Summary
DNA methylation is a conserved epigenetic mark in plants and mammals. In Arabidopsis, DNA methylation can be triggered by small interfering RNAs (siRNAs) through an RNA-directed DNA methylation (RdDM) pathway. Here we report the identification of a new RdDM effector, RDM3/KTF1. Loss-of-function mutations in RDM3/KTF1 reduce DNA methylation and release the silencing of RdDM target loci without abolishing the siRNA triggers. KTF1 has similarity to the transcription elongation factor SPT5 and contains a C-terminal extension rich in GW/WG repeats. KTF1 colocalizes with ARGONAUTE 4 (AGO4) in punctate nuclear foci, and binds AGO4 and RNA transcripts. Our results suggest KTF1 as an adaptor protein that binds scaffold transcripts generated by Pol V and recruits AGO4 and AGO4-bound siRNAs to form an RdDM effector complex. The dual interaction of an effector protein with AGO and small RNA target transcripts may be a general feature of RNA silencing effector complexes.
Keywords: RDM3/KTF1, AGO4, RNA-binding, DNA methylation, gene silencing
Introduction
RNA interference (RNAi) is a conserved gene silencing mechanism in eukaryotic cells (Matzke and Birchler, 2005; Zaratiegui et al., 2007). In RNAi, double-stranded RNAs are processed by the RNaseIII enzyme Dicer into small interfering RNAs (siRNAs) that are then incorporated into an RNA-Induced Silencing Complex (RISC) to direct the cleavage or translational inhibition of complementary RNA (Matzke and Birchler, 2005; Tomari and Zamore, 2005; Filipowicz, 2005). The core component of RISC is the PAZ-and PIWI-domain-containing protein, Argonaute (AGO), which binds to siRNAs and can slice complementary RNAs (Filipowicz, 2005; Qi et al., 2006). Similarly, miRNAs are also generated by Dicers and direct a miRNA RISC to cause degradation or translational inhibition of target mRNAs (Bartel, 2004). In fission yeast, siRNAs are incorporated into the RNA-Induced Transcriptional Silencing (RITS) complex to cause heterochromatin formation (Volpe et al., 2002; Verdel et al., 2004). RITS also contains an AGO that slices transcripts complementary to the bound siRNAs (Verdel et al., 2004; Irvine et al., 2006). The conserved GW182 family of proteins is associated with miRNA RISC by binding to AGOs and is required for miRNA-mediated gene silencing (Ding and Han, 2007; Eulalio et al., 2008). In RITS, the Tas3 protein binds to AGO1 and is necessary for transcriptional gene silencing (Partridge et al., 2007; Till et al., 2007). The GW182 family of proteins and Tas3 all contain the GW/WG repeat sequence motif, which is considered an AGO hook that mediates interaction with AGOs (Ding and Han, 2007).
In plants, the overwhelming majority of small RNAs are 24-nt siRNAs corresponding to transposons and other repetitive elements (Zhang et al., 2007; Mosher et al., 2008). The 24-nt siRNAs cause epigenetic silencing by directing de novo DNA methylation through the RNA-directed DNA methylation (RdDM) pathway (Matzke and Birchler, 2005; Chan et al., 2005). In the RdDM pathway, siRNAs are generated by the action of the putative DNA-directed RNA polymerase Pol IV, RDR2 (RNA-DEPENDENT RNA POLYMERASE 2), and DCL3 (DICER-LIKE 3) (Matzke et al., 2009). The siRNAs are loaded into AGO4 and AGO6 to direct DNA methylation by the de novo DNA methyltransferase DRM2 (Matzke et al., 2009). The functioning of the siRNAs also requires another putative DNA-dependent RNA polymerase, Pol V; the chromatin remodeling protein, DRD1; and a structural-maintenance-of-chromosomes hinge domain-containing protein (Matzke et al., 2009). Pol IV and Pol V have distinct largest subunits, NRPD1 and NRPE1, respectively, but share with Pol II and/or with each other numerous additional subunits (Ream et al., 2009; Huang et al., 2009; He et al., 2009). NRPE1 contains a long C-terminal domain that is very rich in GW/WG repeats (El-Shami et al., 2007). The GW/WG repeats are required for Pol V function and are both sufficient and necessary for interaction with AGO4 (El-Shami et al., 2007). Recently, Pol V was found to generate uncapped and non-polyadenylated transcripts from several non-coding sequences that are targeted by RdDM (Wierzbicki et al., 2008). It was proposed that AGO4/6-bound siRNAs may find target DNA by binding to nascent scaffold transcripts generated by Pol V (Wierzbicki et al., 2008). The de novo DNA methyltransferase DRM2, which is presumably in the RdDM effector complex, is responsible for catalyzing cytosine methylation in CG, CHG, and CHH (H represents A, T, or C) sequence contexts (Cao and Jacobsen, 2002).
Active DNA demethylation mediated by the ROS1 family of DNA glycosylases is important for counteracting the activity of RdDM to prevent or attenuate hypermethylation and transcriptional silencing of transgene repeats, certain endogenous genes, transposons, and other repetitive sequences (Gong et al., 2002; Zhu et al., 2007; Penterman et al., 2007; Lister et al., 2008; He et al., 2009). In the DNA demethylase mutant, ros1, these sequences show enhanced transcriptional silencing (Gong et al., 2002; Zhu et al., 2007; Lister et al., 2008). To identify RdDM pathway components, we carried out a genetic screen for second-site suppressors of the ros1 mutant. Here, we report two allelic ros1 suppressor mutants, rdm3-1 and rdm3-2. The rdm3 mutations release the silencing of an RD29A promoter-driven LUCIFERASE (LUC) transgene, and the endogenous RD29A gene, in ros1 mutant plants. In the rdm3 mutants, DNA methylation is reduced at RdDM target loci such as 5S rDNA, MEA-ISR, AtSN1, AtGP1, and AtMU1. The rdm3 mutations do not affect the levels of siRNAs corresponding to these loci, suggesting that RDM3 may function with Pol V in the effector step of RNA-directed DNA methylation. Like ago4 mutations, however, rdm3 does not block production of Pol V transcripts. RDM3 encodes a protein that was annotated as KTF1 (KOW domain-containing Transcription Factor 1). RDM3/KTF1 has similarity to SPT5, a conserved transcription elongation factor for RNA polymerase II (Wada et al., 1998; Winston, 2001). We found that KTF1 and AGO4 interact in vitro and in vivo, and the two proteins are colocalized in discrete nucleoplasmic foci. These results suggest that KTF1 may physically link Pol V transcription with AGO4-mediated transcript cleavage and epigenetic regulation. KTF1 contains a C-terminal region rich in WG repeats, and these repeats are sufficient for interaction with AGO4. Importantly, an RNA-binding site was identified in the C-terminal region of KTF1. We hypothesize that WG repeats have co-evolved with an RNA-binding site in AGO-interacting proteins to facilitate the formation of a tight protein-transcript-siRNA effector complex for gene silencing.
Results
The rdm3 mutations suppress transcriptional gene silencing in the ros1 mutant
Loss-of-function mutations in the DNA demethylase ROS1 cause transcriptional gene silencing (TGS) of the stress-responsive RD29A promoter driven LUCIFERASE (RD29A-LUC) transgene, the endogenous RD29A gene, and the CaMV 35S promoter driven NPTII (35S-NPTII) kanamycin resistance transgene that is physically linked to the RD29A-LUC transgene (Gong et al., 2002). The silencing of the 35S-NPTII and RD29A-LUC transgenes is indicated by plant sensitivity to kanamycin and loss of luminescence, respectively. To identify components mediating TGS in ros1, we screened a T-DNA mutagenized population in the ros1 background, based on re-activation of luminescence from RD29A-LUC (He et al., 2009). Two allelic mutants, rdm3-1 and rdm3-2 (for RNA-directed DNA Methylation 3), were characterized in this study.
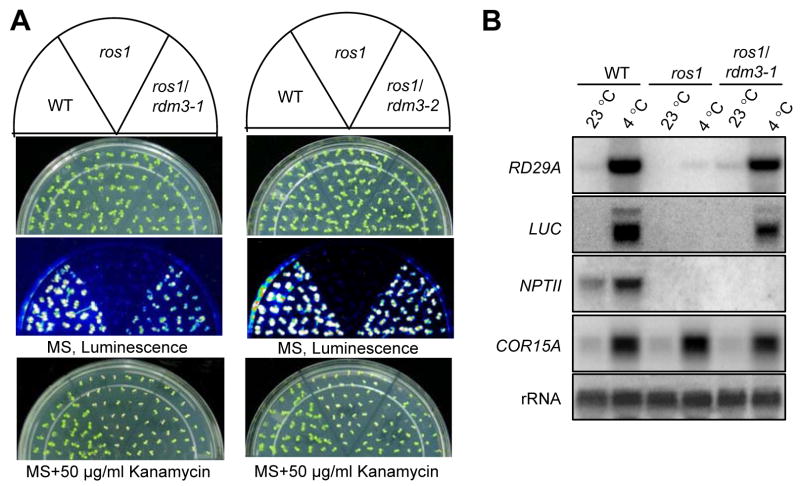
Figure 1A shows the luminescence phenotypes of the wild type, ros1, ros1rdm3-1, and ros1rdm3-2. Like the wild type, both ros1rdm3-1 and ros1rdm3-2 emitted strong luminescence after cold treatment, whereas ros1 emitted little or no luminescence. The result shows that the silencing of RD29A-LUC in ros1 was suppressed by the rdm3 mutations. However, the kanamycin sensitivity of ros1rdm3-1 and ros1rdm3-2 was similar to that of ros1, which indicated that the silencing of 35S-NPTII in ros1 was not suppressed by the rdm3 mutations. The ros1rdm3-1 and ros1rdm3-2 double mutants were crossed to ros1 (Figure 1A). The F1 plants emitted as little luminescence as the ros1 plants, but the F2 progenies segregated approximately 3:1 for ros1: ros1rdm3 luminescence phenotypes, suggesting that the rdm3-1 and rdm3-2 mutations were recessive and that each mutation was in a single nuclear gene (data not shown).
Figure 1. Transcriptional gene silencing of RD29A-LUC is suppressed by the rdm3-1 and rdm3-2 mutations.
(A) Effect of rdm3-1 and rdm3-2 on luminescence and kanamycin-resistance phenotypes in the ros1 background. Plants were grown on MS plates and subjected to luminescence imaging after cold treatment (4°C, 24 h). The plants were also grown on MS plates with kanamycin (50 μg/ml) and photographed after 10 days. (B) Northern blot analysis of RNA levels of endogenous RD29A, RD29A-LUC, and 35S-NPTII in wild type, ros1, and ros1rdm3-1. The constitutively expressed 18S rRNA was used as an RNA loading control while COR15A was used as a cold treatment control.
We crossed ros1/rdm3-1 plants to the wild type and identified the rdm3-1 single mutants. Interestingly, rdm3-1 plants emitted stronger luminescence than the wild type (Figures S1A and S1B). The result is consistent with the presence of a low level of TGS of the RD29A-LUC transgene in the wild type (Gong et al., 2002; Agius et al., 2006), and suggests that RDM3 is required for this TGS.
As reported previously (Gong et al., 2002), the mRNA levels of the endogenous RD29A, RD29A-LUC, and NPTII transgenes were dramatically reduced by the ros1 mutation. In ros1rdm3-1, the mRNA levels of both the endogenous RD29A and the RD29A-LUC transgene were substantially higher than those in ros1 (Figure 1B). In contrast, the mRNA level of the NPTII transgene in ros1rdm3-1 mutant was undetectable, as it was in ros1, which is consistent with the kanamycin-sensitive phenotype of the ros1rdm3-1 and ros1 plants (Figure 1B). These results demonstrate that the rdm3 mutations suppress the TGS of the endogenous RD29A gene and the RD29A-LUC transgene but not the NPTII transgene in the ros1 mutant.
The rdm3 mutation reduces DNA methylation at the RD29A promoter and other RdDM targets
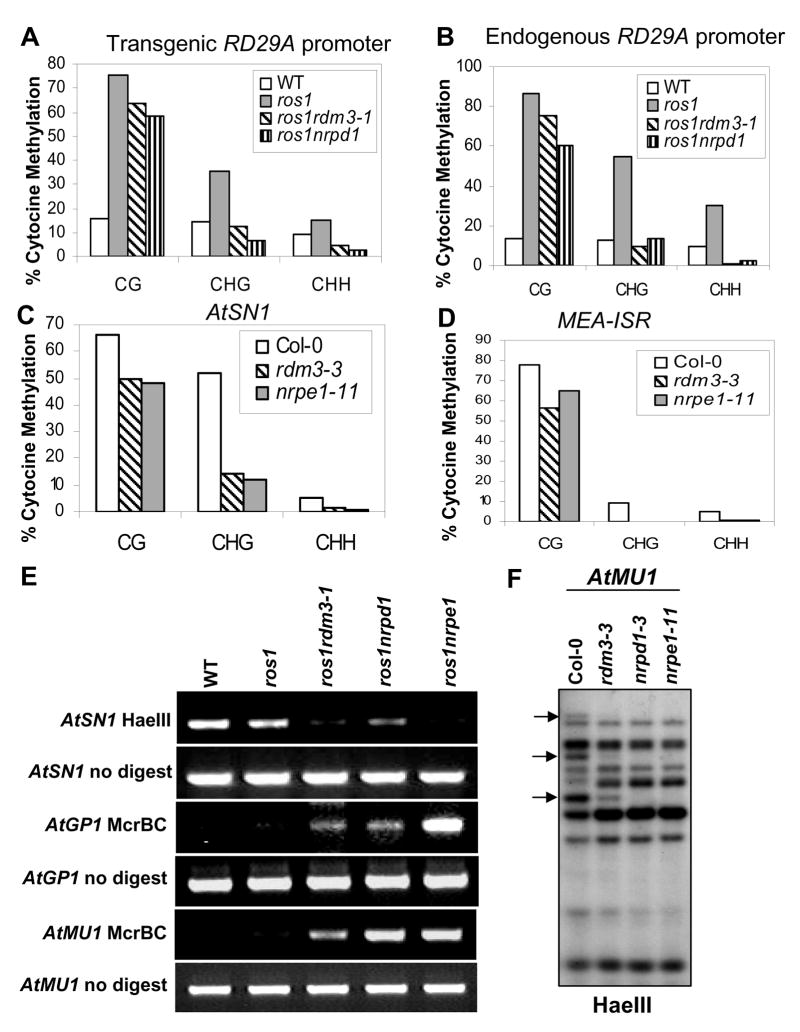
To test whether the suppression of RD29A-LUC transgene silencing in the ros1rdm3-1 mutant correlates with loss of DNA hypermethylation, we analyzed the DNA methylation status of the RD29A promoter by bisulfite sequencing. The results show that both the endogenous and transgenic RD29A promoters were heavily methylated at cytosine residues in all sequence contexts (CG, CHG, and CHH; H represents A, T, or C) in the ros1 mutant, but the methylation was reduced in the ros1rdm3-1 mutant (Figures 2A and 2B). The reductions were comparable to those in ros1nrpd1 and were particularly evident at CHG and CHH sites (Figures 2A and 2B). For example, at the transgenic RD29A promoter the asymmetric CHH methylation was 8.9% in the wild type, 15.2% in ros1, 4.5% in ros1rdm3-1, and 2.8% in ros1nrpd1 (Figure 2A). The methylation change at the endogenous RD29A promoter was also demonstrated by Southern hybridization (Figure S2A). Hypermethylation at the BstUI site of RD29A promoter in ros1 prevented the methylation-sensitive restriction enzyme from cleaving the promoter. The partial cleavage in ros1rdm3-1 and ros1nrpd1 by BstUI thus indicates a reduction in DNA methylation (Figure S2A). The results suggest that like nrpd1, rdm3-1 suppresses the TGS in ros1 by preventing DNA hypermethylation at the RD29A promoter.
Figure 2. The rdm3 mutations reduce DNA methylation at RdDM target loci.
The percentage of cytosine methylation was determined by bisulfite sequencing at transgenic (A) and endogenous (B) RD29A promoters, AtSN1 (C) and MEA-ISR (D). The percentage of cytosine methylation on CG, CHG, and CHH sites is shown. H represents A, T, or C. (E) The rdm3-1 mutation suppressed DNA methylation in AtSN1, AtGP1, and AtMU1. After the indicated genomic DNA was digested with the methylation-sensitive restriction enzyme HaeIII, it was used for amplification of AtSN1. After the genomic DNA was digested with the methylated DNA-specific restriction enzyme McrBC, it was used for amplification of AtGP1 and AtMU1. The amplifications of non-digested genomic DNA were used as controls. (F) The rdm3-3 mutation reduced AtMU1 methylation at CHH sites. Genomic DNA from the indicated genotypes was digested with HaeIII, followed by Southern blot analysis. The three undigested bands (arrows) that are present in the Col-0 wild type were mostly digested in rdm3-3, nrpd1-3 and nrpe1-11.
The DNA methylation status of the highly repetitive 180-bp centromeric repeat, which is not controlled by RdDM, was analyzed by Southern hybridization. No difference in DNA methylation of the centromeric repeat was detected among wild type, rdm3-3 (a T-DNA allele from Stock Center, in the Col-0 background; Figure S3A), and nrpe1-11 (nrpd1b-11) (Figure S2B). However, DNA methylation of 5S rDNA, an RdDM target locus, was reduced by the rdm3 mutations at all cytosine contexts, similar to the effects of nrpd1 and nrpe1 (Figures S2C and S2D).
We then examined the methylation status of several other well-characterized RdDM target loci including AtSN1, MEA-ISR, AtMU1 and AtGP1. Bisulfite sequencing was used to examine DNA methylation at AtSN1, a retroelement, and at MEA-ISR, a sub-telomeric repeat sequence present downstream of the MEA gene. The results show that in the wild-type plants, AtSN1 was heavily methylated with 66.1% of cytosine methylation at CG sites, 52.0% at CpXG, and 5.2% at CHH, but that the methylation levels were reduced to 50.0%, 14.3%, and 1.2%, respectively, in the rdm3-3 mutant. This effect of the rdm3-3 mutation on AtSN1 methylation was similar to that of nrpe1 (Figure 2C). Our bisulfite sequencing results also revealed that the DNA methylation level at MEA-ISR was reduced in rdm3-3, as it was in nrpe1, compared to that in the wild type (Figure 2D).
The reduced methylation in rdm3 mutants at CHH sites of AtSN1 was further tested by digestion with the methylation-sensitive enzyme HaeIII, followed by PCR. Figure 2E shows that AtSN1 was heavily methylated in wild type and ros1, and was thus resistant to HaeIII cleavage, but the methylation was much reduced in ros1rdm3-1, ros1nrpd1, and ros1nrpe1 (Figure 2E). For analysis of the DNA methylation of AtGP1 and AtMU1, the methylated DNA-digesting enzyme McrBC was applied, followed by PCR (Lippman et al., 2003). The results show that like the nrpd1 and nrpe1 mutations, rdm3-1 reduced the DNA methylation of AtGP1 and AtMU1 (Figure 2E). The effect of rdm3 on DNA methylation at AtSN1, AtGP1, and AtMU1 was confirmed by examining the rdm3-3 allele (Figure S4A). Moreover, the methylation status of AtMU1 at CHH sites was further tested by Southern hybridization, which showed that three HaeIII undigested bands in the wild type were mostly digested in rdm3-3, nrpd1, and nrpe1, confirming the reduced AtMU1 methylation in rdm3 (Figure 2F). Taken together, our results show that rdm3 mutations reduce DNA methylation at RdDM target sites, and that the effect was similar to that of mutations in known RdDM components, such as nrpd1, nrpe1, rdr2, dcl3, and ago4.
The effect of rdm3 mutations on 24-nt siRNAs and TGS of transposons
The above results suggest that RDM3 is required for DNA methylation at specific genomic loci. By semi-quantitative RT-PCR, we tested whether the reduction of DNA methylation at AtSN1, AtGP1, and AtMU1 in rdm3 mutant plants resulted in elevated transcript levels from these loci. The AtSN1 transcript level was compared among the wild-type Col-0, rdm3-3, nrpd1, and nrpe1 mutant plants. The results show that a very low level of AtSN1 transcript was detected in the Col-0 wild type but that the transcript level was drastically increased in rdm3-3, as well as in nrpd1 and nrpe1 (Figure 3A). Similarly, AtGP1 and AtMU1 had very low transcript levels in the wild type and ros1, but the transcript levels were substantially increased in ros1rdm3-1, as well as in ros1nrpd1 and ros1nrpe1 (Figure 3A). The results suggest that loss of DNA methylation caused by the rdm3 mutations leads to a release of TGS at the RdDM target loci.
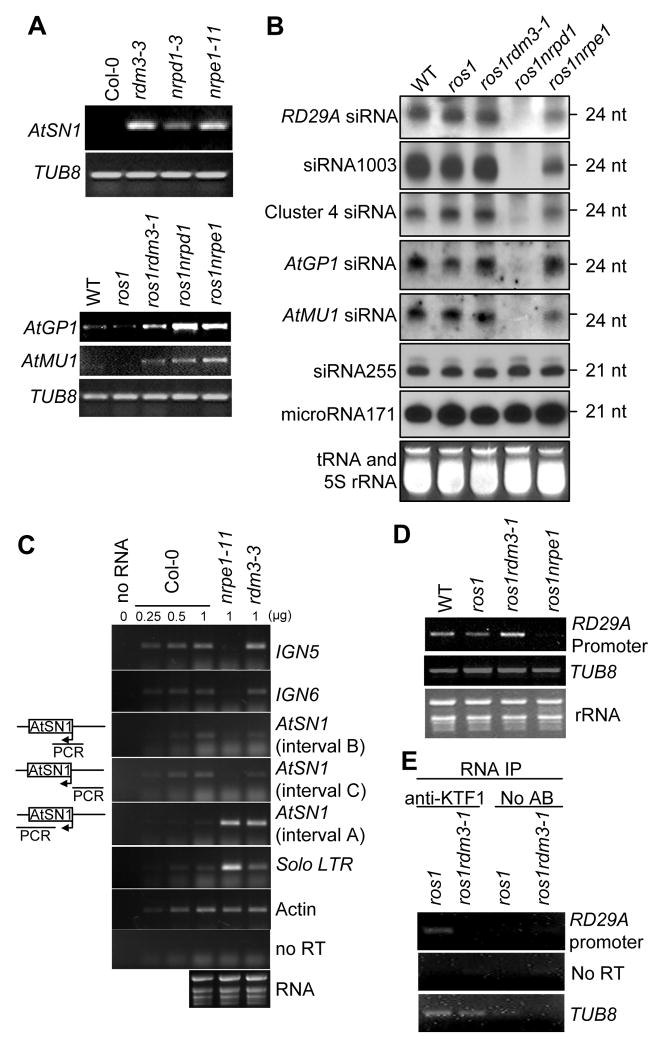
Figure 3. Effect of the rdm3 mutations on RNA and siRNA levels from the RdDM target loci.
(A) The rdm3 mutations increase the RNA expression levels of AtSN1, AtGP1, and AtMU1. Semi-quantitative RT-PCR was used to detect the transcript levels of AtSN1 (interval A, see diagram in panel C), AtGP1, and AtMU1 in the indicated genotypes. TUB8 was amplified as an internal control. (B) Small RNA blot analysis of 24-nt siRNAs, 21-nt ta-siRNAs, and microRNAs in the various genotypes. The positions of size markers (21 nt and 24 nt) are indicated. The ethidium bromide-stained small RNA gel is shown as a loading control. (C) Strand-specific RT-PCR analysis of IGN5, IGN6, AtSN1 and solo LTR transcripts in the Col-0 wild type, nrpe1-11 and rdm3-3. Actin PCR products and total RNA resolved by agarose gel electrophoresis serve as loading controls. Reactions without reverse transcriptase (no RT) were performed to control for background DNA contamination. The positions of the different AtSN1 intervals tested by RT-PCR are indicated in the diagram on the left. (D) RT-PCR analysis of RD29A promoter transcript. TUB8 and ethidium bromide-stained gel are shown as controls. (E) RT-PCR detection of RD29A promoter transcript in KTF1 immunoprecipitates. The background signal from TUB8 was used as an internal control, which indicated no difference between the RNA amounts from ros1 and ros1rdm3-1. No AB, controls without using anti-KTF1 antibody.
Our previous studies suggested that 24-nt siRNAs from the RD29A promoter are the trigger for TGS of RD29A-LUC and endogenous RD29A in ros1 (Gong et al., 2002; Zheng et al., 2007; He et al., 2009). To determine whether rdm3 mutations affect siRNA accumulation, we carried out small RNA blot analysis and found that the rdm3-1 mutation has no effect on the accumulation of 24-nt RD29A promoter siRNAs, whereas nrpd1 blocks the siRNAs and nrpe1 partially reduces them (Figure 3B). The results indicate a role for RDM3 that is downstream of siRNA production in the RdDM pathway. We also tested the effect of rdm3 on endogenous siRNAs from several other RdDM target loci including AtSN1, 5S rDNA, Cluster 4, AtGP1, and AtMU1 (Figure 3B and Figure S4B). The siRNA1003 from 5S rDNA was not affected in ros1rdm3-1, although it was blocked in ros1nrpd1 and partially reduced in ros1nrpe1. The siRNAs from AtGP1, AtMU1, and Cluster 4 were mostly unaffected in ros1rdm3-1 and ros1nrpe1, although they were eliminated in ros1nrpd1 (Figure 3B). The AtGP1, AtMU1, 5S rDNA, and Cluster 4 siRNA results were further confirmed in rdm3-3 (Figure S4B). In addition, AtSN1 siRNAs and siRNA02 were not reduced by the rdm3-3 mutation (Figure S4B). The 21-nt miRNA171 and ta-siRNA siRNA255 were also tested, and the results show that the rdm3 mutations had no effect on either of them (Figure 3B and Figure S4B). These results suggest that RDM3 acts in the RdDM pathway downstream of siRNA production, and thus like NRPE1 (Pol V), RDM3 can be considered as an effector of RdDM.
The rdm3-3 mutation does not block the accumulation of Pol V-dependent non-coding transcripts
To further delineate the role of RDM3 in the RdDM pathway, we tested whether RDM3 is required for the accumulation of Pol V-dependent transcripts (Wierzbicki et al., 2008). Pol V-dependent transcripts at IGN5, IGN6 and AtSN1 loci (intervals B and C) were still detectable in the rdm3-3 mutant (Figure 3C). In fact, the IGN5 transcript level was slightly increased in rdm3-3 (Figure 3C). Pol II and Pol III transcripts at the solo LTR and AtSN1 (interval A) loci, respectively, were upregulated in nrpe1, were also upregulated in the rdm3-3 mutant (Figure 3C).
The RD29A promoter generates 24-nt siRNAs and is targeted by the RdDM pathway for silencing (Gong et al., 2002; He et al., 2009). We found that the RD29A promoter also generated an RNA transcript that requires NRPE1 (Figure 3D). This Pol V-dependent RD29A promoter transcript was present in ros1rdm3-1, as well as in ros1 and the wild type (Figure 3D). Collectively, these data suggest that RDM3 affects RdDM at a step downstream, or independent, of Pol V transcription.
RDM3 encodes the KTF1 protein, a SPT5-like transcription elongation factor with a C-terminal extension rich in WG/GW repeats
To clone the RDM3 gene, we used TAIL-PCR to determine the T-DNA insertion site in the ros1rdm3-1 mutant, and found a single T-DNA insertion in the first intron of AT5G04290 (Figure S3A). In the ros1rdm3-2 mutant, we found a 61-bp deletion in the seventh exon of AT5G04290 (Figure S3A). We obtained the rdm3-3 mutant from the SALK collection (Salk_001254), which has a T-DNA insertion in the tenth exon of AT5G04290 (Figure S3A). In rdm3-3, DNA methylation was dramatically reduced at several tested genomic loci including AtSN1, AtGP1, AtMU1, 5S rDNA, and MEA-ISR (Figures 2C, 2D and 2F; Figures S2C, S2D, S3C and S4A). These multiple mutations indicate that AT5G04290 is the RDM3 gene. AT5G04290 was annotated as KTF1 (KOW domain-containing Transcription Factor 1) (http://www.arabidopsis.org/servlets/TairObject?id=136799&type=gene). The identity of the RDM3 gene was confirmed by complementation tests where introduction of a KTF1 genomic fragment from the wild type restored the silencing of RD29A-LUC and AtSN1 methylation in ros1rdm3-1 and rdm3-3 (Figures S3B and S3C).
A cDNA containing the entire open reading frame of KTF1 was cloned. Based on the sequence of the cloned cDNA, the KTF1 gene contains 18 exons (Figure S3A) and encodes a protein of 1476 amino acid residues (Figure 4A). This differs from the computational gene structure prediction on the TAIR website (http://www.arabidopsis.org/servlets/TairObject?id=136799&type=gene). RT-PCR assays showed that the three rdm3 mutations blocked the accumulation of full length KTF1 transcripts (Figure S5A). Western blot analysis revealed a lack of the KTF1 protein in ros1rdm3-1 and rdm3-3 mutant plants (Figure S5B). These results suggest that the three rdm3 mutants are null alleles.
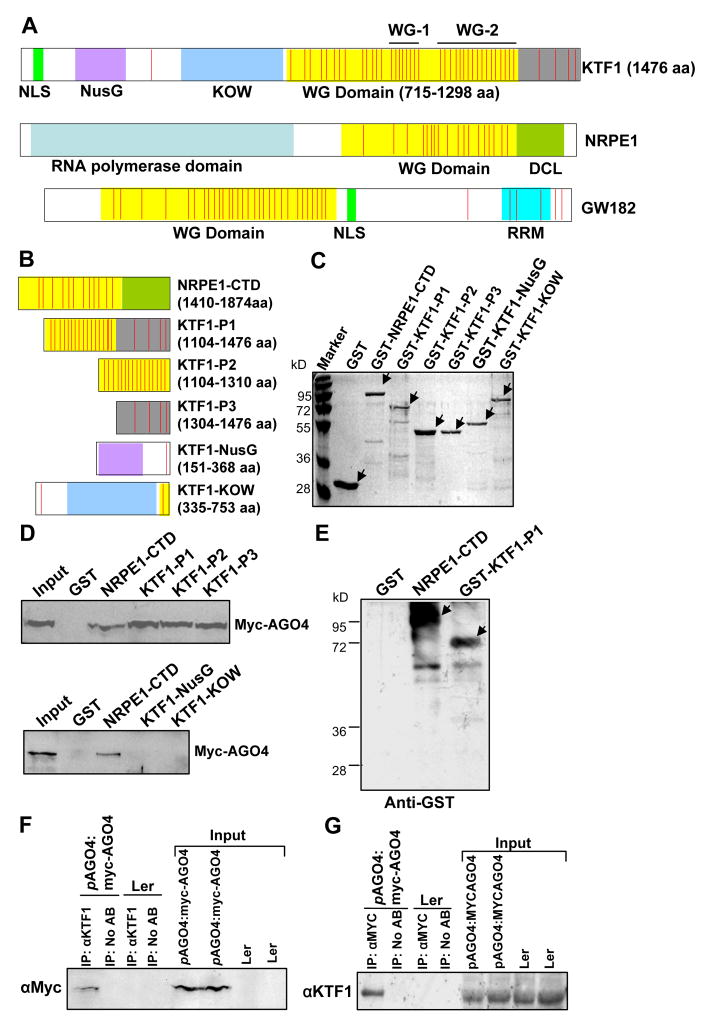
Figure 4. The WG/GW repeats in KTF1 C-terminal domain interact with AGO4.
(A) KTF1, NRPE1, HsGW182 and proteins are shown schematically. All three proteins are characterized by the reiterated WG/GW repeats-containing domains (in yellow). The red stripes represent each of the WG/GW repeat. WG-1 and WG-2 represent two highly conserved WG repeat regions in KTF1. (B) Diagram of the bacterially expressed NRPE1-CTD and truncated KTF1 proteins. (C) The purified proteins were subjected to SDS-PAGE and gels were stained with Coomassie. Arrows point to the proteins of interest. (D) Western blot analysis showing that the GST-fused truncated KTF1 and NRPE1-CTD interact with Myc-AGO4 from plant extracts. Ten percent of the input was used in the “Input” lane. (E) Anti-Myc antibody-conjugated beads captured truncated KTF1 proteins and NRPE1-CTD from a mixture of the proteins with extract from Myc-AGO4 plants. Arrows point to the proteins of interest. (F) and (G) Western blot analysis showing coimmunoprecipitation of KTF1 and Myc-AGO4. Ler wild-type plants without the Myc-AGO4 transgene were used as controls. No AB, control precipitation without using antibodies.
The KTF1 protein is highly similar to the SPT5 family of transcription elongation factors in its N-terminal region (Figure S6A). SPT5 is an essential gene conserved in all eukaryotes and is known to play both positive and negative roles in transcription elongation (Winston, 2001; Yamada et al., 2006). SPT5 proteins contain an acidic domain at the N-terminal region in addition to NusG and KOW domains and C-terminal repeats (Ivanov et al., 2000). KTF1 lacks an acidic N-terminus, and its C-terminal region is different from that of SPT5 proteins, so its similarity to SPT5 proteins is restricted to the NusG and KOW domains. Phylogenetic analysis shows that KTF1 and its orthologs from other plants including rice and grape are more closely related to SPT5 orthologs from plants and animals than to SPT5 proteins from yeasts (Figure S6B). Consistent with a role of KTF1 in RdDM, KTF1 contains a putative nuclear localization signal (NLS) near the N-terminus (Figure 4A). The long C-terminal region of KTF1 is characterized by more than 40 WG-containing repeats (Figure 4A). A large number of WG/GW repeats are also present in the C-terminal region of the largest subunit of Pol V, NRPE1, and in GW182 and related proteins in metazoans (El-Shami et al., 2007; Ding and Han, 2007; Eulalio et al., 2008). The fission yeast Tas3 protein also contains several WG/GW repeats (Till et al., 2007; Patridge et al., 2007). WG/GW-containing sequences are not conserved beyond the defining WG/GW core pattern, even between orthologous proteins. However, part of the WG repeat region of KTF1 is similar to the WG repeat region in Tas3, including residues surrounding the WG sequences (Figure S6C). There are 42 WG repeats in KTF1, and most of them are clustered in the C-terminal region of the protein as diagrammed in Figure 4A. As in NRPE1 (El-Shami et al., 2007), the WG repeat domain in KTF1 is highly hydrophilic and rich in G, S, D, K, and W (Table S1). The WG repeats are concentrated in two sub-domains in the C-terminal region of KTF1, which are designated WG-1 and WG-2 (Figure 4A); WG-1 has seven highly conserved WG repeats while WG-2 has 15 such repeats (Figure S6D).
KTF1 interacts with AGO4 via its WG repeat domain
Recently, it was discovered that WG/GW repeats in Arabidopsis NRPE1, animal GW182 (Figure 4A) and its paralogs, and the fission yeast Tas3 serve as AGO-binding motifs (El-Shami et al., 2007; Eulalio et al., 2008; Ding and Han, 2007; Till et al., 2007; Patridge et al., 2007). To determine whether the C-terminal WG repeat region of KTF1 can provide binding sites for AGO4, we generated GST-KTF1 fusion constructs (GST-KTF1-P1, P2, P3) and produced truncated versions of KTF1 (Figure 4B). The C-terminal region of NRPE1 was used as a positive control (NRPE1-CTD; Figure 4B). The bacterially produced GST fusion proteins were purified (Figure 4C) and used to capture Myc-AGO4 from the crude protein extracts from Myc-AGO4 transgenic plants. Myc-AGO4 was captured by all three versions of truncated KTF1 proteins from the C-terminal region, as well as by NRPE1-CTD, but was not captured by the two truncated KTF1 proteins (KTF1-NusG and KTF1-KOW) from the conserved NusG and KOW domains (Figure 4D). As expected, Myc-AGO4 was not captured by the negative control protein, GST (Figure 4D). Because RNase was present in the binding assay, the capture of Myc-AGO4 by the GST fusion proteins (GST-KTF1-P1, P2 and P3) was probably due to direct directions between AGO4 and the truncated KTF1 proteins rather than indirect interactions mediated by scaffold RNA transcripts. We also used anti-Myc antibodies to capture bacterially purified GST-KTF1-P1 and GST-NRPE1-CTD after mixing the recombinant proteins with crude protein extracts from Myc-AGO4 transgenic plants. The result shows that GST-KTF1-P1 and GST-NRPE1-CTD but not GST were captured by the Myc antibodies (Figure 4E). These results demonstrate that KTF1 and AGO4 interact in vitro, and that only a few WG repeats from KTF1 are sufficient for the interaction because KTF1-P3 contains only four WG/GW repeats. In the fission yeast Tas3 protein, one or two WG repeats are sufficient for binding to Ago1 (Partridge et al., 2007; Till et al., 2007). Nevertheless, KTF1-NusG and KTF1-KOW did not interact with AGO4, although the truncated proteins have one and two WG motifs, respectively (Figure 4B and 4D). Perhaps the hydrophilic amino acid residues in the WG repeat domain of KTF1 are also required for AGO4 interaction.
We performed co-immunoprecipitation experiments to test whether KTF1 interacts with AGO4 in vivo. Myc-AGO4 was detected in KTF1 immunoprecipitates from Myc-AGO4 transgenic plants (Figure 4F). Likewise, KTF1 was detected in Myc-AGO4 immunoprecipitates (Figure 4G). The result suggests that KTF1 and AGO4 interact in vivo.
Partial co-localization of KTF1 with AGO4 and Pol V in the nucleoplasm
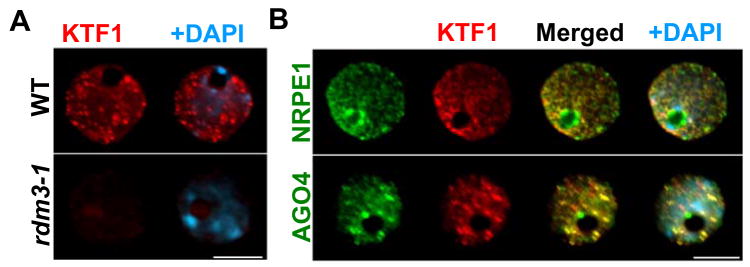
To visualize the sub-nuclear localization of KTF1, we performed immunostaining in interphase nuclei using antibodies specific for KTF1. In 100% of labeled nuclei (n=123), KTF1 immuno signals show distinct nucleoplasmic foci but no nucleolar localization in wild type leaves (Figure 5A). In the rdm3-1 mutant, the signal intensity is strongly reduced (Figure 5A, 100% of the nuclei, n=97), confirming that the KTF1 antibody is specific.
Figure 5. Sub-nuclear localization of KTF1 in interphase Arabidopsis nuclei.
A. Detection of KTF1 (in red) in wild type (WT) and rdm3-1 mutant nuclei by immunofluorescence using anti-KTF1. B. Simultaneous localization of KTF1 and AGO4 or NRPE1. KTF1 (red) was localized using its specific antibody in cells expressing cMyc- and Flag-tagged AGO4 and NRPE1 (in green), respectively. The bright yellow signals due to the overlap of red and green channels in merged images indicate colocalization of two labeled proteins. In all panels DNA was stained with DAPI (blue). Size bar corresponds to 5μm.
In order to determine whether KTF1 might be associated with AGO4 and Pol V in vivo in the nucleus, we performed co-immunolocalization experiments using antibody against the native KTF1 protein and transgenic lines expressing NRPE1-Flag and Myc-AGO4. Both NRPE1 and AGO4 are localized to the nucleoplasm but also display an intense, round-shaped nucleolar signal (Figure 5B) as described previously (Pontes et al., 2006; Li et al., 2006). The merging of KTF1 and AGO4 signals showed a similar distribution pattern and colocalization in the same nucleoplasmic sites but not in the nucleolus, as indicated by the yellow signals (Figure 5B) in most of the cells (81%, n=147). With respect to KTF1 and NRPE1, the majority of cells (63%, n=135) also showed a partial colocalization of the two proteins in the nucleoplasm but not the nucleolus or nucleolar periphery (Figure 5B). These colocalization results are consistent with a physical interaction between KTF1 and AGO4 in vivo in the nucleoplasm. There may also be an interaction between KTF1 and NRPE1, albeit to a lesser extent.
KTF1 is a novel RNA-binding protein
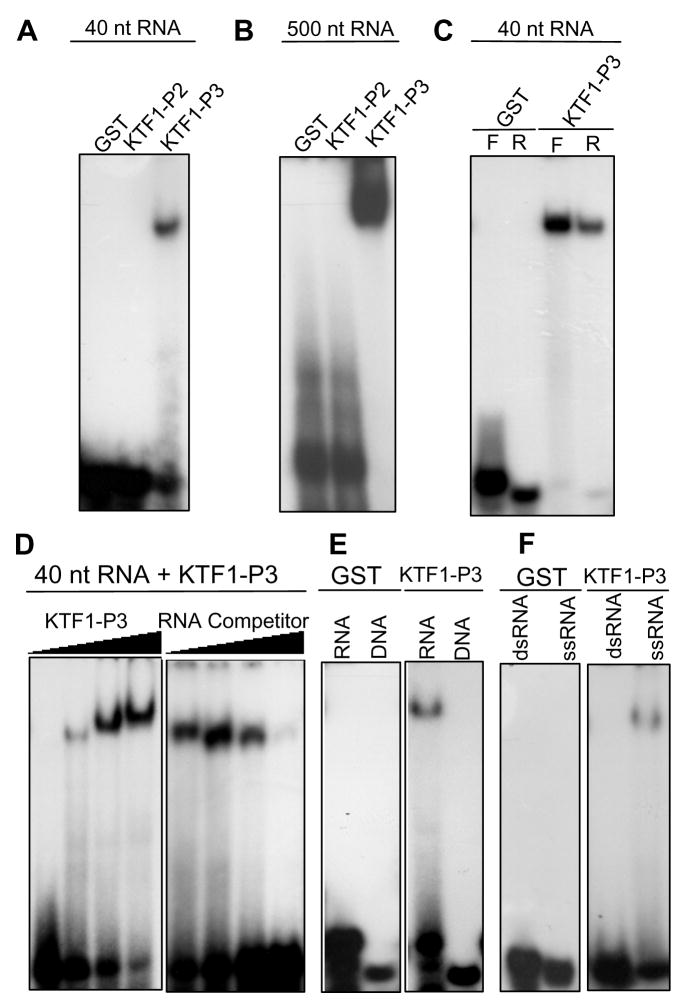
In electrophoretic mobility shift assays (EMSA), the C-terminal region of KTF1 (i.e., KTF1-P1) displays an RNA binding activity. KTF1-P1 but not GST could bind to 40-nt single-stranded RNA corresponding to the RD29A promoter (part of the Pol V-dependent RD29A promoter transcript) (Figure S7A). Interestingly, KTF1-P3 but not KTF1-P2 was also capable of binding to the RNA (Figure 6A). Because KTF1-P2 contains many more WG repeats than KTF1-P3, the results indicate that WG repeats are not responsible for RNA-binding. KTF1-P1 and KTF1-P3 but not GST or KTF1-P2 also bound to a 24-nt RNA, which corresponds to part of the 40-nt RNA (Figure S7B). Similarly, KTF1-P3 but not GST or KTF1-P2 was capable of binding to a 21-nt RNA (Figure S7C). Very strong binding was observed for KTF1-P3 to 500-nt RNA corresponding to the RD29A promoter (Figure 6B). KTF1-P3 was capable of binding to not only the 40-nt RNA but also to its complementary RNA (Figure 6C). These results show that KTF1 can bind RNAs of various sizes and that the binding is not sequence- or strand-specific.
Figure 6. The KTF1 C-terminal domain binds RNAs.
(A) KTF1-P3 but not KTF1-P2 binds to a 40-nt RNA (corresponding to the RD29A promoter) in electrophoretic mobility shift assays. (B) KTF1-P3 but not KTF1-P2 binds to a 500-nt RNA corresponding to the RD29A promoter. (C) KTF1-P3 binds to both the forward (F) and reverse (R) strands of the 40-nt RNA. (D) Protein concentration-dependence of the RNA-binding and competition by unlabeled RNA. The protein-RNA complex increased when an increasing amount of KTF1-P3 protein (0.2, 0.4, 0.8, 1.6 μg) was added to the binding reaction. The protein-RNA complex decreased when an increasing amount of unlabeled 40-nt RNA (1x, 5x, 25x, 125x of labeled RNA) was added to the binding reaction. (E) KTF1-P3 binds to the 40-nt RNA but does not bind to DNA of the same sequence. (F) KTF1-P3 binds to the single-stranded but not double-stranded 40-nt RNA.
KTF1-P3 bound to the 40-nt Pol V transcript in a dose-dependent manner (Figure 6D). The binding was reduced or blocked by competition with un-labeled RNA of the same sequence (Figure 6D). KTF1-P3 did not bind to 40-nt DNA of the same sequence as the RNA (Figure 6E). Furthermore, KTF1-P3 specifically bound to the 40-nt single-stranded RNA, and did not bind to the corresponding double-stranded RNA (Figure 6F).
To test whether KTF1 may be associated with Pol V transcripts in vivo, RNA immunoprecipitation was carried out using anti-KTF1 antibodies. The Pol V-dependent transcript from the RD29A promoter was detected in the KTF1 immunoprecipitate from ros1 but not ros1rdm3-1 (Figure 3E). No signal was detected in ros1 or ros1rdm3-1 following precipitation in the absence of the anti-KTF1 antibody. Taken together, the results suggest that KTF1 can bind to the Pol V-dependent RD29A promoter transcript in vitro and in vivo.
Discussion
Our genetic analysis indicates that KTF1 is a critical effector in the RNA-directed DNA methylation (RdDM) pathway. Like most of the previously identified RdDM components (He et al., 2009), KTF1 is required for transcriptional gene silencing (TGS) of the RD29A-LUC but not of the 35S-NPTII transgene, suggesting that KTF1 functions specifically in siRNA-dependent but not siRNA-independent TGS. KTF1 contains the NusG and KOW domains that are highly conserved in the SPT5 family of proteins from yeast to humans. In humans, SPT5 is involved in transcriptional inhibition mediated by 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB) (Wada et al., 1998). SPT5 and SPT4 form a heterodimeric complex that is also known as DSIF (DRB sensitivity-inducing factor) (Wada et al., 1998). DSIF can regulate transcription elongation in both positive and negative manners by regulating the processivity of RNA polymerase II (Wada et al., 1998; Yamada et al., 2006).
Considering its specific role in the RdDM pathway, KTF1 may regulate Pol V transcription. Recently, Huang et al (2009) reported that KTF1 could be detected in Pol V complexes purified from the inflorescence tissue of cauliflower, and T-DNA insertion mutants in this gene in Arabidopsis had reduced DNA methylation at several RdDM target loci. We found that Pol V transcripts are not blocked by the rdm3 mutation (Figures 3C and 3D). Therefore, unlike Pol V, KTF1 is not required for generating the non-coding transcripts. In Pol II transcription, SPT5 interacts with Pol II and RNA processing factors, and is also associated with the exosome (Lindstrom et al., 2003). Thus, SPT5 is involved in the coordination between transcription elongation and RNA processing and degradation. Our results show that KTF1 interacts with AGO4, another effector of RdDM. AGO4 binds siRNAs and its slicer activity cleaves at least some target RNA transcripts (Qi et al., 2006). This cleavage activity is required for de novo methylation at some RdDM target loci (Qi et al., 2006). Therefore, our results suggest that KTF1 is involved in coordinating Pol V transcription elongation with AGO4-mediated transcript binding or cleavage.
Our data show that KTF1 is an RNA-binding protein. KTF1 is capable of binding Pol V transcripts in vitro and in vivo (Figures 6 and 3E). KTF1 binding to Pol V transcripts is presumably important for the assembly of a functional RISC-like complex containing AGO4, AGO4-bound siRNAs, and Pol V transcripts complementary to the siRNAs. The interaction between an AGO-bound small RNA and its target transcript alone may not be sufficient for the assembly of a functional effector complex, and an adaptor protein like KTF1 that binds to both the AGO and target transcript may be needed. In metazoans, the P-body constituents GW182 and the related TNRC6B and TNRC6C proteins are important for miRNA RISC function (Ding and Han, 2007). These proteins contain not only GW/WG repeats for interacting with AGOs but also RNA recognition motif (RRM) for binding miRNA target mRNAs. This dual interaction of GW182 with AGO and target transcripts of small RNAs may be important for efficient location of target transcripts by AGO-bound small RNAs and for the assembly of a functional RISC to silence the target transcripts. In the RITS complex in fission yeast, Tas3 contains WG repeats and interacts with Ago1 (Verdel et al., 2004; Patridge et al., 2007; Till et al., 2007). The targeting of the Clr4 methyltransferase complex (ClrC) is closely coupled to transcription of the target loci and Tas3 is required for the physical interaction between ClrC and RITS (Chen et al., 2008; Zhang et al., 2008). Although Tas3 has not yet been reported to bind RNA transcripts, artificial tethering of Tas3 but not any other components of RITS to a nascent reporter transcript caused the silencing of the tethered transcript (Buhler et al., 2006). It is possible that Tas3 may contain an as yet unidentified RNA-binding site. We propose that the effector complex of various small RNA-mediated silencing pathways may contain an adaptor protein capable of bridging AGO and target transcripts of the small RNAs. In this regard, an RNA-binding site may have co-evolved with the AGO-binding WG/GW repeats in KTF1, GW182, and other related adaptor proteins in silencing effector complexes.
In the RdDM pathway, the NRPE1 subunit of Pol V also contains many WG/GW repeats in the C-terminal domain that binds to AGO4 (El-Shami et al., 2007). Like KTF1, the C-terminal domain of NRPE1 can also bind RNA transcripts (X.-J. He, Y.-F. Hsu and J.-K. Zhu, unpublished data). The non-coding transcripts produced by Pol V and bound to NRPE1 may be cleaved by the NRPE1-associated AGO4. The cleaved transcripts may be transferred to KTF1, which attracts additional AGO4 with bound siRNAs. This transcript-KTF1-AGO4-siRNA complex may then recruit DRM2 to cause de novo methylation of the non-coding transcripts-generating loci. Future experiments will be able to test this speculative model.
Experimental Procedures
Plant growth, mutant screening, and gene cloning
The wild-type C24 and the ros1 mutant plants carry a homozygous stress-responsive RD29A-LUC transgene (He et al., 2009). A T-DNA mutagenized population in the ros1 background was generated as described previously (Kapoor et al., 2005). Screening for ros1 suppressors and isolation of T-DNA tagged gene by TAIL-PCR (rdm3-1) and of untagged gene (rdm3-2) by map-based cloning were as described previously (He et al., 2009). An approximately 8.8 kb KTF1 genomic fragment was amplified from Col-0 wild type plants and cloned into the Gateway vector PMDC164 for complementation assay in ros1rdm3-1 and rdm3-3.
DNA methylation assay
The DNA methylation status was analyzed by Chop-PCR, Southern hybridization, and bisulfite sequencing. For Chop-PCR, genomic DNA (500 ng) was digested with the methylation-sensitive restriction ezyme HaeIII overnight or the methylated DNA-digesting enzyme McrBC for 1 h. The digested DNA was used to amplify the RdDM targets including AtSN1, AtGP1, and AtMU1. The undigested genomic DNA was simultaneously amplified as controls. Southern hybridization and bisulfite sequencing were carried out as described in He et al (2009).
RNA analysis
RNA blot assays were carried out as described in He et al (2009). The sequences of DNA oligo probes and primers for probe amplification are listed in Table S2.
For semi-quantitative RT-PCR analysis, total RNA was extracted from flowers using Trizol reagent (Invitrogen). After contaminating DNA was removed by DNase, 5 μg of total RNA was used for synthesis of cDNA with SuperScript III (Invitrogen). The cDNA reaction mixture was diluted and used for RT-PCR. The PCR conditions were: 95°C for 5 min followed by 28–35 amplification cycles (95°C for 30 s, 56°C for 30 s, and 72°C for 1 min). The constitutively expressed TUB8 was used as an internal control. Primers used in RT-PCR are listed in Table S2. Conditions and primers for RT-PCR analysis of Pol V-dependent transcripts are as described in Wierzbicki et al (2008).
Binding assay of GST fusion proteins and Myc-AGO4
The NRPE1-CTD (1410–1874 aa) and the three truncated forms of KTF1 (1104–1476 aa; 1104–1310 aa; 1304–1476 aa; 151–368 aa; 335–753 aa) were amplified and cloned into bacterial expression vector pGEX4T1 (Invitrogen). The constructs were transformed into E. coli competent cells BL21 for expression. The GST fusion proteins were purified by glutathione Sepharose 4B beads (Amersham) and used for binding assays as described previously (Li et al., 2006). In brief, a total of 150 μl of protein extract from flowers was added to the GST fusion protein beads (in bacterial protein extraction buffer containing DNase and RNase) and incubated at 4°C for 3 h. The bound proteins were eluted with SDS-PAGE sample buffer after the beads were washed with IP buffer three times. Anti-Myc conjugated agarose (Upstate) was used for immunoprecipitating Myc-AGO4 complexes from cell lysates mixed with purified GST fusion proteins at 4°C for 3 h. The immunoprecipitated protein was eluted with SDS-PAGE sample buffer. All the eluted protein was subjected to 10% SDS-PAGE gel for Western blotting.
Coimmunoprecipitation of AGO4 and RDM3
One gram of flowers from Myc-AGO4 transgenic plants (Li et al., 2009) as well as from the wild-type plants was used for preparation of cell lysates with 2 ml of protein extraction buffer. Equal amounts of crude protein extracts were precleared with protein A agarose beads (Sigma), followed by anti-KTF1 or anti-Myc incubation. The immunoprecipitated protein complexes were captured with protein A agarose beads, washed five times with the extraction buffer, and eluted by boiling the beads in SDS-PAGE sample buffer. The eluted sample was resolved by 8% SDS-PAGE for Western blotting.
Anti-KTF1 antibodies were generated by injecting rabbits with purified recombinant KTF1 protein (KTF1-P3), and purified by affinity chromatography using KTF1-conjugated beads.
Immunofluorescence microscopy
Interphase nuclei were isolated as described by Jasencakova et al (2000). Nuclei preparations were post-fixed with 4% paraformaldehyde and incubated overnight at 4ºC with primary antibodies for KTF1 (1:50), anti-Flag (1:200, Sigma) or anti-cMyc (1:200, Chemicon). Secondary antibodies anti-rabbit Alexa 488 (Invitrogen) and anti-mouse Alexa 594 (Invitrogen) were diluted at 1:200 in PBS and incubated for 4 h at 37°C. DNA was counterstained with 1 μg/ml DAPI in Prolong Gold mounting medium (Invitrogen). The nuclei preparations were analyzed with a Nikon Eclipse E80i epifluorescence microscope equipped with a Photometrics Coolsnap ES Mono digital camera. Images were acquired by the Phylum software and pseudocolored and merged in Adobe Photoshop.
Electrophoretic mobility shift assay (EMSA)
NRPE1-CTD and various truncated forms of KTF1 cDNA were cloned into the pGEX4T1 vector (Invitrogen) for GST fusion constructs. The constructs were transformed into E. coli strain BL21 (Invitrogen) for protein expression. The GST fusion protein was purified by glutathione Sepharose 4B beads (Amersham) and used for EMSA. For the binding assay, single-stranded RNA or DNA probes were directly synthesized and end-labeled with T4 polynucleotide kinase and γ-32P-ATP, while the 500-nt single-stranded RNA probe was generated from the RD29A promoter sequence using an in vitro transcription labeling kit (Ambion). All the probes used for binding assays were purified using 50 G columns (Biolab). The binding reaction included 2μl of labeled RNA or DNA probes, 0.5μg of GST fusion protein, 25 mM HEPES (pH 7.6), 50 mM KCl, 0.1 mM EDTA (pH 8.0), 12.5 mM MgCl2, 1 mM DTT, 0.5% (w/v) BSA, and 5% (w/v) glycerol. Reactions were incubated at room temperature for 30 min. The reaction mixtures were resolved on 4% non-denaturing polyacrylamide gel following electrophoresis at 200 V for 2 h. Gels were dried and exposed to X-Ray film for analysis.
RNA-IP
The ros1 and ros1rdm3-1 were subjected to RNA immunoprecipitation with anti-KTF1 antibodies, following the method of Wierzbicki, et al (2008).
Supplementary Material
Acknowledgments
We thank Xuemei Chen and Shou-Wei Ding for helpful discussion. This work was supported by National Institutes of Health grants R01GM070795 and R01GM059138 to J.-K. Zhu, GM077590 to C.S. Pikaard and National Science Foundation grant MCB-0642843 to H. Jin. O. Pontes was supported by an Edward Mallinckrodt Foundation Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci USA. 2006;103:11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- Ding L, Han M. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol. 2007;17:411–416. doi: 10.1016/j.tcb.2007.06.003. [DOI] [PubMed] [Google Scholar]
- El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, Vega D, Hakimi MA, Jacobsen SE, Cooke R, Lagrange T. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- Filipowicz W. RNAi: the nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- Gong Z, Morales-Ruiz T, Ariza RR, Roldan-Arjona T, David L, Zhu JK. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK. NRPD4, a protein similar to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for siRNA production, RNA-directed DNA methylation, and transcriptional gene silencing. Genes Dev. 2009;23:318–330. doi: 10.1101/gad.1765209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Jones AM, Searle I, Patel K, Vogler H, Hubner NC, Baulcombe DC. An atypical RNA polymerase involved in RNA silencing shares small subunits with RNA polymerase II. Nat Struct Mol Biol. 2009;16:91–93. doi: 10.1038/nsmb.1539. [DOI] [PubMed] [Google Scholar]
- Irvine DV, Zaratiegui M, Tolia NH, Goto DB, Chitwood DH, Vaughn MW, Joshua-Tor L, Martienssen RA. Argonaute slicing is required for heterochromatic silencing and spreading. Science. 2006;313:1134–1137. doi: 10.1126/science.1128813. [DOI] [PubMed] [Google Scholar]
- Ivanov D, Kwak YT, Guo J, Gaynor RB. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol Cell Biol. 2000;20:2970–2983. doi: 10.1128/mcb.20.9.2970-2983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasencakova Z, Meister A, Walter J, Turner BM, Schubert I. Histone H4 acetylation of euchromatin and heterochromatin is cell cycle dependent and correlated with replication rather than with transcription. Plant Cell. 2000;12:2087–2100. doi: 10.1105/tpc.12.11.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Agarwal M, Andreucci A, Zheng X, Gong Z, Hasegawa PM, Bressan RA, Zhu JK. Loss-of-function mutations in a conserved replication protein suppress transcriptional gene silencing in a DNA methylation-independent manner in Arabidopsis. Curr Biol. 2005;579:5889–5898. doi: 10.1016/j.cub.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, McCleery JA, Yates JR, Hartzog GA. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol Cell Biol. 2003;23:1368–1378. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, May B, Yordan C, Singer T, Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003;1:E67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009 Feb 23; doi: 10.1016/j.ceb.2009.01.025. [Epub ahead of print] PMID: 19243928. [DOI] [PubMed] [Google Scholar]
- Mosher RA, Schwach F, Studholme D, Baulcombe DC. Pol IVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci USA. 2008;105:3145–3150. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JF, DeBeauchamp JL, Kosinski AM, Ulrich DL, Hadler MJ, Noffsinger VJ. Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Mol Cell. 2007;26:593–602. doi: 10.1016/j.molcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Penterman J, Zilberman D, Huh JH, Ballinger T, Henikoff S, Fischer RL. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Li CF, Nunes PC, Haag J, Ream T, Vitins A, Jacobsen SE, Pikaard CS. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- Ream TS, Haag JR, Wierzbicki AT, Nicora CD, Norbeck A, Zhu JK, Hagen G, Guilfoyle TJ, Pasa-Tolic L, Pikaard CS. Subunit Compositions of the RNA-Silencing Enzymes Pol IV and Pol V Reveal Their Origins as Specialized Forms of RNA Polymerase II. Mol Cell. 2009;33:192–203. doi: 10.1016/j.molcel.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till S, Lejeune E, Thermann R, Bortfeld M, Hothorn M, Enderle D, Heinrich C, Hentze MW, Ladurner AG. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat Struct Mol Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F. Control of eukaryotic transcription elongation. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-2-reviews1006. reviews1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell. 2006;21:227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE. Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci USA. 2007;104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Zhu J, Kapoor A, Zhu JK. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 2007;26:1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Kapoor A, Sridhar VV, Agius F, Zhu JK. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.