for example, can be used to deliver drugs to cells with reduced toxicity and in a targeted manner. However, traditional methods for liposome preparation require postprocessing steps (i.e. sonication or membrane extrusion) to yield formulations of an appropriate size. A method to engineer liposomes, without postprocessing steps, has been reported by changing the flow conditions in a microfluidic channel [90]. A stream of lipids dissolved in alcohol was hydrodynamically-focused between two sheathed aqueous streams in a microfluidic channel. The laminar flow in the microchannel enabled controlled diffusive mixing at the two liquid interfaces where the lipids self-assemble into vesicles. The vesicle sizes were tunable over a mean diameter from 50 to 150 nm by adjusting the ratio of the alcohol-to-aqueous volumetric flow rate. Other microfluidic methods to prepare lipid vesicles were also reported [91–93]. These lipids vesicles can be used to encapsulate drug and cells for pharmaceutical applications.
Human blood sampling and processing
There are several differences between human and animal blood sampling and analysis. In animal studies, in particular for end of study analysis, the animal is often euthanized and the organs removed. On the other hand, human sampling needs to be designed to minimize the trauma for the patient and to be of maximal assistance to nurses and doctors. Microfluidics can help to improve the human blood sampling process by providing microneedle-based sampling devices and integrated metering and sedimentation disks.
A titanium microneedle with the same size as a female mosquito’s labium (60μm outer diameter, 25μm inner diameter) has been recently developed to enable minimally-invasive and pin-free blood sampling [83]. The design was based on the mosquito’s blood extracting mechanism. The device may provide an alternative way for blood sampling by patient themselves. Integrating centrifugal disks for the rapid assay of whole human blood can become a suitable tool for the analysis of minute blood samples from the microneedles. Such devices can provide all the necessary steps involved in the process such as plasma extraction and the final drug estimation [94–96]. For example, a microfluidic disk featuring a plasma extraction structure with a capacity of 500 nl can purify plasma by centrifugation, meter the purified plasma by overflow, and subsequently transfer the plasma into the detection chamber. A significant advantage of these devices is that they can be easily manufactured and used by patients [97]. These microfluidic devices will be an alternative method for blood sampling in the clinical trials with the benefit of reduced cost and alleviated trauma for the patients.
Manufacturing
Clinical trials last for several years before the approval of a drug candidate[4]. Upon approval, there are significant challenges in efficiently manufacturing compounds on a large scale, while complying with guidelines for good manufacturing practice (GMP). Controlling and optimizing conditions of chemical synthesis are important for the drug studies. Miniaturized and automated microfluidic microreactors provide a promising tool for the synthesis of many chemicals, because they can perform chemical reactions with high efficiency and rapid reaction times [98–100]. The channel length scales of microfluidic microreactors for the chemical synthesis ranged between tens and hundreds of micrometers [98,100]. These devices were fabricated from a variety of materials such as glass, quartz, silicon, metals and polymers, with various techniques, such as photolithography, hot embossing, injection molding, laser microforming and powder blasting [100]. Although each microfluidic reactor may not have the same volume or throughput as larger reaction vessels, they can be easily scaled-up by parallelization. For example, the large-scale synthesis of the immunoactivating natural product, pristine, was achieved by using a microfluidic dehydration device [101]. Furthermore, the ability to perform high-throughput experiments can be used to optimize reaction conditions. For example, a continuous flow microfluidic-based microreactor has been used to study the glycosylation reaction in organic transformations[102].
Microfluidics is also useful for studying DNA and gene synthesis, which is potentially useful for biological drugs. A microfluidic DNA oligonucleotide synthesizer containing multiple on-chip valves was made by perfluoropolyether (PFPE)[103]. Using this device, 60 pmol of DNA oligonucleotides was synthesized while consuming less than 500 nl of phosphoramidite solution in each reaction. This device could be useful for screening small interfering RNA (siRNA) sequences and creating DNA nanostructures. Besides DNA synthesis, multiplex genes were also synthesized in a microfluidic device[104]. Genes that encoded the proteins of the Escherichia coli 30S ribosomal subunit (21 in total) were synthesized and optimized in a device. Rapid prototyping of individual genes can be useful for the synthesis of ribosomes in vitro.
Product lifecycle management
After the launch of a new drug product, efforts must be made to extend and modify the drug’s lifecycle by assessing combination therapy, new therapeutic indications, target patient populations, new dosing regimens and modified formulation. Among these strategies, the development of new dosage forms is one of the most effective methods. Dosage forms can be improved by the appropriate selection of excipients. In a study for a cremophor EL-free paclitaxel formulation, a full factorial combination of many compounds at three different concentrations was screened using an automated liquid dispenser[105]. It revealed that of the 9,880 combinations that were initially tested, only 19 were identified as hit combinations. A miniaturized assay has also been developed for solubility and residual solid screening of drug compounds[106]. The concentration of the drug was determined and drug crystals were analyzed. This result revealed the formation of hydrates in aqueous vehicles of drugs such as caffeine, carbamazepine, and piroxicam. These studies demonstrate the power of the high-throughput combinatorial approach for alternative formulations and suggest that this approach can improve the speed and efficiency of drug formulation design. Thus, many microfluidic approaches that were previously described might be also used for these systems to analyze alternative drug formulations.
Finally, microfluidic systems could be useful for a number of other applications such as in vitro dissolution experiments which are routinely used to control the quality of dosage forms and reduce batch to batch variability. Typically, for such devices, long working hours and large volumes of dissolution media are required. Microfluidic devices can be used to overcome these limitations. For example, a self-calibrating microfabricated capillary viscometer has been reported to generate a wide range of shear rates [107]. The measurement of viscosity was based on monitoring the capillary pressure-driven movement of fluid samples whose shear rate varied with time. Such devices could be integrated into the dissolution testers to provide more information about the drug dissolution process.
Conclusion
Microfluidic technologies are powerful tools for various applications for the drug discovery and development process. Microfluidic-based approaches have already made a significant impact in the area of chemical synthesis, protein crystallization, high-throughput drug screening, and drug delivery, since they address a number of limitations imposed by conventional macroscale methods including low throughput, expensive processes, and large volume of reagents. In particular, microfluidic technologies have great potential in high-throughput studies involving target selection, lead compound generation, identification, and dosage design. However, despite exponential growth of microfluidics in the past few years, a number of challenges still need to be addressed. In particular, microfluidic devices must be simple and highly versatile to enable their use in both academic and industrial pharmaceutical laboratories. A standard microfluidic platform should be developed to enable easy coupling of extant microfluidic systems. More studies should be conducted to determine the reliability of microfluidic chips over hundreds of thousands of samples and months of constant use. Thus, much progress remains to further enhance the use of microfluidics in addressing challenges of drug discovery and development studies.
Acknowledgments
This paper was partially supported by the Coulter Foundation, National Institutes of Health (NIH), the Center for Integration of Medicine and Innovative Technology (CIMIT), the US Army Core of Engineers, and the Charles Stark Draper Laboratory. L.Kang is a recipient of the NUS overseas postdoctoral fellowship. Authors thank Dr. Edward Ramon Brown of GSK Singapore and Dr. K.P.P. Prasad of Pfizer Asia Pacific for sharing their knowledge in drug discovery and development. The authors also thank Dr. Marjo Yliperttula of University of Helsinki, Dr. Yanan Du, Mr. Edward Lo and Mr. Alex Bick their proofreading and helpful comments.
Biographies
 Lifeng Kang is a postdoctoral research fellow in the Harvard-MIT Division of Health Sciences and Technology and Harvard Medical School. He received his Ph.D. from the National University of Singapore in 2006 and his undergraduate and master’s degrees in China Pharmaceutical University. His research field is pharmaceutical sciences for drug discovery and development. He is a recipient of the NUS-overseas postdoctoral fellowship.
Lifeng Kang is a postdoctoral research fellow in the Harvard-MIT Division of Health Sciences and Technology and Harvard Medical School. He received his Ph.D. from the National University of Singapore in 2006 and his undergraduate and master’s degrees in China Pharmaceutical University. His research field is pharmaceutical sciences for drug discovery and development. He is a recipient of the NUS-overseas postdoctoral fellowship.
 Bong Geun Chung is a postdoctoral research fellow in the Harvard-MIT Division of Health Sciences and Technology and Harvard Medical School. He received his Ph.D. at University of California, Irvine in 2007. He also received B.S. and M.S. in Hanyang University, Seoul, Korea. His research is focused on developing microfluidic platforms for controlling stem cell fates and drug delivery.
Bong Geun Chung is a postdoctoral research fellow in the Harvard-MIT Division of Health Sciences and Technology and Harvard Medical School. He received his Ph.D. at University of California, Irvine in 2007. He also received B.S. and M.S. in Hanyang University, Seoul, Korea. His research is focused on developing microfluidic platforms for controlling stem cell fates and drug delivery.
 Robert S. Langer is one of 13 Institute Professors (the highest honor awarded to a faculty member) at the Massachusetts Institute of Technology (MIT). Dr. Langer has over 900 articles, nearly 550 patents, licensed or sublicensed to over 180 pharmaceutical, chemical, biotechnology and medical device companies. Dr. Langer served as a chairman of United States Food and Drug Administration’s Science Board from 1999 to 2002. He has received nearly 150 major awards including the Charles Stark Draper Prize (2002), considered the equivalent of the Nobel Prize for engineers and the world’s most prestigious engineering prize, from the National Academy of Engineering.
Robert S. Langer is one of 13 Institute Professors (the highest honor awarded to a faculty member) at the Massachusetts Institute of Technology (MIT). Dr. Langer has over 900 articles, nearly 550 patents, licensed or sublicensed to over 180 pharmaceutical, chemical, biotechnology and medical device companies. Dr. Langer served as a chairman of United States Food and Drug Administration’s Science Board from 1999 to 2002. He has received nearly 150 major awards including the Charles Stark Draper Prize (2002), considered the equivalent of the Nobel Prize for engineers and the world’s most prestigious engineering prize, from the National Academy of Engineering.
 Ali Khademhosseini is an Assistant Professor of Medicine and Health Sciences and Technology at Harvard-MIT’s Division of Health Sciences and Technology and the Harvard Medical School. He has published over 60 papers and 14 issued or pending patents. He received his Ph.D. in bioengineering from MIT, MSc and BSc from University of Toronto. He has received multiple awards including outstanding undergraduate research mentor at MIT (2004) Coulter Foundation Early Career (2006), BMW Group Scientific Award (2007) and TR-35 Top Young Innovator award from Technology Review (2007). His research is based on developing micro- and nanoscale technologies to control cellular behavior for tissue engineering and drug delivery.
Ali Khademhosseini is an Assistant Professor of Medicine and Health Sciences and Technology at Harvard-MIT’s Division of Health Sciences and Technology and the Harvard Medical School. He has published over 60 papers and 14 issued or pending patents. He received his Ph.D. in bioengineering from MIT, MSc and BSc from University of Toronto. He has received multiple awards including outstanding undergraduate research mentor at MIT (2004) Coulter Foundation Early Career (2006), BMW Group Scientific Award (2007) and TR-35 Top Young Innovator award from Technology Review (2007). His research is based on developing micro- and nanoscale technologies to control cellular behavior for tissue engineering and drug delivery.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Editorial. Five years on…and four challenges for the pharmaceutical industry. Nat Rev Drug Discov. 2007;6:3. doi: 10.1038/nrd2229. [DOI] [PubMed] [Google Scholar]
- 2.Editorial. Same old story? Nat Rev Drug Discov. 2007;6:97. doi: 10.1038/nrd2259. [DOI] [PubMed] [Google Scholar]
- 3.Owens J. 2006 drug approvals: finding the niche. Nat Rev Drug Discov. 2007;6:99–101. doi: 10.1038/nrd2247. [DOI] [PubMed] [Google Scholar]
- 4.PhRMA. Drug discovery and development: Understanding the R&D process. 2007 http://www.innovation.org/drug_discovery/objects/pdf/RD_Brochure.pdf.
- 5.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 6.Smith C. Tools for drug discovery: tools of the trade. Nature. 2007;446:219–222. doi: 10.1038/446219a. [DOI] [PubMed] [Google Scholar]
- 7.Psaltis D, et al. Developing optofluidic technology through the fusion of microfluidics and optics. Nature. 2006;442:381–386. doi: 10.1038/nature05060. [DOI] [PubMed] [Google Scholar]
- 8.Yager P, et al. Microfluidic diagnostic technologies for global public health. Nature. 2006;442:412–418. doi: 10.1038/nature05064. [DOI] [PubMed] [Google Scholar]
- 9.Imming P, et al. Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov. 2006;5:821–834. doi: 10.1038/nrd2132. [DOI] [PubMed] [Google Scholar]
- 10.Russ AP, Lampel S. The druggable genome: an update. Drug Discov Today. 2005;10:1607–1610. doi: 10.1016/S1359-6446(05)03666-4. [DOI] [PubMed] [Google Scholar]
- 11.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 12.Mayr LM. Tackling the chemogenomic space by novel screening technologies. Ernst Schering Res Found Workshop. 2006;58:111–173. doi: 10.1007/978-3-540-37635-4_8. [DOI] [PubMed] [Google Scholar]
- 13.Cheng AC, et al. Structure-based maximal affinity model predicts small-molecule druggability. Nat Biotechnol. 2007;25:71–75. doi: 10.1038/nbt1273. [DOI] [PubMed] [Google Scholar]
- 14.Huang B, et al. Counting low-copy number proteins in a single cell. Science. 2007;315:81–84. doi: 10.1126/science.1133992. [DOI] [PubMed] [Google Scholar]
- 15.Wu H, et al. Chemical cytometry on a picoliter-scale integrated microfluidic chip. Proc Natl Acad Sci U S A. 2004;101:12809–12813. doi: 10.1073/pnas.0405299101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao J, et al. Integration of single cell injection, cell lysis, separation and detection of intracellular constituents on a microfluidic chip. Lab Chip. 2004;4:47–52. doi: 10.1039/b310552k. [DOI] [PubMed] [Google Scholar]
- 17.El-Ali J, et al. Cells on chips. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 18.Freire SL, Wheeler AR. Proteome-on-a-chip: mirage, or on the horizon? Lab Chip. 2006;6:1415–1423. doi: 10.1039/b609871a. [DOI] [PubMed] [Google Scholar]
- 19.Zeng Y, Harrison DJ. Self-assembled colloidal arrays as three-dimensional nanofluidic sieves for separation of biomolecules on microchips. Anal Chem. 2007;79:2289–2295. doi: 10.1021/ac061931h. [DOI] [PubMed] [Google Scholar]
- 20.Cooper JW, et al. Recent advances in capillary separations for proteomics. Electrophoresis. 2004;25:3913–3926. doi: 10.1002/elps.200406154. [DOI] [PubMed] [Google Scholar]
- 21.Petricoin EF, et al. Clinical proteomics: translating benchside promise into bedside reality. Nat Rev Drug Discov. 2002;1:683–695. doi: 10.1038/nrd891. [DOI] [PubMed] [Google Scholar]
- 22.Wang YC, et al. Two-dimensional protein separation with advanced sample and buffer isolation using microfluidic valves. Anal Chem. 2004;76:4426–4431. doi: 10.1021/ac0497499. [DOI] [PubMed] [Google Scholar]
- 23.Kuntz ID. Structure-based strategies for drug design and discovery. Science. 1992;257:1078–1082. doi: 10.1126/science.257.5073.1078. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn P, et al. The genesis of high-throughput structure-based drug discovery using protein crystallography. Curr Opin Chem Biol. 2002;6:704–710. doi: 10.1016/s1367-5931(02)00361-7. [DOI] [PubMed] [Google Scholar]
- 25.Lundstrom K. Structural biology of G protein-coupled receptors. Bioorg Med Chem Lett. 2005;15:3654–3657. doi: 10.1016/j.bmcl.2005.05.041. [DOI] [PubMed] [Google Scholar]
- 26.Zheng B, et al. Screening of protein crystallization conditions on a microfluidic chip using nanoliter-size droplets. J Am Chem Soc. 2003;125:11170–11171. doi: 10.1021/ja037166v. [DOI] [PubMed] [Google Scholar]
- 27.Zheng B, et al. Using nanoliter plugs in microfluidics to facilitate and understand protein crystallization. Curr Opin Struct Biol. 2005;15:548–555. doi: 10.1016/j.sbi.2005.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen CL, et al. A robust and scalable microfluidic metering method that allows protein crystal growth by free interface diffusion. Proc Natl Acad Sci U S A. 2002;99:16531–16536. doi: 10.1073/pnas.262485199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lau BT, et al. A complete microfluidic screening platform for rational protein crystallization. J Am Chem Soc. 2007;129:454–455. doi: 10.1021/ja065855b. [DOI] [PubMed] [Google Scholar]
- 30.Li L, et al. Nanoliter microfluidic hybrid method for simultaneous screening and optimization validated with crystallization of membrane proteins. Proc Natl Acad Sci U S A. 2006;103:19243–19248. doi: 10.1073/pnas.0607502103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson MJ, et al. Phase knowledge enables rational screens for protein crystallization. Proc Natl Acad Sci U S A. 2006;103:16746–16751. doi: 10.1073/pnas.0605293103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Copeland RA, et al. Drug-target residence time and its implications for lead optimization. Nat Rev Drug Discov. 2006;5:730–739. doi: 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- 33.Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–237. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 34.Choi CJ, Cunningham BT. A 96-well microplate incorporating a replica molded microfluidic network integrated with photonic crystal biosensors for high throughput kinetic biomolecular interaction analysis. Lab Chip. 2007;7:550–556. doi: 10.1039/b618584c. [DOI] [PubMed] [Google Scholar]
- 35.Gohlke H, Klebe G. Approaches to the description and prediction of the binding affinity of small-molecule ligands to macromolecular receptors. Angew Chem Int Ed Engl. 2002;41:2644–2676. doi: 10.1002/1521-3773(20020802)41:15<2644::AID-ANIE2644>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 36.Golovanov AP, et al. Isotopically discriminated NMR spectroscopy: a tool for investigating complex protein interactions in vitro. J Am Chem Soc. 2007;129:6528–6535. doi: 10.1021/ja070505q. [DOI] [PubMed] [Google Scholar]
- 37.Perez JM, et al. Magnetic relaxation switches capable of sensing molecular interactions. Nat Biotechnol. 2002;20:816–820. doi: 10.1038/nbt720. [DOI] [PubMed] [Google Scholar]
- 38.Burg TP, et al. Weighing of biomolecules, single cells and single nanoparticles in fluid. Nature. 2007;446:1066–1069. doi: 10.1038/nature05741. [DOI] [PubMed] [Google Scholar]
- 39.Bleicher KH, et al. Hit and lead generation: beyond high-throughput screening. Nat Rev Drug Discov. 2003;2:369–378. doi: 10.1038/nrd1086. [DOI] [PubMed] [Google Scholar]
- 40.Bohacek RS, et al. The art and practice of structure-based drug design: a molecular modeling perspective. Med Res Rev. 1996;16:3–50. doi: 10.1002/(SICI)1098-1128(199601)16:1<3::AID-MED1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 41.Mitchell MC, et al. Microchip-based synthesis and analysis: control of multicomponent reaction products and intermediates. Analyst. 2001;126:24–27. doi: 10.1039/b007397k. [DOI] [PubMed] [Google Scholar]
- 42.Watts P, et al. Investigation of racemisation in peptide synthesis within a micro reactor. Lab Chip. 2002;2:141–144. doi: 10.1039/b203977j. [DOI] [PubMed] [Google Scholar]
- 43.Garcia-Egido E, et al. Synthesis and analysis of combinatorial libraries performed in an automated micro reactor system. Lab Chip. 2003;3:73–76. doi: 10.1039/b302381h. [DOI] [PubMed] [Google Scholar]
- 44.O-Charoen S, et al. Simulation and visualization of flow pattern in microarrays for liquid phase oligonucleotide and peptide synthesis. Biotechnol Prog. 2007;23:755–761. doi: 10.1021/bp060363o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou X, et al. Microfluidic PicoArray synthesis of oligodeoxynucleotides and simultaneous assembling of multiple DNA sequences. Nucleic Acids Res. 2004;32:5409–5417. doi: 10.1093/nar/gkh879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marcus JS, et al. Microfluidic single-cell mRNA isolation and analysis. Anal Chem. 2006;78:3084–3089. doi: 10.1021/ac0519460. [DOI] [PubMed] [Google Scholar]
- 47.Kartal M. Intellectual property protection in the natural product drug discovery, traditional herbal medicine and herbal medicinal products. Phytother Res. 2007;21:113–119. doi: 10.1002/ptr.2036. [DOI] [PubMed] [Google Scholar]
- 48.Gramowski A, et al. Functional screening of traditional antidepressants with primary cortical neuronal networks grown on multielectrode neurochips. Eur J Neurosci. 2006;24:455–465. doi: 10.1111/j.1460-9568.2006.04892.x. [DOI] [PubMed] [Google Scholar]
- 49.Yao X, et al. Carbon nanotube/poly(methyl methacrylate) composite electrode for capillary electrophoretic measurement of honokiol and magnolol in Cortex Magnoliae Officinalis. Electrophoresis. 2006;27:3233–3242. doi: 10.1002/elps.200600048. [DOI] [PubMed] [Google Scholar]
- 50.Luckarift HR, et al. Silica-immobilized enzymes for multi-step synthesis in microfluidic devices. Biotechnol Bioeng. 2007;98:701–705. doi: 10.1002/bit.21447. [DOI] [PubMed] [Google Scholar]
- 51.Macarron R. Critical review of the role of HTS in drug discovery. Drug Discov Today. 2006;11:277–279. doi: 10.1016/j.drudis.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 52.Hajduk PJ, Greer J. A decade of fragment-based drug design: strategic advances and lessons learned. Nat Rev Drug Discov. 2007;6:211–219. doi: 10.1038/nrd2220. [DOI] [PubMed] [Google Scholar]
- 53.Thorsen T, et al. Microfluidic large-scale integration. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 54.Quake SR, Scherer A. From micro- to nanofabrication with soft materials. Science. 2000;290:1536–1540. doi: 10.1126/science.290.5496.1536. [DOI] [PubMed] [Google Scholar]
- 55.Hong JW, et al. A nanoliter-scale nucleic acid processor with parallel architecture. Nat Biotechnol. 2004;22:435–439. doi: 10.1038/nbt951. [DOI] [PubMed] [Google Scholar]
- 56.Pregibon DC, et al. Multifunctional encoded particles for high-throughput biomolecule analysis. Science. 2007;315:1393–1396. doi: 10.1126/science.1134929. [DOI] [PubMed] [Google Scholar]
- 57.Khademhosseini A, et al. Microscale technologies for tissue engineering and biology. Proc Natl Acad Sci U S A. 2006;103:2480–2487. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rettig JR, Folch A. Large-scale single-cell trapping and imaging using microwell arrays. Anal Chem. 2005;77:5628–5634. doi: 10.1021/ac0505977. [DOI] [PubMed] [Google Scholar]
- 59.Chin VI, et al. Microfabricated platform for studying stem cell fates. Biotechnol Bioeng. 2004;88:399–415. doi: 10.1002/bit.20254. [DOI] [PubMed] [Google Scholar]
- 60.Khademhosseini A, et al. Cell docking inside microwells within reversibly sealed microfluidic channels for fabricating multiphenotype cell arrays. Lab Chip. 2005;5:1380–1386. doi: 10.1039/b508096g. [DOI] [PubMed] [Google Scholar]
- 61.Di Carlo D, et al. Single-cell enzyme concentrations, kinetics, and inhibition analysis using high-density hydrodynamic cell isolation arrays. Anal Chem. 2006;78:4925–4930. doi: 10.1021/ac060541s. [DOI] [PubMed] [Google Scholar]
- 62.Chen DL, Ismagilov RF. Microfluidic cartridges preloaded with nanoliter plugs of reagents: an alternative to 96-well plates for screening. Curr Opin Chem Biol. 2006;10:226–231. doi: 10.1016/j.cbpa.2006.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adamson DN, et al. Production of arrays of chemically distinct nanolitre plugs via repeated splitting in microfluidic devices. Lab Chip. 2006;6:1178–1186. doi: 10.1039/b604993a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dittrich PS, Manz A. Lab-on-a-chip: microfluidics in drug discovery. Nat Rev Drug Discov. 2006;5:210–218. doi: 10.1038/nrd1985. [DOI] [PubMed] [Google Scholar]
- 65.Pihl J, et al. Microfluidic gradient-generating device for pharmacological profiling. Anal Chem. 2005;77:3897–3903. doi: 10.1021/ac050218+. [DOI] [PubMed] [Google Scholar]
- 66.Chung BG, et al. Human neural stem cell growth and differentiation in a gradient-generating microfluidic device. Lab Chip. 2005;5:401–406. doi: 10.1039/b417651k. [DOI] [PubMed] [Google Scholar]
- 67.FDA. 2007 http://www.fda.gov/cder/handbook/preclin.htm.
- 68.Hojelse F. Preclinical safety assessment: in vitro -- in vivo testing. Pharmacol Toxicol. 2000;86(Suppl 1):6–7. doi: 10.1034/j.1600-0773.2000.d01-2.x. [DOI] [PubMed] [Google Scholar]
- 69.Viravaidya K, et al. Development of a microscale cell culture analog to probe naphthalene toxicity. Biotechnol Prog. 2004;20:316–323. doi: 10.1021/bp0341996. [DOI] [PubMed] [Google Scholar]
- 70.Baudoin R, et al. Trends in the development of microfluidic cell biochips for in vitro hepatotoxicity. Toxicol in Vitro. 2007;21:535–544. doi: 10.1016/j.tiv.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 71.Khamsi R. Labs on a chip: meet the stripped down rat. Nature. 2005;435:12–13. doi: 10.1038/435012a. [DOI] [PubMed] [Google Scholar]
- 72.Wu HM, et al. In vivo quantitation of glucose metabolism in mice using small-animal PET and a microfluidic device. J Nucl Med. 2007;48:837–845. doi: 10.2967/jnumed.106.038182. [DOI] [PubMed] [Google Scholar]
- 73.Cellar NA, et al. Microfluidic chip for low-flow push-pull perfusion sampling in vivo with on-line analysis of amino acids. Anal Chem. 2005;77:7067–7073. doi: 10.1021/ac0510033. [DOI] [PubMed] [Google Scholar]
- 74.Kim SM, et al. Low-power concentration and separation using temperature gradient focusing via Joule heating. Anal Chem. 2006;78:8028–8035. doi: 10.1021/ac061194p. [DOI] [PubMed] [Google Scholar]
- 75.Striemer CC, et al. Charge- and size-based separation of macromolecules using ultrathin silicon membranes. Nature. 2007;445:749–753. doi: 10.1038/nature05532. [DOI] [PubMed] [Google Scholar]
- 76.Kralj JG, et al. Integrated continuous microfluidic liquid-liquid extraction. Lab Chip. 2007;7:256–263. doi: 10.1039/b610888a. [DOI] [PubMed] [Google Scholar]
- 77.Neal G, et al. Separation of immunoglobulin G precipitate from contaminating proteins using microfiltration. Biotechnol Appl Biochem. 2004;39:241–248. doi: 10.1042/BA20030129. [DOI] [PubMed] [Google Scholar]
- 78.Valaskovic GA, et al. Ultra-low flow nanospray for the normalization of conventional liquid chromatography/mass spectrometry through equimolar response: standard-free quantitative estimation of metabolite levels in drug discovery. Rapid Commun Mass Spectrom. 2006;20:1087–1096. doi: 10.1002/rcm.2414. [DOI] [PubMed] [Google Scholar]
- 79.Dove A. Taking on chromatography’s hard cases. Nat Methods. 2007;4:289–293. [Google Scholar]
- 80.DeVoe DL, Lee CS. Microfluidic technologies for MALDI-MS in proteomics. Electrophoresis. 2006;27:3559–3568. doi: 10.1002/elps.200600224. [DOI] [PubMed] [Google Scholar]
- 81.Csajka C, Verotta D. Pharmacokinetic-pharmacodynamic modelling: history and perspectives. J Pharmacokinet Pharmacodyn. 2006;33:227–279. doi: 10.1007/s10928-005-9002-0. [DOI] [PubMed] [Google Scholar]
- 82.Abdel-Rahman SM, Kauffman RE. The integration of pharmacokinetics and pharmacodynamics: understanding dose-response. Annu Rev Pharmacol Toxicol. 2004;44:111–136. doi: 10.1146/annurev.pharmtox.44.101802.121347. [DOI] [PubMed] [Google Scholar]
- 83.Tsuchiya K, et al. Development of blood extraction system for health monitoring system. Biomed Microdevices. 2005;7:347–353. doi: 10.1007/s10544-005-6077-8. [DOI] [PubMed] [Google Scholar]
- 84.Nugroho AK, et al. Pharmacokinetics and pharmacodynamics analysis of transdermal iontophoresis of 5-OH-DPAT in rats: in vitro-in vivo correlation. J Pharm Sci. 2006;95:1570–1585. doi: 10.1002/jps.20528. [DOI] [PubMed] [Google Scholar]
- 85.Langer R. Drug delivery. Drugs on target Science. 2001;293:58–59. doi: 10.1126/science.1063273. [DOI] [PubMed] [Google Scholar]
- 86.Santini JT, Jr, et al. A controlled-release microchip. Nature. 1999;397:335–338. doi: 10.1038/16898. [DOI] [PubMed] [Google Scholar]
- 87.Groisman A, et al. Microfluidic memory and control devices. Science. 2003;300:955–958. doi: 10.1126/science.1083694. [DOI] [PubMed] [Google Scholar]
- 88.Brazil M. Drug delivery - Controlling the flow. Nat Rev Drug Discov. 2003;2:510–510. [Google Scholar]
- 89.McAllister DV, et al. Microfabricated microneedles for gene and drug delivery. Annu Rev Biomed Eng. 2000;2:289–313. doi: 10.1146/annurev.bioeng.2.1.289. [DOI] [PubMed] [Google Scholar]
- 90.Jahn A, et al. Microfluidic directed formation of liposomes of controlled size. Langmuir. 2007;23:6289–6293. doi: 10.1021/la070051a. [DOI] [PubMed] [Google Scholar]
- 91.Tan YC, et al. Controlled microfluidic encapsulation of cells, proteins, and microbeads in lipid vesicles. J Am Chem Soc. 2006;128:5656–5658. doi: 10.1021/ja056641h. [DOI] [PubMed] [Google Scholar]
- 92.Xu S, et al. Generation of monodisperse particles by using microfluidics: control over size, shape, and composition. Angew Chem Int Ed Engl. 2005;44:724–728. doi: 10.1002/anie.200462226. [DOI] [PubMed] [Google Scholar]
- 93.Okushima S, et al. Controlled production of monodisperse double emulsions by two-step droplet breakup in microfluidic devices. Langmuir. 2004;20:9905–9908. doi: 10.1021/la0480336. [DOI] [PubMed] [Google Scholar]
- 94.Hatch A, et al. A rapid diffusion immunoassay in a T-sensor. Nat Biotechnol. 2001;19:461–465. doi: 10.1038/88135. [DOI] [PubMed] [Google Scholar]
- 95.Steigert J, et al. Integrated siphon-based metering and sedimentation of whole blood on a hydrophilic lab-on-a-disk. Biomed Microdevices. 2007;9:675–679. doi: 10.1007/s10544-007-9076-0. [DOI] [PubMed] [Google Scholar]
- 96.Steigert J, et al. Fully integrated whole blood testing by real-time absorption measurement on a centrifugal platform. Lab Chip. 2006;6:1040–1044. doi: 10.1039/b607051p. [DOI] [PubMed] [Google Scholar]
- 97.Peoples MC, et al. A capillary-based microfluidic instrument suitable for immunoaffinity chromatography. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;848:200–207. doi: 10.1016/j.jchromb.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 98.Brivio M, et al. Miniaturized continuous flow reaction vessels: influence on chemical reactions. Lab Chip. 2006;6:329–344. doi: 10.1039/b510856j. [DOI] [PubMed] [Google Scholar]
- 99.Geyer K, et al. Microreactors as tools for synthetic chemists - The chemists’ round-bottomed flask of the 21st century? Chem-Eur J. 2006;12:8434–8442. doi: 10.1002/chem.200600596. [DOI] [PubMed] [Google Scholar]
- 100.Watts P, Haswell SJ. Microfluidic combinatorial chemistry. Curr Opin Chem Biol. 2003;7:380–387. doi: 10.1016/s1367-5931(03)00050-4. [DOI] [PubMed] [Google Scholar]
- 101.Tanaka K, et al. Large-scale synthesis of immunoactivating natural product, pristane, by continuous microfluidic dehydration as the key step. Org Lett. 2007;9:299–302. doi: 10.1021/ol062777o. [DOI] [PubMed] [Google Scholar]
- 102.Ratner DM, et al. Microreactor-based reaction optimization in organic chemistry--glycosylation as a challenge. Chem Commun (Camb) 2005:578–580. doi: 10.1039/b414503h. [DOI] [PubMed] [Google Scholar]
- 103.Huang Y, et al. Solvent resistant microfluidic DNA synthesizer. Lab Chip. 2007;7:24–26. doi: 10.1039/b613923j. [DOI] [PubMed] [Google Scholar]
- 104.Tian J, et al. Accurate multiplex gene synthesis from programmable DNA microchips. Nature. 2004;432:1050–1054. doi: 10.1038/nature03151. [DOI] [PubMed] [Google Scholar]
- 105.Chen H, et al. A high-throughput combinatorial approach for the discovery of a cremophor EL-free paclitaxel formulation. Pharm Res. 2003;20:1302–1308. doi: 10.1023/a:1025021603288. [DOI] [PubMed] [Google Scholar]
- 106.Wyttenbach N, et al. Miniaturized assay for solubility and residual solid screening (SORESOS) in early drug development. Pharm Res. 2007;24:888–898. doi: 10.1007/s11095-006-9205-0. [DOI] [PubMed] [Google Scholar]
- 107.Srivastava N, Burns MA. Analysis of non-Newtonian liquids using a microfluidic capillary viscometer. Anal Chem. 2006;78:1690–1696. doi: 10.1021/ac0518046. [DOI] [PubMed] [Google Scholar]