Abstract
Among secreted phospholipases A2 (sPLA2s), human group X sPLA2 (hGX sPLA2) is emerging as a novel attractive therapeutic target due to its implication in inflammatory diseases. To elucidate whether hGX sPLA2 plays a causative role in coronary artery disease (CAD), we screened the human PLA2G10 gene to identify polymorphisms and possible associations with CAD end-points in a prospective study, AtheroGene. We identified eight polymorphisms, among which, one non-synonymous polymorphism R38C in the propeptide region of the sPLA2. The T-512C polymorphism located in the 5′ untranslated region was associated with a decreased risk of recurrent cardiovascular events during follow-up. The functional analysis of the R38C polymorphism showed that it leads to a profound change in expression and activity of hGX sPLA2, although there was no detectable impact on CAD risk. Due to the potential role of hGX sPLA2 in inflammatory processes, these polymorphisms should be investigated in other inflammatory diseases.
Keywords: Secreted phospholipase A2, hGX sPLA2, PLA2G10, Polymorphisms, CAD
Introduction
Secreted phospholipases A2 (sPLA2) form a growing family of enzymes that hydrolyze the sn-2 position of glycerophospholipids to generate proinflammatory lipid mediators, lysophospholipids, and free fatty acids. Ten enzymatically active sPLA2s have been identified in mammals. Increased sPLA2 activity has been shown to be an independent risk factor for coronary artery disease (CAD) [1], and group IIA, III, V, and X sPLA2s have been detected in human atherosclerotic lesions [2–5]. The tissue-specific expression pattern and the enzymatic properties of the various sPLA2s suggest that the cellular role of each of these enzymes is different [6]. Studies with transgenic and knockout mice for different sPLA2s support the above hypothesis [4, 7–10].
Among sPLA2s, the role of human group X sPLA2 (hGX sPLA2) is emerging as several studies show its implication in various inflammatory diseases [8, 9, 11–13]. The gene encoding hGX sPLA2 (PLA2G10) maps to human chromosome 16p13.1-p12. A 1.5-Kb transcript was found in the thymus, spleen, and leukocytes, indicating a possible role for hGX sPLA2 in immunity and/or inflammation [14]. One of the key features of hGX sPLA2 is its high catalytic activity towards phosphatidylcholine (PC), the major phospholipid of cell membranes and lipoproteins. This substrate preference is due to the presence of a tryptophan residue on the interfacial binding surface of the enzyme [15]. On the basis of the structural similarities between platelet-activating factor (PAF) and PC, we demonstrated that hGX sPLA2, but not other sPLA2s, can efficiently hydrolyze PAF when it is incorporated into large unilamellar PC phospholipid vesicles and when it is present in PC-rich lipoproteins [16]. This suggests that hGX sPLA2 may be a novel player in PAF regulation during inflammatory processes. hGX sPLA2 can also efficiently hydrolyze lipoproteins (low-density lipoprotein (LDL) and high-density lipoprotein (HDL)), preferentially releasing arachidonic and linoleic acids [17, 18]. Finally, we have recently shown that hGX sPLA2 is present in human atherosclerotic lesions and that the hydrolysis of LDL by hGX sPLA2 results in a modified LDL particle that induces lipid accumulation in human monocyte-derived macrophages [2].
Recent studies using PLA2G10-deficient mice in a model of ischemia reperfusion showed that deficiency in GX sPLA2 leads to a reduction in myocardial ischemia reperfusion injury as neutrophil functions were suppressed [11]. Several studies implicated hGX sPLA2 in the pathogenesis of inflammatory diseases such as asthma [9], and recent data support the idea that hGX sPLA2 may be an important therapeutic target in diseases where inflammation plays an important role, including atherosclerosis.
The aim of the present study was to examine the variability of the PLA2G10 gene and to determine whether it plays a role in CAD and its complications. In addition, we examined the potential functional impact of the non-synonymous R38C polymorphism on the expression and activity of the enzyme.
Materials and methods
Study populations
The SIPLAC study The SIPLAC study was specifically designed to investigate the variability of candidate genes for myocardial infarction (MI) in groups highly contrasted for disease risk and to further characterize the structure of linkage disequilibrium (LD) within these genes in order to select the polymorphisms that will be genotyped in larger studies. It is derived from Etude Cas-Témoins de l’Infarctus du Myocarde (ECTIM), a case–control study of MI based on the multinational monitoring of trends and determinants in cardiovascular disease project registers in the UK and France [19]. The entire ECTIM study comprised 1,332 MI patients and 1,490 controls (62% from UK, 21% female). For the SIPLAC sub-study, cases with MI and a parental history of MI (n = 312, 35% females, mean age: 56 ± 16 years) and controls without CAD and without parental history of MI (n = 317, 42% females, mean age: 59 ± 15 years) were selected from the samples recruited in Belfast and Glasgow. All participants were of European descent and gave an informed consent.
The AtheroGene study Baseline characteristics are provided in Table 1; whereas, detailed description of the study has been provided elsewhere [20, 21]. Briefly, between November 1996 and June 2000, 1,303 CAD patients (75% males, mean age: 61.7 ± 0.3 years) were recruited at the Department of Medicine II of the Johannes Gutenberg University Mainz and the Bundeswehrzentralkrankenhaus Koblenz at the occasion of a diagnostic coronary angiography. A priori inclusion criterion was the presence of a diameter stenosis >30% in at least one major coronary artery. Exclusion criteria were evidence of significant concomitant diseases, in particular, hemodynamically significant valvular heart disease, known cardiomyopathy, and malignant diseases, as well as febrile conditions. Patients were followed-up during a median period of 6.2 years. Follow-up information was obtained about death from cardiovascular causes and non-fatal MI (n = 235). Information about the cause of death or clinical events was obtained by hospital or general practitioner. Healthy control subjects (n = 484, 73% males, mean age 60.0 ± 0.4 years) were recruited either from general practitioners’ offices in the course of a routine check-up or by newspaper announcement. Study participants had German nationality, were inhabitants of the Rhein-Main area, and were of Caucasian origin. The study was approved by the ethics committee of the University of Mainz. Each participant gave written informed consent.
Table 1.
Baseline characteristics of patients with coronary artery disease in the AtheroGene study according to occurrence of cardiovascular event (cardiovascular death or non-fatal myocardial infarction) during follow-up
| Cardiovascular event | |||
|---|---|---|---|
| No (n = 1,039) | Yes (n = 235) | p value | |
| Age, years | 61.0 (0.3) | 63.8 (0.6) | <0.0001 |
| Females, % | 25.1 | 25.5 | 0.90 |
| Body mass index | 27.2 (0.1) | 27.3 (0.2) | 0.68 |
| Current smoker, % | 12.3 | 16.6 | 0.08 |
| Diabetes, % | 14.6 | 22.1 | 0.005 |
| Hypertension, % | 72.6 | 74.0 | 0.65 |
| Statin, % | 36.5 | 29.4 | 0.04 |
| Beta-blocker, % | 59.9 | 54.5 | 0.13 |
| Angiotensin-converting enzyme inhibitors, % | 45.9 | 53.2 | 0.04 |
| Total cholesterol, mg/dL | 220.9 (1.4) | 214.5 (2.9) | 0.05 |
| LDL cholesterol, mg/dL | 141.6 (1.2) | 138.3 (2.6) | 0.27 |
| HDL cholesterol, mg/dL | 49.0 (0.4) | 45.1 (0.9) | 0.0002 |
| Triglyceride, mg/dLa | 167.8 (3.2) | 172.7 (6.9) | 0.15 |
| Fibrinogen, mg/dLa | 354.2 (3.8) | 386.1 (8.0) | 0.0002 |
| hs-CRP, mg/La | 13.8 (1.0) | 19.5 (2.1) | 0.01 |
Categorical variables are presented as percentages; continuous variables are presented as age- and sex-adjusted mean (SEM)
aTest performed on log-transformed variable
Molecular screening of the PLA2G10 gene and genotyping of the polymorphisms
We screened the PLA2G10 gene using the genomic sequences retrieved from public depositories. The screening of the gene was performed using genomic DNA from 62 unrelated MI patients selected from the ECTIM study (http://www.genecanvas.org). Using polymerase chain reaction (PCR) and direct sequencing, we explored 1 kb of the promoter, the entire exonic sequences with their corresponding flanking intronic sequences, and 1 kb of the 3′ region after the codon stop. The sequences were aligned and analyzed with the SeqScape software (Applied Biosystems) to determine the potential polymorphisms. The genotyping of the polymorphisms was performed using a modified PCR-restriction fragment length polymorphism method or the TaqMan (5′ nuclease assay) technology. The description of polymorphisms and genotyping conditions can be found at our web site (http://www.genecanvas.org, genes: PLA2G10).
Statistical analysis
Differences in genotype frequencies between cases and controls were tested by a Chi2 test with 2 degrees of freedom (df), adjusted for center in the SIPLAC study (Mantel–Haenszel test). Allele frequencies were deduced from genotype frequencies. Departure from Hardy–Weinberg equilibrium was tested for each polymorphism in controls by Chi2 testing with 1 df. P values < 0.05 were taken as significant.
LD and haplotype analyses were performed using the Testing Haplotype EffectS In Association Studies (THESIAS) program available online (http://www.genecanvas.org) [22]. Differences in haplotypic frequency distributions between cases and controls in the AtheroGene study were tested by means of the likelihood ratio test. Associations between polymorphisms and cardiovascular risk factors (lipids and inflammatory markers) were tested by a general linear model adjusted for age and sex. Association of genotypes with prospective cardiovascular outcome was tested by a Cox proportional hazards regression analysis adjusted for age and sex.
Functional analysis of the PLA2G10/R38C polymorphism
DNA construct
Active human GX sPLA2 complementary DNA (cDNA) starting at the second methionine initiator site, containing additional nucleotide sequences for expressing the hemagglutinin epitope tag (HA tag, nine amino acids YPYDVPDYA) at the C terminus of human GX sPLA2 protein, were generated by PCR, using the forward oligonucleotide (5′_TGTCGAATTCTGCAGATATCCCGCCATGCTGCTCCTGCTACTGCC_3′) with EcoRI and EcoRV sites (italics), and reverse oligonucleotide (5′_CTTAGCGGCCGCTCAAGCGTAATCTGGAACATCGTATGGGTAGATATCGTCACACTTGGGCGAGTC_3′) with EcoRI and NotI sites (italics), stop codon (underlined), and additional nucleotide sequences (bold) coding for the HA tag sequence. The amplified fragments were purified, digested with EcoRI and NotI, and inserted into the EcoRI and NotI sites of pcDNA3 vector to generate the expression plasmid pcDNA-hGX-HA. All constructs were confirmed by sequencing. Inserting the HA tag at the C terminus of hGX sPLA2 protein has no effect on hGX sPLA2 activity (data not shown).
Site-directed mutagenesis
The pcDNA-hGX-HA construct was used as a template in the Quick-change site-directed mutagenesis procedure according to manufacturer’s instructions. Forward mutagenic primer used to create the R38C point mutation was 5′_GGCGAGGCCTCCAGGATATTAtGTGTGCACCGGCGTGGGATCC -3′. Text in bolded lowercase represents the mismatch base introduced to obtain the desired mutation. Reverse mutagenic primer was of the same length and complementary to the forward mutagenic primer. Following amplification, the reaction mixture was treated with DpnI to eliminate the template DNA. Each of the mutated amplification products were transformed into max efficiency DH5α Escherichia coli strain (Invitrogen), and mutations were confirmed by DNA sequencing of the resulting constructs.
Constructs containing the R38 or the C38 allele
COS-7 cells were cultured in Dulbecco’s modified essential medium containing 10% fetal calf serum, 100 units/ml penicillin, and 0.1 mg/ml streptomycin. Confluent cells (75%) were harvested with trypsin and transfected with the above-described constructs containing the R38 or C38 harboring cDNA and pmax green fluorescent protein (GFP; Amaxa) using the Cell line nucleofector kit V and Amaxa nucleofector (Amaxa Inc.) according to the manufacturer’s instructions. Transfection efficiency was estimated 24 h after electroporation by fluorescence activated cell sorter analysis. The number of GFP positive cells in a sample of pmaxGFP (Amaxa)-transfected cells varied between 75% and 79%. After transfection, cells were cultured in 6-well plates for 24 h additionally at 37°C. Selected experiments were performed at both 37°C and 30°C.
Time-resolved fluoroimmunoassay (TR-FIA) and phospholipase A2 activity
Cell lysates extracted in radio immunoprecipitation assay (RIPA) buffer and cell supernatants were directly used for time-resolved fluoroimmunoassays (TR-FIA) to detect hGX sPLA2 protein concentration as described [23]. hGX sPLA2 enzymatic activity was determined using radiolabeled E. coli membranes as previously described [24].
Fluorescence immunocytochemistry and confocal microscopy
COS-7 cells were transiently transfected with HA-tagged either wild-type or mutant hGX sPLA2 as described below. 24 h after transfection, the phosphate buffer solution (PBS)-washed cells were fixed in 4% paraformaldehyde. Cells were permeabilized with methanol for 10 min. Washed cells were incubated in PBS containing 1% bovine serum albumin (BSA; PBS/BSA) for 1 h. Incubation with primary antibodies (rabbit polyclonal hGX sPLA2, mouse monoclonal protein disulphide isomerase (PDI; Stressgen) for endoplasmic reticulum (ER) staining, and mouse monoclonal Golgi 58 K protein (Sigma) for Golgi apparatus) were carried out in PBS/BSA for 1 h at reverse transcription (RT). Cover slips were subsequently washed four times in PBS/BSA and incubated with secondary antibodies (fluorescein isothiocyanate-conjugated anti-mouse, Cy5-conjugated anti-rabbit (Dako)) and 4′-6-diamidino-2-phenylindole (DAPI; 0.5 mg/ml) for 1 h at RT. Cover slips were washed in PBS and mounted in Moviol. Confocal analysis was performed on a Leica SP2-AOBS confocal microscope, and the images were taken using alternating mode to minimize the channel interference (Leica confocal software).
Results
Screening of the gene
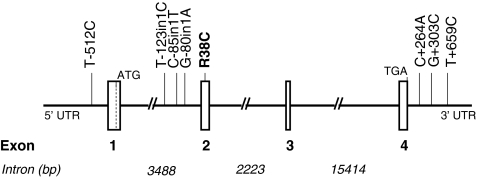
The organization of the PLA2G10 gene is shown in Fig. 1. It consists of four exons (537, 151, 107, and 143 bp, respectively) and three introns (3,488, 2,223, and 15,414 bp, respectively). We screened all exonic sequences with their corresponding flanking intronic sequences, as well as 1 kb in the putative promoter region and 1 kb after the stop codon. From our molecular screening in 124 chromosomes from 62 unrelated subjects, we identified eight polymorphisms (Table 2) which were first genotyped in the SIPLAC study in order to determine their allelic and haplotypic frequencies. Their description can be found at our web site (http://www.genecanvas.org, genes: PLA2G10).
Fig. 1.
Schematic representation of the human PLA2G10 gene and the position of SNPs. Exons are shown as vertical bars. Intron size is indicated. Exons 1–4 encode for the 123 amino acid hGX sPLA2 protein. The polymorphism that resulted in arginine to cysteine substitution is shown in bold
Table 2.
Polymorphisms identified in the PLA2G10 gene (chromosome 16)
| Polymorphism | SNP ID | Base substitution on reverse strand | Gene location | Chromosomal position |
|---|---|---|---|---|
| T-512C | ss#115456686 | T/c | 5′ | 14696099 |
| T-123/in1C | rs#4003232 | T/c | Intron 1 | 14692126 |
| C-85/in1Ta | rs#35822154 | C/t | Intron 1 | 14692088 |
| G-80/in1A | rs#34782548 | G/a | Intron 1 | 14692083 |
| R38C | rs#4003228 | C/t | Exon 2 | 14691989 |
| C+264A | ss#105111473 | C/a | 3′ | 14673705 |
| G+303C | ss#105111474 | G/c | 3′ | 14673666 |
| T+659C | ss#105111475 | T/c | 3′ | 14673310 |
aNot genotyped owing to technical difficulties
PLA2G10 polymorphisms in the SIPLAC study
Seven polymorphisms were investigated in SIPLAC. C-85/in1T was not typed because of technical difficulties. The T+659C substitution, which had been found in a single chromosome among the 124 chromosomes screened, was not observed among the SIPLAC subjects. Accordingly, pairwise LD coefficients were estimated among the six remaining polymorphisms. All polymorphisms exhibited strong LD one with each other, and the six polymorphisms generated eight common haplotypes (frequency > 0.01). Genotype frequencies did not significantly differ between cases and controls for any polymorphism in the SIPLAC study (Table 3). The LD coefficients and haplotypic frequencies estimated in the SIPLAC study are accessible at (http://www.genecanvas.org, genes: PLA2G10).
Table 3.
Genotype and minor allele frequencies in myocardial infarction cases (n = 312) and controls (n = 317) of the SIPLAC study (Belfast and Glasgow pooled, UK)
| Genotype frequency (%) | p value adjusted for center | Minor allele frequency | ||||
|---|---|---|---|---|---|---|
| 11 | 12 | 22 | ||||
| T-512C | Cases | 36.2% | 48.3% | 15.5% | 0.40 | |
| Controls | 41.8% | 46.6% | 11.6% | 0.31 | 0.35 | |
| T-123/in1C | Cases | 61.0% | 34.2% | 4.8% | 0.22 | |
| Controls | 57.6% | 35.5% | 6.9% | 0.55 | 0.25 | |
| G-80/in1Aa | Cases | 54.9% | 38.5% | 6.6% | 0.26 | |
| Controls | 49.3% | 41.5% | 9.2% | 0.40 | 0.30 | |
| R38C | Cases | 97.0% | 3.0% | 0.0% | 0.02 | |
| Controls | 95.2% | 4.8% | 0.0% | 0.30 | 0.02 | |
| C+264A | Cases | 87.5% | 12.1% | 0.4% | 0.06 | |
| Controls | 89.3% | 10.3% | 0.4% | 0.82 | 0.06 | |
| G+303C | Cases | 92.1% | 7.9% | 0.0% | 0.04 | |
| Controls | 92.3% | 7.7% | 0.0% | 0.97 | 0.04 | |
11 homozygotes for the major allele, 12 heterozygotes, 22 homozygotes for the minor allele
aNearly complete concordance with T-123/in1C (85% of subjects concordant for the 2 polymorphisms)
Association of the PLA2G10 polymorphisms with CAD risk in the AtheroGene study
The AtheroGene study was genotyped for five polymorphisms: T-512C, T-123/in1C, R38C, C+264A, and G+303C. There was no significant departure from Hardy–Weinberg equilibrium in controls. In single-locus analysis, none of the polymorphisms was significantly associated with CAD risk (Table 4). Haplotype frequencies did not differ between cases and controls (Table 5). Further subdivision of CAD cases into patients with stable angina pectoris and those with an acute coronary syndrome did not provide significant associations.
Table 4.
Genotype and minor allele frequencies in coronary artery disease cases (n = 1,299) and controls (n = 484) of the AtheroGene study (Mainz, Germany)
| Genotype frequency (%) | p value | Minor allele frequency | ||||
|---|---|---|---|---|---|---|
| 11 | 12 | 22 | ||||
| T-512C | Cases | 46.6% | 43.6% | 9.8% | 0.32 | |
| Controls | 46.0% | 43.4% | 10.6% | 0.88 | 0.32 | |
| T-123/in1C | Cases | 60.4% | 33.9% | 5.7% | 0.23 | |
| Controls | 57.7% | 36.9% | 5.4% | 0.50 | 0.24 | |
| R38C | Cases | 96.4% | 3.5% | 0.1% | 0.02 | |
| Controls | 95.4% | 4.6% | 0.0% | 0.50 | 0.02 | |
| C+264A | Cases | 90.0% | 9.8% | 0.2% | 0.05 | |
| Controls | 90.4% | 9.6% | 0.0% | 0.68 | 0.05 | |
| G+303C | Cases | 96.0% | 4.0% | 0.0% | 0.02 | |
| Controls | 94.8% | 5.2% | 0.0% | 0.29 | 0.03 | |
11 homozygotes for the major allele, 12 heterozygotes, 22 homozygotes for the minor allele
Table 5.
Main haplotypic frequencies estimated in coronary artery disease cases and controls of the AtheroGene study
| Haplotypes | Frequency | |||||
|---|---|---|---|---|---|---|
| T-512C | T-123/in1C | R38C | C+264A | G+303C | Cases (2,426 alleles) | Controls (940 alleles) |
| T | T | R | C | G | 0.399 | 0.390 |
| c | T | R | C | G | 0.297 | 0.293 |
| T | c | R | C | G | 0.208 | 0.217 |
| T | T | R | a | G | 0.054 | 0.048 |
| c | T | R | C | c | 0.018 | 0.026 |
| T | c | C | C | G | 0.014 | 0.017 |
Global difference between cases and controls: p = 0.72
Association of polymorphisms with cardiovascular risk factors and prospective cardiovascular outcome in the AtheroGene study
No significant association was found with baseline lipids or inflammatory markers (data not shown). In CAD patients, the minor allele of T-512C was associated with a decreased risk of death from cardiovascular causes and non-fatal MI during the follow-up (hazard ratio [95% CI] for one copy of the C allele assuming an additive allele effect: 0.76 [0.62–0.94] p < 0.01) and Table 6.
Table 6.
Hazard ratio for cardiovascular event at follow-up associated with the -512C allele
| Cardiovascular event | T-512C genotype | |||
|---|---|---|---|---|
| TT | TC | CC | ||
| No | Number | 469 | 461 | 109 |
| Percent | 45.1% | 44.4% | 10.5% | |
| Yes | Number | 125 | 94 | 16 |
| Percent | 53.2% | 40.0% | 6.8% | |
| Hazard ratio (95% CI) | 0.76 [0.62–0.94] p < 0.01 | |||
Cardiovascular event: cardiovascular death or non-fatal myocardial infarction
Median time at follow-up: 6.2 years
Hazard ratio for one copy of the C allele, assuming an additive allele effect adjusted on age and sex
Functional study of the R38C polymorphism
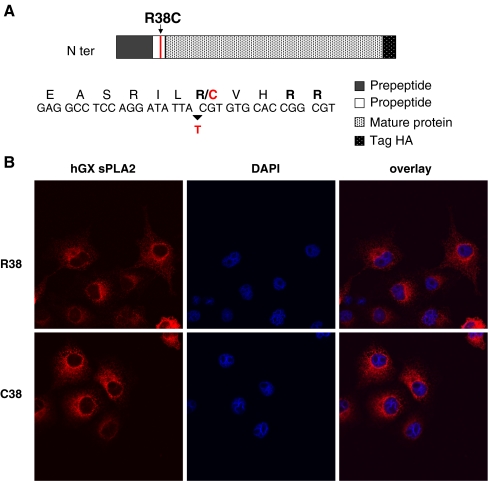
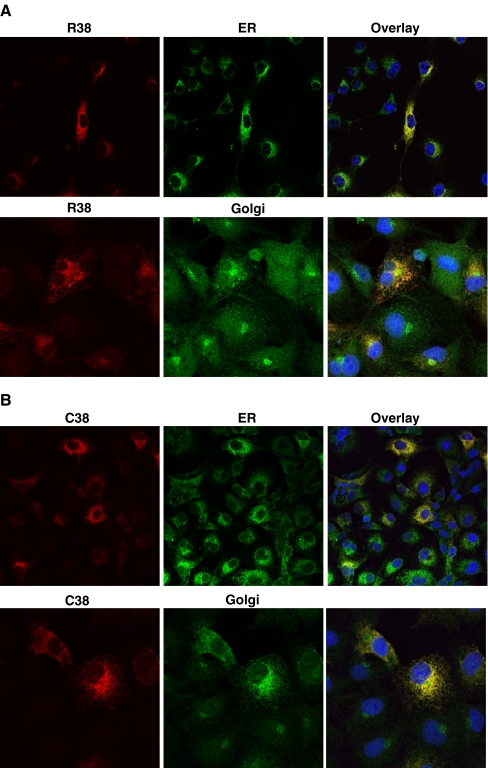
Because of the potential role of hGX sPLA2 in inflammatory processes, we investigated whether the non-synonymous R38C polymorphism might have a functional impact on the protein. The R38C polymorphism, by substituting an arginine to a cysteine at codon 38 (Fig. 2a), leads to an odd number of cysteines in the hGX sPLA2 protein, which may result in misfolding and/or mistargeting and subsequent protein degradation. To analyze the effect of the mutation, we prepared plasmid constructs for HA-tagged hGX sPLA2 with R38 or C38 in the propeptide segment by site-directed mutagenesis and analyzed their expression after transient transfection into COS-7 cells. Fluorescence immunocytochemistry performed 24 h after transfection showed that both R38 and C38 proteins are present intracellularly (Fig. 2b). To determine the subcellular location of the C38 protein, cells were fixed and immunostained with antibodies to PDI and 58 K Golgi proteins, which are well established markers of the ER and the Golgi apparatus, respectively. No difference was observed in their subcellular localization as both R38 and C38 proteins colocalized with ER and Golgi apparatus (Fig. 3a, b).
Fig. 2.
a Schematic representation of the hGX sPLA2 protein containing the R38C polymorphism in the propeptide region. The arginine doublet preceding the mature protein sequence is indicated in bold. The arrow indicates the allelic substitution, and the amino acid change (arginine R to cysteine C) is indicated in red. b Immunofluorescence localization of hGX sPLA2 in COS-7 cells transfected with vectors encoding the R38 and the C38 proteins. The hGX sPLA2 protein is labeled with red. Nucleus is labeled in blue with DAPI. Confocal analysis was performed on a Leica SP2-AOBS confocal microscope as described in “Materials and methods”. Magnification: 50
Fig. 3.
Subcellular immunofluorescence localization of hGX sPLA2 in COS-7 cells transiently transfected with vectors encoding either (a) R38 or (b) C38 hGX sPLA2. Twenty hours after transfection cells were fixed and stained with rabbit polyclonal against hGX sPLA2 (red) and either a mouse monoclonal against the ER marker PDI (green), or a mouse monoclonal against the Golgi apparatus marker 58 K protein (green). Nucleus is labelled in blue with DAPI Confocal analysis was performed on a Leica SP2-AOBS confocal microscope as described in “Materials and methods”. Magnification 50 for ER and 100 for Golgi
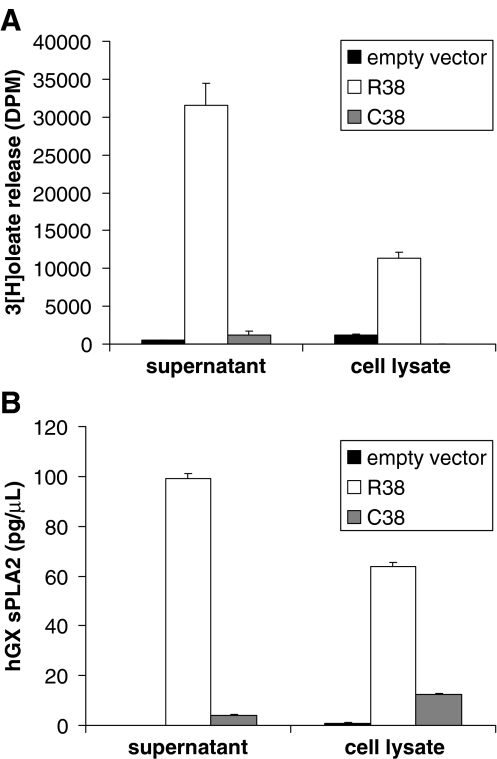
We next determined the level of expression of hGX sPLA2 in cell lysates and cell supernatants by measuring catalytic activity with the sensitive E. coli assay [25] and total protein amount by TR-FIA [23]. In both cell lysate and supernatant, the level of sPLA2 catalytic activity measured for the C38 protein was dramatically lower than that measured for the R38 protein (Fig. 4a). TR-FIA analyses confirmed the lower expression of the C38 sPLA2 protein as compared to R38 (Fig. 4b). Together, our results indicate that the R38C polymorphism has a profound impact on protein activity and expression. In order to test whether the mutation has an impact on the expression of hGX sPLA2 at the messenger RNA (mRNA) level, we performed real-time quantitative PCR (RT-QPCR) analysis of COS-7 transfected cells with either the mutant or the wild type protein. The expression of hGX sPLA2 mRNA in cells transfected with the two expression plasmids was similar (data not shown), suggesting that the polymorphism has no effect on the mRNA production and stability.
Fig. 4.
a hGX sPLA2 enzymatic activity measured with radiolabeled Escherichia coli membranes in supernatant and cell lysates of COS-7 cells transfected either with an empty vector or with vectors encoding the R38 or C38 proteins as described in “Materials and methods”. A representative experiment of at least three experiments with similar results is shown. b Concentration of hGX sPLA2 protein quantified in supernatant and cell lysates of COS-7 cells transfected either with an empty vector or with vectors encoding the R38 or C38 proteins using TR-FIA assay as described in “Materials and methods”. A representative experiment of at least three experiments with similar results is shown
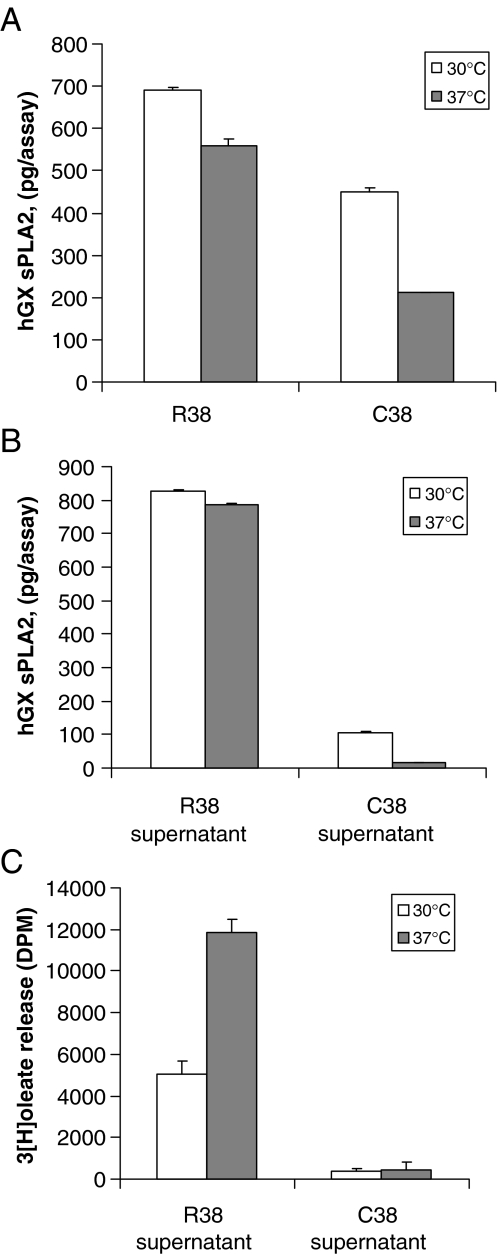
Reduction of cell growth temperature to 30°C has been shown to rescue protein expression and folding of different proteins [26]. To validate our hypothesis that the mutation affects the folding of the hGX sPLA2, we performed parallel transfections of COS-7 cells with the C38 and R38 expression constructs and then incubated the cells at either 30°C or 37°C. Results of TRF-IA assays showed that incubation of cells at 30°C dramatically increased the amount of the C38 protein in both cell lysate and supernatant when compared to cells incubated at 37°C (Fig. 5a, b). As expected, no major effect was observed for the R38 construct (Fig. 5a, b). However, the increased amount of C38 protein was not accompanied by an increased level of sPLA2 activity in the cell supernatant (Fig. 5c), indicating that the overexpressed protein produced at 30°C is not catalytically active. Together, these results suggest that the C38 mutation results in the production of a defective enzyme, which is likely to be misfolded and rapidly degraded.
Fig. 5.
Effect of temperature cell growth on the expression of hGX sPLA2. COS-7 cells transfected with either R38- or C38-containing expression vectors were grown at 30°C or 37°C for 72 h. The total amount of hGX sPLA2 protein was then quantified in the cell lysate (a) and supernatant (b) using the TR-FIA assay as described in “Materials and methods”. c COS-7 cells transfected with either the R38- or C38-containing or empty expression vectors were grown at 30°C or 37°C for 72 h, and hGX sPLA2 activity was measured in cell supernatants as described in “Materials and methods”. Cells transfected with the empty vector showed no detectable hGX sPLA2 protein or enzyme activity (not shown)
Discussion
Growing evidence suggests that sPLA2s IIA, III, V, and X are involved in various inflammatory diseases including CAD and cancer [6, 27]. Circulating levels of sPLA2 activity is an independent predictor of death and new or recurrent MI in patients with acute coronary syndrome [28]. Serum levels of sPLA2 GIIA are linked to variations in the PLA2G2A gene while tagging single nucleotide haplotypes reveal a strong association of the PLA2G5 gene with lipid levels (LDL and oxidized LDL) [29, 30]. Considering the simultaneous expression of IIA, III, V, and X enzymes in the atherosclerotic plaque, it becomes challenging to elucidate the pathophysiological roles of each enzyme in the arterial wall. Using a genetic approach, we identified polymorphisms in the gene coding for hGX sPLA2 and examined whether these polymorphisms play an etiological role in CAD. Our study identified eight polymorphisms, one of which results in the substitution of arginine 38 to cysteine (R38C) in the propeptide sequence of the sPLA2. The full open-reading frame of the hGX cDNA encodes for a signal peptide of 21 amino acids followed by a propeptide sequence of 11 amino acids ending with an arginine doublet and a mature acidic protein of 123 amino acids [14]. The R38C polymorphism is located in the propeptide region of the protein near the putative arginine doublet cleavage site. Overexpression of GX sPLA2 in transgenic mice showed that the zymogen form of the enzyme predominates in most tissues, and that certain inflammation-associated proteases may play a role in activating the enzyme [10]. Conversely, recombinant expression of hGX sPLA2 in various eukaryotic cells leads to the predominant release of the active enzyme, making unclear whether the enzyme is secreted in an active form after propeptide removal before secretion or whether the enzyme is secreted as a proenzyme and then matured extracellularly [8, 31]. The fact that R38C mutation may alter the formation of disulfide bonds and thus the folding of hGX sPLA2, led us to examine its impact on sPLA2 protein expression. Fluorescence immunocytochemistry experiments in COS-7 cells transiently transfected with wild-type and mutant hGX sPLA2 cDNAs showed that both proteins are present in ER and Golgi compartments. Analysis of protein expression and enzymatic activity to reveal an active secretion clearly showed that the mutation lowered the total amount of protein produced by COS-7 cells, prevented its efficient secretion, and resulted in low or no detectable enzymatic activity. When transfected cells were grown at 30°C, a temperature that rescue the folding of various mutated proteins [26], we observed an increased production of the C38 protein in cell lysate and supernatant of transfected cells, but no increase in sPLA2 enzymatic activity. Together, these results support our hypothesis that the R38C mutation produces a misfolded protein that is catalytically inactive and rapidly degraded.
Despite the functional effects observed for the R38C polymorphism at the protein level, no association was observed between this polymorphism and CAD risk or cardiovascular risk factors in the AtheroGene study and the SIPLAC study. This lack of association might be explained by the low frequency of the C38 allele (approximately 3%) and, consequently, the low power of the present studies to detect an association. Furthermore, due to the unavailability of suitable substrates to distinguish between different sPLA2s, the circulating level of GX sPLA2 could not be investigated as an intermediate phenotype in the AtheroGene study. In addition to the R38C polymorphism, we identified other polymorphisms in the putative regulatory region of the PLA2G10 gene. The T-512C was associated with the risk of future cardiovascular event during the prospective follow-up of the AtheroGene cohort. However, this polymorphism was not associated with CAD or MI risk in a case/control setting and did not correlate with any cardiovascular risk factor. This result has thus to be taken with caution and requires further replication in other cohorts. As this polymorphism is located in the putative promoter region of the gene, it might influence the expression level of the enzyme; however, the promoter of the PLA2G10 is still unknown, and thus will be interesting to explore.
Recent studies implicate GX sPLA2 in myocardial ischemia reperfusion injury [11], in allergen-induced airway inflammation [9], and in lung diseases [8]. A possible role of hGX sPLA2 was also suggested in lung and colon carcinogenesis [12, 13]. In this context, functional genetic mutations leading to an alteration of protein expression and/or enzymatic activity may have a profound impact on the pathophysiological role of the enzyme, in particular, by modulating the concentration of lipid mediators produced locally and thus modifying the inflammatory process in diseases like atherosclerosis and cancer.
Acknowledgements
This study was supported by the Institut National de la Santé Et de la Recherche Médicale and by the fellowships from the Fondation pour la Recherche Medicale (SPF20080512033) to SAK and (FDT2000910244) to SG. The study was also supported by an ANR program (ANR 2006 Physiopathologie des maladies humaines: sPLA2/ATHEROSCLEROSE) to GL and EN, and in part by the CNRS and the Association pour la Recherche sur le Cancer to GL. EN and GL are directors of research of the Centre National de la Recherche Scientifique. We also thank the “Plate-forme d’Imagerie Cellulaire Pitié Salpêtrière” for confocal imaging, and the “Plate-forme Postgénomique P3S”.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Claire Perret and Ikram Jemel contributed equally to this paper.
References
- 1.Kugiyama K, Ota Y, Takazoe K, Moriyama Y, Kawano H, Miyao Y, Sakamoto T, Soejima H, Ogawa H, Doi H, Sugiyama S, Yasue H (1999) Circulating levels of secretory type II phospholipase A(2) predict coronary events in patients with coronary artery disease. Circulation 100:1280–1284 [DOI] [PubMed]
- 2.Karabina SA, Brocheriou I, Le Naour G, Agrapart M, Durand H, Gelb M, Lambeau G, Ninio E (2006) Atherogenic properties of LDL particles modified by human group X secreted phospholipase A2 on human endothelial cell function. FASEB J 20:2547–2549 [DOI] [PubMed]
- 3.Rosengren B, Jonsson-Rylander AC, Peilot H, Camejo G, Hurt-Camejo E (2006) Distinctiveness of secretory phospholipase A2 group IIA and V suggesting unique roles in atherosclerosis. Biochim Biophys Acta 1761:1301–1308 [DOI] [PubMed]
- 4.Sato H, Kato R, Isogai Y, Saka GI, Ohtsuki M, Taketomi Y, Yamamoto K, Tsutsumi K, Yamada J, Masuda S, Ishikawa Y, Ishii T, Kobayashi T, Ikeda K, Taguchi R, Hatakeyama S, Hara S, Kudo I, Itabe H, Murakami M (2008) Analyses of group III secreted phospholipase A2 transgenic mice reveal potential participation of this enzyme in plasma lipoprotein modification, macrophage foam cell formation, and atherosclerosis. J Biol Chem 283:33483–33497 [DOI] [PMC free article] [PubMed]
- 5.Wooton-Kee CR, Boyanovsky BB, Nasser MS, de Villiers WJ, Webb NR (2004) Group V sPLA2 hydrolysis of low-density lipoprotein results in spontaneous particle aggregation and promotes macrophage foam cell formation. Arterioscler Thromb Vasc Biol 24:762–767 [DOI] [PubMed]
- 6.Lambeau G, Gelb MH (2008) Biochemistry and physiology of mammalian secreted phospholipases A2. Annu Rev Biochem 77:495–520 [DOI] [PubMed]
- 7.Bostrom MA, Boyanovsky BB, Jordan CT, Wadsworth MP, Taatjes DJ, de Beer FC, Webb NR (2007) Group V secretory phospholipase A2 promotes atherosclerosis: evidence from genetically altered mice. Arterioscler Thromb Vasc Biol 27:600–606 [DOI] [PubMed]
- 8.Curfs DM, Ghesquiere SA, Vergouwe MN, van der Made I, Gijbels MJ, Greaves DR, Verbeek JS, Hofker MH, de Winther MP (2008) Macrophage secretory phospholipase A2 group X enhances anti-inflammatory responses, promotes lipid accumulation, and contributes to aberrant lung pathology. J Biol Chem 283:21640–21648 [DOI] [PubMed]
- 9.Henderson WR Jr, Chi EY, Bollinger JG, Tien YT, Ye X, Castelli L, Rubtsov YP, Singer AG, Chiang GK, Nevalainen T, Rudensky AY, Gelb MH (2007) Importance of group X-secreted phospholipase A2 in allergen-induced airway inflammation and remodeling in a mouse asthma model. J Exp Med 204:865–877 [DOI] [PMC free article] [PubMed]
- 10.Ohtsuki M, Taketomi Y, Arata S, Masuda S, Ishikawa Y, Ishii T, Takanezawa Y, Aoki J, Arai H, Yamamoto K, Kudo I, Murakami M (2006) Transgenic expression of group V, but not group X, secreted phospholipase A2 in mice leads to neonatal lethality because of lung dysfunction. J Biol Chem 281:36420–36433 [DOI] [PubMed]
- 11.Fujioka D, Saito Y, Kobayashi T, Yano T, Tezuka H, Ishimoto Y, Suzuki N, Yokota Y, Nakamura T, Obata JE, Kanazawa M, Kawabata K, Hanasaki K, Kugiyama K (2008) Reduction in myocardial ischemia/reperfusion injury in group X secretory phospholipase A2-deficient mice. Circulation 117:2977–2985 [DOI] [PubMed]
- 12.Masuda S, Murakami M, Mitsuishi M, Komiyama K, Ishikawa Y, Ishii T, Kudo I (2005) Expression of secretory phospholipase A2 enzymes in lungs of humans with pneumonia and their potential prostaglandin-synthetic function in human lung-derived cells. Biochem J 387:27–38 [DOI] [PMC free article] [PubMed]
- 13.Morioka Y, Ikeda M, Saiga A, Fujii N, Ishimoto Y, Arita H, Hanasaki K (2000) Potential role of group X secretory phospholipase A(2) in cyclooxygenase-2-dependent PGE(2) formation during colon tumorigenesis. FEBS Lett 487:262–266 [DOI] [PubMed]
- 14.Cupillard L, Koumanov K, Mattei MG, Lazdunski M, Lambeau G (1997) Cloning, chromosomal mapping, and expression of a novel human secretory phospholipase A2. J Biol Chem 272:15745–15752 [DOI] [PubMed]
- 15.Bezzine S, Bollinger JG, Singer AG, Veatch SL, Keller SL, Gelb MH (2002) On the binding preference of human groups IIA and X phospholipases A2 for membranes with anionic phospholipids. J Biol Chem 277:48523–48534 [DOI] [PubMed]
- 16.Gora S, Lambeau G, Bollinger JG, Gelb M, Ninio E, Karabina SA (2006) The proinflammatory mediator platelet activating factor is an effective substrate for human group X secreted phospholipase A2. Biochim Biophys Acta 1761:1093–1099 [DOI] [PubMed]
- 17.Hanasaki K, Yamada K, Yamamoto S, Ishimoto Y, Saiga A, Ono T, Ikeda M, Notoya M, Kamitani S, Arita H (2002) Potent modification of low density lipoprotein by group X secretory phospholipase A2 is linked to macrophage foam cell formation. J Biol Chem 277:29116–29124 [DOI] [PubMed]
- 18.Pruzanski W, Lambeau L, Lazdunsky M, Cho W, Kopilov J, Kuksis A (2005) Differential hydrolysis of molecular species of lipoprotein phosphatidylcholine by groups IIA, V and X secretory phospholipases A(2). Biochim Biophys Acta 1736:38–50 [DOI] [PubMed]
- 19.Parra HJ, Arveiler D, Evans AE, Cambou JP, Amouyel P, Bingham A, McMaster D, Schaffer P, Douste-Blazy P, Luc G et al (1992) A case-control study of lipoprotein particles in two populations at contrasting risk for coronary heart disease. The ECTIM study. Arterioscler Thromb 12:701–707 [DOI] [PubMed]
- 20.Rupprecht HJ, Blankenberg S, Bickel C, Rippin G, Hafner G, Prellwitz W, Schlumberger W, Meyer J (2001) Impact of viral and bacterial infectious burden on long-term prognosis in patients with coronary artery disease. Circulation 104:25–31 [DOI] [PubMed]
- 21.Tiret L, Godefroy T, Lubos E, Nicaud V, Tregouet DA, Barbaux S, Schnabel R, Bickel C, Espinola-Klein C, Poirier O, Perret C, Munzel T, Rupprecht HJ, Lackner K, Cambien F, Blankenberg S (2005) Genetic analysis of the interleukin-18 system highlights the role of the interleukin-18 gene in cardiovascular disease. Circulation 112:643–650 [DOI] [PubMed]
- 22.Tregouet DA, Barbaux S, Escolano S, Tahri N, Golmard JL, Tiret L, Cambien F (2002) Specific haplotypes of the P-selectin gene are associated with myocardial infarction. Hum Mol Genet 11:2015–2023 [DOI] [PubMed]
- 23.Nevalainen TJ, Eerola LI, Rintala E, Laine VJ, Lambeau G, Gelb MH (2005) Time-resolved fluoroimmunoassays of the complete set of secreted phospholipases A2 in human serum. Biochim Biophys Acta 1733:210–223 [DOI] [PubMed]
- 24.Rouault M, Le Calvez C, Boilard E, Surrel F, Singer A, Ghomashchi F, Bezzine S, Scarzello S, Bollinger J, Gelb MH, Lambeau G (2007) Recombinant production and properties of binding of the full set of mouse secreted phospholipases A2 to the mouse M-type receptor. Biochemistry 46:1647–1662 [DOI] [PubMed]
- 25.Rouault M, Rash LD, Escoubas P, Boilard E, Bollinger J, Lomonte B, Maurin T, Guillaume C, Canaan S, Deregnaucourt C, Schrevel J, Doglio A, Gutierrez JM, Lazdunski M, Gelb MH, Lambeau G (2006) Neurotoxicity and other pharmacological activities of the snake venom phospholipase A2 OS2: the N-terminal region is more important than enzymatic activity. Biochemistry 45:5800–5816 [DOI] [PMC free article] [PubMed]
- 26.Bernier V, Lagace M, Bichet DG, Bouvier M (2004) Pharmacological chaperones: potential treatment for conformational diseases. Trends Endocrinol Metab 15:222–228 [DOI] [PubMed]
- 27.Mounier CM, Wendum D, Greenspan E, Flejou JF, Rosenberg DW, Lambeau G (2008) Distinct expression pattern of the full set of secreted phospholipases A2 in human colorectal adenocarcinomas: sPLA2-III as a biomarker candidate. Br J Cancer 98:587–595 [DOI] [PMC free article] [PubMed]
- 28.Mallat Z, Steg PG, Benessiano J, Tanguy ML, Fox KA, Collet JP, Dabbous OH, Henry P, Carruthers KF, Dauphin A, Arguelles CS, Masliah J, Hugel B, Montalescot G, Freyssinet JM, Asselain B, Tedgui A (2005) Circulating secretory phospholipase A2 activity predicts recurrent events in patients with severe acute coronary syndromes. J Am Coll Cardiol 46:1249–1257 [DOI] [PubMed]
- 29.Wootton PT, Arora NL, Drenos F, Thompson SR, Cooper JA, Stephens JW, Hurel SJ, Hurt-Camejo E, Wiklund O, Humphries SE, Talmud PJ (2007) Tagging SNP haplotype analysis of the secretory PLA2-V gene, PLA2G5, shows strong association with LDL and oxLDL levels, suggesting functional distinction from sPLA2-IIA: results from the UDACS study. Hum Mol Genet 16:1437–1444 [DOI] [PubMed]
- 30.Wootton PT, Drenos F, Cooper JA, Thompson SR, Stephens JW, Hurt-Camejo E, Wiklund O, Humphries SE, Talmud PJ (2006) Tagging-SNP haplotype analysis of the secretory PLA2IIa gene PLA2G2A shows strong association with serum levels of sPLA2IIa: results from the UDACS study. Hum Mol Genet 15:355–361 [DOI] [PubMed]
- 31.Masuda S, Murakami M, Takanezawa Y, Aoki J, Arai H, Ishikawa Y, Ishii T, Arioka M, Kudo I (2005) Neuronal expression and neuritogenic action of group X secreted phospholipase A2. J Biol Chem 280:23203–23214 [DOI] [PubMed]