Abstract
The current standard interferon-alpha (IFN-α)-based therapy for chronic hepatitis C virus (HCV) infection is only effective in approximately half of the patients, prompting the need for alternative treatments. RNA interference (RNAi) represents novel approach to combat HCV by sequence-specific targeting of viral or host factors involved in infection. Monotherapy of RNAi, however, may lead to therapeutic resistance by mutational escape of the virus. Here, we proposed that combining lentiviral vector-mediated RNAi and IFN-α could be more effective and avoid therapeutic resistance. In this study, we found that IFN-α treatment did not interfere with RNAi-mediated gene silencing. RNAi and IFN-α act independently on HCV replication showing combined antiviral activity when used simultaneously or sequentially. Transduction of mouse hepatocytes in vivo and in vitro was not effected by IFN-α treatment. In conclusion, RNAi and IFN-α can be effectively combined without cross-interference and may represent a promising combinational strategy for the treatment of hepatitis C.
Keywords: RNAi, IFN-α, Gene therapy, Lentiviral vector, HCV
Introduction
The hepatitis C virus (HCV) infection remains a major cause of chronic liver disease with an estimated 170 million carriers worldwide. The current standard therapy, pegylated interferon-alpha (IFN-α) in combination with ribavirin, has achieved substantial success [1, 2]. However, still half of the patients fail to develop a sustained virologic response after this therapy. To improve treatment outcomes, novel monotherapies or alternative combination of IFN-α based therapies are urgently required.
RNA interference (RNAi), the degradation of cognate mRNA by small interfering RNA (siRNA), has emerged as a novel therapeutic entity for viral infections. Since the HCV genome is a single-stranded RNA that functions as both a template for transcription and template for a negative strand replication intermediate, it is a prime candidate for RNAi [3]. Instead of a 5′ cap, the internal ribosome entry site (IRES), located at the 5′ noncoding region of the viral genome, plays an essential in initiating translation [4]. Because of the most conserved sequence within the viral genome, IRES seems an ideal target for RNAi, and indeed, several studies have demonstrated inhibition of HCV replication by targeting this region [5–7]. HCV replication is mediated by NS5B, an RNA-dependent RNA-polymerase that lacks proofreading abilities. As a result of extremely high mutation rate (103 per nucleotide per generation) and replication rate (1012 virions per day) in patients, HCV quasispecies are generated [8]. NS5b region has been shown to be very effective for RNAi-induced suppression of HCV replication [9]. Host cellular factors involved in the viral entry, such as CD81, Claudin-1, Occludin, or SR-B1, could also be candidate targets for RNAi antiviral therapy [10–12]. Knockdown of CD81 by RNAi significantly prevented the binding of Huh7 cells to E2 protein [13]. A definitive proof of the therapeutic value of CD81 in vivo was provided by a recent study showing that treatment with anti-CD81 antibodies completely protected human liver-uPA-SCID mice from a subsequent challenge with HCV consensus strains of different genotypes [14].
The success of RNAi in therapeutic application also depends on an efficient delivery system, which can support long-term siRNA production and continuous gene silencing. Integrating self-inactivating lentiviral vector (LV) can achieve these criteria by encoding small hairpin RNA (shRNA), a precursor of siRNA that is cleaved into biologically active siRNA by host cell enzyme, Dicer [15, 16].
Similar to existing antiviral monotherapies, a new monotherapy based on a single RNAi target will likely fail due to the development of resistance. HCV can potentially overcome the antiviral effects of RNAi through several mechanisms, including genomic diversity, mutational escape, and attenuation of RNAi machinery [17–20]. RNA viruses like HIV and HCV are highly adaptive in this context, and mutational escape from therapy has been documented against single RNAi treatments [18, 21]. Thus, a combined strategy would be necessary for eliminating HCV infection. Previous studies reported by our group and others have shown that simultaneously targeting both viral and host cell elements by RNAi could increase the potency of antiviral therapies [13, 21–23]. IFN-α possesses indirect antiviral activity by stimulating genes that can lead to a non-virus-specific antiviral response, whereas RNAi can directly interfere with viral entry and replication through targeting viral RNA genome or mRNA of cellular factors. Based on their complementary antiviral mechanisms, we propose that combining RNAi with IFN-α may prevent therapeutic resistance and exhibit enhanced antiviral activity. Moreover, the additional combination of ribavirin to RNAi and IFN-α may further improve the therapeutic effects in the treatment of chronic hepatitis C.
To date, very little is known about the interaction of exogenous IFN-α with lentiviral vector delivery and the antiviral action of RNAi. Reports have suggested that the administration of lentiviral vectors to mice triggers a rapid and transient type I IFN response (i.e., IFN-α and IFN-β). In animals that lack the capacity to produce type I IFN, dramatic increases in transduction to hepatocytes were seen, indicating that endogenous IFNs may interfere [24].
In the current study, we investigated the effect of combining lentiviral-mediated RNAi with IFN-α in an in vitro HCV replication model. The results indicate that IFN treatment does not affect the transduction efficiency of hepatocytes in vitro and in vivo. Moreover, IFN did not interfere with RNAi-mediated knockdown of host cell target genes, including CD81, and showed combined antiviral activity with shRNAs directed against the HCV genome. The combined efficacy of RNAi and IFN-α may provide new opportunities for highly effective antiviral therapy for hepatitis C.
Materials and methods
Cell culture
Cell monolayers of the human embryonic kidney epithelial cell line HEK293 and human hepatoma cell line Huh7 and Huh6 were maintained in Dulbecco's modified Eagle medium (DMEM; Invitrogen–Gibco, Breda, The Netherlands) and complemented with 10% v/v fetal calf serum (Hyclone, Logan, Utah), 100 IU/ml penicillin, 100 mg/ml streptomycin, and 2 mM l-glutamine (Invitrogen–Gibco; cDMEM). Huh7 cells containing a subgenomic HCV bicistronic replicon (I389/NS3-3V/LucUbiNeo-ET, Huh7-ET) were maintained with 250 μg/ml G418 (Sigma, Zwijndrecht, The Netherlands) [25].
Construction and production of lentiviral vectors
The vectors, LV expressing green fluorescent protein (LV-GFP), LV-shCD81, LV-shIRES, and LV-shNS5b containing shRNA cassettes were constructed and produced as previously reported [13]. LV-shIRES targets HCV IRES region (AGGUCUCGUAGACCGUGCA 321-340). LV-shNS5b targets the viral NS5b region (GACACUGAGACACCAAUUGAC 6367-6388). LV-shCD81 targets human CD81 mRNA (GGAUGUGAAGCAGUUCUAU 594-614). Briefly, a third-generation lentiviral packaging system pND-CAG/GFP/WPRE containing shRNA expression cassette targeting HCV IRES, NS5b region, or viral receptor CD81 was used to produce high-titer VSV-G-pseudotyped lentiviral vectors in HEK293 cells. Vector supernatants were removed 36 and 48 h post-transfection, passed through a 0.45-μM filter, and concentrated 1,000-fold by ultracentrifugation. Plasmid SHC005 containing both shGFP cassette and puromycin selectable marker was obtained from Erasmus Center for Biomics (Rotterdam, The Netherlands). LV-shGFP was produced in the third-generation lentiviral packaging system as mentioned above.
In vitro LV transductions, GFP, and CD81 analysis
Concentrated LV stocks were titrated using HEK293 cells 24 h after infection, with transduction efficiency based on the number of GFP-positive cells as determined by flow cytometry (FACSCalibur; BD BioSciences, Mountain View, CA, USA). Transduction efficiency of LV-GFP in the presence of IFN-α was tested using this method in Huh7 cells 3 days post-infection. Transduction efficiency of LV-shIRES and LV-shNS5b in Huh7-ET cells was also tested by this method. Pegylated IFN-α 2a (dissolved in water) was provided by Roche (Basel, Switzerland). CD81 expression was determined using flow cytometry by staining with phycoerythrin conjugated mouse antihuman CD81 monoclonal antibody (BD Pharmingen, San Diego, USA). Mouse IgG1 was used as isotype-matched control antibody (BD Pharmingen).
In vivo LV transductions in mice
Eleven-week-old female NOD/LtSz-scid/scid (NOD/SCID) mice were injected intravenously with suboptimal dose of LV-GFP (5 × 106 transducing units in 0.25 ml PBS) in the tail vein. Three mice were injected subcutaneously with pegylated IFN-α 2b (Intron A, Schering-Plough, Kenilworth, NJ, USA) at 6 h before and 72 h after intravenous administration of LV-GFP. The dose of peg-IFN-α per injection was 30 μg/kg body weight, as previously described [26]. Two mice were injected with vector alone and one animal served as negative control. After 7 days, mice were killed and livers were harvested. Mouse liver tissue was digested by collagenase type IV (Sigma, Zwijndrecht, The Netherlands) for 30 min at 37°C in order to generate a single-cell suspension. Transduction efficiency was determined by the percentage of GFP-positive hepatocytes using flow cytometric analysis (FACSCalibur, BD BioSciences).
Luciferase assay
Luciferin potassium salt (100 mM; Sigma) was added to Huh7-ET cells and incubate for 30 min at 37°C. Luciferase activity was quantified using a LumiStar Optima luminescence counter (BMG LabTech, Offenburg, Germany).
Statistical analysis
Statistical analysis was performed by using either matched pair nonparametric test (Wilcoxon signed-rank test) or the nonpaired, nonparametric test (Mann–Whitney test; GraphPad Prism software). P values less than 0.05 were considered as statistically significant.
Results
Potent inhibition of HCV replication by lentiviral vector-mediated RNAi
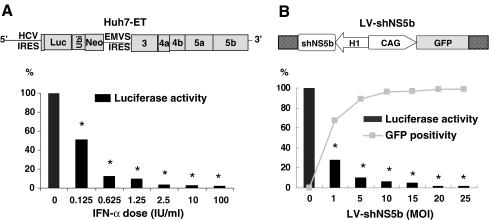
A subgenomic replicon cell line (Huh7-ET), containing HCV nonstructural sequences and a luciferase reporter gene [25, 27], was used to determine the antiviral potency of IFN-α and RNAi (Fig. 1a). HCV replication was monitored by measuring luciferase activity. Robust reduction of luciferase activity (97 ± 2% inhibition, mean ± SD, n = 9) was observed from 2.5 to 100 IU/ml of IFN-α.
Fig. 1.
Inhibition of HCV replication by IFN-α or LV-shNS5B. a Huh7-ET replicon was used for testing HCV replication by monitoring luciferase activity. IFN-α treatment inhibits viral replication in a dose-dependent manner. Profound reduction of luciferase activity (97 ± 2% inhibition, mean ± SD, n = 9) was observed from 2.5 to 100 IU/ml concentration of IFN-α. b LV-shNS5b contains both GFP reporter gene and shRNA targeting HCV were tested. Huh7-ET treated with increasing dose of LV-shNS5b resulted in higher levels of transduction efficiency and inhibition of HCV replication, monitored by GFP-positive population and luciferase activity, respectively. Maximum inhibition of HCV replication was observed at high dose (20 or 25 MOI) by 98 ± 3% (mean ± SD, n = 6). *P < 0.01 (Wilcoxon test) significantly different from untreated conditions
LV-shNS5b vector containing both GFP and shRNA targeting HCV NS5b sequence was tested. Huh7-ET treated with increasing dose of LV-shNS5b resulted in higher levels of transduction efficiency and inhibition of HCV replication as monitored by GFP reporter gene expression and luciferase activity, respectively. Maximum inhibition of HCV replication was observed at multiplicity of infection (MOI) of 20 and 25 by 98 ± 3% (mean ± SD, n = 6; Fig. 1b). The results indicate that RNAi-mediated inhibition of HCV replication can be as effective as IFN-α in Huh7-ET model.
Exogenous IFN-α has no negative impact on lentiviral transduction in vitro and in vivo
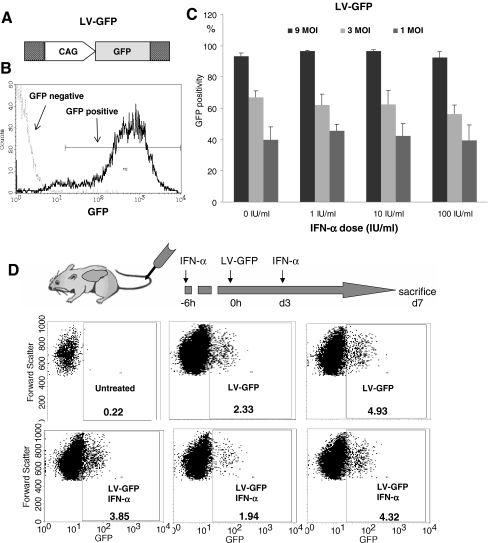
To address whether exogenous IFN-α has a negative impact on lentiviral transduction, a vector transduction assay was performed by infecting Huh7 cells with LV-GFP vector (Fig. 2a) in the absence or presence of 1, 10, or 100 IU/ml of IFN-α. At day 3, transduction efficiency was assessed by flow cytometry for GFP-positive cells (Fig. 2b). When compared with nontreated control group (93 ± 2%, mean ± SD, n = 4), 1, 10, or 100 IU/ml concentration of IFN-α had no significant effect on vector transduction at MOI of 9 (97 ± 0.4%, 97 ± 0.9%, and 93 ± 4%, respectively; P > 0.05). Similar results were observed at medium (3 MOI) and low (1 MOI) vector concentrations (Fig. 2c). Also, pretreatment of cells with IFN-α for 24 h did not influence vector transduction (data not shown).
Fig. 2.
Effect of exogenous IFN-α on lentiviral vector transductions in vitro and in mice. a LV-GFP vector expressing GFP under control of CAG promoter was used for transduction of Huh7 cells. b Representative histogram of GFP expression determined by flow cytometry of control cells and transduced cells 3 days after culture. c No significant (P > 0.05) differences of transduction efficiency, compared with nontreated control group, were observed with 1, 10, or 100 IU/ml of IFN-α for high (9 MOI), intermediate (3 MOI), and low (1 MOI) vector concentrations. Shown is the mean ± SD of four independent experiments. MOI multiplicity of infection. d Mice treated with exogenous IFN-α 6 h before and 3 days after administration of LV-GFP (suboptimal dose, 5 × 106 transducing units) showed comparable transduction efficiency in the liver (1.7% to 4.1% transduced hepatocytes), compared with the group injected with vector only (2.1% and 4.7% transduced hepatocytes). Percentage of GFP-positive cells was indicated in the FACS picture for each individual mouse
To further investigate the effects of exogenous IFN-α on lentiviral transductions in vivo, mice were treated with peg-IFN-α 6 h before and 72 h after to intravenous administration of LV-GFP. As shown in Fig. 2d, there was no significant effect of peg-IFN-α treatment on vector transduction in mice livers (1.7% to 4.1% transduced hepatocytes) compared to untreated transduction group injected with vector only (2.1% and 4.7% transduced hepatocytes).
IFN-α does not interfere with RNAi-mediated host gene silencing
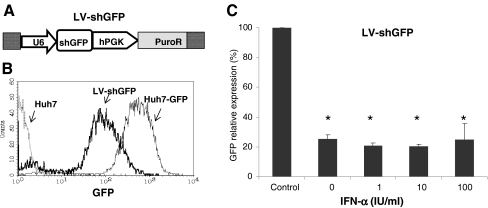
In the setting of combining RNAi with IFN-α, a critical concern would be the influence of exogenous IFN-α on RNAi gene silencing efficiency. To evaluate this issue, a lentiviral vector expressing shRNA to target report gene GFP (LV-shGFP; Fig. 3a) was produced, and gene-silencing efficacy was determined in the absence or presence of IFN-α. First, a stable GFP expressed cell line (Huh7-GFP) was created by infecting naive Huh7 cells with LV-GFP and expanded for several passages. Huh7-GFP cells were treated with LV-shGFP with or without IFN-α. LV-shGFP significantly inhibited GFP mean fluorescence intensity in Huh7-GFP cells (Fig. 3b) by 74.7 ± 2.6% (mean ± SD, n = 3, P < 0.01). Treatment of 1, 10, or 100 IU/ml IFN-α did not significantly interfere with LV-shGFP-mediated GFP knockdown (79.1 ± 1.9%, 79.5 ± 1.4%, and 74.9 ± 10%, respectively, P > 0.05; Fig. 3c).
Fig. 3.
IFN-α does not interfere with RNAi-mediated knockdown of GFP. a LV-shGFP vector containing a shGFP cassette driven by U6 promoter were used. b Control (Huh7) and stable GFP expressing cell line (Huh7-GFP) were treated with LV-shGFP. GFP expression was measured by flow cytometry 3 days post- transductions, clearly showing inhibition of GFP expression by LV-shGFP in Huh7-GFP cells. c The relative GFP expression based on the mean fluorescence intensity. LV-shGFP significantly inhibited GFP expression by 74.7 ± 2.6% (P < 0.01), and treatment of 1, 10, or 100 IU/ml of IFN-α did not interfere with the knockdown by LV-shGFP. Shown is the mean ± SD of three independent experiments (*P < 0.01)
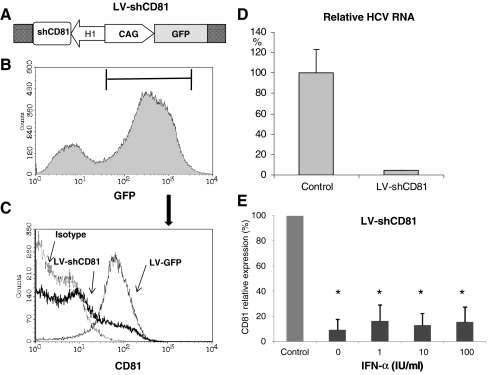
To further confirm these findings, LV-shCD81 was used to target HCV entry receptor CD81. The LV-shCD81 vector contains a GFP reporter gene to track transduction efficiencies (Fig. 4a). CD81 knockdown was determined by flow cytometry based on the GFP-positive population (Fig. 4b, c). LV-shCD81, compared with LV-GFP control vector, significantly reduced CD81 expression by 90.8 ± 8.1% (mean ± SD, n = 3, P < 0.01). Importantly, knockdown of CD81 in Huh7 cells dramatically reduced infection by the JFH1-derived infectious HCV particles (Fig. 4d). As shown in Fig. 4e, LV-shCD81 retained robust gene silencing efficacy in the IFN-α (1, 10, or 100 IU/ml) treatment groups (83.9 ± 12.9%, 87.2 ± 9.6%, and 84.3 ± 11.6%, respectively). Similar results were obtained using other hepatoma cell line, Huh6 (data not shown). Taken together, these results indicate that exogenous IFN-α does not interfere with RNAi-mediated gene silencing.
Fig. 4.
IFN-α does not interfere with RNAi-mediated silencing of CD81. a LV-shCD81 vector containing a GFP reporter gene and shRNA targeting CD81 was used to transduce Huh7 cells. LV-GFP vector without shRNA cassette was used as control. b Three days after transfection, GFP-positive transduced cells were gated to determine CD81 expression. c Representative CD81 expression histogram of GFP-positive cells is shown. Isotype-matched control antibody staining of LV-shCD81 cells was included as negative control. d Knockdown of CD81 in Huh7 cells profoundly reduced HCV infection. Three days after LV-shCD81 transduction, Huh7 cells were exposed to JFH1-derived infectious HCV particles for 6 h. Three days after infection, the LV-shCD81 treated cells showed a clear reduction in intracellular viral RNA levels as determined by quantitative RT-PCR. e Relative CD81 expression was calculated based on mean fluorescence intensity. LV-shCD81 significantly reduced CD81 expression by 90.8 ± 8.1% (P < 0.01), compared to LV-GFP cells. LV-shCD81 retained robust gene silencing efficacy at different concentrations of IFN-α. Shown is the mean ± SD of three independent experiments (*P < 0.01)
Simultaneous and sequential combined antiviral activity of IFN-α and RNAi
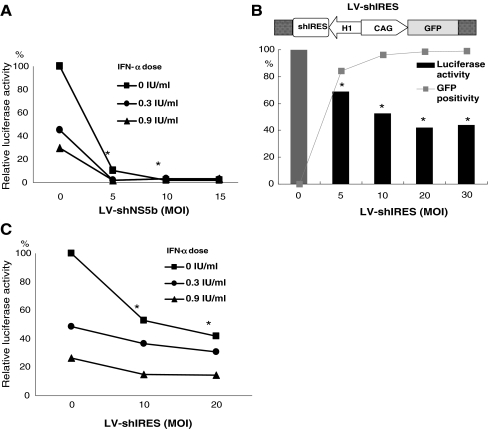
The finding that IFN-α did not affect transduction efficacy and gene silencing prompted the question whether IFN and RNAi have combined antiviral effects on HCV replication. As shown in Fig. 1b, LV-shNS5b has potent antiviral activity. Combination of low-dose IFN-α (<1 IU/ml) with low vector dose (MOI ≤ 5) resulted in a significant enhanced antiviral effect (Fig. 5a). For example, combination of 1 MOI vector with 0.9 IU/ml IFN-α showed 92.1 ± 8.1% (mean ± SD, n = 6, P < 0.01) inhibition versus 72.2 ± 9.4% with vector alone or 71.5 ± 4.1% with IFN-α alone. Real-time quantitative PCR confirmed the results in this group of treatments (data not shown). At high-dose vector (MOI ≥ 10), inhibition of HCV replication was nearly complete, and therefore, no significant combinational effect with IFN could be observed.
Fig. 5.
Enhanced inhibition of HCV replication by simultaneous treatment with IFN-α and RNAi. a The structure and action of LV-shNS5b on HCV replication have been shown in Fig. 1b. The combination of low-dose IFN-α (<1 IU/ml) with low-dose vector (≤10 MOI) resulted in enhanced antiviral effects. For example, combination of 1 MOI vector with 0.9 IU/ml IFN-α showed 92.1% ± 8.1 inhibition, versus 72.2% ± 9.4 with vector alone or 71.5 ± 4.1% with IFN-α alone (mean ± SD, P < 0.01). Also, with 5 MOI, a significant combinational effect was observed (P < 0.05), but at higher MOIs, inhibition of HCV replication was nearly complete, and no significant additive effect of IFN-α was observed. b LV-shIRES vector containing GFP reporter gene and shRNA targeting HCV IRES was used to transduce Huh7-ET cells. Increasing dose of LV-shIRES resulted in higher levels of transduction efficiency and inhibition of HCV replication, shown by GFP positivity and luciferase activity, respectively. c Combining low-dose IFN-α with LV-shIRES resulted in enhanced inhibition of HCV replication at each combined condition. Shown is the mean ± SD of six independent experiments (*P < 0.05)
In order to more clearly show the combined antiviral effect of IFN-α and RNAi, a less potent vector containing both GFP and shRNA targeting HCV IRES was used (Fig. 5b). Huh7-ET treated with increasing dose of LV-shIRES vector resulted in higher levels of transduction efficiency and inhibition of HCV replication, monitored by GFP-positive population and luciferase activity, respectively. There was a clear threshold for the level of HCV silencing observed at vector dose of MOI 20 and higher (Fig. 5b). Similar to the results with LV-shNS5B vector, a combination of low-dose IFN-α with LV-shIRES resulted in enhanced inhibition of HCV replication at each combined condition. For example, the combination of LV-shIRES (MOI 20) with 0.9 IU/ml IFN-α enhanced inhibition of HCV replication to 86.8% ± 3.4 compared to vector alone (56.4%, n = 6, P < 0.001) or IFN-α alone (72.5%, P < 0.05; Fig. 5c). Real-time quantitative PCR confirmed the results in this group of treatments (data not shown).
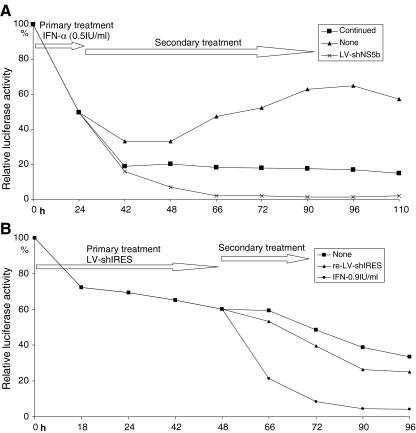
To further investigate this combined antiviral activity, Huh7-ET cells were sequentially treated with IFN-α and RNAi. Cells were treated with suboptimal dose of IFN-α (0.5 IU/ml) for 24 h followed by a secondary treatment with either LV-shNS5b or again with IFN-α. As shown in Fig. 6a, cells were more sensitive to subsequent treatment with RNAi than to re-treatment with IFN-α. Maximal inhibition of HCV replication with IFN-α was 81.3% ± 1.3 (n = 7) versus 98.2% ± 0.2 inhibition (n = 6) with LV-shNS5b (P < 0.001). This indicates that HCV treated with IFN-α remains sensitive to RNAi, more so than to additional IFN-α. Conversely, treatment of LV-shIRES led to partial inhibition of viral replication and was more sensitive to secondary treatment with IFN-α than re-treatment of the same vector (Fig. 6b). At 48 h after the initial treatment of LV-shIRES, additional treatment of IFN-α (0.9 IU/ml) resulted in 96.0% ± 0.6 inhibition of viral replication versus 75.1% ± 3.7 inhibition after re-treatment with LV-shIRES (n = 6, P < 0.001).
Fig. 6.
Subsequential treatment of IFN-α and RNAi reciprocally enhances inhibition of HCV replication. a Huh7-ET cells were treated with low dose (0.5 IU/ml) of IFN-α 24 h after which medium was replaced, and cells were treated a second time for an additional 86 h. Secondary treatment with IFN-α resulted in a maximum inhibition of 81.3% ± 1.3 (n = 7, P < 0.001) from t = 42 h onward. However, subsequently switching IFN-α to LV-shNS5b resulted in a significantly greater inhibition of viral replication (98.2% ± 0.2 inhibition, n = 6, P < 0.001). In Huh7-ET cells without secondary treatment, HCV replication was partially restored to approx. two third of baseline levels at 2 days after switching. b Conversely, Huh7-ET cells primarily treated with LV-shIRES were more sensitive to a secondary treatment with IFN-α than re-treatment with the same LV-shIRES vector. Secondary treatment with 0.9 IU/ml IFN-α resulted in profound inhibition of viral replication (96% ± 0.6 inhibition at t = 96 h, n = 6, P < 0.001) as compared to secondary treatment with vector (75.1% ± 3.7 inhibition, n = 6). In Huh7-ET, cells without additional treatment HCV replication was (66.4% ± 5.4 inhibition, n = 6). Overall, these findings indicate that cells treated with IFN-α are more sensitive to subsequential treatment with RNAi than re-treatment with IFN-α. Equally, cells treated with RNAi are more sensitive to subsequential treatment with IFN-α than re-treatment with RNAi
Taken together, these results indicate that RNAi and IFN-α act independently on HCV replication and that combination of these agents resulted in an enhanced antiviral effect. Furthermore, IFN-α and RNAi appear complementary, suggesting that viral resistance to treatment of IFN-α remains sensitive to treatment with RNAi and, reversely, that viral resistance to RNAi remains sensitive to inhibition by IFN-α.
Discussion
Peg-IFN-α in combination with ribavirin is currently the standard therapy for chronic HCV leading to a sustained viral response in approximately half of patients with the common genotypes [28]. In order to improve treatment outcomes and to provide opportunities to treat previous nonresponders, it is urgent to exploit alternative therapies. RNAi represents as a novel antiviral strategy that could be effective for HCV. To be applicable in therapeutic setting, it is critical that RNAi induces long-term and stable gene silencing. Since integrating lentiviral vectors have shown great advantages in transgenic delivery [29], third-generation LV are currently the best option for stable shRNA delivery. We found that LV and RNAi can be applied in combination with IFN-α without effecting transduction to hepatocytes or interfering with RNAi-mediated gene silencing.
The action of RNAi technology is clearly different from conventional antiviral compounds. Particularly when delivered by a HIV derived lentiviral vector, the first concern would be whether the vector itself could have complicated interaction with innate immune system, especially IFN response. The induction of an endogenous IFN response by shRNA expressing vectors is dependent on the cell type and the sequence. Studies have demonstrated that human dendritic cells can mount an IFN response after exposure to wild-type HIV or lentiviral vectors [30–32]. To our knowledge, our current study is the first report on the effect of exogenous IFN-α on lentiviral transduction and RNAi. A recent study has shown that lentiviral vector triggers a type I interferon (IFN-α/β) response that restricts hepatocyte gene transfer and promotes vector clearance in mice [24]. These mouse experiments contradict our experiment in NOD/SCID mice where no negative effect of exogenous peg-IFN-α treatment was observed on transduction of hepatocytes (Fig. 2d). Also, previous studies by others [33, 34] and our group [13] showed no in vitro evidence of endogenous type I interferon responses after transduction with lentiviral vectors. The fact that exogenous IFN-α does not have an apparent impact on LV transductions makes a combined approach feasible.
In the setting of combining RNAi with IFN-α, the most critical issue would be the influence of exogenous IFN-α on gene silencing efficiency. This study has clearly shown that IFN-α does not have significant negative effects on RNAi-mediated silence of both GFP report gene and native host gene CD81. Down regulation of viral-binding co-receptor CD81 expression was previously shown to reduce HCV E2 envelope binding [35]. Now, we (Fig. 4d) and others [36] have shown RNAi-mediated reduction of CD81 also inhibit infection of replication competent HCV virus, JFH1. Therefore, it is conceivable that lentiviral-mediated RNAi has the potential not only to inhibit viral replication but also to prevent infection. Most importantly, a simultaneous combination of IFN-α with RNAi directly targeting viral genome (IRES or NS5b region) significantly enhanced their individual antiviral effects. Furthermore, IFN-α and RNAi clearly acted complementary shown by the fact that cells treated with IFN-α were more sensitive to subsequent treatment with RNAi than to re-treatment with IFN-α. Conversely, treatment of LV-shIRES led to partial inhibition of viral replication and was more sensitive to secondary treatment with IFN-α than re-treatment of the same vector (Fig. 6b). These findings could suggest that HCV that develops resistance to treatment with IFN-α would still be sensitive to RNAi therapy and, reversely, that potential viral resistance developed against RNAi would still be susceptible to inhibition by IFN-α.
Owing to the high risk of escape and resistance development, combination of RNAi with IFN-α may be necessary to completely cure HCV infection by attacking the virus in two distinct ways. In particular, this approach possesses unique advantages in preventing and treating HCV recurrence after liver transplantation. Lentiviral-mediated RNAi could be used to modify a donor graft and prevent or slow HCV recurrence after transplantation; meanwhile, low-dose IFN-α can be used to systemically treat HCV enhancing the therapeutic effects of both. Additional combinations with ribavirin are conceivable in order to further enhance therapeutic effects. Our preliminary results show that ribavirin has no negative effects on the action of RNAi (data not shown).
In conclusion, in this study, we found that exogenous IFN-α had no significant negative influence on lentiviral transduction. In the presence of IFN-α, RNAi-mediated gene knockdown was unaffected, and moreover, a combination of viral genome targeted RNAi with IFN-α achieved enhanced antiviral effects. Therefore, this novel combination strategy may offer the potential to eliminate HCV infection in chronically infected patients.
Acknowledgments
The authors would like to thank Prof. Ralf Bartenschlager and Dr. Volker Lohmann (University of Heidelberg, Germany) for generously providing the Huh7 and Huh6 subgenomic HCV replicon cells, Prof. Takaji Wakita (National Institute of Infectious Diseases, Japan) for providing the full-length JFH1-derived infectious HCVcc, Prof. Gerard Wagemaker (Erasmus MC, Rotterdam) for providing the mice, and Pascal van der Wegen (Erasmus MC, Rotterdam) for technical support. We thank the Erasmus MC Translational Research Fund, the Liver Research Foundation (SLO) Rotterdam, for funding this research.
Conflict of interest statement The authors declare that they have no competing financial interests.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- HCV
Hepatitis C virus
- IFN-α
Interferon-alpha
- RNAi
RNA interference
- siRNA
Small interfering RNA
- shRNA
Small hairpin RNA
- LV
Lentiviral vector
- GFP
Green fluorescent protein
References
- 1.Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK (2001) Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958–965 [DOI] [PubMed]
- 2.Keating GM, Plosker GL (2005) Peginterferon alpha-2a (40KD) plus ribavirin: a review of its use in the management of patients with chronic hepatitis C and persistently ‘normal’ ALT levels. Drugs 65:521–536 [DOI] [PubMed]
- 3.Pan QW, Henry SD, Scholte BJ, Tilanus HW, Janssen HL, van der Laan LJ (2007) New therapeutic opportunities for hepatitis C based on small RNA. World J Gastroenterol 13:4431–4436 [DOI] [PMC free article] [PubMed]
- 4.Prabhu R, Garry RF, Dash S (2006) Small interfering RNA targeted to stem-loop II of the 5′ untranslated region effectively inhibits expression of six HCV genotypes. Virol J 3:100 [DOI] [PMC free article] [PubMed] [Retracted]
- 5.Hamazaki H, Ujino S, Abe E, Miyano-Kurosaki N, Shimotohno K, Takaku H (2004) RNAi expression mediated inhibition of HCV replication. Nucleic Acids Symp Ser 48:307–308 [DOI] [PubMed]
- 6.Ilves H, Kaspar RL, Wang Q, Seyhan AA, Vlassov AV, Contag CH, Leake D, Johnston BH (2006) Inhibition of hepatitis C IRES-mediated gene expression by small hairpin RNAs in human hepatocytes and mice. Ann N Y Acad Sci 1082:52–55 [DOI] [PubMed]
- 7.Kanda T, Steele R, Ray R, Ray RB (2007) Small interfering RNA targeted to hepatitis C virus 5′ nontranslated region exerts potent antiviral effect. J Virol 81:669–676 [DOI] [PMC free article] [PubMed]
- 8.Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, Perelson AS (1998) Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103–107 [DOI] [PubMed]
- 9.Wilson JA, Jayasena S, Khvorova A, Sabatinos S, Rodrigue-Gervais IG, Arya S, Sarangi F, Harris-Brandts M, Beaulieu S, Richardson CD (2003) RNA interference blocks gene expression and RNA synthesis from hepatitis C replicons propagated in human liver cells. Proc Natl Acad Sci U S A 100:2783–2788 [DOI] [PMC free article] [PubMed]
- 10.Koutsoudakis G, Herrmann E, Kallis S, Bartenschlager R, Pietschmann T (2007) The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J Virol 81:588–598 [DOI] [PMC free article] [PubMed]
- 11.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM (2007) Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801–805 [DOI] [PubMed]
- 12.Bartosch B, Verney G, Dreux M, Donot P, Morice Y, Penin F, Pawlotsky JM, Lavillette D, Cosset FL (2005) An interplay between hypervariable region 1 of the hepatitis C virus E2 glycoprotein, the scavenger receptor BI, and high-density lipoprotein promotes both enhancement of infection and protection against neutralizing antibodies. J Virol 79:8217–8229 [DOI] [PMC free article] [PubMed]
- 13.Henry SD, van der Wegen P, Metselaar HJ, Tilanus HW, Scholte BJ, van der Laan LJ (2006) Simultaneous targeting of HCV replication and viral binding with a single lentiviral vector containing multiple RNA interference expression cassettes. Mol Ther 14:485–493 [DOI] [PubMed]
- 14.Meuleman P, Hesselgesser J, Paulson M, Vanwolleghem T, Desombere I, Reiser H, Leroux-Roels G (2008) Anti-CD81 antibodies can prevent a hepatitis C virus infection in vivo. Hepatology 48:1761–1768 [DOI] [PubMed]
- 15.Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L (1998) A third-generation lentivirus vector with a conditional packaging system. J Virol 72:8463–8471 [DOI] [PMC free article] [PubMed]
- 16.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Zhang M, Ihrig MM, McManus MT, Gertler FB, Scott ML, Van Parijs L (2003) A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet 33:401–406 [DOI] [PubMed]
- 17.Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N, Feinstone S, Halfon P, Inchauspe G, Kuiken C, Maertens G, Mizokami M, Murphy DG, Okamoto H, Pawlotsky JM, Penin F, Sablon E, Shin IT, Stuyver LJ, Thiel HJ, Viazov S, Weiner AJ, Widell A (2005) Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962–973 [DOI] [PubMed]
- 18.Wilson JA, Richardson CD (2005) Hepatitis C virus replicons escape RNA interference induced by a short interfering RNA directed against the NS5b coding region. J Virol 79:7050–7058 [DOI] [PMC free article] [PubMed]
- 19.Ji J, Glaser A, Wernli M, Berke JM, Moradpour D, Erb P (2008) Suppression of short interfering RNA-mediated gene silencing by the structural proteins of hepatitis C virus. J Gen Virol 89:2761–2766 [DOI] [PubMed]
- 20.Wang Y, Kato N, Jazag A, Dharel N, Otsuka M, Taniguchi H, Kawabe T, Omata M (2006) Hepatitis C virus core protein is a potent inhibitor of RNA silencing-based antiviral response. Gastroenterology 130:883–892 [DOI] [PubMed]
- 21.ter Brake O, Konstantinova P, Ceylan M, Berkhout B (2006) Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol Ther 14:883–892 [DOI] [PubMed]
- 22.Grimm D, Kay MA (2007) Combinatorial RNAi: a winning strategy for the race against evolving targets? Mol Ther 15:878–888 [DOI] [PMC free article] [PubMed]
- 23.Ter Brake O, Legrand N, von Eije KJ, Centlivre M, Spits H, Weijer K, Blom B, Berkhout B (2009) Evaluation of safety and efficacy of RNAi against HIV-1 in the human immune system (Rag-2(-/-)(c)(-/-)) mouse model. Gene Ther 16:149–153 [DOI] [PubMed]
- 24.Brown BD, Sitia G, Annoni A, Hauben E, Sergi LS, Zingale A, Roncarolo MG, Guidotti LG, Naldini L (2007) In vivo administration of lentiviral vectors triggers a type I interferon response that restricts hepatocyte gene transfer and promotes vector clearance. Blood 109:2797–2805 [DOI] [PubMed]
- 25.Frese M, Schwarzle V, Barth K, Krieger N, Lohmann V, Mihm S, Haller O, Bartenschlager R (2002) Interferon-gamma inhibits replication of subgenomic and genomic hepatitis C virus RNAs. Hepatology 35:694–703 [DOI] [PubMed]
- 26.Inoue K, Umehara T, Ruegg UT, Yasui F, Watanabe T, Yasuda H, Dumont JM, Scalfaro P, Yoshiba M, Kohara M (2007) Evaluation of a cyclophilin inhibitor in hepatitis C virus-infected chimeric mice in vivo. Hepatology 45:921–928 [DOI] [PubMed]
- 27.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R (1999) Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113 [DOI] [PubMed]
- 28.Feld JJ, Hoofnagle JH (2005) Mechanism of action of interferon and ribavirin in treatment of hepatitis C. Nature 436:967–972 [DOI] [PubMed]
- 29.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D (1996) In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267 [DOI] [PubMed]
- 30.Beignon AS, McKenna K, Skoberne M, Manches O, DaSilva I, Kavanagh DG, Larsson M, Gorelick RJ, Lifson JD, Bhardwaj N (2005) Endocytosis of HIV-1 activates plasmacytoid dendritic cells via Toll-like receptor-viral RNA interactions. J Clin Invest 115:3265–3275 [DOI] [PMC free article] [PubMed]
- 31.Fonteneau JF, Larsson M, Beignon AS, McKenna K, Dasilva I, Amara A, Liu YJ, Lifson JD, Littman DR, Bhardwaj N (2004) Human immunodeficiency virus type 1 activates plasmacytoid dendritic cells and concomitantly induces the bystander maturation of myeloid dendritic cells. J Virol 78:5223–5232 [DOI] [PMC free article] [PubMed]
- 32.Breckpot K, Emeagi P, Dullaers M, Michiels A, Heirman C, Thielemans K (2007) Activation of immature monocyte-derived dendritic cells after transduction with high doses of lentiviral vectors. Hum Gene Ther 18:536–546 [DOI] [PubMed]
- 33.Gonzalez-Alegre P, Bode N, Davidson BL, Paulson HL (2005) Silencing primary dystonia: lentiviral-mediated RNA interference therapy for DYT1 dystonia. J Neurosci 25:10502–10509 [DOI] [PMC free article] [PubMed]
- 34.Cave E, Weinberg MS, Cilliers T, Carmona S, Morris L, Arbuthnot P (2006) Silencing of HIV-1 subtype C primary isolates by expressed small hairpin RNAs targeted to gag. AIDS Res Hum Retroviruses 22:401–410 [DOI] [PubMed]
- 35.McKeating JA, Zhang LQ, Logvinoff C, Flint M, Zhang J, Yu J, Butera D, Ho DD, Dustin LB, Rice CM, Balfe P (2004) Diverse hepatitis C virus glycoproteins mediate viral infection in a CD81-dependent manner. J Virol 78:8496–8505 [DOI] [PMC free article] [PubMed]
- 36.Molina S, Castet V, Pichard-Garcia L, Wychowski C, Meurs E, Pascussi JM, Sureau C, Fabre JM, Sacunha A, Larrey D, Dubuisson J, Coste J, McKeating J, Maurel P, Fournier-Wirth C (2008) Serum-derived hepatitis C virus infection of primary human hepatocytes is tetraspanin CD81 dependent. J Virol 82:569–574 [DOI] [PMC free article] [PubMed]