Abstract
Background
The genetics of spikelet formation, a feature unique to grasses such as rice and maize, is yet to be fully understood, although a number of meristem and organ identity mutants have been isolated and investigated in Arabidopsis and maize. Using a two-element Ac/Ds transposon tagging system we have isolated a rice mutant, designated branched floretless 1 (bfl1) which is defective in the transition from spikelet meristem to floret meristem.
Results
The bfl1 mutant shows normal differentiation of the primary rachis-branches leading to initial spikelet meristem (bract-like structure equivalent to rudimentary glumes) formation but fails to develop empty glumes and florets. Instead, axillary meristems in the bract-like structure produce sequential alternate branching, thus resulting in a coral shaped morphology of the branches in the developing panicle. The bfl1 mutant harbours a single Ds insertion in the upstream region of the BFL1 gene on chromosome 7 corresponding to PAC clone P0625E02 (GenBank Acc No. message URL http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=nucleotide&list_uids=34395191&dopt=GenBank&term=ap004570AP004570). RT-PCR analyses revealed a drastic reduction of BFL1 transcript levels in the bfl1 mutant compared to that in the wild-type. In each of the normal panicle-bearing progeny plants, from occasional revertant seeds of the vegetatively-propagated mutant plant, Ds was shown to be excised from the bfl1 locus. BFL1 contains an EREBP/AP2 domain and is most likely an ortholog of the maize transcription factor gene BRANCHED SILKLESS1 (BD1).
Conclusions
bfl1 is a Ds-tagged rice mutant defective in the transition from spikelet meristem (SM) to floret meristem (FM). BFL1 is most probably a rice ortholog of the maize ERF (EREBP/AP2) transcription factor gene BD1. Based on the similarities in mutant phenotypes bfl1 is likely to be an allele of the previously reported frizzy panicle locus.
Background
The orderly production of meristems with specific fates is crucial for the proper development of plant architecture. The shoot apical meristem (SAM) is the ultimate source of all aerial structures of the plant, including inflorescences and flowers. The identity of a given meristem is defined by the types of structures it produces [21]. The SAM produces leaves in its vegetative developmental stage, but begins to generate an inflorescence meristem after switching to the reproductive developmental stage [23].
The large number of genes involved in this complex process can be classified into three categories, namely flowering time genes, meristem identity genes and organ identity genes [18]. The flowering time genes are those involved in the transition from a vegetative meristem to a reproductive meristem. Many mutations that affect flowering time have been described in Arabidopsis [3,6,18,27] and some in rice [12,38]. Once this transition is achieved, the inflorescence meristem (IM) produces intermediate meristems that give rise to floret meristems (FMs) to produce floral organs [21].
A number of meristem and organ identity genes have been investigated in Arabidopsis and Antirrhinum [28]. Although monocots are expected to have molecular and genetic mechanisms of flowering similar to those of dicots, little is known about the genetics of spikelet formation – a feature unique to grasses such as rice and maize. Many developmental mutants that affect the elaboration of the inflorescence have been isolated in maize [21] but, to date, only a few of the corresponding genes have been cloned. The maize KNOTTED1 (KN1) is one such spikelet meristem (SM) identity gene and encodes a homeodomain-containing protein which acts as a transcription factor [33]. The kn1 mutant develops fewer branches and spikelet pairs because of defects in inflorescence meristem maintenance [14]. Another maize gene LIGULELESS2 (LG2) has been shown to encode a basic-leucine zipper (bZIP) protein, and the lg2 mutant plants have reduced long tassel branches [34]. One of the well characterized maize SM identity genes is INDETERMINATE SPIKELET1 (IDS1) that encodes an AP2-like transcription factor, and the mutant gene ids1 specifies determinate fate by suppressing indeterminate growth within the spikelet meristem [8]. Recently, yet another maize SM identity gene BRANCHED SILKLESS1 (BD1), encoding a transcription factor containing an ERF (EREBP/AP2) domain has been cloned and characterized [9]. BD1 specifies SM identity by repressing indeterminate branch fate within the lateral domain of the SM [9] and plays a crucial role in mediating the transition from spikelet to floret meristem during maize ear development [10]. The bd1 mutant shows highly branched ears [9,10].
Although more than 20 genes that control panicle morphology have been reported in rice [17], few have been analyzed in detail [15,22] and none has been cloned to date. However, some rice orthologs of floret meristem identity genes of Arabidopsis and Antirrhinum have been cloned and their expression patterns investigated. For instance, in Arabidopsis and Antirrhinum, the indeterminate state of the IM is maintained by TERMINAL FLOWER1 (TFL1) and CENTRORADIALIS (CEN), respectively. Mutations in these genes result in the IM being converted into a terminal flower [7,25]. Over-expression of RCN1 and RCN2, two rice TFL1/CEN orthologs, resulted in a delayed transition to the reproductive phase and more branched panicles due to a delayed switch from the branch shoot to FM [23]. The expression pattern of RFL, the rice ortholog of the Antirrhinum FLORICAULA (FLO) and Arabidopsis LEAFY (LFY) genes, indicates that it plays a distinctly-different role from that of LFY in Arabidopsis and FLO in Antirrhinum [16]. More recently, a BD1-like gene has also been cloned from rice but has yet to be functionally characterized [9].
Here we report the isolation of a Ds-tagged rice mutant designated branched floretless 1 (bfl1) and identification of the BRANCHED FLORETLESS 1 (BFL1) gene. The bfl1 mutant showed normal differentiation of the primary rachis-branches leading to initial SM (bract-like structure equivalent to rudimentary glumes) formation but failed to develop empty glumes and florets. Instead, axillary meristems in the rudimentary glumes produced sequential alternate branching resulting in a highly-branched panicle. BFL1 encodes a transcription factor containing an EREBP/AP2 domain, identical to the rice BD1-like gene cloned recently [9]. Genotypic and phenotypic analyses of the bfl1 mutant and BFL1 revertants suggest that the BFL1 is essential for the transition from SM to FM in rice.
Results
bfl1 is a mutant defective in the transition from SM to FM
Out of 20 progeny of a mutagenic plant B2-8A-2-18 (F2 generation) derived from a cross between iAc and DsG transgenic lines [31], we found a mutant plant (B2-8A-2-18-16) with panicle characteristics similar to a previously reported EMS-induced frizzy panicle mutant fzp [20] and a γ-ray induced frizzy panicle mutant fzp2 [15]. We have designated this mutant branched floretless 1 (bfl1).
The panicle of a bfl1 mutant looks like a stick and its rachis-branches never stretch because of tangling (Fig. 1B). Development of the primary rachis-branches appeared to be normal (Fig. 1C). The panicle length of wild-type and the bfl1 mutant were 15.5 cm and 14.9 cm, respectively and the number of primary rachis-branches of wild-type and the mutant were 6.1 and 6.4, respectively. However, lateral and terminal spikelets of primary and secondary order rachis-branches failed to produce florets. Instead, they continued to produce next-order rachis branches in alternate axis (Fig. 1G, 2E) most likely after initiation of bract-like structures (Fig. 1F, 1G, 2E, 2F). As observed in the scanning electron mirgrographs (SEMs), the bract-like structure in the mutant (Fig. 2F,2G,2H) corresponds to rudimentary glumes of wild-type (Fig. 2B,2C,2D) because they have comparable positions and surface appearances. The mutant showed no difference from the wild-type plant with respect to vegetative growth, or to the time of transition from vegetative phase to reproductive phase.
Figure 1.
Inflorescence morphology of wild-type and bfl1 mutant. A, Mature panicle of wild-type. B, Mature panicle of bfl1 mutant. C, Manually stretched mature panicle of bfl1 mutant. D, Primary rachis-branch (PB) of wild-type at the stage between spikelet primordium and spikelet organ differentiation, which is composed of secondary rachis-branch (SB), lateral spikelets (LS) and terminal spikelet (TS). E, Secondary rachis-branch of wild-type at microspore developmental stage, which is composed of lateral spikelets and terminal spikelet. F, Primary rachis-branch of bfl1 mutant at the developmental stage equivalent to D, alternating higher level rachis-branches are continuously developed instead of spikelets. G, Secondary rachis-branch of bfl1 mutant at the developmental stage equivalent to E. Abbreviations: DP: degenerate point; PB: primary rachis-branch; SB: secondary rachis-branch; LS: lateral spikelet; TS: terminal spikelet; TB: tertiary rachis-branch; BM: branch meristem; RG: rudimentary glume (indicated by triangles).
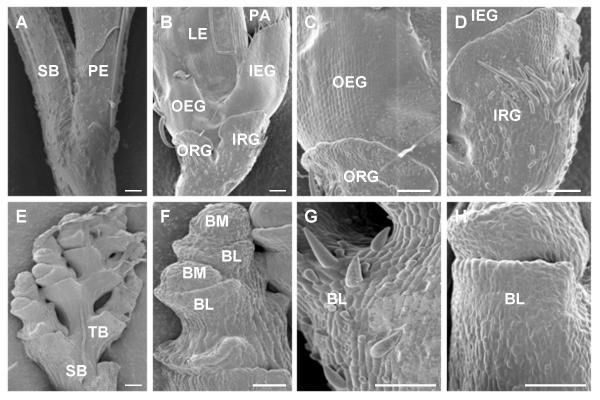
Figure 2.
Scanning electron mircrographs (SEMs) of spikelet structures of wild-type and bfl1 mutant plants. A, Junction of secondary rachis-branch (SB) and pedicel (PE) of spikelet in wild-type. B, A close-up of the basal part of a spikelet showing all the spikelet organs of wild-type. C, A close-up of the surface of outer rudimentary glume (ORG) and outer empty glume (OEG) of a wild-type spikelet. The cells of empty glume are orderly arranged and have a smooth flat surface, whereas the cells of rudimentary glume have irregular shapes and a rugose surface bearing hairy structures. D, Surface of inner rudimentary glume (IRG) of wild-type, having short or long hairs. E, Secondary rachis-branches of bfl1 and subsequently formed tertiary and higher order rachis-branches which develop continuously in alternating axes. F, Bract-like structure (BL) and rachis-branch meristem (BM) of bfl1. Following differentiation BL, BM either arrests its growth or continues to produce new BL. Most likely BL is equivalent to rudimentary glume because of the resemblance of the surface structures. G and H, Close-up view of the surface of BL of bfl1. Bars in all diagrams represent 100 μm. Abbreviations: SB: secondary rachis-branch; TB: tertiary rachis-branch; PE: pedicel of spikelet; ORG: outer rudimentary glume; IRG: inner rudimentary glume; OEG: outer empty glume; IEG: inner empty glume; PA: palea; LE: lemma; BL: bract-like structure; BM: branch meristem.
bfl1 is a Ds-tagged mutant
PCR analyses showed that the original bfl1mutant contained both iAc and Ds (Table 2) and was negative for GFP (the T-DNA or Ds launching pad selection marker), indicating that the transposed Ds and iAc have not segregated but the original Ds launching pad has segregated away. As iAc was present, excision of Ds and possible reversion to wild-type phenotype could be expected. The mutant plant was therefore propagated vegetatively to recover seeds produced after reversion events.
Table 2.
PCR characterization of bfl1 mutant and its putative revertants (PR)
| Plant | Ac PCR | Ds PCR | PCR1a | PCR2a | PCR3a | Genotype of Ds insertion | Phenotype |
| bfl1 | + | + | - | + | + | Ds/Ds | Mutant |
| PR1 | + | - | - | + | - | Ds/Ds | Mutant |
| PR2 | + | + | + | - | - | +/+ | Normal |
| PR3 | + | + | - | + | + | Ds/Ds | Mutant |
| PR4 | + | + | - | + | + | Ds/Ds | Mutant |
| PR5 | - | + | + | - | - | +/+ | Normal |
| PR6 | + | + | - | + | + | Ds/Ds | Mutant |
| PR7 | + | + | + | + | + | Ds/+ | Normal |
| PR8 | + | + | - | + | + | Ds/Ds | Mutant |
| PR9 | + | + | - | + | + | Ds/Ds | Mutant |
| PR10 | - | + | - | + | + | Ds/Ds | Mutant |
| PR12 | ND | + | + | - | - | +/+ | Normal |
| WT | - | - | + | - | - | +/+ | Normal |
a): PCR1, PCR2 and PCR3 were done with primers LW1125_For and LW1125_Rev, LW1125_Rev and Ds5_112-, and LW1125_For and Ds3_6587+, respectively.
In such vegetatively-propagated plants, several revertant sectors were found within a tiller, or in different tillers (Fig. 3A, 3B). In total, 15 normal mature seeds (Fig. 3E), 11 sterile and abnormal seeds (Fig. 3F, 3G) and 6 incomplete spikelets (Fig. 3H) were obtained. Out of these 15 putative revertant seeds, 10 germinated (PR1 to PR10) and were grown to maturity. PR2, PR5 and PR7 showed normal panicle development and partial sterility (Fig. 3C; Table 2), whereas the other 7 plants (PR1, PR3, PR4, PR6, PR8, PR9 and PR10) still displayed the bfl1 phenotype (Fig. 3D; Table 2). Ds-specific PCR analysis showed positive results for PR2 to PR10 and a negative result for PR1 (Table 2). The latter was attributed to a possible Ds3' end truncation as indicated in the subsequent Southern blot hybridization analyses (see below).
Figure 3.
Revertants of bfl1 mutant. A, Heading panicle with revertant spikelets. B, Mature panicle with revertant seeds. C, Panicles of PR2 showing normal developed spikelets. D, Panicles of PR3 showing bfl1 mutant phenotype. E, Normal looking revertant seed. F, Abnormal sterile revertant seed. G, Sterile revertant spikelet with an extra sterile spikelet inside. H, Sterile revertant spikelet with just lemma.
Ds3' (LW1125; GenBank Acc. No. message URL http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=nucleotide&list_uids=33771666&dopt=GenBank&term=bfl1AY343496) and Ds5' (LW1455; GenBank Acc. No. message URL http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=nucleotide&list_uids=33771665&dopt=GenBank&term=bfl1AY343495) flanking sequences were rescued from the bfl1 mutant and PR1 using TAIL-PCR [19] and plasmid rescue system, respectively. Initial searches (February 2002) failed to identify any homologous sequences from GenBank (NCBI) database but the China Rice Genome database http://btn.genomics.org.cn/rice/ contained one entry (contig 239) showing homology to bfl1 flanking sequences. PCR characterization of the bfl1 locus and gene prediction were therefore based on contig 239. Subsequently, the Ds insertion in the bfl1 mutant was mapped to rice chromosome 7 corresponding to PAC clone P0625E02 (GenBank Acc No. message URL http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=nucleotide&list_uids=34395191&dopt=GenBank&term=ap004570AP004570).
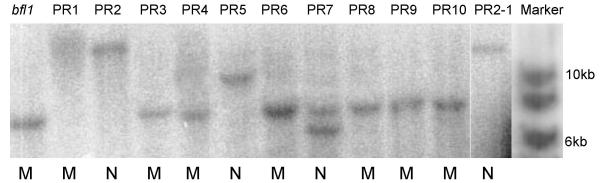
Southern blot hybridization analyses with the gus probe (which hybridizes with the 3' end of the Ds insertion) showed that the bfl1 mutant had a single-copy of Ds showing one 7.2 kb positively-hybridizing band (Fig. 4). PR3, PR4, PR6, PR8, PR9 and PR10 also had the same 7.2 kb band and showed mutant panicles, indicating that they were homozygous Ds insertion plants resulting from self-pollination of gametes with Ds in the bfl1 locus. PR1, however, produced mutant panicles but did not show any positively-hybridizing band, suggesting a deletion of the 3' region of Ds comprising the gus gene sequences. PR2 and PR5 did not have the same 7.2 kb band, but had a band of a different size indicating that they were homozygous revertants with Ds excised from the bfl1 locus and re-inserted into other locations in the rice genome. PR7 had one extra positively-hybridizing band besides the 7.2 kb band, indicating that it was a heterozygous revertant resulting from fertilization of a gamete with Ds in the bfl1 locus by a gamete with a re-transposed Ds. Thus, normal panicle-development appeared to result from excision of Ds from the bfl1 locus. Based on the sequences of contig 239 and the PAC clone P0625E02, the expected size of the positively-hybridizing band in the Southern blot analyses of HindIII-digested bfl1 mutant plant DNA with thegus probe is 9.3 kb. However, the observed band size was ~7.2 kb. A close observation of the published sequence of this fragment revealed that a single nucleotide substitution could result in an additional HindIII restriction site, and a shorter positively-hybridizing band. We tried locating the possible nucleotide substitution in various sources of genomic DNA including a freshly amplified DNA but failed to see any nt transition. Most likely this particular transition varies between different samples of Nipponbare. The position of the HindIII RE recognition site formed due to possible transition is indicated in Fig. 6. Nevertheless, our genomic sequences flanking both Ds3 (GenBank Acc. No. message URL http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=nucleotide&list_uids=33771666&dopt=GenBank&term=bfl1AY343496) and Ds5 (GeneBank Acc. No. message URL http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=nucleotide&list_uids=33771665&dopt=GenBank&term=bfl1AY343495) ends unambigously maps the location of the Ds insertion.
Figure 4.
Southern blot hybridization analyses of Ds insertion in the bfl1 mutant and Ds excision in revertants. Genomic DNA (~10 μg) was extracted from putative revertants, digested with HindIII, fractionated on a 0.7% agarose gel, and blotted to a Hybond-N+ membrane. The membrane was hybridised with a gus probe (see Figure 6). PR1 to PR10 are revertants of the original bfl1 mutant and PR2-1 is one of the progenies of PR2. Abbreviations: N, wild-type phenotype; M, mutant phenotype.
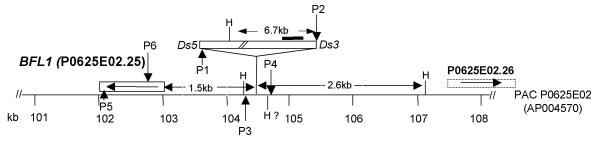
Figure 6.
Genomic location of BFL1 and Ds insertion in bfl1 mutant. Schematic representation of the positions of the BFL1 gene and the Ds insertion in bfl1 mutant in the Rice genomic PAC clone P0625E02 (GenBank Acc. No.AP004570). The orientation of the BFL1 gene (P0625E02.25) is indicated by arrow. Binding sites of primers Ds5-112- (P1), Ds3-6587+ (P2), LW1125_Rev (P3), LW1125_For (P4) used in PCR1, PCR2 and PCR3, and OsBfl1_Rev2 (P5) and OsBfl1_For (P6) used in RT-PCR (see Table 2 for details) are indicated. Solid line represents gus probe used for Southern blot hybridization analysis. HindIII restriction sites are indicated (H).
Three sets of PCRs were performed to confirm the presence or absence of a Ds insertion in the bfl1 locus: PCR1 to amplify the region flanking the Ds insertion; PCR2 and PCR3 to amplify part of the 5' and 3' ends of the Ds element and their flanking genomic regions, respectively (Table 1; Fig. 6). PR2 and PR5 were positive for PCR1 and negative for PCR2 and PCR3 confirming that they were homozygous revertants. PR7 was positive for PCR1, PCR2 and PCR3 confirming that it was a heterozygous revertant (Table 2). The genotypes of PR2, PR5 and PR7 were confirmed by subsequent analyses of progeny plants. The PCR results of PR1 confirmed that it had a Ds3' truncation (Table 2). All other plants were negative for PCR1 and positive for PCR2 and PCR3, suggesting that there had been no excision of the Ds element from the bfl1 locus.
Table 1.
Primers used in this study.
| Primer | Sequence | Target |
| Ds5_112- | ATCGGTTATACGATAACGGTC | Ds5' |
| Ds3_1 | ACCCGACCGGATCGTATCGGT | Ds3' |
| Ds3_3 | GTATTTATCCCGTTCGTTTTCGT | Ds3' |
| Ds3_6587+ | CCGTCCCGCAAGTTAAATATG | Ds3' |
| AD2 | NGTCGA(G/C)(A/T)GANA(A/T)GAA | |
| GPAInt | TCCAAGTCCACAAGGAAAATTG | Ds |
| GUS_313- | TCACTTCCTGATTATTGACCCAC | Ds |
| Ac_1931+ | CAGCTCCAAAGACAAAGACAAC | Ac |
| Ac_2382- | TGCAGCAGCAATAACAGAGTC | Ac |
| LW1125_For | TGTGGAGGAGAAATTAGACAGG | Ds insertion flank (P0625E02) |
| LW1125_Rev | CAGTGTGAAATGTGTAGAAGGG | Ds insertion flank (P0625E02) |
| OsBfl1_For | GCACCAACTTCGTCTACACCCA | BFL1 (P0625E02) |
| OsBfl1_Rev2 | TGAATGGAGAGTAGGAGTCGGAGC | BFL1 (P0625E02) |
| RSs1_F | TGCCTTGATCGAAGCTGAC | RSs1 |
| RSs1_R | AGCAAGGGGTAGAGGCTCTC | RSs1 |
Taken together, the above analyses strongly suggest that the insertion of the Ds is the cause of the bfl1 mutation and the excision of the Ds from the bfl1 locus is associated with the reversion of the mutation.
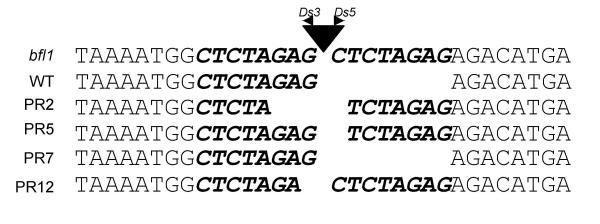
It is a well known fact that during Ds insertion it creates 8 bp host sequence duplication. However, when the Ds is re-excised it often leaves footprints of this direct repeat sequences. Ds excision footprints of PR2, PR5, PR7 and PR12 showed retention of 4, 7, 0 and 7 bp of the duplicated host sequence, respectively (Fig. 5). Although the proper reading frame could be restored with the excision of Ds in PR7, the reading frame would not be restored for PR2, PR5 and PR12, suggesting that the Ds insertion was not in the coding region of the BFL1 gene, but in its non-coding or regulatory region.
Figure 5.
Identification of Ds excision footprints. Ds excision footprints of PR2, PR5, PR7 and PR12 that showed normal looking panicle. The footprints were amplified using primers LW1125_For and LW1125_Rev, and sequenced with primer LW1125_For (see Table 2 and Fig. 6 for details)
Subsequently, we obtained additional revertants, including a whole normal panicle with 34 seeds. When 26 seeds of this panicle were germinated and grown to maturity, they showed segregation of homozygous un-excised Ds (Ds/Ds), heterozygous Ds excision (Ds/+) and homozygous Ds excision (+/+) in the ratio of 1:2:1 (6:13:7). All Ds excision plants had the same excision footprint as PR12 (Table 2 and Fig. 5), indicating that there had been germinal excision of Ds during the transition from vegetative to reproductive stage.
BFL1 is a transcription factor gene with an EREBP/AP2 domain
Analyses of 10-kb sequence of the ~30-kb contig 239 from the China Rice Genome database harboring the BFL1 locus by gene prediction programs FGENESH http://www.softberry.com/berry.phtml/ and GENSCAN http://genes.mit.edu/GENSCAN.html identified a single-exon gene capable of encoding a protein with the DNA binding domain of the EREBP/AP2 family of plant transcription factors [26,36], 1515 bp downstream from the Ds insertion. Subsequently, the same gene was predicted in the rice PAC clone P0625E02 sequences (Fig. 6). The predicted message URL http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=nucleotide&list_uids=34899319&dopt=GenBank&term=P0625E02.25P0625E02.25 protein corresponds to our predicted protein of 318 amino acids. In view of the function of EREBP/AP2 domain genes, and the excision of Ds from the bfl1 locus resulting in revertants with normally-developed panicles, we concluded that this EREBP/AP2-domain gene is likely to be the BFL1 gene. Furthermore, the loss or reduction of the BFL1 expression in mutant plants was confirmed by gene-specific RT-PCR using total RNA isolated from leaf and developing panicles (Fig. 7). BFL1 was expressed in wild-type leaves and developing panicles but was greatly reduced in the bfl1 mutant. The expression of BFL1 in revertant panicles was evident in the RT-PCR analysis (Fig. 7). However, we could not backup the RT-PCR result with Northern blot analysis possibly due to very low expression level.
Figure 7.
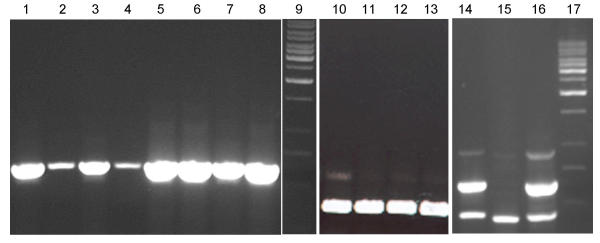
BFL1 transcript analysis by RT-PCR. Semi-quantitative detection of BFL1 transcripts in total RNA samples isolated from wild-type leaf (lane 1) and developing panicle (lane 3) and mutant leaf (lane 2) and developing panicle (lane 4), and same samples without DNase treatment (lanes 5–8) amplified using BFL1 gene-specific primers. Lanes 10–13 are same samples amplified using rice RSs1 gene specific primers (binding to two exons flanking an intron) to serve as internal controls for RNA quantity and DNA contamination. Semi-quantitative detection of BFL1 transcripts along with internal control (RSs1) in total RNA samples isolated from wild-type (lane 14), bfl1 mutant (lane 15) and revertant (lane 16) panicles. Lanes 9 and 17 are the molecular wt. markers.
The EREBP/AP2 domain of BFL1 was found to be identical to the ERF domain of a recently-isolated maize BD1 gene and that of a rice BD1-like gene [9]. The EREBP/AP2 domain of BFL1 also shared a high level of similarity with those of class II ERFs, such as LEAFY PETIOLE and TINY of Arabidopsis [32,37] and, as in LEAFY PETIOLE, the EREBP/AP2 domain of BFL1 was located close to the N-terminus of the protein (Fig. 8).
Figure 8.
Comparison of EREBP/AP2 domain of BFL1 with that of other ERF transcription factors. Multiple sequence alignment of BFL1, the maize BD1, the tobacco EREBP3, and the Arabidopsis LEAFY PETIOLE, TINY and ERF4 proteins using PILEUP program of GCG [11] with gap weight of 8 and gap length weight of 2. Consensus sequence was derived with PRETTY program of GCG with the minimum plurality of 2. EREBP/AP2 (ERF) domain is boxed. Note that BFL1 and BD1 have 80% identity at the predicted amino acid sequence and identical EREBP/AP2 domains.
Discussion
BFL1 mediates the transition from SM to FM during the development of the rice panicle
In rice, immediately after the transition to the reproductive stage, the SAM switches into an IM, from which primary rachis-branch meristems (PBMs) initiate in a spiral phyllotaxy. The IM degenerates after a given number of PBMs have been initiated. Concurrently, primary rachis-branches differentiate into secondary rachis-branch meristems (SBMs) or lateral spikelet meristems (LSMs) alternatively, and are terminated by terminal spikelet meristems (TSMs). SMs produce two pairs of glumes, rudimentary and empty glumes, as well as a single floret composed of lemma, palea, lodicules, stamens and carpel [29]. Therefore, the rice inflorescence or panicle represents a determinate branched structure composed of a main spike bearing primary and secondary rachis-branches, lateral and terminal spikelets (Fig. 1A). Each rice spikelet contains a single floret, a pair of empty glumes and a pair of rudimentary glumes in contrast to the pair of glumes and two florets present in maize but it is still to be resolved whether empty glumes or rudimentary glumes are analogous to maize glumes [5,13,29].
In the bfl1 mutant, although the development of the SAM is normal until primary rachis-branches (PBs) are produced, lateral and terminal spikelets are replaced by secondary rachis-branches (SBs) which then continually initiate higher order branches in a distichous phyllotaxy resulting in an indeterminate branched panicle. Based on our observation, the rudimentary glumes of spikelets seemed to be initiated normally as bract-like structures in the bfl1 mutant (Fig. 1G, 2F), but subsequent SMs ceased development and failed to produce empty glumes and FM, instead, their identity was converted to that of a branch meristem (BM), indicating that SM identity is acquired but fails to transit from SM to FM. Therefore, we propose that the function of BFL1 is to repress BM identity within the spikelet, or to positively regulate the initiation of FM. Similarly, in the maize bd1 mutant, ear SMs could not transit to FMs and continue to produce glumes and spikelet-like structure [10]. As we believe that the spikelet initiation has occurred in bfl1 it is logical to assume that the bract-like structure (rudimentary glume) are in fact actual glumes as discussed previously [29].
BFL1 is a rice ortholog of the maize BD1 gene
Public availability of the near-complete rice genome sequences (http://www.ncbi.nlm.nih.gov/;http://rgp.dna.affrc.go.jp/; http://btn.genomics.org.cn/rice; http://portal.tmri.org/rice/) allowed us to locate the single Ds insertion in the bfl1 mutant on rice chromosome 7 corresponding to the PAC clone P0625E02 and indicated that the gene most likely to have been affected by this Ds insertion is one which encodes an EREBP/AP2 domain-like protein (message URL http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=nucleotide&list_uids=34899319&dopt=GenBank&term=P0625E02.25P0625E02.25). Since this domain is identical to the ERF domains of the recently reported maize BD1 gene and the rice BD1 – like gene [9] we conclude that BFL1 is the rice ortholog of maize BD1.
Although ERF proteins are known as plant-specific transcription factors involved in the ethylene response [24], they are also implicated in plant development [4,32,37]. The direct evidence for the role of ERF proteins in organogenesis comes from the characterization of the maize BD1. The BD1 gene mediates the transition from a spikelet to a floret meristem during maize ear development [9,10] and its expression is temporally and spatially regulated during spikelet and floret initiation [9]. Strong phenotypic resemblance between the maize bd1 mutant and our rice bfl1 mutant further supports our claim that BFL1 is the rice ortholog of BD1.
Failure to detect BFL1 transcript by Northern blot analysis was most probably due to its low expression. It is not uncommon to have such a low expression levels of transcription factor genes such as BFL1. However, expression studies using RT-PCR under optimal conditions indicated substantial reduction in the BFL1 transcript levels in the bfl1 mutant compared to wild-type and was restored in the revertant (Fig. 7). Further studies on spatial and developmental expression patterns of BFL1 in wild-type, mutant, and revertant plants are needed to define the regulation of BFL1 expression.
The Ds insertion in bfl1 blocks the formation of the transcription initiation complex of the BFL1
Based on our prediction, the transcription of BFL1 starts at 121 nt upstream of the translation start site (TSS). A TATA box and the Ds insertion are located 157 and 1515 nt upstream of the TSS, respectively, suggesting that the Ds insertion is not in the core promoter region of BFL1 gene. The observed mutant phenotype and the RT-PCR data, however, suggest that the Ds insertion severely affects the expression of the BFL1 gene, thus suggesting that the region upstream of the core promoter is required for efficient transcription initiation of the BFL1 gene. The Ds insertion in bfl1 is most likely to block assembly of the transcription initiation complex, which includes not only the core promoter but also proximal and distal enhancer elements. Further investigation into such regulatory elements is required to unravel the transcriptional control of BFL1. It is obvious that there are several other genes involved in this complex regulatory pathway and identification of these genes is crucial to understand the three-step transition unique to grasses i.e. branch to spikelet to florets.
Conclusion
Phenotypic, genotypic and expression analyses of a Ds tagged mutant and its Ds-excised revertants and/or wild-type plants in conjunction with publicly-available rice genome sequences have allowed us to identify a rice gene, BRANCHED FLORETLESS 1 (BFL1), involved in the transition from SM to FM. BFL1 is most likely an ortholog of the maize transcription factor gene BD1 and encodes a transcription factor protein containing an EREBP/AP2 domain identical to that of BD1 and its orthologs in other cereals [9]. Because of the phenotypic similarities bfl1 is most likely to be an allele of the previously reported fzp [20] and fzp2 [15] mutants.
Methods
Plant materials and growth conditions
All rice lines used in this study were derived from the japonica cultivar Nipponbare. The bfl1 mutant described in this paper was recognised initially among F3 generation plants derived from a cross between the iAc (pSK300, TT3-24-1-1) and Ds gene trap (DsG, pSK200, TT2-10-1-1) transgenic lines described previously by Upadhyaya et al. (2002). The iAc and DsG binary vector constructs, tissue culture and Agrobacterium transformation procedures used have been described previously [30,31]. Plants were grown under controlled glasshouse conditions with 25 ± 3°C day and 21°C night temperatures with 16 h day length.
DNA extraction and Ds flanking sequence rescue
Genomic DNA was extracted from plants using the PureGene nucleic acid isolation kit (Gentra Systems Inc. Minneapolis, MN, USA) according to the manufacturer's instructions. Ds5' and Ds3' flanking sequences were rescued using the built-in plasmid rescue system [31] and TAIL-PCR [19], respectively. For plasmid rescue, ~2 μg of genomic DNA was digested with SacI, then extracted with phenol/chloroform, precipitated and self-ligated in 500 μl of ligation mix containing 5 Weiss units of T4 DNA ligase at 16°C overnight. The ligated DNA was used for electroporation after ethanol precipitation. Plasmid clones were analysed by appropriate restriction enzyme analyses before being selected for sequencing. TAIL-PCR was performed using three nested primers Ds3_1, Ds3_3, Ds3_6587+ and an arbitrary degenerate primer AD2 (Table 1) according to Liu and others [19] with minor modifications. The purified PCR product was re-amplified using Ds3_6587+ and AD2 to ensure success of the sequencing reaction. Sequencing was performed with the reagents of the ABI Prism BigDye termination cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions.
Sequence analysis
DNA sequence comparisons were performed using the BLAST program [1,2] searching against the NCBI GenBank http://www.ncbi.nlm.nih.gov/ and China's indica rice sequence database http://btn.genomics.org.cn/rice. The ORF was identified by using FGENESH http://www.softberry.com/berry.phtml/ and GENSCAN http://genes.mit.edu/GENSCAN.html. Alignment of EREBP/AP2 domains was performed using programs of Genetics Computer Group Wisconsin software suit [11].
Southern blot hybridization
For Southern blot hybridization analysis, genomic DNA (~10 μg) was digested with HindIII, fractionated on a 0.7% agarose gel, and blotted onto a Hybond-N+ membrane (Amersham Life Science, England) according to the manufacture's instructions. The gus gene of the DsG construct was used to prepare radioactively-labelled probes with the MegaprimeTM DNA labelling system (Amersham Life Science, England) according to the manufacturer's instructions. The membrane was hybridized at 42°C for 6 h, washed at 60°C with 0.1 × SSC and 0.1% SDS and visualized by autoradiography using a phosphor imager (Molecular Dynamics, Sunnyvale, CA, USA).
PCR conditions
PCR primers (Table 1) were designed using the 'prime' program of the Genetics Computer Group (GCG) Wisconsin software suit [11]. Ds insertion plants were analysed by Ac- (primers Ac_1931+ and Ac_2382-) and Ds- (primers GUS_313- and GPAInt) specific PCR amplification. The following PCR program was used: 30 cycles of 94°C for 2 minutes, 58°C for 30 seconds, and 72°C for 1 minute, followed by a final 72°C for 5 minutes. To identify the presence or absence of Ds insertion in the bfl1 locus, a set of primers annealing to the flanking region of the Ds insertion were designed based on the published rice genomic sequences (contig 239 of China Genome database at http://btn.genomics.org.cn/rice. Three sets of PCRs were then performed: PCR1 with primers LW1125_For and LW1125_Rev amplified the region flanking the Ds insertion. PCR2 with primers LW1125_Rev and Ds5_112-and PCR3 with LW1125_For and Ds3_6587 +amplified part of the 5' and 3' ends of the Ds element and their flanking genomic regions, respectively (Table 1; Fig. 6). The PCR conditions were 30 cycles of 94°C for 2 minutes, 60°C for 30 seconds, and 70°C for 1 minute, followed by a final 70°C for 5 minutes.
BFL1 transcript levels in RNA samples of mutant and wild-type were visualized by RT-PCR with BFL1 specific primers (Table 1) using OneStep RT-PCR kit (QIAGEN Inc, California, USA) reagents according to manufacture's instructions. Primers specific to the rice sucrose synthase gene RSs1 [35] was used as an internal control for RNA integrity and DNA contamination.
Authors' contributions
The mutant was isolated among the screening population which was a combined effort of Plant Industry Rice Functional Genomics group http://www.pi.csiro.au/fgrttpub/members.htm. Most of the plant analyses were performed by QHZ and some by MSH. Sequence analyses were performed by MNU and QHZ. QHZ, MNU and ESD participated in the design, conception and coordination of the study. All authors read and approved the final manuscript.
Note added while this manuscript was under review
The FRIZZY PANICLE gene has also been shown to be an ortholog of maize BD1 in the recently published article "Komatsu M, Chujo A, Nagato Y, Shimamoto K, Kyozuka J (2003) FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development, 130(16):3841-50" and hence bfl1 is an allele of frizzy panicle.
Acknowledgments
Acknowledgements
The authors thank Kerrie Ramm, Ramani Shivakkumar, Celia Miller, Shu-Ting Pan, and Kathryn Smith for their invaluable technical assistance. We also thank Drs John Watson, Ming-Bo Wang and Andrew Eamens for their critical reading and discussions. This work was supported by GrainGene, Rural Industries Research and Development Corporation (RIRDC) and NSW Agricultural Genomics Centre. We also wish to acknowledge Dr Junko Kyozuka for information on frizzy panicle mutants.
Contributor Information
Qian-Hao Zhu, Email: qianhao.zhu@csiro.au.
Mohammad Shamsul Hoque, Email: mohammad.hoque@csiro.au.
Elizabeth S Dennis, Email: liz.dennis@csiro.au.
Narayana M Upadhyaya, Email: narayana.upadhyaya@csiro.au.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1006/jmbi.1990.9999. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki T. Transition from vegetative to reproductive phase. Curr Opin Plant Biol. 2001;4:63–68. doi: 10.1016/S1369-5266(00)00137-0. [DOI] [PubMed] [Google Scholar]
- Banno H, Ikeda Y, Niu QW, Chua NH. Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell. 2001;13:2609–2618. doi: 10.1105/tpc.13.12.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AD. Plant form: an illustrated guide to flowering plant morphology. New York:Oxford University Press; 1991. [Google Scholar]
- Blasquez M. Flower development pathways. J Cell Sci. 2000;113:3547–3548. doi: 10.1242/jcs.113.20.3547. [DOI] [PubMed] [Google Scholar]
- Bradley D, Carpenter R, Copsey L, Vincent C, Rothstein S, Coen E. Control of inflorescence architecture in Antirrhinum. Nature. 1996;379:791–797. doi: 10.1038/379791a0. [DOI] [PubMed] [Google Scholar]
- Chuck G, Meeley RB, Hake S. The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1 . Genes Dev. 1998;12:1145–1154. doi: 10.1101/gad.12.8.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Muszynski M, Kellogg E, Hake S, Schmidt RJ. The control of spikelet meristem identity by the BRANCHED SILKLESS 1 gene in maize. Science. 2002;298:1238–1241. doi: 10.1126/science.1076920. [DOI] [PubMed] [Google Scholar]
- Colombo L, Marziani G, Masiero S, Wittich PE, Schmidt RJ, Gorla MS, Enrico PèM. BRANCHED SILKLESS mediates the transition from spikelet to floral meristem during Zea mays ear development. Plant J. 1998;16:355–363. doi: 10.1046/j.1365-313x.1998.00300.x. [DOI] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A Comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–396. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature. 2003;422:719–722. doi: 10.1038/nature01549. [DOI] [PubMed] [Google Scholar]
- Hoshikawa K. The Growing Rice Plant. Tokyo: Nobunkyo; 1989. [Google Scholar]
- Kerstetter RA, Laudencia-Chingcuanco D, Smith LG, Hake S. Loss-of-function mutations in the maize homeobox gene, knotted1, are defective in shoot meristem maintenance. Development. 1997;124:3045–3054. doi: 10.1242/dev.124.16.3045. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Maekawa M, Shimamoto K, Kyozuka J. The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev Biol. 2001;231:364–373. doi: 10.1006/dbio.2000.9988. [DOI] [PubMed] [Google Scholar]
- Kyozuka J, Konishi S, Nemoto K, Izawa T, Shimamoto K. Down-regulation of RFL, the FLO/LFY homolog of rice, accompanied with panicle branch initiation. Proc Natl Acad Sci U S A. 1998;95:1979–1982. doi: 10.1073/pnas.95.5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyozuka J. Flower development of rice. In: Shimamoto K, editor. In Molecular Biology of Rice. Tokyo: Springer-Verlag; 1999. pp. 101–118. [Google Scholar]
- Levy YY, Dean C. The transition to flowering. Plant Cell. 1998;10:1973–1990. doi: 10.1105/tpc.10.12.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313X.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Mackill DJ, Pinson SRM, Rutger JN. Frizzy panicle, an EMS-induced mutant in the Japonica cultivar M-201. Rice Genet Newsl. 1992;9:100–102. [Google Scholar]
- McSteen P, Laudencia-Chingcuanco D, Colasanti J. A floret by any other name: control of meristem identity in maize. Trends Plant Sci. 2000;5:61–66. doi: 10.1016/S1360-1385(99)01541-1. [DOI] [PubMed] [Google Scholar]
- Murai M, Izawa M. Effects of major genes controlling morphology of panicle in rice. Breed Sci. 1994;44:247–255. [Google Scholar]
- Nakagawa M, Shimamoto K, Kyozuka J. Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 2002;29:743–750. doi: 10.1046/j.1365-313X.2002.01255.x. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi M, Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 1995;7:173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima S, Murata M, Sakamoto W, Ogura Y, Motoyoshi F. Cloning and molecular analysis of the Arabidopsis gene Terminal Flower1. Mol Gen Genet. 1997;254:186–194. doi: 10.1007/s004380050407. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcription factors. Biol Chem. 1998;379:633–646. doi: 10.1515/bchm.1998.379.6.633. [DOI] [PubMed] [Google Scholar]
- Simpson GG, Gendall AR, Dean C. When to switch to flowering. Annu Rev Cell Dev Biol. 1999;15:519–550. doi: 10.1146/annurev.cellbio.15.1.519. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Albert VA, Oppenheimer DG, dePamphilis CW, Ma H, Frohlich MW, Theissen G. Missing links: the genetic architecture of flower and floral diversification. Trends Plant Sci. 2002;7:22–31. doi: 10.1016/S1360-1385(01)02098-2. [DOI] [PubMed] [Google Scholar]
- Takeoka Y, Shimizu M, Wada T. Panicles. In: Hoshikawa TM, editor. In Science of the Rice Plant. I. Tokyo: Nobunkyo; 1993. pp. 295–338. [Google Scholar]
- Upadhyaya NM, Surin B, Ramm K, Gaudron J, Schünmann PHD, Taylor W, Waterhouse PM, Wang M-B. Agrobacterium -mediated transformation of Australian rice cultivars Jarrah and Amaroo using modified promoters and selectable markers. Aust J Plant Physiol. 2000;27:201–210. doi: 10.1071/PP99078. [DOI] [Google Scholar]
- Upadhyaya NM, Zhou X-R, Zhu Q-H, Ramm K, Wu L, Eamens A, Sivakumar R, Kato T, Yun D-W, Santhoshkumar C, Narayanan KK, Peacock JW, Dennis ES. An iAc/Ds gene and enhancer trapping system for insertional mutagenesis in rice. Funcl Plant Biol. 2002;29:547–559. doi: 10.1071/PP01205. [DOI] [PubMed] [Google Scholar]
- van der Graaff E, Dulk-Ras AD, Hooykaas PJ, Keller B. Activation tagging of the LEAFY PETIOLE gene affects leaf petiole development in Arabidopsis thaliana. Development. 2000;127:4971–4980. doi: 10.1242/dev.127.22.4971. [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Veit B, Sinha N, Hake S. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature. 1991;350:241–243. doi: 10.1038/350241a0. [DOI] [PubMed] [Google Scholar]
- Walsh J, Freeling M. The liguleless2 gene of maize functions during the transition from the vegetative to the reproductive shoot apex. Plant J. 1999;19:489–495. doi: 10.1046/j.1365-313X.1999.00541.x. [DOI] [PubMed] [Google Scholar]
- Wang M-B, Boulter D, Gatehouse JA. A complete sequence of the rice sucrose syntahse-1 (RSs1) gene. Plant Mol Biol. 1992;19:881–885. doi: 10.1007/BF00027086. [DOI] [PubMed] [Google Scholar]
- Weigel D. The APETALA2 domain is related to a novel type of DNA binding domain. Plant Cell. 1995;7:388–389. doi: 10.1105/tpc.7.4.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K, Long D, Swinburne J, Coupland G. A Dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell. 1996;8:659–671. doi: 10.1105/tpc.8.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Kojima S, Takahashi Y, Lin H, Sasaki T. Genetic control of flowering time in rice, a short-day plant. Plant Physiol. 2001;127:1425–1429. doi: 10.1104/pp.127.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]