It has been known for decades that the Y chromosome carries genes required for spermatogenesis and male fertility. But it has been frustratingly difficult to pin down the relevant genes, to such an extent that a finding reported by Luddi et al.1 in this issue of the Journal — that the ubiquitin-specific peptidase 9, Y-linked gene (USP9Y) is not essential for normal spermatogenesis — represents a noteworthy advance in the field.
Approximately 15% of couples have difficulty conceiving; the causes are many and include genetic factors. The most common known genetic causes of spermatogenic failure are karyotype anomalies and Y-chromosome microdeletions. Karyotype analysis and screening for Yq deletions are now routine diagnostic tests that are able to help elucidate both the cause of spermatogenic perturbations and their clinical consequences. It is therefore of some interest to know which genes are essential to spermatogenesis; with this information, a more specific diagnosis might be possible, and the clinical management — including genetic counseling — of infertile couples would be greatly improved.
The search for these genes began in 1976, when Tiepolo and Zuffardi2 reported cytogenetically visible microdeletions in Yq in six azoospermic men. Such deletions must be very large to be visible and are therefore likely to involve the absence of many genes (and unlikely to reveal the causal gene), but this observation provided a starting point for a classic approach to identifying the gene or genes responsible for this phenotype. Smaller deletions within the Yq interval would be sought using molecular methods, leading to the identification of one or a few candidate genes, and inactivating mutations would then be used to confirm which of the candidates was responsible. This strategy has proved successful in the identification of the causal genes underlying countless single-gene disorders since its initial application to chronic granulomatous disease in 1986.3
At first, the application of this approach for Y-chromosomal spermatogenic abnormalities seemed promising. Molecular analysis did indeed identify smaller deletions within Yq that were specific to men with spermatogenic failure. These were classified, on the basis of chromosomal location, as three azoospermia factor (AZF) regions: AZFa, AZFb, and AZFc.4 AZFb and AZFc were later shown to contain large segments of duplicated sequence and, in fact, to overlap each other. Given the challenge of tracking down specific loci in chromosomal regions enriched in sequence duplications, it is perhaps understandable that the gene or genes responsible for the AZFb-AZFc spermatogenesis phenotype have still not been identified.
In contrast, the AZFa interval consists of regular single-copy DNA (i.e., DNA that does not contain a supranormal level of repeated sequence) that should not present unusual complications for homing in on the key gene. And in 1999, Sun et al.5 described what seemed to be a definitive result. They sequenced the two genes in the AZFa region (USP9Y and the DEAD [Asp–Glu–Ala–Asp] box polypeptide 3, Y-linked gene, DDX3Y) in 576 infertile men and discovered exactly the kind of mutation they were looking for, in a man with spermatogenic failure: a de novo deletion of 4 bp within the USP9Y gene that destroyed a splicedonor site, resulting in the skipping of the next exon and truncation of approximately 90% of the peptide sequence (Fig. 1). Thus, it appeared that the loss of USP9Y was responsible for the observed azoospermia and that one gene underlying spermatogenic failure had been successfully found.
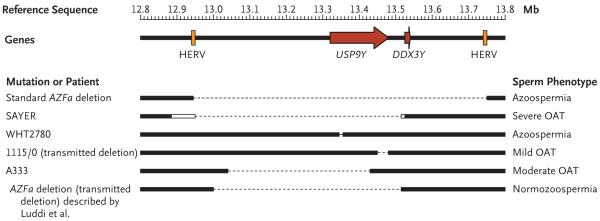
Figure 1. Known Deletions of the Azoospermia Factor Region a (AZFa) of the Human Y chromosome and Their Associated Spermatogenic Phenotypes.
Part of the reference sequence, 1 Mb in length, of the Y chromosome is shown (top), highlighting ubiquitin-specific peptidase 9, Y-linked gene (USP9Y) and the DEAD (Asp–Glu–Ala–Asp) box polypeptide 3, Y-linked gene (DDX3Y) (red arrows) and the two human endogenous retrovirus sequences (HERVs) (orange boxes). Along the same sequence, regions are shown (bottom) as retained (black boxes) or missing (thin dotted line) in patients, including the standard AZFa deletion resulting from nonallelic homologous recombination between the two HERV sequences, as well as confirmed gene-specific deletions. Note the variability in phenotype associated with USP9Y-only deletions and the lack of known DDX3Y-only deletions. The white boxes in SAYER (described by Brown et al.6) indicate the extent of uncertainty in the location of the breakpoint in this patient, and the size of the 4-bp deletion in WHT2780 (described by Sun et al.5) has been exaggerated to make it visible at this scale. The 1115/0 and A333 deletions are described by Krausz et al.7 “Transmitted” indicates a deletion that has been passed naturally from father to son on at least one occasion. OAT denotes oligoasthenoteratozoospermia.
But subsequent findings complicated this simple conclusion. The phenotype associated with the loss of USP9Y turned out to be variable, ranging from normospermia in chimpanzees and bonobos, in which the gene became inactivated millions of years ago,8 to moderate oligoasthenoteratozoospermia in humans,7 to azoospermia.5 The report by Luddi et al. now adds normospermia in humans to this list of phenotypes. These observations have bearing on two related questions: Is the inactivation of USP9Y or DDX3Y responsible for the AZFa azoospermic phenotype? and What conclusions should we draw from the variation in the USP9Y-associated phenotype?
There are a number of reasons why the first, apparently simple, question has been so difficult to resolve. First, AZFa deletions are themselves rare. In literature reports, they represent less than 5% of all AZF deletions.9 Second, AZFa deletions leading to azoospermia obviously cannot be inherited, and so all are de novo mutations. They usually arise by the same mechanism: homologous recombination between two misaligned human endogenous retrovirus sequences (HERVs, elements of viral origin inserted into the genome and inherited as part of the chromosome) spaced about 790 kb apart (Fig. 1), encompassing both genes.10 Mutations due to HERV misalignment are therefore not informative for distinguishing between the contributions of the two genes. Years of study have identified just a handful of patients who carry nonstandard deletions that affect only one of the genes and are thus informative (Fig. 1). Third, all of the confirmed single-gene mutations affect USP9Y; we have no information about the phenotype associated with the loss of DDX3Y only.
The study by Luddi et al. clearly shows that complete USP9Y deletion is compatible with normal spermatogenesis and fertility. But previous studies5-7 show, equally clearly, that its loss can disrupt spermatogenesis to varying degrees and that natural transmission (fertility) is possible when the phenotype is mild.7 So we have to conclude that the phenotype associated with the loss of USP9Y varies according to the genetic or other background of the carrier, although the relevant modifying factors remain unknown. This variation in phenotype contrasts sharply with the effect of losing both genes, which invariably results in the Sertoli-cell–only syndrome (characterized by the lack of germ cells) and, consequently, azoospermia. This consistent effect strongly suggests a key role of DDX3Y in spermatogenesis, a conclusion supported by expression of the gene in premeiotic spermatogonia (USP9Y is expressed in postmeiotic spermatids).11 We predict that the loss of DDX3Y alone is associated with a more severe testicular phenotype, most likely azoospermia.
What are the clinical implications of these conclusions? Screening for USP9Y deletion should not be performed routinely until there is a better understanding of the factors that modify the phenotype. Luddi et al. suggest that revision of the European Academy of Andrology–European Molecular Genetics Quality Network screening guidelines may be warranted to ensure that DDX3Y-only deletions are detected. The lack of confirmed deletions of this type after many years of worldwide screening in research laboratories9 means that the cost of such a step may currently outweigh the clinical benefit, but we can look forward to a time when the thousand-dollar genome will make such information, and much more, readily available. In the meantime, the article by Luddi et al. focuses our attention even more strongly on understanding the role of DDX3Y.
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.Luddi A, Margollicci M, Gambera L, et al. Spermatogenesis in a man with complete deletion of USP9Y. N Engl J Med. 2009;360:881–5. doi: 10.1056/NEJMoa0806218. [DOI] [PubMed] [Google Scholar]
- 2.Tiepolo L, Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Hum Genet. 1976;34:119–24. doi: 10.1007/BF00278879. [DOI] [PubMed] [Google Scholar]
- 3.Royer-Pokora B, Kunkel LM, Monaco AP, et al. Cloning the gene for an inherited human disorder — chronic granulomatous disease — on the basis of its chromosomal location. Nature. 1986;322:32–8. doi: 10.1038/322032a0. [DOI] [PubMed] [Google Scholar]
- 4.Vogt PH, Edelmann A, Kirsch S, et al. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet. 1996;5:933–43. doi: 10.1093/hmg/5.7.933. [DOI] [PubMed] [Google Scholar]
- 5.Sun C, Skaletsky H, Birren B, et al. An azoospermic man with a de novo point mutation in the Y-chromosomal gene USP9Y. Nat Genet. 1999;23:429–32. doi: 10.1038/70539. [DOI] [PubMed] [Google Scholar]
- 6.Brown GM, Furlong RA, Sargent CA, et al. Characterisation of the coding sequence and fine mapping of the human DFFRY gene and comparative expression analysis and mapping to the Sxrb interval of the mouse Y chromosome of the Dffry gene. Hum Mol Genet. 1998;7:97–107. doi: 10.1093/hmg/7.1.97. [DOI] [PubMed] [Google Scholar]
- 7.Krausz C, Degl’Innocenti S, Nuti F, et al. Natural transmission of USP9Y gene mutations: a new perspective on the role of AZFa genes in male fertility. Hum Mol Genet. 2006;15:2673–81. doi: 10.1093/hmg/ddl198. [DOI] [PubMed] [Google Scholar]
- 8.Perry GH, Tito RY, Verrelli BC. The evolutionary history of human and chimpanzee Y-chromosome gene loss. Mol Biol Evol. 2007;24:853–9. doi: 10.1093/molbev/msm002. [DOI] [PubMed] [Google Scholar]
- 9.Krausz C, Degl’Innocenti S. Y chromosome and male infertility: update, 2006. Front Biosci. 2006;11:3049–61. doi: 10.2741/2032. [DOI] [PubMed] [Google Scholar]
- 10.Kamp C, Hirschmann P, Voss H, Huellen K, Vogt PH. Two long homologous retroviral sequence blocks in proximal Yq11 cause AZFa microdeletions as a result of intrachromosomal recombination events. Hum Mol Genet. 2000;9:2563–72. doi: 10.1093/hmg/9.17.2563. [DOI] [PubMed] [Google Scholar]
- 11.Vogt PH, Falcao CL, Hanstein R, Zimmer J. The AZF proteins. Int J Androl. 2008;31:383–94. doi: 10.1111/j.1365-2605.2008.00890.x. [DOI] [PubMed] [Google Scholar]