Abstract
Phospholamban (PLN) is an essential regulator of cardiac muscle contractility. The homopentameric assembly of PLN is the reservoir for active monomers that, upon deoligomerization form 1:1 complexes with the sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA), thus modulating the rate of calcium uptake. In lipid bilayers and micelles, monomeric PLN exists in equilibrium between a bent (or resting) T state and a more dynamic (or active) R state. Here, we report the high-resolution structure and topology of the T state of a monomeric PLN mutant in lipid bilayers, using a hybrid of solution and solid-state NMR restraints together with molecular dynamics simulations in explicit lipid environments. Unlike the previous structural ensemble determined in micelles, this approach gives a complete picture of the PLN monomer structure in a lipid bilayer. This hybrid ensemble exemplifies the tilt, rotation, and depth of membrane insertion, revealing the interaction with the lipids for all protein domains. The N-terminal amphipathic helical domain Ia (residues 1–16) rests on the surface of the lipid membrane with the hydrophobic face of domain Ia embedded in the membrane bilayer interior. The helix comprised of domain Ib (residues 23–30) and transmembrane domain II (residues 31–52) traverses the bilayer with a tilt angle of ≈24°. The specific interactions between PLN and lipid membranes may represent an additional regulatory element of its inhibitory function. We propose this hybrid method for the simultaneous determination of structure and topology for membrane proteins with compact folds or proteins whose spatial arrangement is dictated by their specific interactions with lipid bilayers.
Keywords: hybrid method, membrane proteins, oriented solid-state NMR, molecular modeling, PISEMA
Structure and topology are central to membrane protein function (1). Recently determined high-resolution structures reveal the compact folds for several membrane proteins, such as electron and proton-conducting proteins involved in photosynthesis and respiration (http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html). However, a significant population of membrane proteins does not possess a compact tertiary fold, but has its fold space defined through interactions of secondary structure elements (helices, turns, and loops) with the lipid membrane, i.e., the topology (1). This is the case for phospholamban (PLN), a mammalian protein that is essential in the regulation of cardiac muscle contractility (2), and that has recently become a major target for gene therapy to ameliorate cardiomyopathies (3, 4). PLN is located in the sarco(endo)plasmic reticulum (SR) of cardiac myocytes, inhibiting the SR Ca2+-ATPase (SERCA) by shifting its relative Ca2+ affinity (5). In vitro and in vivo experiments have shown PLN to exist as a homopentamer that deoligomerizes into active monomers that bind SERCA in a 1:1 molar ratio (6). The monomeric form of PLN exists in equilibrium between a dynamically disordered R state and a more restricted T state (7, 8) and has 3 structural domains (helix–loop–helix) and 4 dynamic domains: Ia (residues 1–16), loop (17–22), Ib (residues 23–30), and II (residues 31–52) (9). As measured by NMR (10, 11) and EPR spectroscopies (7, 8, 12, 13), the T state is predominant (≈84%) in both micelles and lipid bilayers (14).
The lack of PLN tertiary structure (noncompact fold) and the semiflexible loop that connects domain Ia to Ib represent major obstacles for structural studies of PLN by solution NMR. In particular, the paucity of NOEs and the degeneracy of residual dipolar coupling solutions prevented the complete characterization of the structural topology (15, 16). Here, we show how solution NMR angular and distance restraints derived from detergent micellar studies, together with solid-state NMR orientational restraints obtained in mechanically aligned lipid bilayers, are used to simultaneously define the high-resolution structure and topology of monomeric PLN. The various restraints are combined into a single energy objective function and used jointly with a membrane immersion depth potential (17). The average structure from the calculated conformational ensemble is then embedded into an explicit 1,2-dioleoyl-glycero-3-sn-phosphocholine (DOPC) lipid bilayer and equilibrated, revealing the high-resolution T state structure of monomeric PLN (L-shaped) and the protein–lipid interactions at the atomic level. The resulting fold of monomeric PLN is defined by its strong interactions with lipid membranes. Unlike the previously determined structural ensemble (15), this ensemble more precisely specifies the position of each structural domain with respect to the lipid membrane, with domain Ib protruding toward the membrane/water interface and ready to make hydrogen bonds and salt bridges with the juxtamembrane region of SERCA. Domain II matches the hydrophobicity of the DOPC bilayer, ready to interact with transmembrane helices M2, M5, and M9 of SERCA's binding groove. The amphipathic domain Ia is helical and adsorbed on the surface of the membrane bilayer. Taken with the recent findings from several other groups (18, 19), we propose that the reversible binding of this domain with the lipid bilayer represents a further regulatory element of PLN function.
Results
Solution and Solid-State NMR Restraints.
A previous solution NMR structure of AFA-PLN (C36A, C36F, C41A; fully functional monomeric mutant) was determined using protein solubilized in 600 mM DPC, pH 4.2, 50 °C (15). Here, we repeated these experiments on AFA-PLN solubilized in 300 mM DPC, pH 6.0, 37 °C, which are conditions that preserve SERCA's enzymatic activity. The NOE patterns at 37 °C and 50 °C were essentially the same. We also performed H(CCO)NH and C(CO)NH TOCSY experiments to fully assign the side chains, which enabled interresidue NOEs to Pro-21 to be identified and incorporated into the structural calculations. In addition, we measured 3J-coupling constants to restrain the χ1 rotamers (Table S2).
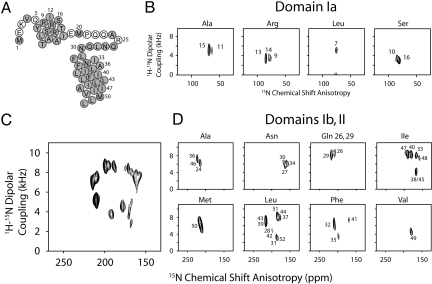
For solid-state NMR spectroscopy, the AFA-PLN samples were reconstituted into DOPC lipids at a lipid/protein ratio of ≈160/1. The solid-state NMR restraints and residue assignments for the 15N chemical-shift anisotropy (CSA) and 1H-15N dipolar coupling (DC) of domain Ib and II were obtained from PISEMA experiments conducted at 30 °C (20). For smaller 1H-15N DC values (i.e., for residues located in the PLN cytoplasmic domain), we carried out SAMPI4 experiments (21) with selectively labeled PLN protein. A higher level of hydration and temperature optimization led to a substantial increase in resolution and sensitivity with respect to our previous sample preparations, without jeopardizing the macroscopic alignment of the lipid membranes (Fig. S1). A uniformly 15N-labeled [U-15N] AFA-PLN sample now gives a complete PISA-wheel spectrum for the helix comprised of domains Ib and II (Fig. 1C). To complete the resonance assignments, we also collected PISEMA spectra on several selectively labeled PLN samples. Fig. 1 shows PISEMA and SAMPI4 spectra for [15N-Leu, Ile, Ala, Met, Phe, Gln-26/Gln-29, Ser, Arg (15Nα), and Asn] AFA-PLN. The orientational restraint dataset consists of 44 assigned residues (82 total restraints), which correspond to ≈85% assignment of the backbone.
Fig. 1.
Solid-state NMR spectra of AFA-PLN in lipid bilayers. (A) Primary sequence of AFA-PLN showing assigned residues in gray (CSA and DC assignments in Table S1). (B) Assigned PISEMA (Leu, Ala) and SAMPI4 spectra (Ser, Arg) of domain Ia. (C) [U-15N] PISEMA spectrum of PLN domain Ib and II displaying a uniform intensity across the PISA wheel. (D) Selectively labeled PLN PISEMA spectra of domain Ib and II. Overlay shown in Fig. S3.
Evidence for the dynamic N terminus and loop region is seen in the [15N-Met] AFA-PLN PISEMA spectrum that shows 2 resonances with 15N CSA values of 123 and 101 ppm, corresponding to Met-1 and Met-20 (Fig. S2). These 15N chemical shifts appear at essentially isotropic values, indicating that the observed anisotropic chemical shifts are dynamically averaged for both the loop and N terminus. Based on these results and the dynamic characterization of PLN by solution NMR (10), the DC and CSA values for residues located in the most dynamic regions of PLN (Glu-2, Lys-3, Glu-19, and Pro-21) were not determined.
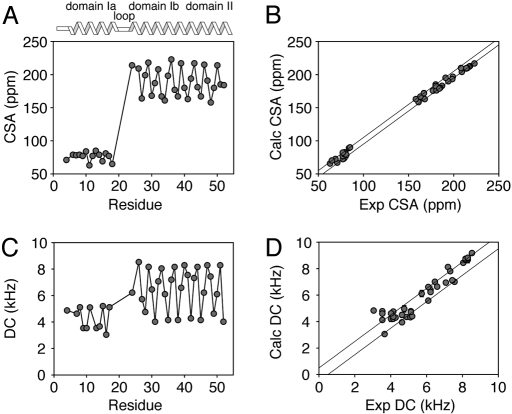
The solid-state NMR assignments (Table S1) were carried out by using extensive selective labeling and a combinatorial algorithm for minimizing the resonance positions based on the periodic nature of the helices (22). Fig. 2 shows the CSA and DC plotted vs. the residue number in AFA-PLN. As expected for helical proteins, both CSA and DC oscillate with a period of ≈3.6 (Fig. S4). The amplitude of the oscillation is larger for domains Ib and II than for domain Ia, reflecting the different orientations with respect to the external magnetic field. These oscillations can be interpreted in terms of tilt (θ) and rotation (ρ) angles with respect to the membrane bilayer normal (parallel to B0) for the different protein domains. An estimate of alignment using a static ideal helix to fit the PISA-wheel patterns (16, 23, 24) gives θ and ρ angles of ≈24° and ≈198°, respectively, for the helix comprised of domains Ib and II and a θ angle of ≈97° for domain Ia (Fig. S5). These angles are in close agreement with those published using CSA and DC for selectively labeled [15N-Leu, Ala, and Ile] AFA-PLN alone (25), and reinforce the previously proposed L-shaped topology for monomeric PLN (15, 26).
Fig. 2.
Experimental CSA (A) and DC (C) values from Fig. 1 plotted vs. residue number. (B and D) show correlation plots of calculated vs. experimental CSA and DC for the hybrid ensemble, respectively. The parallel lines in B and D indicate the experimental errors used in the structural calculations.
Structure Calculations.
The technical details of the structure determination protocols and the validation methods for the hybrid approach are reported in ref. 27. In the first stage of the structure calculation, we started with PLN in an extended configuration, and carried out simulated annealing using only the solution NMR restraints [NOEs, 3J-coupling, dihedral angles from TALOS (28) and hydrogen bonds]. Because there are still very few restraints in the loop region (residues 17–22), this step generated 200 PLN conformers that converged into well defined secondary structure elements but lacked precision in the 3D structure (Fig. S6B). In the second stage of the refinement, we introduced the solid-state NMR orientational restraints (total of 82) using torsion angle dynamics and simulated annealing calculations. This protocol allowed for the secondary structural elements to be maintained during the overall orientation of the protein according to the alignment tensor (Bo, the membrane normal is fixed along the z axis), avoiding possible integration problems of the simulated annealing algorithm. The third stage consisted of rigid-body minimization of the conformational ensemble into a virtual membrane potential (i.e., the depth of insertion potential, Ez) according to the energy function described by DeGrado and coworkers (17). A final step of Cartesian molecular dynamics refinement was used to relax the PLN conformers.
Convergence and Validation of the Hybrid Conformational Ensemble.
The conformers were selected according to the following criteria: NOE violations <0.5 Å, covalent bond violations <0.01 Å, and bond angle violations <5° (29). Fig. 2 shows the agreement between calculated and experimental CSA and DC, which is excellent given the large number of structural restraints (solution and solid-state NMR restraints). Some small deviations (≈5 ppm) that were observed could be due to imperfect CSA tensors for each amino acid residue as shown by Wang et al. (23); however, these deviations do not substantially affect the structure or domain orientations (see ref. 27). A summary of the parameters analyzed to assess the quality of the structures is reported in Table 1.
Table 1.
Summary of structural ensemble statistics
| RMSD from solution NMR restraints | |
| NOEs, Å (422 total) | 0.06 ± 0.006 |
| Torsion angles,° (99 total) | 0.41 ± 0.22 |
| PISEMA R-factors | |
| CSA (43 total) | 0.87 ± 0.02 |
| DC (39 total) | 0.99 ± 0.03 |
| RMSD from idealized covalent geometry | |
| Bond, Å | 0.006 ± 0.0004 |
| Angle, ° | 0.70 ± 0.03 |
| Impropers, ° | 0.21 ± 0.02 |
| Measure of structural quality, % residues in | |
| Most Favored Region | 90 ± 2 |
| Additional Allowed Region | 8 ± 3 |
| Generously Allowed Region | 2 ± 2 |
| Disallowed Regions | 0 |
| Precision of atomic coordinates, Å | |
| Backbone Atoms (4–50) | 2.3 |
| Domain Ia (residues 4–18) | 0.6 |
| Domains Ib/II (residues 24–50) | 0.6 |
| Protein Topology (domain in subscript) | |
| θIa, ° | 102 ± 2 |
| ρIa, ° | 92 ± 3 |
| InsertionIa, Å | 16.2 ± 0.8 |
| θIb,II, ° | 24 ± 2 |
| ρIb,II, ° | 204 ± 4 |
| InsertionIb,II, Å | 5.5 ± 0.5 |
Comparison of the Hybrid Conformational Ensemble with the Conformational Ensemble Obtained from Solution NMR Data Alone.
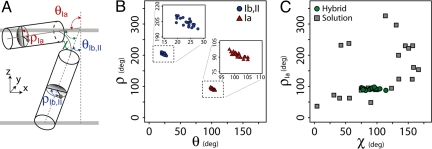
Because our hybrid ensemble has an angular dependence, it is not appropriate to perform a simple overlay of the backbone atoms as the criterion to calculate the precision of the ensemble. Rather, the 2 ensembles were compared on the basis of 3 angles (Fig. 3A): χ (the angle between the helix comprising domains Ib and II and the helix comprising domain Ia), ρIa (the rotation angle for domain Ia), and ρIb,II (rotation angle for domains Ib and II). Fig. 3C shows a plot of ρIa vs. the χ angle for the solution NMR ensemble alone and the final ensemble generated with the introduction of the orientational restraints of solid-state NMR. It is apparent that the solution NMR ensemble has a broad distribution of angles. Among the different conformers obtained in detergent micelles, we originally chose the L-shaped conformations on the basis of paramagnetic quenching experiments (15). Using the hybrid method, we do not have to select the structures manually; rather the orientational restraints derived from CSA and DC reduce the conformational space allowed, resulting in a structural ensemble with precise structures (Fig. S6A) and topologies (Fig. 3) with respect to the lipid bilayer. It is important to note that the deviations of the angles (the errors reported for ρ and θ in Table 1) reflect the quality of the fit and not the mosaic spread of the sample.
Fig. 3.
Comparison between the solution NMR ensemble and the conformational ensemble generated with the hybrid protocol. (A) Angles used to describe the topology of monomeric AFA-PLN. (B) Rotation (ρ) versus tilt angle (θ) for the 20 structures in the deposited ensemble. The Insets better allow for variations to be seen in the angles describing the topology with respect to the membrane normal for domains Ib and II (Left, circles) and domain Ia (Right, triangles). (C) Comparison of the ensemble of structures generated from solution NMR restraints alone (squares) with the hybrid solution and solid-state NMR method (circles).
Equilibration of DOPC Lipids Around the Average Hybrid Structure.
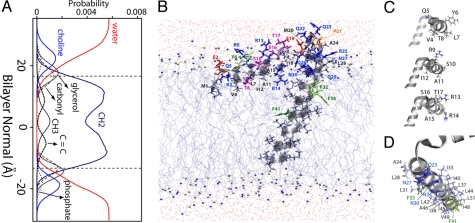
To fully describe the atomic-level interactions of PLN with lipids, we placed the average hybrid structure into an equilibrated DOPC lipid bilayer using CHARMM version 33a2 (30). We then performed 30 ns of molecular dynamics simulation using NAMD version 2.6 (31) to allow the lipids to equilibrate around the PLN structure, which was harmonically restrained with a force constant of 20 kcal mol−1Å−2 throughout the simulation. This ensured that the experimental restraints would be satisfied, and provided a detailed picture of the specific interactions the AFA-PLN monomer makes with DOPC lipids (Fig. 4B). The setup of the molecular dynamics calculations is reported in SI Text.
Fig. 4.
Structural topology of monomeric PLN in a DOPC lipid bilayer. (A) Probability distribution profile for the chemical groups of DOPC in the molecular dynamics simulation of PLN within the bilayer. (B) Structure of monomeric AFA-PLN in a DOPC lipid bilayer. (C) Detailed images of the residues within the amphipathic domain Ia helix show that the hydrophobic residues (Val-4, Leu-7, Ala-11, Ile-12, Ala-15) face the interior of the bilayer. (D) Structure highlighting the face of PLN within domains Ib and II that have been shown by mutagenesis and cross-linking data to interact with SERCA (56). Residues in D shown with side chains in sticks and labeled on the structure reside on the same helix of PLN, i.e., they are positioned for binding SERCA. In both B and D, the colors reflect the hydrophobicity: light gray, hydrophobic; blue/red/purple, hydrophilic; green, aromatic (Phe); specifically Ser and Thr residues are shown in purple, Glu is shown in red, and Asn and Gln are shown in blue.
Discussion
The complexity and challenges presented by membrane protein structure determination call for interdisciplinary approaches (32). X-ray crystallography and solution NMR have been widely used to determine high-resolution structures for several membrane proteins (33–36). These techniques are able to give atomic resolution information about the backbone and side chains but fall short in the elucidation of membrane protein topology. In contrast, solid-state NMR using oriented membrane protein samples can give molecular details regarding both backbone structure and topology. Because a number of membrane protein structures recently determined show similar folds in micelles, lipid bilayers, and crystals (34, 37–41), we propose to combine the restraints from these techniques into a unique protocol with the goal of obtaining the high-resolution structure (backbone and side chains) and topology within the lipid bilayer. In the literature, there are several examples of backbone structure determination of membrane protein using solid-state NMR data alone (42, 43) and a few examples reported for the combined use of solution and solid-state NMR information in a qualitative fashion (39, 44, 45). However, there is no precedent for the combination of these restraints into a total energy function, which includes distance, torsion angle, and orientational restraints with a semiempirical depth of insertion potential.
We applied this hybrid approach to the monomeric active form of PLN, which, despite its size, is a challenging system. In fact, PLN does not possess a compact fold, but instead, has its 3D architecture dictated by interactions with the lipid bilayer. Several in vivo and in vitro studies have shown that PLN adopts a remarkably similar structure both in micelles and lipid bilayers (8, 12, 15, 18, 19, 25, 46, 47), justifying the combination of restraints. In the event that restraints obtained in lipid bilayers and detergent micelles were incompatible, it is recommended that those from lipids be weighted more heavily, due to potential limitations of micelles.
Our structural ensemble [Protein Data Bank (PDB) ID code 2KB7], generated from simulated annealing procedures, shows PLN to exist in an L-shaped conformation (T state), while giving a qualitative view of its immersion depth within the lipid bilayer (Fig. 4). In close agreement with the static fit carried out on the initial solid-state NMR data (25, 48), the structural ensemble of PLN shows that the membrane embedded helix (domains Ib and II) crosses the membrane bilayer with a tilt angle of 24 ± 2° and an azimuthal (rotation) angle of 204 ± 4°. This indicates that a specific face of the helix, i.e., residues Arg-25, Gln-29, Phe-32, Ala-36, Leu-43, Ile-47, and Met-50, is on average oriented toward the SR lumen. Domain Ia lies on the surface of the lipid bilayer (Fig. 4), adopting an angle of 102 ± 2° with respect to the membrane plane. Although the PISA wheel pattern for a helix at 102° is exactly the same as that for 78°, the CSA and DC plotted vs. residue are able to distinguish these angles within the data. The rotation angle of this domain is challenging to resolve using PISEMA or SAMPI4 experiments, because of the small dispersion of CSA and DC values for helices positioned at ≈90° with respect to the bilayer normal. However, the inclusion of the Ez potential (17) helped overcome these ambiguities by identifying 2 energetically different ensembles. One ensemble had higher energy, with the hydrophobic face of the helix pointed toward the bulk solvent, and a second more energetically favorable ensemble, which had the hydrophobic face toward the hydrocarbon region. The latter agrees with several topological studies that we have carried out by using various hydrophilic and hydrophobic paramagnetic probes (15), and the direct observation of NOEs between the detergent and the methyl groups of Val-4 and Leu-7 in domain Ia (49).
For the final stage of refinement, we chose one of the structures from the conformational ensemble (model 6 from PDB ID code 2KB7) and embedded it into an explicit DOPC lipid bilayer for molecular dynamics calculations. Fig. 4 shows a snapshot from the simulation, which allows for appreciation of the atomic interactions of PLN within the lipid bilayer. Specifically, the hydrophobic side chains of residues Val-4, Leu-7, Ala-11, Ile-12, and Ala-15 are in contact with the hydrophobic interior (methylene region) of the membrane. Glu-2, Lys-3, and Tyr-6 are positioned in the interfacial region of the bilayer, and make hydrogen bonds with the DOPC lipid headgroup. Of the 3 arginine residues within domain Ia, Arg-9 and Arg-13 are solvent-exposed and accessible for interaction with PLN binding partners, whereas Arg-14 snorkels into the lipid bilayer headgroup region. Preliminary studies also indicate that the Nε of Arg-14 has the highest-order parameter of all arginines within AFA-PLN, which is consistent with increased solvent protection. This is noteworthy, because mutations at Arg-9 and Arg-14 have been linked to the abnormal regulation of SERCA, resulting in dilated cardiomyopathy (50, 51). Ser-16 and Thr-17 (the 2 phosphorylatable sites) are exposed to the bulk solvent, prone to interact with their respective kinases (i.e., protein kinase A and CamKII kinase, respectively). The amphipathic interactions of domain Ia with the lipid bilayer are also present in the oligomeric state of PLN, as measured by solution NMR in detergent micelles (14, 50), solid-state NMR in lipid bilayers (13, 52), EPR saturation transfer experiments in lipid bilayers and detergent micelles (8, 12, 13), and fluorescence resonance energy transfer in intact cells (46) and detergent reconstituted systems (53).
The topology of PLN determined with this approach also reveals that part of domain Ib extends outside the lipid bilayer. This reconciles the structural model of PLN with several datasets derived from solution NMR (11, 15, 54) and saturation transfer EPR measurements (8). Specifically, Gln-23 and Ala-24 are solvent exposed (15), whereas Arg-25, Gln-26 and Asn-27 are located within the choline/glycerol interface of the lipid headgroup region, in agreement with paramagnetic quenching and hydrogen/deuterium exchange experiments (15). Leu-28, Gln-29, and Asn-30 reside in the glycerol/carbonyl region, making hydrogen bonds with the carbonyl groups. Because this domain forms key inhibitory interactions with SERCA via hydrogen bonds and salt bridges (55), the depth of insertion may be an important mechanism regulating SERCA and PLN function.
Domain II is the most hydrophobic and rigid part of PLN. Hydrogen/deuterium exchange experiments show that the amide protons of this region exchange with the solvent on the timescale of days (15). This domain crosses the membrane bilayer with an angle of 24°, with the proposed SERCA binding face pointing away from the cytoplasmic domain Ia and prone to the interaction with the enzyme (Fig. 4D). Remarkably, domain II does not cross the 2 membrane leaflets completely; rather the C-terminal leucine (Leu-52) stops short in the luminal leaflet with its side chain in the methylene region of the bilayer and the carboxyl terminus snorkeling into the carbonyl region of the DOPC headgroups in contact with transient water molecules. Unlike the N-terminal domain Ia, the interactions of the C-terminal carboxyl group constitute the only anchor of domain II in the luminal leaflet of the membrane, an element that may confer the necessary mobility (piston-like motion) for PLN to be lifted up from the membrane to interact with SERCA (12).
The presence of amphipathic cytoplasmic domains is emerging as a common motif for single-pass regulatory and viral membrane proteins. In fact, the L-shaped membrane architecture of PLN is reminiscent of the structures and topologies of the FXYD proteins (a family of membrane associated proteins that serve as subunits to Na,K-ATPases) (40), the fd coat protein (responsible for viral assembly) (56), the VpU protein from HIV-1 (57), and the M2 channel from the influenza A virus (58). Amphipathic helix motifs are known to be functionally important in a number of capacities, ranging from stabilizing protein structures to sensing changes in the physical properties of the membrane (59, 60).
In our allosteric model of regulation (7), the amphipathic helix of PLN drives the conformational equilibrium toward the T state (the resting state), whereas the transition to the more pliable R-state is necessary for PLN to mold into SERCA's binding groove. The dynamic nature of the R-state was underscored by EPR analysis (7, 8, 12) and probably detected by magic angle spinning solid-state NMR experiments (47). Based on these data, it is likely that the interaction between lipids and the amphipathic domain Ia is modulated by the composition of the lipid membrane, and that these interactions may constitute an additional mechanism of SERCA regulation. Because PLN has at least 6 different binding partners (SERCA, protein kinase A, CamKII kinase, protein phosphatase I, A-kinase anchoring protein, and the antiapoptotic HS-1 associated protein X-1), the reversible binding to the membrane surface and the interplay between the T and R states may give the necessary flexibility in binding these proteins.
Materials and Methods
Sample Preparation.
[U-15N] AFA-PLN was expressed in Escherichia coli bacteria and purified as described (61). Most of the selectively labeled samples were also expressed in E. coli (Table S3). PLN samples that showed isotopic scrambling, [15N-Ser] and [15N-Gln], were synthesized by using Fmoc solid-phase peptide synthesis as reported (62, 63). For solution NMR experiments, AFA-PLN was solubilized in 300 mM DPC, 120 mM NaCl, and 20 mM NaH2PO4 (pH 6.0). Solid-state NMR samples were prepared in either 4/1 DOPC/1,2-dioleoyl-glycero-3-sn-phosphoethanolamine (DOPE) or DOPC lipid bilayers as described (no difference in spectra was discernable with or without DOPE) (25). Samples were hydrated for 4–5 days at 40 °C in a saturated solution of Na2HPO4 (the [U-15N] AFA-PLN in D2O).
NMR Spectroscopy.
Solution NMR experiments were collected in 300 mM DPC (pH 6) and at 37 °C. Solid-state NMR experiments PISEMA (20) and SAMPI4 (21) were acquired at 30 °C. Spectra of [U-15N] samples were acquired with 1 k scans and 30-t1 increments, whereas selectively labeled samples required 4- to 12 k scans and ≈8–16 increments. Experiments were performed at 1H frequencies of 600 and 700 MHz using Bruker (DMX) and Varian (VNMRS) spectrometers, respectively, equipped with low electric field flat-coil probes (64). Additional details are given in the SI Text.
Structural Calculations.
The structure calculations were carried out by using X-PLOR-NIH software (29). We defined a hybrid solution and solid-state NMR target function (ET), which is formulated as a combination of geometrical (Echem), solution NMR (EsolNMR), and solid-state NMR (EssNMR) terms:
The potential energy functions (E) are expressed in the form wE, where w represents the relative weight for each energy term, which has been optimized as reported by Shi et al. (27). All of the restraints are approximated by harmonic functions. Specifically, both CSA and DC potentials were implemented as flat-well potential functions according to Bertram and coworkers (65). The experimental errors for CSA and DC were set to 5 ppm and 0.5 kHz, respectively. A total of 500 monomers were generated. One hundred conformers displayed no violations and were selected for statistical analysis (Table 1). Further details regarding the calculations and molecular dynamics simulations in DOPC are described in the SI Text.
Supplementary Material
Acknowledgments.
We thank J. Hoch for helpful discussions, R. Bertram for sharing the DC and CSA python modules, C. Schwieters for help with the X-PLOR-NIH code, and P. Gor'kov, W. Brey, R. Fu, Z. Gan, K. Shetty, and T. Cross at the National High Magnetic Field Laboratory. This work was supported by the National Institutes of Health (Grants GM64742, HL80081, and GM072701, to G.V.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2KB7).
This article contains supporting information online at www.pnas.org/cgi/content/full/0904290106/DCSupplemental.
References
- 1.von Heijne G. Membrane-protein topology. Nat Rev Mol Cell Biol. 2006;7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 2.Tada M, Kirchberger MA, Katz AM. Phosphorylation of a 22,000-dalton component of the cardiac sarcoplasmic reticulum by adenosine 3′:5′-monophosphate-dependent protein kinase. J Biol Chem. 1975;250:2640–2647. [PubMed] [Google Scholar]
- 3.Hoshijima M, et al. Chronic suppression of heart-failure progression by a pseudophosphorylated mutant of phospholamban via in vivo cardiac rAAV gene delivery. Nat Med. 2002;8:864–871. doi: 10.1038/nm739. [DOI] [PubMed] [Google Scholar]
- 4.Kaye DM, et al. Percutaneous cardiac recirculation-mediated gene transfer of an inhibitory phospholamban peptide reverses advanced heart failure in large animals. J Am Coll Cardiol. 2007;50:253–260. doi: 10.1016/j.jacc.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 5.Simmerman HK, Jones LR. Phospholamban: Protein structure, mechanism of action, and role in cardiac function. Physiol Rev. 1998;78:921–947. doi: 10.1152/physrev.1998.78.4.921. [DOI] [PubMed] [Google Scholar]
- 6.Kimura Y, Kurzydlowski K, Tada M, MacLennan DH. Phospholamban inhibitory function is activated by depolymerization. J Biol Chem. 1997;272:15061–15064. doi: 10.1074/jbc.272.24.15061. [DOI] [PubMed] [Google Scholar]
- 7.Zamoon J, Nitu F, Karim C, Thomas DD, Veglia G. Mapping the interaction surface of a membrane protein: Unveiling the conformational switch of phospholamban in calcium pump regulation. Proc Natl Acad Sci USA. 2005;102:4747–4752. doi: 10.1073/pnas.0406039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karim CB, Kirby TL, Zhang Z, Nesmelov Y, Thomas DD. Phospholamban structural dynamics in lipid bilayers probed by a spin label rigidly coupled to the peptide backbone. Proc Natl Acad Sci USA. 2004;101:14437–14442. doi: 10.1073/pnas.0402801101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Traaseth NJ, et al. Structural and dynamic basis of phospholamban and sarcolipin inhibition of ca(2+)-ATPase. Biochemistry. 2008;47:3–13. doi: 10.1021/bi701668v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metcalfe EE, Zamoon J, Thomas DD, Veglia G. 1H/15N heteronuclear NMR spectroscopy shows four dynamic domains for phospholamban reconstituted in dodecylphosphocholine micelles. Biophys J. 2004;87:1–10. doi: 10.1529/biophysj.103.038844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metcalfe EE, Traaseth NJ, Veglia G. Serine 16 phosphorylation induces an order-to-disorder transition in monomeric phospholamban. Biochemistry. 2005;44:4386–4396. doi: 10.1021/bi047571e. [DOI] [PubMed] [Google Scholar]
- 12.Karim CB, Zhang Z, Howard EC, Torgersen KD, Thomas DD. Phosphorylation-dependent conformational switch in spin-labeled phospholamban bound to SERCA. J Mol Biol. 2006;358:1032–1040. doi: 10.1016/j.jmb.2006.02.051. [DOI] [PubMed] [Google Scholar]
- 13.Traaseth NJ, et al. Spectroscopic validation of the pentameric structure of phospholamban. Proc Natl Acad Sci USA. 2007;104:14676–14681. doi: 10.1073/pnas.0701016104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nesmelov YE, Karim CB, Song L, Fajer PG, Thomas DD. Rotational dynamics of phospholamban determined by multifrequency electron paramagnetic resonance. Biophys J. 2007;93:2805–2812. doi: 10.1529/biophysj.107.108910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zamoon J, Mascioni A, Thomas DD, Veglia G. NMR solution structure and topological orientation of monomeric phospholamban in dodecylphosphocholine micelles. Biophys J. 2003;85:2589–2598. doi: 10.1016/s0006-3495(03)74681-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mascioni A, Eggimann BL, Veglia G. Determination of helical membrane protein topology using residual dipolar couplings and exhaustive search algorithm: Application to phospholamban. Chem Phys Lipids. 2004;132:133–144. doi: 10.1016/j.chemphyslip.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 17.Senes A, et al. E(z), a depth-dependent potential for assessing the energies of insertion of amino acid side-chains into membranes: Derivation and applications to determining the orientation of transmembrane and interfacial helices. J Mol Biol. 2007;366:436–448. doi: 10.1016/j.jmb.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 18.Karp ES, Tiburu EK, Abu-Baker S, Lorigan GA. The structural properties of the transmembrane segment of the integral membrane protein phospholamban utilizing (13)C CPMAS, (2)H, and REDOR solid-state NMR spectroscopy. Biochim Biophys Acta. 2006;1758:772–780. doi: 10.1016/j.bbamem.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 19.Clayton JC, Hughes E, Middleton DA. The cytoplasmic domains of phospholamban and phospholemman associate with phospholipid membrane surfaces. Biochemistry. 2005;44:17016–17026. doi: 10.1021/bi0511383. [DOI] [PubMed] [Google Scholar]
- 20.Wu CH, Ramamoorthy A, Opella SJ. High-resolution heteronuclear dipolar solid-state NMR spectroscopy. J Mag Reson. 1994;109:270–272. [Google Scholar]
- 21.Nevzorov AA, Opella SJ. Selective averaging for high-resolution solid-state NMR spectroscopy of aligned samples. J Magn Reson. 2007;185:59–70. doi: 10.1016/j.jmr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 22.Buffy JJ, et al. Two-dimensional solid-state NMR reveals two topologies of sarcolipin in oriented lipid bilayers. Biochemistry. 2006;45:10939–10946. doi: 10.1021/bi060728d. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, et al. Imaging membrane protein helical wheels. J Magn Reson. 2000;144:162–167. doi: 10.1006/jmre.2000.2037. [DOI] [PubMed] [Google Scholar]
- 24.Marassi FM, Opella SJ. A solid-state NMR index of helical membrane protein structure and topology. J Magn Reson. 2000;144:150–155. doi: 10.1006/jmre.2000.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Traaseth NJ, Buffy JJ, Zamoon J, Veglia G. Structural dynamics and topology of phospholamban in oriented lipid bilayers using multidimensional solid-state NMR. Biochemistry. 2006;45:13827–13834. doi: 10.1021/bi0607610. [DOI] [PubMed] [Google Scholar]
- 26.Lambeth S, et al. NMR solution structure of phospholamban. Helv Chim Acta. 2000;83:2141–2152. [Google Scholar]
- 27.Shi L, et al. A refinement protocol to determine the structure, topology, and depth of insertion of membrane proteins using hybrid solution and solid-state restraints. J Biomolec NMR. 2009 doi: 10.1007/s10858-009-9328-9. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 29.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 30.Brooks BR, et al. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 31.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowieson NP, Kobe B, Martin JL. United we stand: Combining structural methods. Curr Opin Struct Biol. 2008;18:617–622. doi: 10.1016/j.sbi.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 33.Liang B, Tamm LK. Structure of outer membrane protein G by solution NMR spectroscopy. Proc Natl Acad Sci USA. 2007;104:16140–16145. doi: 10.1073/pnas.0705466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hiller S, et al. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321:1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baker KA, Tzitzilonis C, Kwiatkowski W, Choe S, Riek R. Conformational dynamics of the KcsA potassium channel governs gating properties. Nat Struct Mol Biol. 2007;14:1089–1095. doi: 10.1038/nsmb1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chill JH, Louis JM, Delaglio F, Bax A. Local and global structure of the monomeric subunit of the potassium channel KcsA probed by NMR. Biochim Biophys Acta. 2007;1768:3260–3270. doi: 10.1016/j.bbamem.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bayrhuber M, et al. Structure of the human voltage-dependent anion channel. Proc Natl Acad Sci USA. 2008;105:15370–15375. doi: 10.1073/pnas.0808115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grishaev A, Tugarinov V, Kay LE, Trewhella J, Bax A. Refined solution structure of the 82-kDa enzyme malate synthase G from joint NMR and synchrotron SAXS restraints. J Biomol NMR. 2008;40:95–106. doi: 10.1007/s10858-007-9211-5. [DOI] [PubMed] [Google Scholar]
- 39.Franzin CM, Teriete P, Marassi FM. Structural similarity of a membrane protein in micelles and membranes. J Am Chem Soc. 2007;129:8078–8079. doi: 10.1021/ja0728371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bogusky MJ, Leo GC, Opella SJ. Comparison of the dynamics of the membrane-bound form of fd coat protein in micelles and in bilayers by solution and solid-state nitrogen-15 nuclear magnetic resonance spectroscopy. Proteins. 1988;4:123–130. doi: 10.1002/prot.340040205. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Y, et al. NMR solution structure of the integral membrane enzyme DsbB: Functional insights into DsbB-catalyzed disulfide bond formation. Mol Cell. 2008;31:896–908. doi: 10.1016/j.molcel.2008.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu J, et al. Backbone structure of the amantadine-blocked trans-membrane domain M2 proton channel from influenza A virus. Biophys J. 2007;92:4335–4343. doi: 10.1529/biophysj.106.090183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J, Chen J, Brooks CL, 3rd, Im W. Application of solid-state NMR restraint potentials in membrane protein modeling. J Magn Reson. 2008;193:68–76. doi: 10.1016/j.jmr.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Opella SJ, et al. Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy. Nat Struct Biol. 1999;6:374–379. doi: 10.1038/7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Werner K, et al. Combined solid state and solution NMR studies of alpha,epsilon-15N labeled bovine rhodopsin. J Biomol NMR. 2007;37:303–312. doi: 10.1007/s10858-007-9143-0. [DOI] [PubMed] [Google Scholar]
- 46.Kelly EM, Hou Z, Bossuyt J, Bers DM, Robia SL. Phospholamban oligomerization, quaternary structure, and sarco(endo)plasmic reticulum calcium ATPase binding measured by fluorescence resonance energy transfer in living cells. J Biol Chem. 2008;283:12202–12211. doi: 10.1074/jbc.M707590200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andronesi OC, et al. Determination of membrane protein structure and dynamics by magic-angle-spinning solid-state NMR spectroscopy. J Am Chem Soc. 2005;127:12965–12974. doi: 10.1021/ja0530164. [DOI] [PubMed] [Google Scholar]
- 48.Mascioni A, Karim C, Zamoon J, Thomas DD, Veglia G. Solid-state NMR and rigid body molecular dynamics to determine domain orientations of monomeric phospholamban. J Am Chem Soc. 2002;124:9392–9393. doi: 10.1021/ja026507m. [DOI] [PubMed] [Google Scholar]
- 49.Traaseth NJ, Verardi R, Veglia G. Asymmetric methyl group labeling as a probe of membrane protein homo-oligomers by NMR spectroscopy. J Am Chem Soc. 2008;130:2400–2401. doi: 10.1021/ja711499r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmitt JP, et al. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science. 2003;299:1410–1413. doi: 10.1126/science.1081578. [DOI] [PubMed] [Google Scholar]
- 51.Haghighi K, et al. A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy. Proc Natl Acad Sci USA. 2006;103:1388–1393. doi: 10.1073/pnas.0510519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abu-Baker S, et al. The structural topology of wild-type phospholamban in oriented lipid bilayers using 15N solid-state NMR spectroscopy. Protein Sci. 2007;16:2345–2349. doi: 10.1110/ps.072977707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robia SL, Flohr NC, Thomas DD. Phospholamban pentamer quaternary conformation determined by in-gel fluorescence anisotropy. Biochemistry. 2005;44:4302–4311. doi: 10.1021/bi0478446. [DOI] [PubMed] [Google Scholar]
- 54.Traaseth NJ, Thomas DD, Veglia G. Effects of Ser16 phosphorylation on the allosteric transitions of phospholamban/Ca(2+)-ATPase complex. J Mol Biol. 2006;358:1041–1050. doi: 10.1016/j.jmb.2006.02.047. [DOI] [PubMed] [Google Scholar]
- 55.Toyoshima C, et al. Modeling of the inhibitory interaction of phospholamban with the Ca2+ ATPase. Proc Natl Acad Sci USA. 2003;100:467–472. doi: 10.1073/pnas.0237326100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marassi FM, Opella SJ. Simultaneous assignment and structure determination of a membrane protein from NMR orientational restraints. Protein Sci. 2003;12:403–411. doi: 10.1110/ps.0211503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marassi FM, et al. Correlation of the structural and functional domains in the membrane protein vpu from HIV-1. Proc Natl Acad Sci USA. 1999;96:14336–14341. doi: 10.1073/pnas.96.25.14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tian C, Gao PF, Pinto LH, Lamb RA, Cross TA. Initial structural and dynamic characterization of the M2 protein transmembrane and amphipathic helices in lipid bilayers. Protein Sci. 2003;12:2597–2605. doi: 10.1110/ps.03168503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Drin G, et al. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 60.Cornell RB, Taneva SG. Amphipathic helices as mediators of the membrane interaction of amphitropic proteins, and as modulators of bilayer physical properties. Curr Protein Pept Sci. 2006;7:539–552. doi: 10.2174/138920306779025675. [DOI] [PubMed] [Google Scholar]
- 61.Buck B, et al. Overexpression, purification, and characterization of recombinant ca-ATPase regulators for high-resolution solution and solid-state NMR studies. Protein Expr Purif. 2003;30:253–261. doi: 10.1016/s1046-5928(03)00127-x. [DOI] [PubMed] [Google Scholar]
- 62.Vorherr T, Wrzosek A, Chiesi M, Carafoli E. Total synthesis and functional properties of the membrane-intrinsic protein phospholamban. Protein Sci. 1993;2:339–347. doi: 10.1002/pro.5560020306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karim CB, Marquardt CG, Stamm JD, Barany G, Thomas DD. Synthetic null-cysteine phospholamban analogue and the corresponding transmembrane domain inhibit the ca-ATPase. Biochemistry. 2000;39:10892–10897. doi: 10.1021/bi0003543. [DOI] [PubMed] [Google Scholar]
- 64.Gor'kov PL, et al. Using low-E resonators to reduce RF heating in biological samples for static solid-state NMR up to 900 MHz. J Magn Reson. 2007;185:77–93. doi: 10.1016/j.jmr.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 65.Bertram R, et al. Atomic refinement with correlated solid-state NMR restraints. J Magn Reson. 2003;163:300–309. doi: 10.1016/s1090-7807(03)00147-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.