Abstract
Plant microspores can be reprogrammed from their normal pollen development to an embryogenic route in a process termed microspore embryogenesis or androgenesis. Stress treatment has a critical role in this process, inducing the dedifferentiation of microspores and conditioning the following androgenic response. In this study, we have used three barley doubled haploid lines with similar genetic background but different androgenic response. The Barley1 GeneChip was used for transcriptome comparison of these lines after mannitol stress treatment, allowing the identification of 213 differentially expressed genes. Most of these genes belong to the functional categories “cell rescue, defense, and virulence”; “metabolism”; “transcription”; and “transport”. These genes were grouped into clusters according to their expression profiles among lines. A principal component analysis allowed us to associate specific gene expression clusters to phenotypic variables. Genes associated with the ability of microspores to divide and form embryos were mainly involved in changes in the structure and function of membranes, efficient use of available energy sources, and cell fate. Genes related to stress response, transcription and translation regulation, and degradation of pollen-specific proteins were associated with green plant production, while expression of genes related to plastid development was associated with albino plant regeneration.
Electronic supplementary material
The online version of this article (doi:10.1007/s10142-009-0113-3) contains supplementary material, which is available to authorized users.
Keywords: Barley, Dedifferentiation, Microspore embryogenesis, Stress treatment
Introduction
Plant microspores can switch from their normal pollen developmental pathway towards an embryogenic route in a process termed microspore embryogenesis or androgenesis. Androgenesis has proven to be an essential tool in plant breeding and genetics due to the ability to generate homozygous lines (doubled haploids, DH) in a single generation (Forster and Thomas 2005).
Barley (Hordeum vulgare L.) is used as a model system for microspore embryogenesis studies in cereals and the use of this method has resulted in many new barley cultivars (COST Action 851 2005). However, microspore embryogenesis is very genotype-dependent and there are several agronomically important genotypes that are recalcitrant, primarily due to low embryogenesis rate and/or a high albino plant regeneration (Li and Devaux 2001; Muñoz-Amatriaín et al. 2008).
The application of a stress treatment is necessary for the reprogramming of microspores (Touraev et al. 1997). Stress treatment represses the normal gametophytic pathway of microspores to fertile pollen, which leads to an intermediate stage of dedifferentiation and cell totipotency. This transitional stage allows microspores, under appropriate culture conditions, to divide, develop into embryos, and regenerate complete plants. A variety of stresses are known to trigger androgenesis, but the type of stress applied depends on the plant species or even the genotype (Shariatpanahi et al. 2006). In barley, the highest regeneration efficiency is obtained with uninucleated microspores subjected to starvation and osmotic stress, triggered by incubating anthers in a medium containing mannitol (Hoekstra et al. 1992; Cistué et al. 1994). Stress treatment is not only needed for switching the developmental fate, but it also conditions the numbers of divisions and embryos, green and albino plant regeneration, and spontaneous doubling (Cistué et al. 1994, 1999; Hoesktra et al. 1997; Kasha et al. 2001; Li and Devaux 2003; Wojnarowiez et al. 2004; Oleszczuk et al. 2006; Shariatpanahi et al. 2006).
Many studies have described the morphological changes that take place in microspores upon stress, such as a cellular enlargement, vacuole regression, and nuclear migration (for review, see Touraev et al. 2001 and Maraschin et al. 2005). However, the molecular mechanisms underlying microspore dedifferentiation are largely unknown. Several labs have recently used functional genomics tools to study transcriptional changes during the microspore embryogenesis process (Kyo et al. 2003; Maraschin et al. 2006; Muñoz-Amatriaín et al. 2006; Hosp et al. 2007; Joosen et al. 2007; Malik et al. 2007; Tsuwamoto et al. 2007). Studies focused on the stress treatment stage of tobacco microspore embryogenesis identified genes associated with metabolism, chromosome remodelling, transcription and translation, and signalling (Kyo et al. 2003; Hosp et al. 2007). In barley, two studies characterized the stress-induced gene expression. In a previous study, we used the 22 k Barley1 GeneChip to analyze the transcriptome of anthers before and after 4 days of mannitol treatment (Muñoz-Amatriaín et al. 2006). This study revealed large changes in the expression of genes related to central metabolism, stress response, and suppression of the gametophytic developmental pathway. Maraschin et al. (2006), using optimal and sub-optimal stress treatments of androgenesis induction, revealed that metabolic changes and proteolysis could have a critical role in the dedifferentiation phase of microspore embryogenesis.
In this report, we have selected three barley doubled haploid lines with a very different response to microspore embryogenesis, but a similar genetic background [chromosomes 1H, 2H, 4H and 7H, and 80% of 3H, 5H and 6H were common among them (Muñoz-Amatriaín et al. 2008)]. Moreover, QTLs for different components of the androgenic response were found in the polymorphic regions (Muñoz-Amatriaín et al. 2008). Transcriptome comparison of these lines after 4 days of mannitol stress treatment has allowed the identification of those genes defining, at the time of microspore dedifferentiation, their specific response to microspore embryogenesis.
Materials and methods
Plant material and anther culture
Barley (Hordeum vulgare L.) doubled haploid lines DH46, DH6188, and DH6004 were selected for this study. DH46 was obtained via anther culture from a cross between the winter two-rowed Igri and the facultative six-rowed Dobla (Chen et al. 2007). A population of 100 DH lines was developed from the cross between DH46 and Igri in order to identify QTLs for green plant percentage in barley anther culture (Muñoz-Amatriaín et al. 2008). Lines DH6188 and DH6004 belonged to this population and were selected for this study based on their similar genetic background, the presence of QTLs for different components of the androgenic response on the polymorphic regions, and their extreme maximum and minimum values for variables associated with the microspore embryogenesis response.
Donor plants were grown in growth chambers as described by Cistué et al. (2003). The three DH lines were evaluated for anther culture response, also following the protocol described by Cistué et al. (2003). Ten replications of 15 anthers were used for characterization. Each replication consisted of one spike harvested from the second and third tiller of the plants. Variables recorded were the numbers of dividing microspores (nDM), embryos (nEMB), green plants (nGP), and albino plants (nAP), all referred to 100 cultured anthers, as well as the percentage of green plants (number of green plants per total plants; pGP). Analysis of variance was performed with the generalized linear model (GLM) procedure from the SAS/STAT statistics software package (SAS Institute, Cary, NC, USA). Variable values for each line were compared by the Duncan’s test (P < 0.05) using data from each replication.
Microarray analysis was carried out with anthers harvested after 4 days of mannitol treatment. Three biological replicates of each line were generated to assess the reproducibility of microarray analysis.
Cytological studies
Protocols for cytological studies, including DAPI staining of isolated microspores and semithin sections of anthers, are described in detail in Supplementary Materials and Methods S1.
RNA isolation and array hybridization
Total RNA was isolated using TRIzol Reagent (Gibco BRL) and passed through RNeasy columns (Qiagen) for further clean up, following the manufacturer’s instructions in both cases. All starting total RNA samples were quality-assessed prior to beginning target preparation/processing steps by loading 25–250 ng/well onto a RNA Lab-On-A-Chip (Caliper Technologies Corp., Mountain View, CA, USA) and evaluated on an Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA, USA). Double-stranded cDNA was synthesized from the poly(A) + RNA present in the isolated total RNA (8.5 g total RNA starting material each sample reaction) using the SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen, Carlsbad, CA, USA) and poly (T)-nucleotide primers that contained a sequence recognized by T7 RNA polymerase. Biotin-tagged cRNA, generated from the cDNA, was fragmented and hybridized to Affymetrix Barley1 GeneChips, followed by washing, staining (SAPE, streptavidin-phycoerythrin), and scanning on a GeneChip® Scanner 3000, according to standard protocols (Affymetrix GeneChip® Expression Analysis Technical Manual available at http://www.affymetrix.com).
Microarray data analysis
Scanned images were analyzed with GCOS 1.2 (Affymetrix, Santa Clara, CA, USA). Expression estimates was calculated using gcRMA implemented in GeneSpring 7.1 (Silicon Genetics, Redwood City, CA, USA). We used the flag “present” as an indicator of whether or not a gene was expressed. Filtering was done for probe sets with a present call in all three replicates in at least one line and with an expression difference between two lines (≥2). To find statistically significant changes we used one-way ANOVA, together with the Benjamini and Hochberg method to adjust the false discovery rate (FDR adjusted p value cutoff was set to 0.05). Hierarchical clustering (Pearson correlation) was carried out on genes with statistically significant changes using the mean value for each line. For annotation purposes, blastx (e value cutoff = e−10) data was exported from HarvEST:Barley version 1.50 (http://harvest.ucr.edu). The Munich Information Center for Proteins Sequences Arabidopsis thaliana Database (MIPS) (http://mips.gsf.de/proj/funcatDB/search_main_frame.html) was used for functional classification.
Real-time and semiquantitative RT-PCR experiments
Real-time and semiquantitative reverse transcription polymerase chain reaction (RT-PCR) experiments are described in Supplementary Materials and Methods S1. Primer sequences for selected genes are shown in Supplementary Table S2.
Principal component analysis
A graphical representation of the association between genotypes, microarray expression data (mean values), and the mean values of the anther culture variables for each line was achieved using principal component analysis (PCA). The analysis was based on the correlation matrix, standardized and centred data, and was carried out using standard SAS/STAT procedures (SAS Institute, Cary, NC, USA). As the analysis was based on just three genotypes, the PCA biplot should explain 100% of the total variability.
Results
Characterization of the response to microspore embryogenesis
Microspore embryogenesis response of the three DH lines (DH46, DH6188, and DH6004) was characterized by recording five phenotypic variables representing different stages of the process (Table 1). DH6188 had the highest values for all traits, except the number of dividing microspores (nDM) and number of albino plants (nAP). DH6188 produced a number of green plants significantly higher than the other two lines. DH6004 had lower numbers of divisions and embryos (nEMB) than DH6188 and DH46, which determined its low productivity (nGP). Even though DH46 had the highest nDM and high nEMB, this line had the lowest nGP due to its high regeneration of albino plants.
Table 1.
Response of the doubled haploid lines DH46, DH6004, and DH6188 to microspore embryogenesis
| Line | nDM | nEMB | pGP | nAP | nGP |
|---|---|---|---|---|---|
| DH46 | 2644.78 (a) | 459.13 (a) | 2.35 (a) | 213.47 (a) | 8.26 (b) |
| DH6004 | 380.00 (c) | 81.43 (b) | 93.6 (b) | 5.00 (b) | 50.71 (b) |
| DH6188 | 1678.13 (b) | 579.14 (a) | 88.44 (b) | 10.45 (b) | 440.91 (a) |
Values followed by the same letter are not significantly different (P = 0.05) as described by Duncan’s test
nDM number of dividing microspores, nEMB number of embryos, pGP percentage of green plants, nAP number of albino plants, nGP number of green plants
Cytological studies
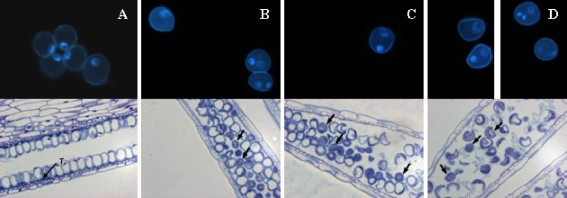
Before stress treatment, microspores were distributed in the periphery of the anther in direct contact with the tapetum (Fig. 1a—bottom). Most microspores were at late uninucleate stage, corresponding to G2 phase according to Shim and Kasha (2003) (Fig. 1a—top). Moreover, after 4 days of mannitol treatment, tapetum was degraded and microspores had a random distribution inside the anther loculus (Fig. 1b, c, d—bottom). A slow progression of the cell cycle was observed in all lines along mannitol treatment (Fig. 1b, c, d—top) as described by Muñoz-Amatriaín et al. (2006). However, differences among lines could be observed: whereas most of the microspores in DH6188 and DH46 had undergone the first mitosis ending with two asymmetric nuclei (Fig. 1b, c—top), DH6004 presented many signs of degradation and a very high degree of asynchrony, with microspores at uninucleate, binucleate or trinucleate stage (Fig. 1d).
Fig. 1.
DAPI staining of barley isolated microspores (top) and semithin sections of anthers stained with toluidine blue (bottom). a Anthers before mannitol treatment. b, c, d Anthers after 4 days of mannitol stress treatment of DH46 (b), DH6188 (c), and DH6004 (d). (T tapetum, Black arrows indicate binucleate microspores)
Identification and clustering of the differentially expressed genes after the stress treatment
The identification of genes differentially expressed between the three DH lines after 4 days of mannitol treatment was accomplished using the Barley1 GeneChip (Close et al. 2004). Using a false discovery rate adjusted p value of 0.05, ANOVA resulted in 213 differentially expressed genes. Annotation of these genes to functional categories was done using the functional categories described at MIPS website (for details, see “Materials and methods”). Of the 213 genes, 22% (47) had no homologue in any Uniprot, Rice, or Arabidopsis protein databases and 16% (35) belonged to the functional category “unclassified proteins.” Functional categories with the highest number of genes were “cell rescue, defense, and virulence” (32 genes) and “metabolism” (22 genes). These categories were followed by “transcription” and “transport”, with 12 and ten genes, respectively.
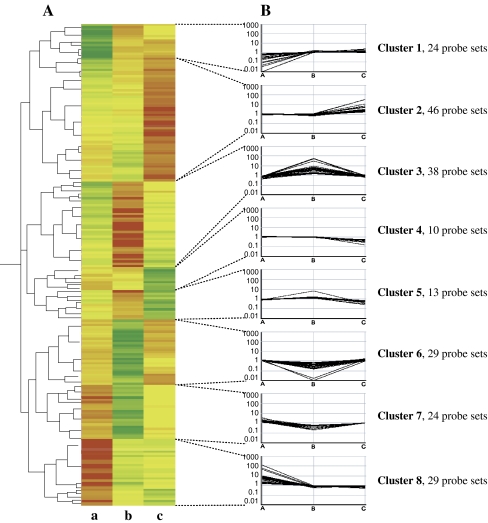
The 213 differentially expressed genes were subjected to hierarchical clustering based on their relative transcript level in each line and each gene was assigned to one of eight expression clusters (Fig. 2). Of eight clusters, six included genes differentially expressed in one line compared to the other two lines (clusters 1, 2, 3, 4, 6, and 8) and two clusters, 5 and 7, comprised genes whose expression profile was not associated with the change of a unique line. Expression data and annotation are available in the Supplementary Table S1.
Fig. 2.
Hierarchical clustering and expression profiles of differentially expressed genes. a Hierarchical cluster analysis of the 213 genes in DH46 (a), DH6004 (b), and DH6188 (c). b Expression profiles for the genes in each cluster presented in a graph format
Expression levels of seven differentially expressed genes and one invariant gene were analyzed using real-time RT-PCR for validation of microarray data. The correlation coefficient was 0.86 and the concordance was 100%. These results confirmed the reliability of microarray data. For more information, see Supplementary Table S3 and Figure S1.
Association between clusters and microspore embryogenesis response
To establish associations between clusters and microspore embryogenesis variables, a principal component analysis was carried out. Only genes with differences in the expression level over 2.5-fold were chosen for the PCA.
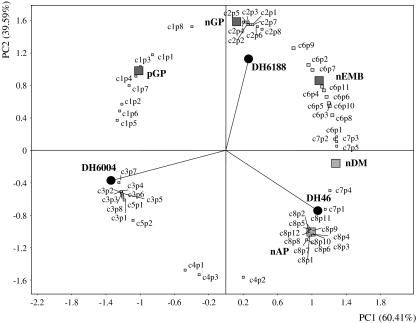
The well-separated distribution of the DH lines reflected the great differences in their anther culture response (Fig. 3). First principal component (PC1) accounted for 60.41% of the total variation and was mainly determined by the number of dividing microspores (nDM). The second axis (PC2) accounted for 39.59% of the total variation and was particularly associated with the number of green plants.
Fig. 3.
Principal component analysis of the five androgenic variables (grey squares) and the genes representative of each cluster (white squares). The three lines DH46, DH6004, and DH6188 are also identified (black circles). Each gene was designated as p (probe set), preceded by its cluster number and followed by its position in the cluster, according to the classified genes of Supplemental Table S1. nDM number of dividing microspores, nEMB number of embryos, pGP percentage of green plants, nAP number of albino plants, nGP number of green plants
Early stages of microspore embryogenesis are represented by the numbers of dividing microspores (nDM) and embryos (nEMB). Variable nDM gives a measure of the number of microspores that are able to divide and form a multicellular structure, which is the first evidence of a change in the developmental pathway, whereas nEMB indicates the number of microspores that develop into a complete embryo. For nDM, no close association with a specific cluster was revealed by PCA and only three genes of cluster 7 (beta-1.3-glucanase, c7p2; type 1 non-specific lipid transfer protein precursor, c7p3; and EF hand family protein, c7p5) and one of cluster 6 (TMS membrane family protein, c6p1) could be related with this variable. Variable nEMB was found to be associated with genes of cluster 3 (hAT dimerization domain-containing protein, c3p1; elongation factor 1-alpha, c3p3; F-box domain-containing protein, c3p5; mitochondrial glycoprotein, c3p6; and protein csAtPR5, c3p8), one cluster 5 gene (endonuclease, c5p1) and, in lesser extent, with cluster 6 genes (sucrose transporter, c6p4; aldose reductase, c6p3; oxidoreductase, c6p7; and histone H2B, c6p11) (Table 2).
Table 2.
Genes associated with early stages of microspore embryogenesis
| PCA name | Probe set | Expression level | E-value | Description | ||
|---|---|---|---|---|---|---|
| DH46 | DH6004 | DH6188 | ||||
| Cluster 3 | ||||||
| c3p1 | rbags22p06_s_at | 0.834 | 57.65 | 0.604 | 900E-76 | hAT dimerization domain-containing protein |
| c3p2 | Contig9177_at | 0.793 | 56.88 | 0.881 | 100E-100 | Shikimate kinase |
| c3p3 | Contig679_s_at | 0.707 | 51.97 | 0.997 | 4.00E-71 | Elongation factor 1-alpha (EF-1-alpha) |
| c3p4 | Contig16560_at | 0.724 | 31.48 | 1 | 2.00E-17 | D111/G-patch domain-containing protein |
| c3p5 | HU11B09u_s_at | 0.965 | 7.243 | 0.876 | 1.00E-126 | F-box domain containing protein |
| c3p6 | Contig18336_at | 0.921 | 6.356 | 0.764 | 4.00E-87 | Mitochondrial glycoprotein family protein |
| c3p7 | HVSMEa0004F18f2_s_at | 0.65 | 4.279 | 0.976 | 0.0 | Chloroplastic glutamine synthetase (GS2) |
| c3p8 | Contig7247_at | 0.952 | 3.404 | 0.837 | 4.00E-40 | Protein csAtPR5 |
| Cluster 5 | ||||||
| c5p1 | Contig4113_at | 0.845 | 7.032 | 0.366 | 1.00E-168 | Endonuclease |
| Cluster 6 | ||||||
| c6p1 | Contig3707_at | 1.295 | 0.01 | 0.999 | 5.00E-88 | TMS membrane family protein |
| c6p2 | Contig20828_at | 1.045 | 0.163 | 1.591 | 8.00E-59 | Cytochrome P450 |
| c6p3 | Contig8224_at | 1.101 | 0.199 | 1.128 | 1.00E-162 | Aldose reductase |
| c6p4 | Contig14939_at | 1.175 | 0.27 | 1.341 | 8.00E-81 | Sucrose transporter |
| c6p5 | Contig6294_at | 1.051 | 0.33 | 1.061 | 1.00E-167 | Brix domain containing protein |
| c6p6 | Contig21733_at | 1.126 | 0.396 | 1.207 | 7.00E-16 | Indole-3-acetate beta-glucosyltransferase |
| c6p7 | Contig3564_s_at | 1.23 | 0.399 | 1.65 | 1.00E-128 | Oxidoreductase. 2OG-Fe oxygenase family protein |
| c6p8 | Contig22536_at | 1.149 | 0.417 | 1.098 | 3.00E-48 | FAD-dependent oxidoreductase family protein |
| c6p9 | Contig7032_at | 1.037 | 0.424 | 1.778 | 0.0 | Beta-D-xylosidase |
| c6p10 | Contig7897_at | 1.11 | 0.439 | 1.139 | 2.00E-88 | Proton-dependent oligopeptide transport (POT) family protein |
| c6p11 | Contig1154_s_at | 1.227 | 0.475 | 1.402 | 1.00E-56 | Histone H2B |
| Cluster 7 | ||||||
| c7p2 | Contig17372_at | 1.251 | 0.252 | 1.007 | 5.00E-39 | Beta-1.3-glucanase |
| c7p3 | Contig2043_s_at | 1.393 | 0.258 | 1.09 | 4.00E-20 | Type 1 non-specific lipid transfer protein precursor |
| c7p5 | Contig25034_at | 1.358 | 0.33 | 1.048 | 2.00E-38 | EF hand family protein |
The number of green plants variable (nGP) measures the final efficiency of the microspore embryogenesis process. This variable was strongly linked to cluster 2 genes (glucan endo-1.3-beta-glucosidase, c2p1 and c2p3; NADPH:quinone reductase, c2p2; transcription initiation factor IIE, c2p4; and arsenite transport subunit B, c2p5) and was also associated, although not as closely, with genes of cluster 4 (mitochondrial processing peptidase, c4p1; and transport protein particle, c4p3) (Table 3).
Table 3.
Genes associated with the efficiency of microspore embryogenesis
| PCA name | Probe set | Expression level | E-value | Description | ||
|---|---|---|---|---|---|---|
| DH46 | DH6004 | DH6188 | ||||
| Cluster 2 | ||||||
| c2p1 | HVSMEl0010O18r2_s_at | 0.815 | 1.077 | 31.87 | 1.00E-180 | Glucan endo-1.3-beta-glucosidase |
| c2p2 | Contig18638_at | 0.957 | 0.895 | 7.024 | 4.00E-52 | NADPH:quinone reductase |
| c2p3 | Contig11921_x_at | 0.996 | 0.835 | 6.925 | 1.00E-180 | Glucan endo-1.3-beta-glucosidase |
| c2p4 | Contig8499_at | 0.887 | 1.04 | 5.262 | 0.0 | Transcription initiation factor IIE (TFIIE) alpha subunit |
| c2p5 | Contig25746_at | 1.01 | 0.993 | 4.741 | 5.00E-16 | Arsenite transport subunit B |
| c2p6 | HVSMEm0004L13r2_s_at | 1.103 | 0.749 | 3.12 | 1.00E-10 | Splicing factor PWI domain-containing protein |
| c2p7 | Contig4260_at | 1.069 | 0.948 | 2.68 | 1.00E-129 | Mannitol dehydrogenase (ELI3–1) |
| c2p8 | Contig9094_at | 0.911 | 0.706 | 2.644 | 1.00E-124 | Osmotin-like protein |
| Cluster 4 | ||||||
| c4p1 | Contig8352_at | 0.965 | 1.167 | 0.156 | 0.0 | Mitochondrial processing peptidase alpha subunit |
| c4p2 | Contig1781_s_at | 1.268 | 0.939 | 0.284 | 1.00E-106 | Histone H1 |
| c4p3 | Contig8849_at | 1.022 | 1.07 | 0.33 | 2.00E-99 | Transport protein particle (TRAPP) |
Variables number of albino plants (nAP) and percentage of green plants (pGP) are a measure of albinism, a phenomenon that limits the potential yield of microspore embryogenesis. A close association of these two variables was observed with genes of cluster 8 (RNA polymerase II 15.9 kDa subunit, c8p2; 40S ribosomal protein S23, c8p3; histone H4, c8p4; pumilio/Puf RNA binding domain-containing protein, c8p5; S1 RNA binding domain-containing protein, c8p8; and a protein similar to DAG, c8p9) and, to a lesser extent, with genes of cluster 1 (histone H2B, c1p3; and heterogeneous nuclear ribonucleoprotein, c1p4). A gene of cluster 7 (alpha-glucosidase, c7p1) was also found to be related to albinism (Table 4).
Table 4.
Genes associated with albinism
| PCA name | Probe set | Expression level | E-value | Description | ||
|---|---|---|---|---|---|---|
| DH46 | DH6004 | DH6188 | ||||
| Cluster 1 | ||||||
| c1p1 | Contig8733_at | 0.01 | 1.072 | 1.233 | 7.00E-55 | RNA polymerase II 15.9 kDa subunit |
| c1p2 | Contig24810_at | 0.0341 | 1.466 | 0.975 | 3.00E-23 | CG-1 domain containing protein |
| c1p3 | rbags21h11_at | 0.0849 | 1.256 | 1.242 | 3.00E-52 | Histone H2B |
| c1p4 | Contig2602_at | 0.102 | 1.21 | 1.096 | 1.00E-147 | Heterogeneous nuclear ribonucleoprotein |
| c1p5 | Contig12222_at | 0.178 | 1.708 | 0.997 | 0.0 | NBS-LRR disease resistance protein |
| c1p6 | Contig11423_at | 0.326 | 1.399 | 0.975 | 1.00E-153 | Endoribonuclease Dicer |
| c1p7 | Contig6316_s_at | 0.363 | 1.164 | 0.998 | 1.00E-25 | Retrotransposon protein |
| c1p8 | Contig8886_at | 0.363 | 1.222 | 2.142 | 0.0 | GMP synthase |
| Cluster 7 | ||||||
| c7p1 | Contig12639_at | 3.521 | 0.196 | 0.999 | 1.00E-147 | Alpha-glucosidase (AGLU) |
| Cluster 8 | ||||||
| c8p1 | Contig869_x_at | 126.1 | 0.901 | 0.811 | 4.00E-60 | Histone H2A |
| c8p2 | Contig8732_at | 44.37 | 0.679 | 0.781 | 6.00E-55 | RNA polymerase II 15.9 kDa subunit |
| c8p3 | Contig2098_at | 9.913 | 0.786 | 0.968 | 1.00E-74 | 40S ribosomal protein S23 |
| c8p4 | Contig724_at | 8.776 | 0.882 | 0.878 | 8.00E-54 | Histone H4 |
| c8p5 | Contig7020_at | 8.26 | 0.556 | 0.932 | 4.00E-60 | Pumilio/Puf RNA-binding domain-containing protein |
| c8p6 | Contig12770_at | 7.162 | 0.909 | 0.585 | 7.00E-98 | Wall-associated kinase |
| c8p7 | HVSMEg0011L24r2_at | 6.232 | 0.966 | 0.958 | 2.00E-12 | Zinc finger (C3HC4-type RING finger) family protein |
| c8p8 | Contig9928_at | 6.129 | 0.81 | 0.811 | 1.00E-102 | S1 RNA binding domain containing protein |
| c8p9 | Contig4384_at | 4.908 | 0.828 | 0.97 | 1.00E-105 | Similar to DAG protein |
| c8p10 | EBro03_SQ004_L04_at | 4.34 | 0.889 | 0.975 | 8.00E-26 | Peroxidase family protein |
| c8p11 | HZ50B24r_s_at | 2.547 | 0.812 | 0.864 | 1.00E-149 | Cinnamate 4-hydroxylase CYP73 |
| c8p12 | Contig2607_at | 2.287 | 0.888 | 0.942 | 1.00E-147 | Heterogeneous nuclear ribonucleoprotein |
Analysis of gene expression in different stages of microspore development
To study the expression patterns of some of the differentially expressed genes at different of stages of both pollen and androgenic development, semiquantitative RT-PCRs were done. Developmental stages included: uninucleated microspores before stress treatment (UM); microspores after 4 days of mannitol treatment (s-4d); microspores after 4 days of culture, following protocol described by Cistué et al. (2003) (c-4d); and young pollen grains (uninucleated microspores developed for 4 days in the mother plant; P).
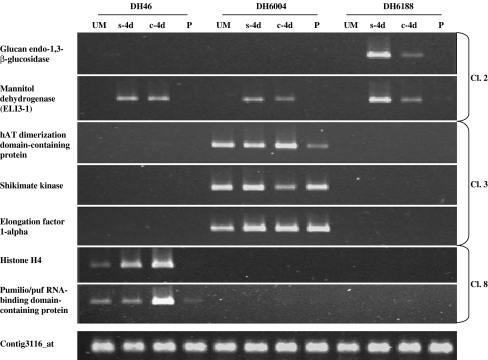
We selected a total of seven genes representing each of the different associations with microspore embryogenesis variables given by the PCA. These genes included: a glucan endo-1,3-β-glucosidase (HVSMEl0010O18r2_s_at, c2p1) and a mannitol dehydrogenase ELI3-1 (contig4260_at, c2p7) from cluster 2, the hAT dimerization domain-containing protein (rbags22p06_s_at, c3p1), a shikimate kinase (contig9177_at, c3p2) and an elongation factor 1-alpha (contig679_s_at, c3p3) from cluster 3, and a histone H4 (contig724_at, c8p4) and pumilio/Puf RNA binding domain-containing protein (contig7020_at, c8p5) from cluster 8 (Tables 2, 3, and 4).
Cluster 2 genes were found to be expressed not only at the dedifferentiation stage (s-4d) but also during early stages of microspore embryogenesis (c-4d). Both genes showed an expression specific of the androgenic development, being the highest expression associated with the fourth day of stress treatment (Fig. 4). Moreover, the gene coding for the glucan endo-1,3-β-glucosidase had expression only in line DH6188. Cluster 3 genes were only expressed in line DH6004, but in all stages of both androgenic and pollen development (Fig. 4). Finally, cluster 8 genes were expressed exclusively in DH46. These two genes were expressed at both stages of the androgenic development, being the highest expression at 4 days of culture. Regarding pollen development, expression of these genes at uninucleated microspores was observed, whereas no or almost no expression was found in young pollen grains (Fig. 4).
Fig. 4.
Expression analyses by semiquantitative RT-PCR of seven genes belonging to cluster 2 (Cl. 2), cluster 3 (Cl. 3), and cluster 8 (Cl. 8). Four different stages were assayed in each of the three DH lines: uninucleated microspores before stress treatment (UM), microspores after 4 days of stress treatment (s-4d), microspores after 4 days of culture (c-4d), and young pollen grains (P). Contig3116_at was used as control
Discussion
Stress treatment has a decisive role in microspore embryogenesis because it is not only needed for switching the dedifferentiation of microspores, but it also conditions the following response to microspore embryogenesis. Despite its critical role, the stress treatment stage has been little studied at the molecular level and there is no information currently available concerning the association between gene expression in this stage and the response to microspore embryogenesis.
The selection of three genetically similar DH lines with very different androgenic response and their transcriptome comparison at the time of dedifferentiation allowed the identification of 213 differentially expressed transcripts. It was not possible to assess the function of nearly 40% of the transcripts. Some of these genes, with a high differential expression level, could be good candidates to carry on further analysis. Functional categories with the highest number of genes were coincident with those of previous studies: “cell rescue, defense, and virulence”, “metabolism”, “transcription”, and “transport” (Kyo et al. 2003; Hosp et al. 2007; Maraschin et al. 2006; Muñoz-Amatriaín et al. 2006).
Genes related to early stages of microspore embryogenesis
After dedifferentiation, some microspores start to divide and form multicellular structures that can develop into complete embryos. Two variables, nDM and nEMB, are used as a measure of the early stages of microspore embryogenesis.
Only four genes from clusters 7 and 6 were positively associated with nDM. The gene with the highest difference in expression level encodes a TMS membrane family protein. Although the function of these proteins is unknown, a member of this family, TDE1, has been related to apoptosis inhibition and tumorigenesis in humans (Bossolasco et al. 2006). Two other genes encoded a beta-1,3-glucanase and a non-specific lipid transfer protein. Genes encoding these proteins were previously found to be expressed in early stages of microspore embryogenesis (Vrinten et al. 1999; Kyo et al. 2003; Borderies et al. 2004; Joosen et al. 2007; Malik et al. 2007). The fourth gene positively related with nDM encodes an EF-hand domain protein, often found in calcium-binding proteins, indicating an important role of calcium in early stages of microspore embryogenesis. This result is in accordance with previous data, since the introduction of Ca2+ in the stress treatment medium increased the number of divisions, embryos, and total plants (Hoesktra et al. 1997; Cistué et al. 2004). These four genes are related to changes in the structure and function of membranes. It is known that membranes are the primary target of plant stresses (Hasegawa et al. 2000). Our results indicate that the microspore membrane plays an important role in the stress response, conditioning the early stages of microspore embryogenesis.
Several genes belonging to clusters 6 and 3 were, respectively, positively and negatively associated with the ability of microspores to form a complete embryo (nEMB). High nEMB was associated with genes involved in carbohydrate and lipid degradation and transport, such as a beta-D-xylosidase, an alpha-galactosidase, a lipase, and a sucrose transporter, and genes related to energy production such as two oxidoreductases and a proton-dependent oligopeptide (POT) transporter. However, the expression of genes involved in reassimilation of ammonia from amino acid degradation, like glutamine synthetase 2 (GS2) and asparagine synthetase (AS), and a shikimate kinase involved in the biosynthesis of aromatic amino acids, was negatively associated with nEMB. As is known, during mannitol treatment there is a reorganization of the central carbon metabolism to a more flexible use of carbon skeletons from different sources (Muñoz-Amatriaín et al. 2006). Results of this study indicated that lines producing high numbers of embryos have active carbohydrate and lipid degradation, which increases the sugar content, whereas low responding lines are characterized by increased proteolysis and catabolism of amino acids.
Other genes of clusters 6 and 3 associated with nEMB are related to different cell fates. It is known that cytoskeleton rearrangements are involved in the induction of microspore embryogenesis (Maraschin et al. 2005). In this analysis, actin gene ACT7 was positively related to nEMB. The actin cytoskeleton plays an active role in cell division, cell shape determination, and cell-polarity establishment (McDowell et al. 1996). At the same time, the expression of genes related to different stages of pollen development was associated to low values of nEMB. These genes included: a ribose-phosphate pyrophosphokinase, which is involved in nucleotide biosynthesis, a fibrillarin, rRNA biogenesis protein RRP5, and elongation factor 1-alpha (EF-1-α). Maraschin et al. (2006) found that the expression of nucleotide biosynthetic genes and rRNA genes like fibrillarin was associated with uninucleate microspores before mannitol treatment, at the same time that EF-1-α was associated with pollen development. Other genes negatively associated with this variable are related to programmed cell death, such as senescence-associated gene SAG102 and an endonuclease.
All these data suggest that many microspores of the low-responding genotypes are not able to dedifferentiate, maintaining the uninucleated initial stage, following the pollen developmental pathway, or undergoing programmed cell death. Cytological studies of recalcitrant line DH6004 are in agreement with these expression data, since microspores in process of degradation, together with uninucleated microspores and even trinucleate pollen grains, were observed after mannitol treatment (Fig. 1d). Moreover, expression analysis of the three genes of the cluster 3 by semiquantitative RT-PCR (Fig. 4) showed that all were already expressed in uninucleated microspores. These expression data suggested that the reprogramming problems in recalcitrant genotypes could originate in earlier stages of microspore development at the time of sampling.
Genes related to the efficiency of microspore embryogenesis
The final efficiency of the microspore embryogenesis process is measured by the number of green plants obtained (nGP). The expression of genes belonging to clusters 2 and 4 at the stress treatment stage was related with high and low values of green plant production, respectively.
Many genes of cluster 2 had a role in stress response, including PR proteins, oxidative stress-related proteins, and heat-shock proteins (HSPs), which is in accordance with the multidimensional stress response described as a consequence of mannitol treatment (Muñoz-Amatriaín et al. 2006). Among the PR proteins, two glucan endo-1,3-beta-glucosidases, three mannitol dehydrogenases (ELI3-1), and an osmotin-like protein were found. The activation of many PR proteins, like beta-glucanases and ELI3 proteins, in a system devoid of pathogens could be related with their metabolic roles (Stoop et al. 1996). Beta-glucanases could degrade cell wall carbohydrates for the mobilization of storage material, while ELI3 proteins could oxidize mannitol to mannose (Williamson et al. 1995). The expression of two of these PR-genes (one beta-1,3-glucanase and one mannitol dehydrogenase) was analyzed by semiquantitative RT-PCR (Fig. 4). Both genes were specifically expressed in the two stages of the androgenic development and their highest expression occurred in the most efficient line (DH6188) at the time of dedifferentiation. Although both genes are good candidates for further study, the gene encoding a beta-1,3-glucanase is of special interest since it could be used as a bio-marker for high green plant production.
High values of nGP were also related to protection against oxidative stress and detoxification, since genes coding for a NAD(P)H:quinone reductase (NQR), a cytochrome P450, riboflavin biosynthesis protein ribA, and a glutathione S-transferase (GST) were present in cluster 2. The induction of GST family members during the initial stages of microspore embryogenesis is well documented (Vrinten et al. 1999; Maraschin et al. 2006; Muñoz-Amatriaín et al. 2006; Joosen et al. 2007; Tsuwamoto et al. 2007). Two HSPs (HSC70 and HSP81-2) were also positively related to nGP. Both proteins were not heat-inducible molecular chaperones. The role of HSP in the induction of microspore embryogenesis has been discussed. It has been suggested that their involvement in the androgenic switch could be indirect, having a role more directly related to stress tolerance (for review, see Seguí-Simarro and Nuez 2008). In our study, the presence of two HSPs suggests that, whether their involvement is direct or indirect, their expression is important for the final production of green plants.
The high number of stress-response genes associated with nGP indicates that microspores best protected against stress during dedifferentiation have more chances to successfully conclude the androgenic process.
Many reports have revealed that stressed microspores show an overall decrease in the protein levels, leading to the hypothesis that down-regulation of pollen-specific proteins or increased protein breakdown might play an important role in the dedifferentiation of microspores (for review, see Maraschin et al. 2005). Further studies have shown that the induction of proteolytic genes was associated with the androgenic potential of microspores (Maraschin et al. 2006). In this study, proteolytic genes such as aspartic protease, subtilase, and 26S proteasome regulatory subunit required for proper proteosome assembly were found to be positively associated with nGP.
Regulation of transcription and translation plays an important role in the final efficiency of the process since some of the genes showing the highest difference in expression level that were associated with high nGP belong to this category. One of them encodes the alpha subunit of the transcription initiation factor TFIIE that plays a central role in the formation of pre-mRNA (Forget et al. 2004). Another had a PWI motif that is important for pre-mRNA splicing (Blencowe and Ouzounis 1999). Finally, a protein factor IF2, which is essential for promoting translation initiation, was also found. The expression of the response regulator ARR3, involved in a His-to-Asp phosphorelay signal transduction system (Suzuki et al. 1998), was also related with high green plant production.
Few genes were found to be associated with a low nGP. Among them, the gene showing the highest difference in the expression level encoded a mitochondrial processing peptidase (MPP), which is part of the cytochrome c reductase complex of the respiratory chain and is expressed in male gametophyte (Noir et al. 2005). In the same way, a caleosin gene and the two histones H1 and H3 could also be related to pollen development, as caleosins are related to the storage of lipid bodies (Murphy et al. 2000), which are known to be accumulated in the cytoplasm of the pollen vegetative cell (Maraschin et al. 2005), and H1 has been associated to pollen differentiation (Tanaka et al. 1998).
Genes related to albinism
Regeneration of chlorophyll-deficient plants is one of the major obstacles for the efficient use of microspore embryogenesis in the production of homozygous plants, since these albino plants only survive for relatively short periods in vitro. Genes belonging to clusters 1 and 8 and one gene of cluster 7 were associated with the occurrence of albinism during androgenesis.
When plastid differentiation during microspore embryogenesis has been studied in albino barley genotypes, abnormal features mainly affecting plastid size and structure (Caredda et al. 1999, 2000) have been found. It has been shown that, after the stress treatment, microspore plastids had differentiated exclusively into amyloplasts, accumulating starch and losing their thylakoids as well as their capacity to divide (Caredda et al. 2000).
High values of nAP were found to be associated with the expression of three genes that could be related to plastid development. One of them had homology to DAG (differentiation and greening), a nuclear gene which encodes a protein targeted to the plastids. Expression of DAG is required for the expression of nuclear genes affecting the chloroplast, such as CAB and RBCS, and for the expression of the gene RPOB encoding the plastidial RNA polymerase β subunit. DAG acts very early in chloroplast development and is essential not only for chloroplast development from proplastids, but also for the formation of other plastid types (Chatterjee et al. 1996). The second plastid development-associated gene encodes a class B ankyrin repeat protein (Becerra et al. 2004). One of the four class B-proteins characterized is known to be involved in crucial events controlling plastid differentiation (Zhang et al. 1992; Garcion et al. 2006). The third gene encodes abscisic acid-insensitive 3 (ABI3), a transcription factor that plays a role in plastid identity and could affect plastid ultrastructure (Rohde et al. 2000).
Starch accumulation in plastids after the stress treatment has been associated with the expression of albino phenotype (Caredda et al. 2000). Surprisingly, we did not find any gene differentially expressed in line DH46 that could be directly related to starch accumulation. The only gene involved in carbohydrate metabolism that was associated to nAP encoded an alpha-glucosidase (AGLU), involved in the last steps of carbohydrate degradation.
Molecular studies of microspore-derived albino plants in wheat have revealed that albino plants seemed to lack plastid ribosomes and showed an altered transcription and translation pattern when compared to green plants (Hofinger et al. 2000). These authors suggested that the translation deficiency in plastids was the primary reason for the expression of the albino phenotype. In this study, two plastid-encoded ribosomal proteins S8 were expressed at lower levels in the albino-producing line DH46, which is consistent with the deficiency of plastid ribosomes in albino plants that leads to the absence of normally abundant plastid translation products (Zubko and Day 2002).
Finally, a gene that could be involved in signal transduction to chloroplast was found to be associated with high pGP and low nAP. This gene encodes a protein with a CG-1 domain that was first identified in parsley as a possible member of the light signal transduction chain (da Costa e Silva 1994).
The relation of the rest of genes associated with nAP and pGP to plastids has not been yet described. Among these, genes involved in processes affecting transcription and translation were found: two isoforms of a RNA polymerase II 15.9 kDa subunit (yeast Rpb4) which is required for transcription and for mRNA export in stress conditions (Farago et al. 2003); two isoforms of a heterogeneous nuclear ribonucleoprotein, with a possible role in pre-mRNA splicing (Martinez-Contreras et al. 2007); and a pumilio/Puf RNA binding domain-containing protein, which is involved in translation repression of specific target mRNAs (Spassov and Jurecic 2003). Low numbers of albino plants were associated with genes encoding the endoribonuclease Dicer, which plays an essential role in RNA interference (Bernstein et al. 2001), and a diacylglycerol kinase. Further studies about the possible involvement of all these genes in albinism would be worthwhile.
The differential expression of plastid-related genes after stress treatment suggested that although albinism is manifested at the time of plant regeneration, it could be previously determined at the stage of microspore dedifferentiation. Semiquantitative RT-PCR analysis showed that the two genes of cluster 8 already showed differential expression in uninucleated microspores at the time of sampling and no or almost no expression in pollen grains (Fig. 4). Our results are in agreement with some studies that indicated that the origin of albinism in some cultivars is determined earlier in microspore embryogenesis or even at the time of sampling (Caredda et al. 2000, 2004). These results also suggest that the mechanisms that lead to plastid disappearance during pollen maturation in albino genotypes are different from those taking place during microspore dedifferentiation.
Electronic supplementary materials
Below is the link to the electronic supplementary materials.
Protocols for cytological studies, real-time and semiquantitative RT-PCR (DOC 33.0 KB)
Data of expression level and annotations of probe sets corresponding to each cluster (XLS 82.5 KB).
PCR primers for semiquantitative and real-time RT-PCR (XLS 16.0 KB).
Validation of microarray results by real-time PCR (DOC 38.5 KB).
Linear regression analysis between microarray and quantitative PCR data (DOC 217 KB).
Acknowledgments
We are grateful to the Dr. J.D. Heck group at the UCI DNA and Protein MicroArray Facility, University of California Irvine, for excellent services. We also thank Prof. I. Romagosa his help with principal component analysis and Dr. Ben Millett for the English correction of the manuscript. María Muñoz-Amatriaín was recipient of a predoctoral fellowship and a short-term scientific mission to the UCR, from Ministry of Education and Science of Spain. This work was supported by Projects AGL2004-03396 and AGL2007-62930/AGR from “Plan Nacional de Recursos y Tecnologías Agroalimentarias” of Spain.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s10142-009-0113-3) contains supplementary material, which is available to authorized users.
References
- Becerra C, Jahrmann T, Puigdomènech P, Vicient CM (2004) Ankyrin repeat-containing proteins in Arabidopsis: characterization of a novel and abundant group of genes coding ankyrin-transmembrane proteins. Gene 340:111–121 [DOI] [PubMed]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363–366 [DOI] [PubMed]
- Blencowe BJ, Ouzounis CA (1999) The PWI motif: a new protein domain in splicing factors. Trends Biochem Sci 24:179–180 [DOI] [PubMed]
- Borderies G, Le Bechec M, Rossignol M, Lafitte C, Le Deunff E, Beckert M, Dumas C, Matthys-Rochon E (2004) Characterization of proteins secreted during maize microspore culture: arabinogalactan proteins (AGPs) stimulate embryo development. Eur J Cell Biol 83:205–212 [DOI] [PubMed]
- Bossolasco M, Veillette F, Bertrand R, Mes-Masson AM (2006) Human TDE1, a TDE1/TMS family member, inhibits apoptosis in vitro and stimulates in vivo tumorigenesis. Oncogene 25:4549–4558 [DOI] [PubMed]
- Caredda S, Devaux P, Sangwan RS, Clément C (1999) Differential development of plastids during microspore embryogenesis in barley. Protoplasma 208:248–256 [DOI]
- Caredda S, Doncoeur C, Devaux P, Sangwan RS, Clément C (2000) Plastid differentiation during androgenesis in albino and non-albino producing cultivars of barley (Hordeum vulgare L.). Sex Plant Reprod 13:95–104 [DOI]
- Caredda S, Devaux P, Sangwan RS, Proult I, Clément C (2004) Plastid ultrastructure and DNA related to albinism in androgenetic embryos of various barley (Hordeum vulgare) cultivars. Plant Cell Tissue Organ Cult 76:35–43 [DOI]
- Chatterjee M, Sparvoli S, Edmunds C, Garosi P, Findlay K, Martin C (1996) DAG, a gene required for chloroplast differentiation and palisade development in Antirrhinum majus. EMBO J 15:4194–4207 [PMC free article] [PubMed]
- Chen X-W, Cistué L, Muñoz-Amatriaín M, Sanz M, Romagosa I, Castillo AM, Vallés MP (2007) Genetic markers for doubled haploid response in barley. Euphytica 158:287–294 [DOI]
- Cistué L, Ramos A, Castillo AM, Romagosa I (1994) Production of large number of doubled haploid plants from barley anthers pretreated with high concentrations of mannitol. Plant Cell Rep 13:709–712 [DOI] [PubMed]
- Cistué L, Ramos A, Castillo AM (1999) Influence of anther pretreatment and culture medium composition on the production of barley doubled haploids from model and low responding cultivars. Plant Cell Tiss Org Cult 55:159–166 [DOI]
- Cistué L, Vallés MP, Echávarri B, Sanz JM, Castillo AM (2003) Barley anther culture. In: Malupszynski M, Kasha K, Foster B (eds) Doubled haploid production in crop plants, a manual. FAO/IAEA Division, Wien, pp 29–35
- Cistué L, Vallés MP, Echávarri B, Sanz JM, Castillo AM (2004) Production of barley doubled haploids by anther and microspore culture. In: Mujib A, Cho M-J, Predieri S, Banerjee S (eds) In vitro application in crop improvement. Science Publishers, Plymouth, pp 1–17
- Close TJ, Wanamaker SI, Caldo RA, Turner SM, Ashlock DA, Dickerson JA, Wing RA, Muehlbauer GJ, Kleinhofs A, Wise RP (2004) A new resource for cereal genomics: 22K barley GeneChip comes of age. Plant Physiol 134:960–968 [DOI] [PMC free article] [PubMed]
- COST Action 851 (2005) http://www.scri.ac.uk/assoc/COST851/DHTable2005.xls
- da Costa e Silva O (1994) CG-1, a parsley light-induced DNA-binding protein. Plant Mol Biol 25:921–924 [DOI] [PubMed]
- Farago M, Nahari T, Hammel C, Cole CN, Choder M (2003) Rpb4p, a subunit of RNA polymerase II, mediates mRNA export during stress. Mol Biol Cell 14:2744–2755 [DOI] [PMC free article] [PubMed]
- Forget D, Langelier MF, Thérien C, Trinh V, Coulombe B (2004) Photo-cross-linking of a purified preinitiation complex reveals central roles for the RNA polymerase II mobile clamp and TFIIE in initiation mechanisms. Mol Cell Biol 24:1122–1131 [DOI] [PMC free article] [PubMed]
- Forster BP, Thomas WTB (2005) Doubled haploids in genetics and plant breeding. Plant Breeding Rev 25:57–88
- Garcion C, Guilleminot J, Kroj T, Parcy F, Giraudat J, Devic M (2006) AKRP and EMB506 are two ankyrin repeat proteins essential for plastid differentiation and plant development in Arabidopsis. Plant J 48:895–906 [DOI] [PubMed]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Ann Rev Plant Physiol Plant Mol Biol 51:463–499 [DOI] [PubMed]
- Hoekstra S, van Zijderveld MH, Louwerse JD, Heidekamp F, van der Mark F (1992) Anther and microspore culture of Hordeum vulgare L. cv. Igri. Plant Sci 86:89–96 [DOI] [PubMed]
- Hoekstra S, van Bergen S, van Brouwershaven IR, Schilperoort RA, Wang M (1997) Androgenesis in Hordeum vulgare L.: effects of mannitol, calcium and abscisic acid on anther pretreatment. Plant Sci 126:211–218 [DOI]
- Hofinger BJ, Ankele E, Gülly Ch, Heberle-Bors E, Pfosser MF (2000) The involvement of the plastid genome in albino plant regeneration from microspores in wheat. In: Bohanec B (ed) Biotechnological approaches for utilization of gametic cells-COST 824. OP-EUR, Luxembourg, pp 215–228
- Hosp J, Tashpulatov A, Roessner U, Barsova E, Katholnigg H, Steinborn R, Melikant B, Lukyanov S, Heberle-Bors E, Touraev A (2007) Transcriptional and metabolic profiles of stress-induced, embryogenic tobacco microspores. Plant Mol Biol 63:137–149 [DOI] [PubMed]
- Joosen R, Cordewener J, Supena EDJ, Vorst O, Lammers M, Maliepaard C, Zeilmaker T, Miki B, America T, Custers J, Boutilier K (2007) Combined transcriptome and proteome analysis identifies pathways and markers associated with the establishment of rapeseed microspore-derived embryo development. Plant Physiol 144:155–172 [DOI] [PMC free article] [PubMed]
- Kasha KJ, Hu TC, Oro R, Simion E, Shim YS (2001) Nuclear fusion leads to chromosome doubling during mannitol pretreatment of barley (Hordeum vulgare L.) microspores. J Exp Bot 52:1227–1238 [DOI] [PubMed]
- Kyo M, Hattori S, Yamaji N, Pechan P, Fukui H (2003) Cloning and characterization of cDNAs associated with the embryogenic dedifferentiation of tobacco immature pollen grains. Plant Sci 164:1057–1066 [DOI]
- Li H, Devaux P (2001) Enhancement of microspore culture efficiency of recalcitrant barley genotypes. Plant Cell Rep 20:475–481 [DOI]
- Li H, Devaux P (2003) High frequency regeneration of barley doubled haploid plants from isolated microspore culture. Plant Sci 164:379–386 [DOI]
- Malik MR, Wang F, Dirpaul JM, Zhou N, Polowick PL, Ferrie AMR, Krochko JE (2007) Transcript profiling and identification of molecular markers for early microspore embryogenesis in Brassica napus. Plant Physiol 144:134–154 [DOI] [PMC free article] [PubMed]
- Maraschin SF, de Priester W, Spaink HP, Wang M (2005) Androgenic switch: an example of plant embryogenesis from the male gametophyte perspective. J Exp Bot 56:1711–1726 [DOI] [PubMed]
- Maraschin SF, Caspers M, Potokina E, Wülfert F, Graner A, Spaink HP, Wang M (2006) cDNA array analysis of stress-induced gene expression in barley androgenesis. Physiol Plantarum 127:535–550 [DOI]
- Martinez-Contreras R, Fisette J F, Cloutier P, Revil T, Shkreta L, Chabot B (2007) hnRNP proteins and splicing control. In: Blencowe B, Graveley B (eds) Alternative splicing in the postgenomic era. Eurekah Biosciences Series, pp 123–147 [DOI] [PubMed]
- McDowell JM, An YQ, Huang S, McKinney EC, Meagher RB (1996) The Arabidopsis ACT7 actin gene is expressed in rapidly developing tissues and responds to several external stimuli. Plant Physiol 111:699–711 [DOI] [PMC free article] [PubMed]
- Muñoz-Amatriaín M, Svensson JT, Castillo AM, Cistué L, Close TJ, Vallés MP (2006) Transcriptome analysis of barley anthers: effect of mannitol treatment on microspore embryogenesis. Physiol Plantarum 127:551–560 [DOI]
- Muñoz-Amatriaín M, Castillo AM, Chen XW, Cistué L, Vallés MP (2008) Identification and validation of QTLs for green plant percentage in barley (Hordeum vulgare L.) anther culture. Mol Breeding 22:119–129 [DOI]
- Murphy DJ, Hernendez-Pinzon I, Patel K, Hope RG, McLauchlan J (2000) New insights into the mechanisms of lipid-body biogenesis in plants and other organisms. Biochem Soc Trans 28:710–711 [DOI] [PubMed]
- Noir S, Bräutigam A, Colby T, Schmidt J, Panstruga R (2005) A reference map of the Arabidopsis thaliana mature pollen proteome. Biochem Biophys Res Commun 337:1257–1266 [DOI] [PubMed]
- Oleszczuk S, Sowa S, Zimny J (2006) Androgenic response to preculture stress in microspore cultures of barley. Protoplasma 228:95–100 [DOI] [PubMed]
- Rohde A, De Rycke R, Beeckman T, Engler G, Van Montagu M, Boerjan W (2000) ABI3 affects plastid differentiation in dark-grown Arabidopsis seedlings. Plant Cell 12:35–52 [DOI] [PMC free article] [PubMed]
- Seguí-Simarro JM, Nuez F (2008) How microspores transform into haploid embryos: changes associated with embryogenesis induction and microspore-derived embryogenesis. Physiol Plantarum 134:1–12 [DOI] [PubMed]
- Shariatpanahi ME, Bal U, Heberle-Bors E, Touraev A (2006) Stresses applied for the re-programming of plant microspores towards in vitro embryogenesis. Physiol Plantarum 127:519–534 [DOI]
- Shim YS, Kasha KJ (2003) The influence of pretreatment on cell stage progression and the time of DNA synthesis in barley (Hordeum vulgare L.) uninucleate microspores. Plant Cell Rep 21:1065–1071 [DOI] [PubMed]
- Spassov DS, Jurecic R (2003) The PUF family of RNA-binding proteins: does evolutionarily conserved structure equal conserved function? IUBMB Life 55:359–366 [DOI] [PubMed]
- Stoop JMH, Williamson JD, Pharr DM (1996) Mannitol metabolism in plants: a method for coping with stress. Trends Plant Sci 1:139–144 [DOI]
- Suzuki T, Imamura A, Ueguchi C, Mizuno T (1998) Histidine-containing phosphotransfer (HPt) signal transducers implicated in His-to-Asp phosphorelay in Arabidopsis. Plant Cell Physiol 39:1258–1268 [DOI] [PubMed]
- Tanaka I, Ono K, Fukuda T (1998) The developmental fate of angiosperm pollen is associated with a preferential decrease in the level of histone H1 in the vegetative nucleus. Planta 206:561–569 [DOI]
- Touraev A, Vicente O, Heberle-Bors E (1997) Initiation of microspore embryogenesis by stress. Trends Plant Sci 2:297–302 [DOI]
- Touraev A, Pfosser M, Heberle-Bors E (2001) The microspore: a haploid multipurpose cell. Adv Bot Res 35:53–109 [DOI]
- Tsuwamoto R, Fukuoka H, Takahata Y (2007) Identification and characterization of genes expressed in early embryogenesis from microspores of Brassica napus. Planta 225:641–652 [DOI] [PubMed]
- Vrinten PL, Nakamura T, Kasha KJ (1999) Characterization of cDNAs expressed in the early stages of microspore embryogenesis in barley (Hordeum vulgare L.). Plant Mol Biol 41:455–463 [DOI] [PubMed]
- Williamson JD, Stoop JMH, Massel MO, Conkling MA, Manson Pharr D (1995) Sequence analysis of a mannitol dehydrogenase cDNA from plants reveals a function for the pathogenesis-related protein ELI3. Proc Natl Acad Sci U S A 92:7148–7152 [DOI] [PMC free article] [PubMed]
- Wojnarowiez G, Caredda S, Devaux P, Sangwan RS, Clément C (2004) Barley anther culture: assessment of carbohydrate effects on embryo yield, green plant production and differential plastid development in relation with albinism. J Plant Physiol 161:747–755 [DOI] [PubMed]
- Zhang H, Schelrer DC, Fowle WH, Goodman HM (1992) Expression of antisense or sense RNA of an ankyrin repeat-containing gene blocks chloroplast differentiation in Arabidopsis. Plant Cell 4:1575–1588 [DOI] [PMC free article] [PubMed]
- Zubko MK, Day A (2002) Differential regulation of genes transcribed by nucleus-encoded plastid RNA polymerase, and DNA amplification, within ribosome-deficient plastids in stable phenocopies of cereal albino mutants. Mol Genet Genomics 267:27–37 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary materials.
Protocols for cytological studies, real-time and semiquantitative RT-PCR (DOC 33.0 KB)
Data of expression level and annotations of probe sets corresponding to each cluster (XLS 82.5 KB).
PCR primers for semiquantitative and real-time RT-PCR (XLS 16.0 KB).
Validation of microarray results by real-time PCR (DOC 38.5 KB).
Linear regression analysis between microarray and quantitative PCR data (DOC 217 KB).