Abstract
Cigarette smoking is the leading preventable cause of death in the US. Although smoking behavior has a significant genetic determination, the specific genes and associated mechanisms underlying smoking behavior are largely unknown. Here, we performed a genome-wide association study on smoking behavior in 840 Caucasians, including 417 males and 423 females, in which we examined ∼380,000 SNPs. We found that a cluster of nine SNPs upstream from the IL15 gene were associated with smoking status in males, with the most significant SNP, rs4956302, achieving a p value (8.80×10−8) of genome-wide significance. Another SNP, rs17354547, that is highly conserved across multiple species, achieved a p value of 5.65×10−5. These two SNPs, together with two additional SNPs (rs1402812 and rs4956396) were selected from the above nine SNPs for replication in an African-American sample containing 1,251 subjects, including 412 males and 839 females. The SNP rs17354547 was successfully replicated in the male subgroup of the replication sample; it was associated with smoking quantity (SQ), the Heaviness of Smoking Index (HSI) and the Fagerstrom Test for Nicotine Dependence (FTND), with p values of 0.031, 0.0046 and 0.019, respectively. In addition, a haplotype formed by rs17354547, rs1402812 and rs4956396 was also associated with SQ, HSI and FTND, achieving p values of 0.039, 0.0093 and 0.0093, respectively. To further confirm our findings, we performed an in silico replication study of the nine SNPs in a Framingham Heart Study sample containing 7,623 Caucasians from 1,731 families, among which, 3,491 subjects are males and 4,132 are females. Again, male-specific association with smoking status was observed, for which seven of the nine SNPs achieved significant p values (p<0.05) and two achieved marginally significant p values (p<0.10) in males. Several of the nine SNPs, including the highly conserved one across species, rs17354547, are located at potential transcription factor binding sites, suggesting transcription regulation as a possible function for these SNPs. Through this function, the SNPs may modulate gene expression of IL15, a key cytokine regulating immune function. As the immune system has long been recognized to influence drug addiction behavior, our association findings suggest a novel mechanism for smoking addiction involving immune modulation via the IL15 pathway.
Keywords: smoking, nicotine addiction, IL15, genomewide association, genetics
Introduction
Cigarette smoking results in an annual death toll of 438,000 in the United States, where one in every five deaths is smoking related (1). Smoking is highly associated with the development of cardiovascular and respiratory diseases, and cancer (2). Most dramatically, men and women who smoke increase their risk of developing lung cancer by 13 and 23 folds, respectively, compared to non-smokers (2).
Despite extensive smoking-control efforts, > 20% (or > 45 million) of American adults continue to smoke (3). There is a substantial genetic component underlying smoking behavior, with a heritability > 50% as demonstrated in studies with twins (4). The specific genes underlying smoking behavior, however, remain largely unknown. To date, more than 20 whole genome linkage studies have identified a number of loci potentially linked to smoking behavior, but few of these loci have been replicated across studies with high statistical significance (for review, see (5)). Genetic association studies have also implicated several genes associated with smoking behavior, such as GABAB2 (6), DOPA decarboxylase (7), and nicotinic acetylcholine receptor α4 subunit (CHRNA4) (8) and α5/α3/β4 cluster on chromosome 15 (9). However each of these studies focused on genes with known significance in neural biology and, consequently, these association studies were not designed to identify potentially novel genes/regulatory mechanisms underlying smoking behavior. Moreover, most of the genes implicated in these association studies await further confirmation from independent studies.
A promising strategy to facilitate identification of genes underlying smoking behavior is genome-wide association studies (GWAS) that take advantage of the knowledge of linkage equilibrium (LD) patterns in humans and the rapid development of high throughput SNP genotyping platforms. With high SNP densities that facilitate detection of culprit DNA changes within a narrow genomic region, the GWAS approach has demonstrated its great power for identifying novel genes associated with human complex diseases/traits (10-14).
Here we conducted one of the first few GWAS investigations to search for novel genetic factors underlying smoking behavior. Using an Affymetrix 500K array, we successfully genotyped and analyzed a total of 379,319 single nucleotide polymorphisms (SNPs) for 840 unrelated Caucasians, including 417 males and 423 females. We identified a cluster of nine SNPs upstream of the IL15 gene, which achieved strong association with smoking status in males. One particular SNP, rs17354547, is highly conserved across multiple species. This SNP, together with several other ones, are located at potential transcription factor binding sites, suggesting their functional importance, possibly by regulating IL15 gene expression. Furthermore, this SNP was replicated in an African-American (AA) cohort, where it was associated with several nicotine dependence (ND) phenotypes, and all of the nine SNPs were replicated in silico for association with smoking status in a Framingham Heart Study (FHS) Caucasian sample. Our findings suggest a novel mechanism for smoking behavior, where the IL15 pathway may play an important role.
Materials and methods
Subjects
GWAS sample
The study was approved by the necessary Institutional Review Boards of all involved institutions. Signed informed-consent documents were obtained from all study participants before they entered the study. For our GWAS, a random sample containing 840 unrelated Caucasians was identified from our established and expanding genetic repertoire currently containing more than 6,000 subjects. All of the chosen subjects were US Caucasians of European origin living in Omaha, Nebraska and its surrounding areas. They were healthy subjects recruited for genetic research of common human complex traits, such as bone mineral density and body mass index. Detailed recruitment and exclusion criteria were published elsewhere (15). Briefly, subjects with chronic diseases and conditions involving vital organs (heart, lung, liver, kidney, and brain) and severe endocrinological, metabolic, and nutritional diseases were excluded from this study. The general relevant characteristics of the study subjects are listed in Table 1.
Table 1.
Characteristics of study subjects
| GWAS sample (Caucasian) |
AA replication sample |
FHS replication sample |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Total sample | Male | Female | Total sample | Male | Female | Total sample | Male | Female | |
| No. of subjects | 840 | 417 | 423 | 1,251 | 412 | 839 | 7,623 | 3,491 | 4,132 |
| Age | 53.8 (16.9) | 54.6 (17.0) | 53.1 (16.2) | 40 (14.6) | 37.2 (14.3) | 41.5 (14.5) | 52.9 (15.1) | 52.5 (16.4) | 53.3 (15.3) |
| No. of smokers | 378 | 224 | 154 | 960 | 352 | 608 | 1,172 | 542 | 630 |
| Indexed SQ | 2.1 (1.08) | 2.3 (1.13) | 1.9 (0.96) | 2.3 (0.96) | 2.3 (0.96) | 2.2 (0.96) | – | – | – |
| HSI | – | – | – | 4.7 (1.5) | 4.8 (1.5) | 4.7 (1.5) | – | – | – |
| FTND score | – | – | – | 7.2 (2.2) | 7.2 (2.2) | 7.3 (2.2) | – | – | – |
Note: Age, SQ, HSI and FTNT score are presented as means (SD).
Smoking-related data from all subjects were recorded in a nurse-administered questionnaire, which also included a detailed medical history. Subjects were categorized as “smokers” based on their answer to the question in the questionnaire “Do/did you smoke cigarettes?” A subject who never smoked is defined as a “non-smoker”. For non-smokers, we intentionally excluded those subjects younger than the age of 25. This exclusion strategy is to ensure that our control subjects (non-smokers) are unlikely to develop smoking behavior if exposed to cigarettes in future since most smokers start smoking in adolescence. For smokers, cigarette consumption information was also collected, which was the number of cigarettes smoked per day. For the purpose of our analyses, the cigarette consumption information was transformed into indexed smoking quantity (SQ) using the criterion from the Fagerström Test for ND (FTND) questionnaire: SQ = 1 if number of cigarettes smoked/day is 10 or less, SQ = 2 if the number is 11-20, SQ = 3 if the number is 21-30, and so on.
AA replication sample
For replication of our GWAS findings, we used a family-based sample containing 402 nuclear families that included a total of 1,251 subjects (412 males and 839 females) (16). All the study subjects are of African-American (AA) origin and were recruited primarily from the Mid-South states of Tennessee, Mississippi, and Arkansas during 1999-2004. Proband smokers were required to be at least 21 years of age, have smoked for at least the last five years, and have consumed an average of 20 cigarettes per day for the last 12 months. Siblings and parents of a smoking proband were recruited whenever possible, regardless of their smoking status. Extensive data were collected on each participant, including demographics (e.g., sex, age, race, biological relationships, weight, height, years of education, and marital status), medical history, smoking history and current smoking behavior, ND, and personality traits assessed by various questionnaires, available at NIDA Genetics Consortium Website (http://zork.wustl.edu/nida). All participants provided informed consent. The study protocol and forms/procedures have been approved by all participating Institutional Review Boards.
In the present replication cohort, ND was ascertained by the three measures most commonly used in the literature: SQ (as defined in the above section), the Heaviness of Smoking Index (HSI: 0-6 scale), which includes SQ and smoking urgency (i.e., how soon after waking up does the subject smoke the first cigarette), and the Fagerström Test for ND (FTND: 0-10 scale) (17). A detailed description of the demographic and clinical characteristics of the sample is presented in Table 1.
FHS replication sample
To further replicate our GWAS findings in Caucasians, we used a sample from the FHS population, which contains 7,623 Caucasians, including 3,491 males and 4,132 females, from 1,731 families. The phenotype and genotype information of the cohort was downloaded from Framingham SHARe (SNP Health Association Resource), accessed through NCBI dbGaP (http://view.ncbi.nlm.nih.gov/dbgap). Appropriate procedures have been taken for the usage of the data, which include approval from UMKC IRB and signatures on the Data Distribution Agreement by all the UMKC investigators who have access to the data.
Self-reported smoking status was available for all the 7,623 subjects according to the Framingham SHARe. For determination of the smoking status, a question “Did you smoke cigarettes regularly in the last year?” was asked to a subject. Those answering “yes” are treated as smokers and “no” as non-smokers in this sample. In total, there are 1,172 smokers, among whom 542 are males and 630 females. To be consistent with our GWAS sample, those non-smokers who are younger than 25 were excluded from the analyses. The basic characteristics of the study subjects are presented in Table 1.
Genotyping
GWAS sample
Genomic DNA was extracted from whole human blood using a commercial isolation kit (Gentra systems, Minneapolis, MN, USA) following the protocols detailed in the kit. Genotyping with Affymetrix Mapping 250k Nsp and Affymetrix Mapping 250k Sty arrays was performed at Vanderbilt Microarray Shared Resource using the standard protocol recommended by Affymetrix. Genotyping calls were determined from the fluorescent intensities using the DM (dynamic model-based) algorithm with a 0.33 P-value setting (18) as well as the B-RLMM algorithm (19). DM calls were used for quality control while the B-RLMM calls were used for all subsequent data analysis. B-RLMM clustering was performed with 94 samples per cluster.
The final average BRLMM call rate across the entire sample reached the high level of 99.14%. However, out of the initial full-set of 500,568 SNPs, we discarded 32,961 SNPs with sample call rates < 95%, another 36,965 SNPs with allele frequencies deviating from Hardy-Weinberg equilibrium (HWE) (P < 0.001) and 51,323 SNPs with minor allele frequencies (MAF) < 1%. Therefore, the final SNP set maintained in the subsequent analyses contained 379,319 SNPs, yielding an average marker spacing of ∼7.9 kb throughout the human genome.
AA replication sample
Based on our GWAS findings and the allowance of the budget, we selected four SNPs for replication analyses (i.e. rs4956302, rs17354547, rs4956396 and rs1402812). Please see the Results section for the detailed justification for selecting these SNPs.
DNA was extracted from peripheral blood samples of each participant using a kit from Qiagen Inc. (Valencia, CA). All SNPs were genotyped using the TaqMan SNP Genotyping Assay in a 384-well microplate format (Applied Biosystems, Foster, CA). Briefly, 15 ng of DNA was amplified in a total volume of 7 μl containing an MGB probe and 2.5 μl of TaqMan universal PCR master mix. Allelic discrimination analysis was performed on the ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster CA). To ensure the quality of genotyping, SNP-specific control samples were added to each 384-well plate.
The final call rate for the 4 genotyped SNPs, rs17354547, rs1402812, rs4956396 and rs4956302 was 99.35%, 99.30%, 99.50% and 99.30%, respectively. To verify the quality of our genotyping, we also checked the SNP data for any significant departure from Hardy-Weinberg equilibrium (HWE). The HWE at each locus was assessed by the χ2 test and the significance level for the test was set at 0.0125 (=0.05/4), adjusting for multiple testing of four SNPs. All the genotyped SNPs were in HWE (p > 0.0125). In particular, for the SNP, rs17354547, the p value was 0.258.
FHS replication sample
Using this FHS sample, we performed in silico replication of the nine interesting SNPs identified in our GWAS cohort (see details for these nine SNPs in the Results section). Genotyping of these nine SNPs in the FHS sample was performed with Affymetrix 500K mapping array plus Affymetrix 50K supplementary array. For details of the genotyping method, please refer to Framingham SHARe at NCBI dbGaP website (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000007.v3.p2). Specifically, for the nine SNPs of interest, the call rates are as follows: 98.98% for rs12505771, 99.10% for rs6838494, 98.58% for rs17354547, 99.14% for rs17354568, 98.84% for rs17007301, 98.85% for rs1402812, 98.41% for rs4956396, 98.60% for rs13133830, and 98.33% for rs4956302. The p values for HWE test at these nine SNPs range from 0.41 to 0.88, suggesting HWE and good genotyping quality at these SNPs.
Statistical Analyses
GWAS
GWAS statistical analyses were performed with software package SAS (SAS Institute Inc., Cary, NC). Genotypic association analysis was conducted with logistic regression for association with “smoking status” and with a Poisson regression analysis for association with “SQ”. Genotypes were treated as independent variables and phenotypes (smoking status and SQ) were treated as dependent variables in each method. To adjust for the effects of “age” or “sex” on the association, these two covariates were included in a model together with the genotype information and the model was compared in terms of likelihood with another restricted model where only the effects of the covariates were estimated. The significance for the covariate-adjusted association, which is the difference in likelihood of the two models, was tested using a chi-square test.
Predisposing risk of each significant SNP was evaluated by an odds ratio (OR) and a corresponding 95% confidence interval (CI), which was calculated using the software package Stata (Stata Corporation, College Station, Texas). To calculate an OR for a certain SNP, subjects were first divided into two genotype groups, carriers of a minor allele (i.e., homozygotes of the minor allele and heterozygotes) and non-carriers of the minor allele (i.e., homozygotes of the major allele). The OR was then calculated by comparing the prevalence of smokers (i.e., ratio of smokers vs. non-smokers) in the two genotype groups.
Block structure of the gene under scrutiny was inferred using Haploview (20) (http://www.broad.mit.edu/mpg/haploview/) based on our own genotype data.
To explore potential functions of the interesting SNPs identified through GWAS, we used the FASTSNP (function analysis and selection tool for SNPs) program (http://fastsnp.ibms.sinica.edu.tw) that analyzes SNP functions based on up-to-date information extracted from 11 external bioinformatic databases at query time (21). In addition, we also evaluated inter-species conservation of the identified SNPs and their flanking sequences using the UCSC Human Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway).
We adopted a GWAS significance threshold at p = 4.2 × 10−7, which was derived by Freimer and Sabatti (22) based on a gene-wise approach, and subsequently modified by Lencz et al. (23) taking into account a more accurate estimate of the total number of genes in the human genome.
To detect population stratification that may lead to spurious association results, we used the software Structure 2.2 (http://pritch.bsd.uchicago.edu/software.html) to investigate the potential substructure of our sample. The program uses a Markov chain Monte Carlo (MCMC) algorithm to cluster individuals into different cryptic sub-populations on the basis of multi-locus genotype data (24). To ensure robustness of our results, we performed independent analyses under three assumed numbers for population strata (k = 2, 3, and 4), using 2,000 un-linked markers that were randomly selected across the entire genome. To confirm the results achieved through Structure 2.2, we further tested population stratification of our GWAS sample using a method of genomic control (25).
To determine whether the association findings in our GWAS could potentially be due to bias (e.g., genotyping error), we examined the distribution of p values for all ∼380,000 SNPs analyzed in our sample using the quantile-quantile (Q-Q) plot.
Replication study
For the genotyped SNPs in our AA replication sample, the PedCheck program was used to determine genotyping consistency for Mendelian inheritance (26). Pair-wise linkage disequilibrium (LD) between all SNP markers was assessed using Haploview (27), with the determination of haplotype blocks based on block definitions proposed by Gabriel and colleagues (28). Association between individual SNPs and the ND measures (SQ, HSI and FTND) was determined by the PBAT program using generalized estimating equations (29), with gender and age entered as covariates. The association test was allele-based. Associations between each ND measure and haplotypes from multiple SNP combinations were examined using the FBAT program, with the computation of p values for the Z statistic based on the Monte Carlo sampling option under the null distribution of no linkage and no association (30). The haplotype analysis was single haplotype-based. Since four major haplotypes were tested, the significance for the analysis was set at 0.0125 (=0.05/4) adjusting multiple testing through Bonferroni correction.
The association analyses of the nine interesting SNPs (see details in the Results section) in the FHS sample were performed in the similar way as for the AA sample, using the FBAT program to examine association of the SNPs with smoking status (29).
Results
GWAS findings
We performed genome-wide genotypic association analyses for smoking status (adjusted for age and sex) and identified a cluster of nine SNPs, upstream (i.e., around 93 kb) from the IL15 gene, which ranked among the most significant 30 SNPs tested genome-wide in the total sample (Appendix I). Of particular interest, three of the nine SNPs, rs4956302, rs17354547 and rs1402812, ranked as the most significant SNPs among all SNPs tested genome-wide (Appendix I). We therefore focused our subsequent analyses on these nine SNPs.
In our total sample containing 840 subjects, the nine SNPs achieved p values at levels of 10−5 to 10−6 for association with smoking status, with the most significant SNP, rs4956302, reaching a p value of 1.19 ×10 −6 (Table 2). We further performed gender-specific association analyses (adjusted for age) for these nine SNPs and found that these SNPs were associated with smoking status much more strongly in males than in females, showing an apparent male-specific association (Table 2). In males, association of these SNPs with smoking status achieved significance levels similar to that seen in the total sample despite the fact that the number of male subjects was approximately one half of the total. In particular, rs4956302 achieved a p value of 8.08 × 10−8 in males, which is significant at the genome-wide level (Table 2).
Table 2.
Male-specific association signals for the 9 smoking behavior- associated SNPs detected upstream from the IL-15 gene in the GWAS sample
| Smoking statusd |
Cigarette consumption (SQ)e |
OR [95% CI]f |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| dbSNP ID | Position | Allelea | MAFb | MAFc | Total sample | Male | Female | Total sample | Male | Female | Male |
| rs12505771 | 142585896 | G/A | 0.178 | 0.190 | 9.53 × 10−6 | 9.62 × 10−5 | 0.02 | 3.30 × 10−3 | 5.72 × 10−3 | 0.22 | 0.42 (0.27-0.65) |
| rs6838494 | 142586854 | C/A | 0.178 | 0.195 | 9.53 × 10−6 | 9.62 × 10−5 | 0.02 | 3.30 × 10−3 | 5.72 × 10−3 | 0.22 | 0.42 (0.27-0.65) |
| rs17354547 | 142610319 | A/C | 0.176 | 0.208 | 5.61 × 10−6 | 5.65 × 10−5 | 0.02 | 1.58 × 10−3 | 2.54 × 10−3 | 0.20 | 0.41 (0.26-0.64) |
| rs17354568 | 142610821 | A/C | 0.177 | 0.200 | 1.72 × 10−5 | 1.51 × 10−4 | 0.02 | 2.50 × 10−3 | 4.32 × 10−3 | 0.20 | 0.43(0.28-0.67) |
| rs17007301 | 142629429 | C/T | 0.175 | 0.200 | 1.39 × 10−5 | 9.71 × 10−5 | 0.02 | 2.22 × 10−3 | 3.48 × 10−3 | 0.21 | 0.42 (0.27-0.65) |
| rs1402812 | 142661116 | C/G | 0.180 | 0.208 | 6.67 × 10−6 | 9.55 × 10−5 | 0.01 | 2.17 × 10−3 | 5.36 × 10−3 | 0.15 | 0.42 (0.27-0.66) |
| rs4956396 | 142662240 | G/A | 0.176 | 0.208 | 1.06 × 10−5 | 1.10 × 10−4 | 0.01 | 1.82 × 10−3 | 3.86 × 10−3 | 0.16 | 0.43 (0.28-0.67) |
| rs13133830 | 142683568 | C/T | 0.151 | 0.158 | 1.22 × 10−5 | 1.12 × 10−5 | 0.07 | 4.25 × 10−3 | 2.07 × 10−3 | 0.47 | 0.36 (0.23-0.58) |
| rs4956302 | 142684172 | T/C | 0.165 | 0.175 | 1.19 × 10−6 | 8.80 × 10−8 | 0.17 | 2.82 × 10−3 | 1.97 × 10−4 | 0.96 | 0.34 (0.21-0.53) |
Note:
The second allele represents the minor allele of each locus.
Minor allele frequency calculated in our own Caucasian sample.
Minor allele frequency reported for Caucasians in the public database of HapMap CEU.
P values for association with smoking status
P values for association with cigarette consumption (SQ)
Odds ratios (OR) with 95% confidence intervals (CI) for smoking status. The OR is for carriers of the minor alleles.
Detailed information for these nine SNPs is shown in Table 2. All SNPs have minor allele frequencies (MAF) of ∼ 0.20, and carriers of minor alleles in the male sample had odds ratios (OR) <0.50, suggesting a protective role against smoking behavior for the minor alleles of these SNPs in males. Genotype frequency distribution at the nine SNPs in smoking vs. non-smoking groups in males, females and the total sample is presented in Appendix II.
To further confirm the significance of these nine SNPs to smoking behavior, we also tested their association with another important phenotype for smoking, cigarette consumption (SQ), in the total sample as well as in males and females. The male-specific association pattern was also detected between these SNPs and SQ (Table 2); association with SQ was non-significant in females but achieved p values at the levels of 10−3-10−4 in both the total and male samples (Table 2).
Haplotype analyses using the Haploview program and our own genotype data indicated that strong LD exists among these nine SNPs, which form a single haplotype block. The haplotype formed by these nine SNPs was also associated with smoking status and cigarette consumption in males, achieving p values of 8.96×10−5 and 6.20×10−3, respectively. Figure 1 presents the structure of the haplotype block formed by these nine SNPs as well as association signals achieved in males for “smoking status” and “cigarette consumption” at these SNPs. (Figure 1 about here)
Figure 1.
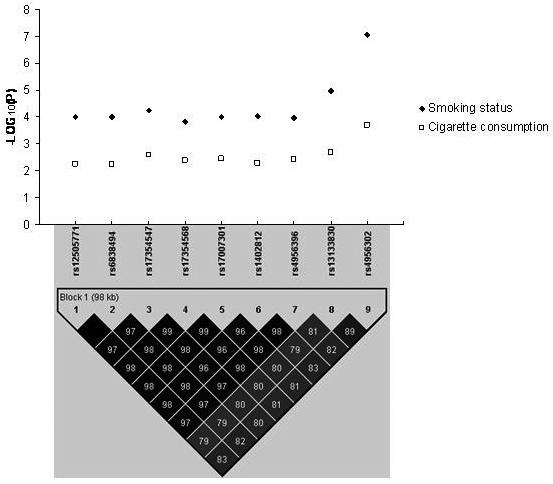
Association signals for the 9 smoking behavior-associated SNPs detected upstream from the IL15 gene in male subjects in the GWAS
Note: The haplotype block map for the 9 SNPs was reconstructed using our own genotype data showing pairwise LD in r2.
Using the FASTSNP program, we analyzed the potential functions of these nine SNPs. According to the analyses, four of the SNPs are located at potential transcription factor (TF) binding sites. For the SNP rs4956302, a polymorphic change of T→C may establish a binding site for the TF, Lyf-1. For the SNP, rs17354547, a polymorphic change of A→C may delete binding sites for the TFs, CdxA and YY1. For the SNP, rs17354568, a polymorphic change of A→C may create a binding site for the TF, Ik-2. For the SNP, rs6838494, a polymorphic change of C→A may create a binding site for TFs, CdxA and Nkx-2 but delete the binding site for Oct-1.
Using the UCSC Human Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway), we determined that rs17354547, and the potential TF binding sequence containing this SNP (as suggested by the FASTSNP program), is highly conserved among 28 vertebrate species (Figure 2). (Figure 2 about here)
Figure 2.
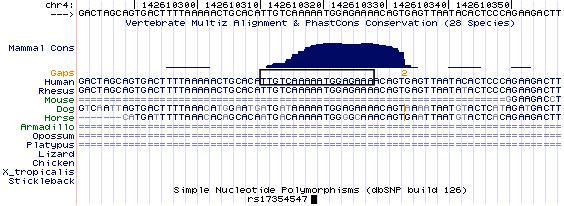
Conservation of the SNP, rs17354547, and a potential transcription factor binding site containing this SNP, across multiple species
Note: The sequence inside the box is a potential transcription factor binding site that may be modulated by the SNP rs17354547, according to FASTSNP analyses (21). The SNP and its flanking sequence are highly conserved across multiple species.
Replication study findings in the AA sample
For this replication study, the SNP rs4956302 was selected due to the fact that it achieved the highest significance for association with smoking status among all the SNPs tested genome-wide. The SNP rs17354547 was selected because it is highly conserved across multiple species. The other two SNPs (rs4956396 and rs1402812) were randomly selected from the remaining seven SNPs since these seven SNPs are in high LD and achieved similar p values (Figure 1).
In the total sample of our AA replication cohort, these four SNPs did not achieve significant p values for association with SQ, HSI and FTND. However, in the male sub-group of our replication sample, the SNP rs17354547 was significantly associated with SQ, HSI and FTND, achieving p values of 0.031, 0.0046 and 0.019, respectively. In addition, a haplotype formed by the SNPs rs17354547, rs1402812 and rs4956396 was also associated with SQ, HSI and FTND, achieving p values of 0.039, 0.0093 and 0.0093, respectively. Since a total of four major haplotypes were tested, to adjust for multiple testing, the significance level for the haplotype association analysis was reset at 0.0125 through Bonferroni correction (0.05/4). Therefore, based on the adjusted significance level, the haplotype association was significant for only two ND phenotypes, which are HSI and FTND. Additional details for the above results are presented in Table 3.
Table 3.
Association signals detected in the AA replication sample
| SNP Name | Allelea | MAFb | MAFc | Total sample |
Male subgroup |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SQ | NSQ | HSI | NHSI | FTND | NFTND | SQ | NSQ | HSI | NHSI | FTND | NFTND | ||||
| rs17354547 | A/C | 0.160 | 0.150 | 0.17 | 149 | 0.17 | 148 | 0.25 | 155 | 0.031 | 94 | 0.0046 | 93 | 0.019 | 97 |
| rs1402812 | C/G | 0.370 | 0.458 | 0.33 | 229 | 0.56 | 228 | 0.59 | 234 | 0.52 | 94 | 0.67 | 95 | 0.39 | 96 |
| rs4956396 | G/A | 0.217 | 0.208 | 0.26 | 184 | 0.23 | 185 | 0.33 | 192 | 0.10 | 111 | 0.24 | 113 | 0.36 | 115 |
| rs4956302 | T/C | 0.356 | 0.449 | 0.69 | 225 | 0.95 | 226 | 0.79 | 231 | 0.26 | 96 | 0.46 | 99 | 0.14 | 100 |
Note:
The second allele represents the minor allele of each locus.
Minor allele frequency calculated in our own AA sample.
Minor allele frequency reported for Sub-Saharan African in the public database of HapMap YRI.
Presented under SQ, HSI and FTND are p values achieved by a certain SNP for each corresponding phenotype.
Presented under N are the numbers of informative families for a certain trait.
Replication study findings in the FHS sample
We performed in silico replication of the nine interesting SNPs identified in our GWAS using the FHS sample. In the total sample and in the female subgroup of the sample, no significant results were achieved. However, in the male subgroup of the sample, seven of the SNPs achieved significant (p<0.05) and two achieved marginally significant p values (p<0.10) for association with smoking status. Additional details of the results are presented in Table 4.
Table 4.
Association signals detected in the FHS replication sample
| SNP | Allelea | MAFb | MAFc | Total sampled | Maled | Femaled |
|---|---|---|---|---|---|---|
| rs12505771 | G/A | 0.202 | 0.190 | 0.53 | 0.028 | 0.82 |
| rs6838494 | C/A | 0.202 | 0.195 | 0.60 | 0.030 | 0.80 |
| rs17354547 | A/C | 0.200 | 0.208 | 0.49 | 0.044 | 0.74 |
| rs17354568 | A/C | 0.199 | 0.200 | 0.48 | 0.044 | 0.76 |
| rs17007301 | C/T | 0.198 | 0.200 | 0.70 | 0.048 | 0.47 |
| rs1402812 | C/G | 0.203 | 0.208 | 0.56 | 0.035 | 0.71 |
| rs4956396 | G/A | 0.200 | 0.208 | 0.77 | 0.083 | 0.44 |
| rs13133830 | C/T | 0.183 | 0.158 | 0.29 | 0.083 | 0.80 |
| rs4956302 | T/C | 0.200 | 0.175 | 0.27 | 0.026 | 0.95 |
Note:
The second allele represents the minor allele of each locus.
Minor allele frequency calculated in the FHS sample.
Minor allele frequency reported for Caucasians in the public database of HapMap CEU.
P values for association with smoking status
Analyses for potential population stratification
To detect potential stratification of our GWAS sample, we analyzed our sample using software Structure 2.2 (24). When 2,000 randomly selected un-linked markers were used to cluster our subjects, all subjects of the sample were tightly clustered together under all assigned values (i.e., 2, 3, or 4) for the assumed number of population strata, k; these results suggest no population stratification. The results are shown in Appendix III.
We further tested our GWAS sample for population stratification using the genomic control method (25). Based on genome-wide SNP information, we estimated the inflation factor (λ), a measure for population stratification. Ideally, for a homogeneous population with no stratification, the value of λ should be equal to or near 1.0. For our total sample, the estimated λ value was 1.009 for smoking status and 1.012 for cigarette consumption (SQ), suggesting essentially no population stratification; these findings further confirm the results achieved through the Structure 2.2 software.
Other analyses
Using the Q-Q plot, we examined the distribution of p values achieved for smoking status in our GWAS for all ∼380,000 SNPs that were analyzed (Appendix IV). As shown in the plot, the observed p values match well with the expected p values over a wide range of values of [−LOG10(p)], which is from 0 to ∼ 4. Observed p values gradually depart from expected p values at the extreme tail, where [−LOG10(p)] is ≥ ∼4. This pattern suggests that our GWAS association findings are more likely to be attributable to true genetic variation than to potential bias, such as genotyping errors.
Discussion
This study reports one of the first few GWAS of smoking behavior in Caucasians. Through this study, we identified a cluster of nine SNPs upstream from the IL15 gene, which ranked among the most significant 30 SNPs associated with smoking status in our GWAS (Appendix I). Gender-specific analyses indicated that these nine SNPs were associated with smoking status in a male-specific manner (Table 2). In particular, the SNP rs4956302 achieved a genome-wide significant p value of 8.80×10−8 in male subjects. Another SNP, rs17354547, is highly conserved across multiple species (Figure 2), suggesting its functional importance. Of note, these nine SNPs were also associated with another important phenotype for smoking behavior, cigarette consumption (SQ), and this association was also male specific (Table 2).
From these nine SNPs, we choose four SNPs (rs4956302, rs17354547, rs1402812 and rs4956396) to replicate our association findings in an AA family-based cohort containing 1,251 subjects, including 412 males and 839 females. We selected rs4956302 due to the fact that it achieved the highest significance among all the SNPs tested genome-wide in our GWAS cohort. We also selected rs17354547 because it is highly conserved across multiple species. Another two SNPs (rs1402812 and rs4956396) were randomly chosen since all seven of the remaining significant SNPs are in high LD and had similar p values. Interestingly, a male-specific association with multiple ND phenotypes was observed for the SNP rs17354547; in male subjects of our AA replication cohort SNP rs17354547 achieved p values of 0.031, 0.0046 and 0.019 for association with SQ, HSI and FTND, respectively (Table 3). Furthermore, in male subjects from our replication sample, a haplotype formed by the SNPs rs17354547, rs1402812 and rs4956396 was also associated with SQ, HSI and FTND, with p values of 0.039, 0.0093 and 0.0093, respectively. The replication results support our GWAS findings for an association between smoking behavior and these SNPs located upstream from the IL15 gene.
To further confirm our GWAS findings, we performed an in silico replication study of the nine interesting SNPs upstream of the IL15 gene using a large FHS sample containing 7,623 Caucasians from 1,731 families. Again, a clear pattern of male-specific association of these SNPs with smoking status was observed; although none of the SNPs achieved p values less than 0.44 in the female subgroup, seven of the SNPs achieved p values less than 0.05 and two achieved p values less than 0.10 in the male subgroup (Table 4). The results provide additional support to our GWAS findings and replication findings achieved in the AA cohort.
It was not until very recently that intergenic transcription has been recognized as an active and common cellular process. Evidence has shown that a significant portion of the transcriptome arises from outside annotated genes (31,32). As an important function, intergenic transcription can regulate expression at nearby genes (33,34). In particular, intergenic transcription was found to be an important mechanism underlying expression of cytokine genes, such as GM-CSF, IL3, IL4, IL5, IL 10, and IL13 (35-38). Given their location at potential TF binding sites, those SNPs identified in our GWAS that are upstream from the cytokine gene IL15, might potentially regulate IL15 gene expression through intergenic transcription. Importantly, the SNP, rs17354547, replicated in both the AA and FHS cohorts, as well as the TF binding site that can be potentially modulated by this SNP, are highly conserved across multiple species (Figure 2), further supporting the functional importance of this SNP in transcription regulation. Overall, our findings suggest that the observed association of the SNPs upstream of the IL15 gene with smoking status and multiple ND phenotypes may be mediated through regulation of IL15 gene expression, and that this appears to represent a novel mechanism underlying smoking behavior.
Multiple lines of evidence demonstrate that the immune system, in particular, lymphoid cells, play an important role in drug addiction. Destruction of the immune system with irradiation or immunosuppressive drugs has been shown to significantly alleviate the opiate-withdrawal syndrome (39,40). In contrast, transfer of lymphoid cells to irradiated rats before morphine administration restores drug-withdrawal signs (41). These findings suggest a mechanism for neuro-immunological interactions, where factors derived from the immune system may regulate functions of the central nervous system, influencing addictive behaviors. This mechanism is supported by the discovery of functional synapses between neurons and lymphocytes (42). Given that IL15 is an important immunoregulatory cytokine influencing activation and proliferation of T lymphocytes and natural killer cells, it appears reasonable to speculate that IL15 influences smoking addiction through its immunoregulatory effects.
Population stratification and/or ethnic admixture can be an important source of spurious association in genetic association studies. However, these factors did not affect our GWAS sample and are therefore unlikely to have interfered with our association results. Our study cohort came from an apparently homogenous US mid-west white population, living in Omaha, Nebraska and its surrounding areas. We found that the allele frequencies for the interesting SNPs in our sample are very similar to those reported in the typical and representative Caucasian samples used in the HapMap CEU (Table 2). Furthermore, using the program Structure 2.2 (24), we analyzed our study subjects thoroughly in order to detect potential sub-populations in our sample. In these analyses, all subjects tightly clustered together as a single group, suggesting no significant population substructure in our sample (Appendix III). Furthermore, the measure for population stratification (λ) for our GWAS sample, calculated through the genomic control method (25), was 1.009 for smoking status and 1.012 for cigarette consumption, suggesting essentially no stratification. For the above reasons, the association results, as detected in our GWAS, are not likely to be plagued by spurious associations due to population admixture/stratification.
In our GWAS discovery cohort, control subjects were defined as never-smokers. This criterion for selecting controls is different from the conventional one, where current non-smoking subjects with a certain degree of previous exposure (e.g., having smoked more than 1 but less than 100 cigarettes in their lifetime) are normally selected as controls. Therefore, a potential problem of our study design is that some “control” subjects in our GWAS sample may in future become smokers if exposed to cigarettes. Depending on the number of such subjects, this potential misclassification problem may undercut the statistical power of our study, leading to false negative results. In our study, we tried to minimize the effects caused by this potential problem by excluding those control (non-smoking) subjects under the age of 25 from our study. Since most smokers initiate smoking behavior in adolescence, non-smoking subjects under the age of 25 may have a much higher chance than older people to develop smoking behavior if exposed to cigarettes. Therefore, after excluding these younger subjects from our control group, the subjects in the group that may later develop smoking behavior due to exposure to cigarettes, if existing, may not be in large numbers. Hence, the potential misclassification problem caused by our control subject selection strategy may have only moderate effects to the overall results of our study. The robustness of our GWAS findings is supported by their replication in both the AA and FHS cohorts.
As another limitation of our study, we did not adjust for multiple testing (for testing multiple smoking behavior-related phenotypes in our GWAS and the AA replication cohorts). However, due to the limited number of different phenotypes (i.e., 2 phenotypes in the GWAS cohort and 3 phenotypes in the AA cohort) and the fact that these phenotypes are correlated smoking behavior traits, adjusting for multiple testing may only have minor effects on the current results. Even with the most stringent correction, Bonferroni correction that does not consider correlation of the multiple traits, the most significant SNP in our GWAS, rs4956302, is still significant at the corrected genome-wide significance level of 2.1×10−7 (= 4.2×10−7/2) for association with smoking status, and the most significant SNP in our AA replication study, rs17354547, is also significant at the corrected significance level of 0.017 (=0.05/3) for association with HSI. Again, replication of our GWAS findings in two different cohorts attests to the findings' robustness and may have attenuated the potential problem due to multiple testing of several phenotypes.
In summary, we identified a group of SNPs, upstream from the IL15 gene, that were associated with both smoking status and quantity of cigarette consumption. Interestingly, a key SNP, rs17354547, which is highly conserved across multiple species, was replicated in an independent AA cohort for association with multiple ND phenotypes. Moreover, all of the nine SNPs were replicated in silico in a FHS cohort for association with smoking status. Remarkably, the association of the SNPs with smoking behavior-related phenotypes in both our GWAS and the two replication samples appeared to be male-specific. Higher prevalence of smoking in males than in females in the US (3) attaches additional importance to our findings. Some of the SNPs, located at potential TF binding sites, may regulate IL15 gene expression and consequently, could have an important regulatory effect on the immune system. The above findings, together with previous data from studies of drug addiction, compel us to propose a novel mechanism for smoking addiction modulated by the immune system, where the IL15 pathway may play a key role. The confirmation and elaboration of this hypothetical mechanism needs further detailed functional studies directed at IL15.
Acknowledgement
Investigators of this work were partially supported by grants from NIH (R01 AR050496-01, R21 AG027110, R01 AG026564, R21 AA015973, P50 AR055081, and R01 DA12844). The study also benefited from grants from National Science Foundation of China, Huo Ying Dong Education Foundation, HuNan Province, Xi'an Jiaotong University, and the Ministry of Education of China.
The Framingham Heart Study and the Framingham SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University. The Framingham SHARe data used for the analyses described in this manuscript were obtained through dbGaP (accession number phs000007.v3.p2). This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or the NHLBI.
Appendix I. The most significant 30 SNPs for association with smoking status identified in the current GWAS
| SNP | P Value | Chromosome | Position | Associated Gene |
|---|---|---|---|---|
| rs4956302 | 1.19×10−6 | 4q31.21 | 142684172 | IL15 |
| rs17354547 | 5.61×10−6 | 4q31.21 | 142610319 | IL15 |
| rs1402812 | 6.67×10−6 | 4q31.21 | 142661116 | IL15 |
| rs2036627 | 7.24×10−6 | 1q21.3 | 152936201 | KCNN3 |
| rs6009041 | 7.71×10−6 | 22q13.31 | 45674541 | TBC1D22A |
| rs2973062 | 7.74×10−6 | 5p13.2 | 37824246 | GDNF |
| rs762145 | 8.70×10−6 | 21q22.13 | 38068188 | DSCR4 |
| rs1995662 | 8.77×10−6 | 1q21.3 | 152946191 | ADAR |
| rs738932 | 9.31×10−6 | 22q13.31 | 45671152 | TBC1D22A |
| rs12505771 | 9.53×10−6 | 4q31.21 | 142585896 | IL15 |
| rs6838494 | 9.53×10−6 | 4q31.21 | 142586854 | IL15 |
| rs484594 | 1.04×10−5 | 6q27 | 165868541 | PDE10A |
| rs4956396 | 1.06×10−5 | 4q31.21 | 142662240 | IL15 |
| rs2715260 | 1.07×10−5 | 3q13.33 | 123276180 | CD86 |
| rs12147616 | 1.07×10−5 | 14q24.1 | 67241093 | RDH11 |
| rs13133830 | 1.22×10−5 | 4q31.21 | 142683568 | IL15 |
| rs9790142 | 1.24×10−5 | 3q22.1 | 133556726 | ACPP |
| rs6580194 | 1.28×10−5 | 5q31.3 | 141014307 | CENTD3 |
| rs10788392 | 1.35×10−5 | 10q23.1 | 86742325 | KIAA1128 |
| rs17007301 | 1.39×10−5 | 4q31.21 | 142629429 | IL15 |
| rs335336 | 1.42×10−5 | 4q13.1 | 62015943 | LPHN3 |
| rs10103840 | 1.46×10−5 | 8p21.1 | 29475632 | unknown |
| rs17354568 | 1.72×10−5 | 4q31.21 | 142610821 | IL15 |
| rs12882315 | 2.00×10−5 | 14q24.1 | 67241481 | RDH11 |
| rs16970398 | 2.03×10−5 | 17q12 | 30155847 | CCT6B |
| rs6595593 | 2.06×10−5 | 5q23.2 | 124572229 | ZNF608 |
| rs2167289 | 2.08×10−5 | 7q32.1 | 127645127 | LEP |
| rs10189390 | 2.35×10−5 | 2q33.2 | 204716926 | PARD3B |
| rs762646 | 2.39×10−5 | 22q13.31 | 45672831 | TBC1D22A |
| rs4887420 | 2.59×10−5 | 15q25.3 | 84585588 | FLJ32310 |
Appendix II. Genotype distribution at the nine SNPs upstream of the IL15 gene
| SNP | Smoking status | Genotypea | N (total)b | N (male)b | N (female)b |
|---|---|---|---|---|---|
| rs12505771 | Nonsmoker | AA | 10 | 7 | 3 |
| AB | 160 | 72 | 88 | ||
| BB | 292 | 114 | 178 | ||
| Smoker | AA | 11 | 5 | 6 | |
| AB | 79 | 45 | 34 | ||
| BB | 288 | 174 | 114 | ||
| rs6838494 | Nonsmoker | AA | 10 | 7 | 3 |
| AB | 160 | 72 | 88 | ||
| BB | 292 | 114 | 178 | ||
| Smoker | AA | 11 | 5 | 6 | |
| AB | 79 | 45 | 34 | ||
| BB | 288 | 174 | 114 | ||
| rs17354547 | Nonsmoker | AA | 9 | 7 | 2 |
| AB | 160 | 72 | 88 | ||
| BB | 292 | 114 | 178 | ||
| Smoker | AA | 11 | 5 | 6 | |
| AB | 78 | 44 | 34 | ||
| BB | 289 | 175 | 114 | ||
| rs17354568 | Nonsmoker | AA | 10 | 7 | 3 |
| AB | 158 | 71 | 87 | ||
| BB | 294 | 115 | 179 | ||
| Smoker | AA | 11 | 5 | 6 | |
| AB | 79 | 45 | 34 | ||
| BB | 288 | 174 | 114 | ||
| rs17007301 | Nonsmoker | AA | 10 | 7 | 3 |
| AB | 157 | 71 | 86 | ||
| BB | 294 | 115 | 179 | ||
| Smoker | AA | 11 | 5 | 6 | |
| AB | 78 | 44 | 34 | ||
| BB | 289 | 175 | 114 | ||
| rs1402812 | Nonsmoker | AA | 10 | 7 | 3 |
| AB | 162 | 73 | 89 | ||
| BB | 287 | 112 | 175 | ||
| Smoker | AA | 11 | 5 | 6 | |
| AB | 80 | 46 | 34 | ||
| BB | 286 | 172 | 114 | ||
| rs4956396 | Nonsmoker | AA | 9 | 7 | 2 |
| AB | 160 | 71 | 89 | ||
| BB | 293 | 115 | 178 | ||
| Smoker | AA | 11 | 5 | 6 | |
| AB | 79 | 45 | 34 | ||
| BB | 287 | 173 | 114 | ||
| rs13133830 | Nonsmoker | AA | 9 | 7 | 2 |
| AB | 143 | 67 | 76 | ||
| BB | 310 | 119 | 191 | ||
| Smoker | AA | 10 | 5 | 5 | |
| AB | 68 | 36 | 32 | ||
| BB | 300 | 183 | 117 | ||
| rs4956302 | Nonsmoker | AA | 9 | 7 | 2 |
| AB | 158 | 73 | 85 | ||
| BB | 294 | 113 | 181 | ||
| Smoker | AA | 10 | 5 | 5 | |
| AB | 73 | 38 | 35 | ||
| BB | 294 | 181 | 113 | ||
Note:
“A” represents the minor allele and “AA” the homozygote of that allele. “B” represents the major allele and “BB” the homozygote of that allele. “AB” represents the heterozygote.
“N (total)” represents the number of a certain genotype in the total sample, “N (male)” the number in the male subgroup, and “N (female)” the number in the female subgroup.
Appendix III. Results of analyses of potential sample stratification
Appendix IV. Q-Q plots for the p values achieved in the GWAS
References
- 1.CDC . Best Practices for Comprehensive Tobacco Control Programs—2007. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health; 2007. http://www.cdc.gov/tobacco/tobacco_control_programs/stateandcommunity/best_practices/index.htm. [Google Scholar]
- 2.CDC . The Health Consequences of Smoking: A Report of the Surgeon General. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health; 2004. http://www.cdc.gov/tobacco/data_statistics/sgr/sgr_2004/index.htm [Google Scholar]
- 3.CDC Cigarette Smoking Among Adults—United States, 2006. Morbidity and Mortality Weekly Report. 2007;56:1157–1161. [PubMed] [Google Scholar]
- 4.Li MD, Cheng R, Ma JZ, Swan GE. A meta-analysis of estimated genetic and environmental effects on smoking behavior in male and female adult twins. Addiction. 2003;98:23–31. doi: 10.1046/j.1360-0443.2003.00295.x. [DOI] [PubMed] [Google Scholar]
- 5.Li MD. Identifying susceptibility loci for nicotine dependence: 2008 update based on recent genome-wide linkage analyses. Hum Genet. 2008;123:119–131. doi: 10.1007/s00439-008-0473-0. [DOI] [PubMed] [Google Scholar]
- 6.Beuten J, Ma JZ, Payne TJ, Dupont RT, Crews KM, Somes G, et al. Single- and multilocus allelic variants within the GABA(B) receptor subunit 2 (GABAB2) gene are significantly associated with nicotine dependence. Am J Hum Genet. 2005;76:859–864. doi: 10.1086/429839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma JZ, Beuten J, Payne TJ, Dupont RT, Elston RC, Li MD. Haplotype analysis indicates an association between the DOPA decarboxylase (DDC) gene and nicotine dependence. Hum Mol Genet. 2005;14:1691–1698. doi: 10.1093/hmg/ddi177. [DOI] [PubMed] [Google Scholar]
- 8.Li MD, Beuten J, Ma JZ, Payne TJ, Lou XY, Garcia V, et al. Ethnic- and gender-specific association of the nicotinic acetylcholine receptor alpha4 subunit gene (CHRNA4) with nicotine dependence. Hum Mol Genet. 2005;14:1211–1219. doi: 10.1093/hmg/ddi132. [DOI] [PubMed] [Google Scholar]
- 9.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007 doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007 doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng HW, Deng H, Liu YJ, Liu YZ, Xu FH, Shen H, et al. A genomewide linkage scan for quantitative-trait loci for obesity phenotypes. Am J Hum Genet. 2002;70:1138–1151. doi: 10.1086/339934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li MD, Payne TJ, Ma JZ, Lou XY, Zhang D, Dupont RT, et al. A genomewide search finds major susceptibility loci for nicotine dependence on chromosome 10 in African Americans. Am J Hum Genet. 2006;79:745–751. doi: 10.1086/508208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br J Addict. 1991;86:1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- 18.Di X, Matsuzaki H, Webster TA, Hubbell E, Liu G, Dong S, et al. Dynamic model based algorithms for screening and genotyping over 100 K SNPs on oligonucleotide microarrays. Bioinformatics. 2005;21:1958–1963. doi: 10.1093/bioinformatics/bti275. [DOI] [PubMed] [Google Scholar]
- 19.Rabbee N, Speed TP. A genotype calling algorithm for affymetrix SNP arrays. Bioinformatics. 2006;22:7–12. doi: 10.1093/bioinformatics/bti741. [DOI] [PubMed] [Google Scholar]
- 20.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 21.Yuan HY, Chiou JJ, Tseng WH, Liu CH, Liu CK, Lin YJ, et al. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34:W635–W641. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freimer N, Sabatti C. The use of pedigree, sib-pair and association studies of common diseases for genetic mapping and epidemiology. Nat Genet. 2004;36:1045–1051. doi: 10.1038/ng1433. [DOI] [PubMed] [Google Scholar]
- 23.Lencz T, Morgan TV, Athanasiou M, Dain B, Reed CR, Kane JM, et al. Converging evidence for a pseudoautosomal cytokine receptor gene locus in schizophrenia. Mol Psychiatry. 2007;12:572–580. doi: 10.1038/sj.mp.4001983. [DOI] [PubMed] [Google Scholar]
- 24.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 26.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 28.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 29.Lange C, Silverman EK, Xu X, Weiss ST, Laird NM. A multivariate family-based association test using generalized estimating equations: FBAT-GEE. Biostatistics. 2003;4:195–206. doi: 10.1093/biostatistics/4.2.195. [DOI] [PubMed] [Google Scholar]
- 30.Horvath S, Xu X, Lake SL, Silverman EK, Weiss ST, Laird NM. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol. 2004;26:61–69. doi: 10.1002/gepi.10295. [DOI] [PubMed] [Google Scholar]
- 31.Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 32.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 33.Hirschman JE, Durbin KJ, Winston F. Genetic evidence for promoter competition in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4608–4615. doi: 10.1128/mcb.8.11.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martens JA, Wu PY, Winston F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 2005;19:2695–2704. doi: 10.1101/gad.1367605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones EA, Flavell RA. Distal enhancer elements transcribe intergenic RNA in the IL-10 family gene cluster. J Immunol. 2005;175:7437–7446. doi: 10.4049/jimmunol.175.11.7437. [DOI] [PubMed] [Google Scholar]
- 36.Urwin DL, Schwenger GT, Groth DM, Sanderson CJ. Distal regulatory elements play an important role in regulation of the human IL-5 gene. Eur J Immunol. 2004;34:3633–3643. doi: 10.1002/eji.200425279. [DOI] [PubMed] [Google Scholar]
- 37.Rogan DF, Cousins DJ, Santangelo S, Ioannou PA, Antoniou M, Lee TH, et al. Analysis of intergenic transcription in the human IL-4/IL-13 gene cluster. Proc Natl Acad Sci U S A. 2004;101:2446–2451. doi: 10.1073/pnas.0308327100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cockerill PN, Shannon MF, Bert AG, Ryan GR, Vadas MA. The granulocyte-macrophage colony-stimulating factor/interleukin 3 locus is regulated by an inducible cyclosporin A-sensitive enhancer. Proc Natl Acad Sci U S A. 1993;90:2466–2470. doi: 10.1073/pnas.90.6.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dafny N, Pellis NR. Evidence that opiate addiction is in part an immune response. Destruction of the immune system by irradiation-altered opiate withdrawal. Neuropharmacology. 1986;25:815–818. doi: 10.1016/0028-3908(86)90003-1. [DOI] [PubMed] [Google Scholar]
- 40.Dafny N, Wagle VG, Drath DB. Cyclosporine alters opiate withdrawal in rodents. Life Sci. 1985;36:1721–1726. doi: 10.1016/0024-3205(85)90554-5. [DOI] [PubMed] [Google Scholar]
- 41.Dafny N, Dougherty PM, Pellis NR. The immune system and opiate withdrawal. Int J Immunopharmacol. 1989;11:371–375. doi: 10.1016/0192-0561(89)90083-0. [DOI] [PubMed] [Google Scholar]
- 42.Felten DL, Felten SY, Bellinger DL, Carlson SL, Ackerman KD, Madden KS, et al. Noradrenergic sympathetic neural interactions with the immune system: structure and function. Immunol Rev. 1987;100:225–260. doi: 10.1111/j.1600-065x.1987.tb00534.x. [DOI] [PubMed] [Google Scholar]


