Abstract
Cadmium is a highly toxic environmental contaminant that has been implicated in various disorders. A major mechanism for cadmium detoxification in the yeast Saccharomyces cerevisiae relies on extrusion via Pca1, a P-type ATPase. While an N-terminal degron targets Pca1 for degradation before its secretion to the plasma membrane, cadmium in the growth media rapidly up-regulates Pca1 by preventing its turnover. Here we show that the endoplasmic reticulum-associated degradation (ERAD) system, known for its role in quality control of secretory proteins, is unexpectedly responsible for the regulation of Pca1 expression by cadmium. Direct cadmium sensing at the ER by a degron in Pca1 leads to an escape of Pca1 from ERAD. This regulated conversion of an ERAD substrate to a secretory competent state in response to a cellular need illustrates a mechanism for expressional control of a plasma membrane protein. Yeast has likely evolved this mode of regulation for a rapid response against cadmium toxicity at the expense of constant synthesis and degradation of Pca1. ERAD of a portion of secretory proteins might occur via signal-dependent regulatory mechanisms as demonstrated for Pca1.
Keywords: ERAD, degron, P-type ATPase, Pca1 yeast
Metal ions are highly toxic, although several metals such as iron, copper, and zinc are essential micronutrients. Organisms possess delicate systems for detoxification and excretion of nonphysiological metals and homeostatic metabolism of nutritional yet toxic metals (1, 2). Pca1 in S. cerevisiae is a multi-spanning transmembrane protein that belongs to the family of P1B-type ATPase heavy-metal transporters widely distributed from bacteria to humans (3). Pca1 functions in the efflux of cadmium across the plasma membrane (4), an extremely toxic environmental pollutant that causes various human diseases, such as cancer, kidney disease, and endocrine disruption (5).
We have previously demonstrated that Pca1 is a short-lived protein that is targeted for ubiquitination and proteasomal degradation via its cytosolic N-terminal domain (amino acids 1-392) before reaching the cell surface (6). However, in the presence of cadmium, Pca1 is rapidly up-regulated due to the prevention of its degradation (6). An autonomous degron encompassing amino acid residues 250-350 within the N-terminal cytosolic domain is necessary and sufficient for both degradation and metal sensing (6). Given that the cell surface expression of several plasma membrane proteins is regulated by ubiquitin-mediated endocytosis followed by vacuolar degradation, Pca1 turnover and cadmium-responsive degradation represents an interesting mode of expressional control in which subcellular trafficking and stability are determined by its substrate during secretion.
To uncover molecular factors involved in the expressional control of Pca1, we have devised a genetic screen to identify mutants that are defective in Pca1 degradation in the absence of cadmium. Unexpectedly, our data presented herein demonstrate that components of the ER-associated degradation (ERAD) system target Pca1 for proteasomal degradation. Secretory proteins in eukaryotes are inserted into the ER lumen or membrane where correct folding and maturation occurs. Numerous factors, such as molecular chaperones, glycosylation enzymes, and protein disulfide isomerases, are involved in this process. Genetic mutations and environmental stresses (e.g., heat, oxidative stress) increase the probability of failure of protein folding and maturation. The unfolded protein response (UPR) pathway enhances the capability of protein folding when cells are challenged by these stresses (7). The ERAD system eliminates terminally misfolded or unassembled proteins, which is critical for the prevention of toxic accumulation of aberrant proteins (8, 9). Several mutant proteins, unassembled subunits of secretory proteins, and heterologously expressed proteins have all been identified as ERAD substrates (8, 9). However, Pca1 is a naturally expressed and functional protein and there is no evidence of multimer assembly for Pca1.
Cadmium is extremely toxic and its levels fluctuate in the environment. Thus, transcriptional and/or translational control of defense factors may not be fast enough for cell protection. Our results suggest that yeast cells have evolved to harness the ERAD system for rapid up-regulation of Pca1 cell surface expression by direct sensing of intracellular cadmium at the ER.
Results
Pca1 Is Targeted for ER-associated Degradation in the Absence of Cadmium.
To identify molecular factors involved in Pca1 turnover, we selected yeast mutants exhibiting defects in Pca1 degradation. An expression construct of a fully functional Pca1 fused with green fluorescent protein (GFP-Pca1) was transformed into a collection of mutant yeast strains (Table S1), each lacking a single nonessential gene (10). Cells emitting a strong GFP signal (as a consequence of a defect in degradation of the GFP-Pca1 fusion) were selected using a flow cytometer, and gene deletions resulting in high Pca1 expression were identified [supporting information (SI) Fig. S1A].
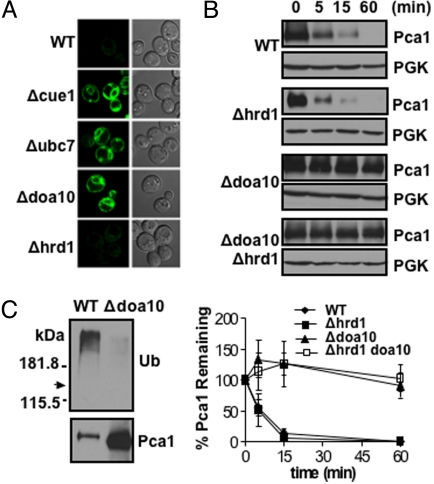
Unexpectedly, Pca1 is stabilized and highly expressed in cells with deletions of the CUE1 gene (Fig. S1 B–D). CUE1 encodes an ER membrane-bound protein that recruits Ubc7, a cytosolic ubiquitin-conjugating enzyme, to the ER surface (11). Given that Cue1 is a crucial factor involved in ERAD, we further examined Pca1 stabilization in yeast strains bearing deletions in factors required for ERAD, including Ubc7 (12) and Ubc6 ubiquitin-conjugating enzymes (13), and ER resident Doa10 (14) and Hrd1 (15) ubiquitin ligases. In yeast, the recognition of integral membrane ERAD substrates is governed by the location of the misfolded domain. Lesions in soluble domains on the cytosolic face of the ER belong to the ERAD-C (cytoplasmic pathway) in which their ubiquitination and degradation are largely dependent on Doa10 (8, 9). On the other hand, Hrd1 is involved in ubiquitination and degradation of substrates with misfolded regions in soluble luminal domains (ERAD-L) or within transmembrane domains (ERAD-M) (8, 9). Pca1 stabilization was evident in Δubc7 or Δdoa10 but not Δhrd1 strains (Fig. 1A). Cycloheximide chase of Pca1 further confirmed no significant role for Hrd1 in Pca1 turnover (Fig. 1B), indicating the specificity of Doa10 in Pca1 turnover. Consistent with ubiquitination by this E3 ligase, Pca1 ubiquitination was dramatically reduced in the Δdoa10 strain (Fig. 1C). Pca1 turnover was also decelerated in Δubc6 strain, but to a lesser extent than in the Δubc7 strain (Fig. S2). Collectively, these data suggest that in the absence of its cadmium substrate, Pca1 is rapidly degraded through the ERAD pathway. Given that our screening method selected only CUE1-null mutants more than 10 times despite the dependence of other ERAD components in Pca1 degradation, our screen was not saturated. It would be expected that more comprehensive screening would identify other factors (e.g., Doa10 and Ubc7) involved in Pca1 turnover.
Fig. 1.
Pca1 is degraded through the ERAD pathway. (A) Confocal microscopy of GFP-fused Pca1 expressed in wild-type (WT), Δcue1, Δubc7, Δdoa10, or Δhrd1 strains. (B) Cycloheximide chase and immunoblotting of HA epitope-tagged Pca1 (HA-Pca1) expressed in wild-type (WT), Δhrd1, Δdoa10, or Δdoa10 Δhrd1 strains at the indicated time points. Each blot was probed for phosphoglycerate kinase (PGK) to determine equal loading. For quantification, pixel densities of HA-Pca1 were normalized to those of PGK. Average ± SD of 3 independent experiments is graphed (Lower). (C) Detection of ubiquitin-conjugated Pca1. Immunoprecipitation of HA-Pca1 in wild-type (WT) or Δdoa10 cells followed by western blot with anti-Ub or HA antibodies. Arrow indicates expected migration of HA-Pca1.
Degradation of Pca1 and Cadmium Sensing Occur at the ER.
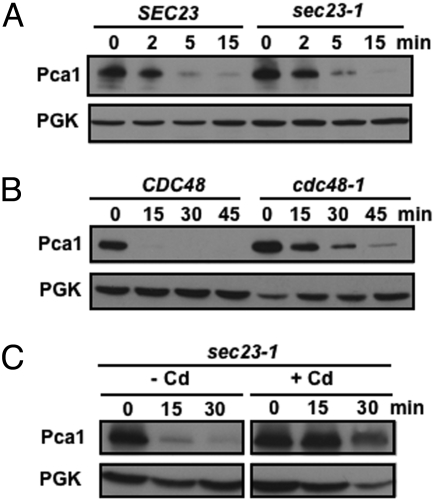
Given that Pca1 degradation and ubiquitination required Doa10, an ER-resident ubiquitin ligase, we reasoned that Pca1 degradation occurs before its secretion from the ER. To demonstrate this hypothesis, we first examined the fate of Pca1 turnover in a sec23-1 mutant, which exhibits a temperature sensitive defect in COPII vesicle-dependent ER to Golgi trafficking (16). As shown in Fig. 2A, trapping Pca1 in the ER had no significant effect on turnover rate, demonstrating that Pca1 degradation does not require its exit from the ER. Second, we determined the stability of Pca1 in a temperature-sensitive mutant of the Cdc48/Np14p/Ufd1 AAA-type ATPase, which is required for efficient ER extraction and degradation of ERAD substrates such as Ste6*, a mutant plasma membrane protein (17). Consistent with a role for Cdc48 in Pca1 turnover, stabilization of Pca1 is apparent in the cdc48-3 strain compared to its isogenic, wild-type strain at restrictive temperature (Fig. 2B). Since Pca1 degradation occurs at the ER, it would be intuitive that cadmium sensing which blocks degradation would also occur at the ER. As expected, addition of cadmium to cell cultures prevented Pca1 degradation when ER exit is blocked (Fig. 2C).
Fig. 2.
Pca1 degradation and cadmium sensing occur at the ER. (A) HA-tagged Pca1 was expressed in SEC23 and a sec23-1 temperature-sensitive mutant. To block ER to Golgi transport, cells were cultured at restrictive temperature (37 °C for 30 min). After cycloheximide addition to culture media, cells were collected at the indicated time points. Pca1 was detected by western blotting using anti-HA antibodies. (B) Cycloheximide chase and western blotting of HA-Pca1 expressed in CDC48 and a cdc48-3 temperature-sensitive mutant cultured at restrictive temperature (37 °C for 30 min). (C) Cadmium stabilizes Pca1 when ER to Golgi transport is blocked. A sec23-1 strain expressing HA-Pca1 was cultured at restrictive temperature (37 °C for 30 min) before the addition of cycloheximide with (+) or without (-) cadmium (Cd) (50 μM CdCl2). Cells were collected for immunoblotting at the indicated time points. Each blot was probed for PGK to determine equal loading.
Inhibition of Pca1 ERAD by Cadmium Is Specific.
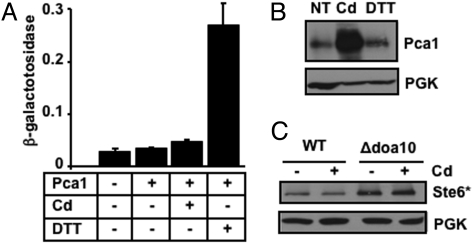
To confirm if the inhibitory effect of cadmium on ERAD is specific to Pca1, we addressed possible toxic effects of cadmium on ER functions, such as inhibition of ERAD machinery, or saturation of ERAD capacity by excess accumulation of damaged and/or misfolded proteins. Accumulation of misfolded proteins in the ER generally leads to induction of the unfolded protein response (UPR) (7, 18). The overall status of ER function was monitored with a UPR reporter construct (UPRE-LacZ) (19). Despite being an ERAD substrate, constitutive expression of Pca1 did not lead to any significant increase in reporter gene expression over basal levels (Fig. 3A). Cadmium concentrations that induce robust expression of Pca1 (Fig. 3B) led to a minimal UPR response (Fig. 3A), whereas DTT strongly enhanced reporter levels (Fig. 3A) without affecting Pca1 expression (Fig. 3B). Furthermore, cadmium did not stabilize Ste6*, a well-studied Doa10 substrate (Fig. 3C). Collectively, these data indicated that the cadmium dependent stabilization of Pca1 is specific and was not due to over-accumulation of misfolded protein intermediates or the inactivation of ERAD machinery.
Fig. 3.
Cadmium-dependent inhibition of Pca1 ERAD is specific. (A) Unfolded protein response (UPR) determined by a UPRE-LacZ reporter construct (19). Empty vector (-) or an expression construct of HA epitope tagged Pca1 (+) was co-transformed with a UPRE-LacZ reporter construct in a wild-type (WT) yeast strain. Cells were cultured in SC selective media with (+) or without (-) supplementation of cadmium (Cd) (50 μM CdCl2, 1 h). As a positive control of UPR response, cells were cultured with DTT (2 mM, 1 h) (19). Each bar represents the average ± SD of β-galactosidase activities of 4 different samples. (B) Western blot analysis of HA-Pca1 expressed in non-treated cells (NT) or cells cultured in media supplemented with cadmium (Cd) (50 μM CdCl2) or DTT (2 mM) for 1 h. (C) Western blot analysis of HA epitope-tagged Ste6* in WT or Δdoa10 cells cultured without (-) or with (+) supplementation of cadmium (Cd) (50 μM CdCl2, 1 h) to culture media. PGK was probed to determine equal loading.
The N Terminus of Pca1 Contains a Targeting Signal for ERAD.
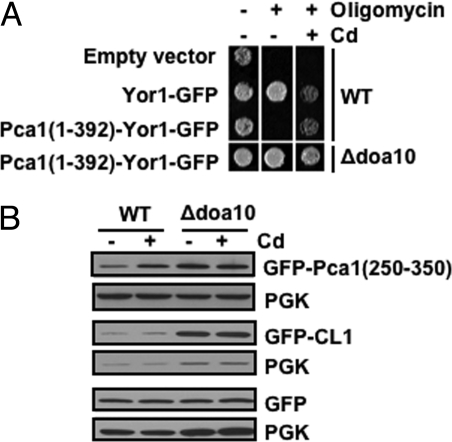
Aberrant secretory proteins typically engage quality control checkpoints, which prevents their exit from the ER. For instance, when the ERAD pathway is inactivated, some substrates are trapped in the ER rather than being secreted to the plasma membrane (17, 20). We took advantage of the ability of the Yor1 ATP-binding cassette (ABC) transporter to extrude oligomycin, a mitochondrial toxin (21). If the Pca1 degron prevents ER exit, then Yor1 fused with this degron would be retained in the ER and no oligomycin resistance would be conferred. Fusion of Pca1 (1-392) with Yor1 leads to rapid turnover of Yor1, which is prevented by cadmium in culture media or deletion of DOA10 (Fig. S3). Consistent with its rapid proteolysis, in the absence of cadmium the Pca1 (1-392)-Yor1-GFP fusion did not confer resistance to oligomycin (Fig. 4A, third row, Middle). However, upon supplementation of cadmium (Fig. 4A, third row, Right) or expression in a Δdoa10 strain (Fig. 4A, fourth row, Middle), the fusion protein confers oligomycin resistance. These results demonstrate that the Pca1 (1-392)-Yor1-GFP fusion reaches the cell surface for oligomycin efflux when its degradation is prevented. Oligomycin resistance in the Δdoa10 strain expressing the Pca1 (1-392)-Yor1-GFP fusion without cadmium suggests that metal sensing is not a prerequisite for ER exit. This is also in agreement with our previous observation where deletion of residues 1-392 did not perturb Pca1 trafficking or function (6).
Fig. 4.
The N-terminal regulatory domain does not function as an ER-retention signal. (A) Oligomycin resistance of wild-type (WT) or Δdoa10 strains expressing vector, Yor1-GFP or Pca1 (1-392)-Yor1-GFP. Cells (5 μL A600 = 0.1) were spotted on YPEG solid media with (+) or without (-) the addition of oligomycin (2.5 μg/mL) and/or cadmium (Cd) (10 μM CdCl2). Cell growth was monitored after 3 days. (B) GFP without or with fusion of Pca1 (250-350) or a CL1 artificial degron were expressed in wild-type (WT) and Δdoa10 strains. Cells were cultured in media with (+) or without (-) supplementation of cadmium (50 μM CdCl2, 1 h). GFP levels in total protein extracts were measured by western blotting using anti-GFP antibodies. Each blot was probed for PGK to determine equal loading.
We next examined whether the Pca1 degron can target a cytosolic protein for ERAD. Expression of a soluble GFP reporter C-terminally fused to Pca1 residues 250-350 [GFP-Pca1 (250-350)] was regulated in a cadmium and Doa10-dependent manner (Fig. 4B). However, cadmium did not prevent Doa10-dependent turnover of GFP fused with a CL1 degron (22) (Fig. 4B). These data indicate that the Pca1 degron can also target a cytosolic protein for ERAD. We conclude that the Pca1 degron does not function as either an ER exit or retention signal but rather serves as a targeting signal for Doa10 dependent ERAD.
Physical Interaction Between Pca1 and Doa10 via the N-Terminal Degron.
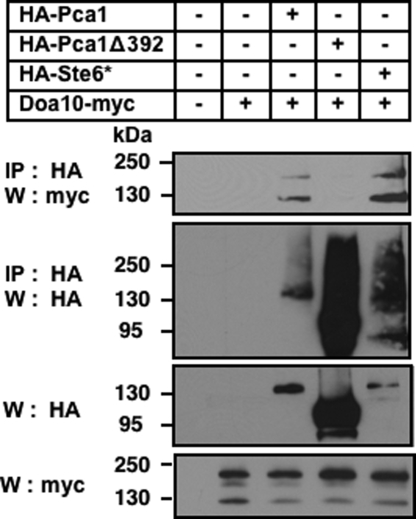
We hypothesized that the N terminus of Pca1 functions as a Doa10-recognition motif. Pca1 and Doa10 interaction was determined by chemical crosslinking. Microsomes were prepared from cells expressing thirteen c-myc epitope-tagged Doa10 (Doa10-myc) (23) and a fully functional HA-tagged Pca1 (HA-Pca1), or with deletion of the N-terminal regulatory domain (HA-Pca1Δ392) (6). Ste6* served as a positive control for physical interaction between an ERAD substrate and Doa10 (23). Microsomes were treated with dimethyl dithiobispropionimidate (DTBP), a thiol reversible and membrane-permeable cross-linker, and HA-tagged proteins were immunoprecipitated by anti-HA conjugated Sepharose beads. Doa10 was co-precipitated from cells expressing Pca1 or Ste6* but not Pca1Δ392 which lacks the N-terminal degron (Fig. 5, first panel). This data demonstrated that the N terminus is required for either direct or indirect interaction of Pca1 with Doa10.
Fig. 5.
The N-terminal regulatory domain is required for physical interaction between Pca1 and Doa10. Expression constructs of HA-Pca1, Pca1 deleted of amino acid residues 1-392 (HA-Pca1Δ392), or Ste6* (HA-Ste6*) were expressed in a strain containing a chromosomal integration of 13 c-myc epitope tagged Doa10 (Doa10-myc). Chemical cross-linking was performed using microsomes prepared from these cells (See Materials and Methods). HA tagged proteins were immunoprecipitated (IP) and denatured under reducing conditions to break crosslinking. Samples were subjected to SDS/PAGE and western blotting (W) using anti-myc antibodies (IP:HA, W:myc), and then the same blot was stripped and re-probed with anti-HA antibodies (IP:HA, W:HA). Total protein extracts were resolved on SDS-PAGE and analyzed by western blotting using anti-HA (W:HA) or anti-myc (W:myc) antibodies.
Metal Binding and Conformational Change of the Pca1 Degron.
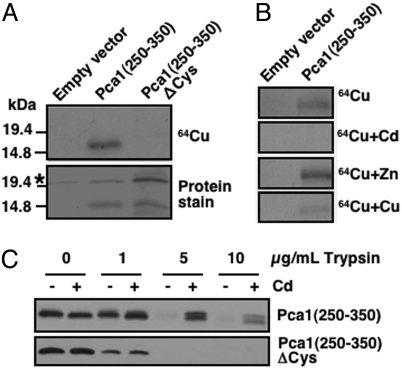
Given that cadmium or copper rescued Pca1 from degradation and that the Pca1 N terminus contains potential metal binding residues (6), we predicted that metal binding to this region would mask a degradation signal. To determine metal binding, we used metal blotting assays (24). HA-Pca1 (250-350) was immunoprecipitated using anti-HA antibody conjugated beads and transferred to a nitrocellulose membrane. Autoradiography of blots incubated with 64Cu(II) showed specific binding of radioactive copper to HA-Pca1 (250-350) as no signal was observed from bands corresponding to the same peptide in which all 7 cysteine residues were mutated to alanine (HA-Pca1 (250-350)ΔCys) (Fig. 6A). Cu(II) or Cd(II) but not Zn(II) effectively competed for 64Cu(II) binding implying that copper and cadmium bind to the same site(s) (Fig. 6B). Cd(II) appeared to compete more effectively than copper at the same concentrations (Fig. 6B and Fig. S4A) suggesting higher-binding affinity of cadmium. These metal-binding specificities were in accordance with previously observed cadmium- and copper-specific regulation of Pca1 stability (4).
Fig. 6.
Metal binding and conformational changes within the Pca1 degron. Pca1 amino acid residues 250-350 with N-terminal triple HA epitope tagging [HA-Pca1 (250-350)] and HA-Pca1 (250-350)ΔCys where all 7 cysteine residues were substituted to alanine were expressed in a Δdoa10 strain. (A) Immunoprecipitated HA-Pca1 (250-350) and HA-Pca1 (250-350)ΔCys were subjected to 64Cu blotting (1 μM CuCl2, 10 μCi) followed by autoradiography (See Material and Methods). Asterisk indicates non-specifically precipitated bands. (B) 64Cu blotting was performed in the presence of 5-fold excess non-radioactive competitor ions. (C) Cell lysates prepared from cells cultured with (+) or without (-) cadmium (CdCl2 50 μM, 1 h) were subjected to increasing concentrations of trypsin proteolysis followed by anti-HA immunoblotting.
We next used a limited trypsin proteolysis assay of HA-Pca1-(250-350) to probe for cadmium-dependent conformational changes, which could be visualized by different trypsin digestion patterns. Yeast cell lysates prepared from cells cultured with or without cadmium in the media were subjected to trypsin proteolysis followed by anti-HA immunoblotting. Indeed, this domain is more resistant to proteolysis when cells were cultured with cadmium (Fig. 6C Upper). No protection of trypsin proteolysis was observed in the same peptide containing site-directed mutations of cadmium-binding cysteine residues (6) (Fig. 6C Lower). Cadmium-dependent protection of trypsin proteolysis was confirmed in vitro using purified Pca1 (250-350) (Fig. S4B). Collectively, these data suggest that cadmium-induced conformational changes within the degron act as a molecular switch allowing Pca1 to circumvent ERAD.
Discussion
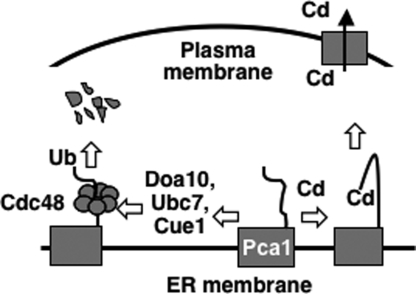
Our data supports a model (Fig. 7) illustrating an unanticipated regulatory mechanism by which cells control the expression of a plasma membrane protein. In the absence of cadmium, newly synthesized Pca1 is targeted for ERAD by an N-terminal degron resulting in rapid turnover at the proteasome. Cadmium sensing by this degron induces a conformational change, which prevents the recognition of Pca1 by the ERAD machinery. Hence, cells are able to rapidly elevate Pca1 expression in response to cadmium.
Fig. 7.
Model of cadmium-regulated expression of Pca1. Cadmium sensing occurs at the ER by metal binding to the cytosolic N-terminal domain of Pca1 followed by conformational changes which prevents recognition for ubiquitination by ERAD factors, including Cue1, Ubc7, and Doa10. Pca1 extraction from the ER is facilitated by Cdc48 after ubiquitination. Black squares indicate Pca1 transmembrane domains. Black arrow indicates cadmium efflux via Pca1 at the plasma membrane.
Given that cadmium is an extremely toxic metal of which environmental levels fluctuate, the constant synthesis of a cadmium exporter would be advantageous for cell survival. ERAD of Pca1 at the early step of synthesis keeps Pca1 levels at a minimum, since expression is not necessary when cells are growing without cadmium stress. It is also possible that Pca1 could play a role other than cadmium efflux, which would be deleterious in the absence of cadmium. For instance, if Pca1 transports nutritional metal ions due to low substrate specificity as shown for other P1B-type ATPases (25), the constant degradation of Pca1 would prevent the loss of essential metals that are limiting for growth.
ERAD eliminates misfolded or unassembled proteins, which is necessary for the prevention of toxic accumulation of aberrant proteins (8, 9). At least 30% of newly synthesized proteins are rapidly degraded by proteasomes (26). It has been suggested that this observation represents the degradation of misfolded proteins and defective ribosomal products resulting from errors in translation. This pool of peptides also provides both host and viral antigenic peptides to be displayed by MHC class I molecules (27). Our study presented herein suggests another unexplored role for ERAD, which occurs in a degron-dependent manner and would allow for fine tuning the expression of a plasma membrane protein in response to cellular cues.
Regulation of Pca1 expression at the ER may represent a conserved and largely uncharacterized system by which cells control the expression levels of secretory proteins. Consistent with this argument, molecular factors in the ERAD pathway are involved in regulated destruction of proteins. Sterol metabolic status regulates ERAD of ER-resident HMG-CoA reductase, the rate-limiting enzyme of sterol synthesis (28). Co-translational degradation of apolipoprotein B at the ER is enhanced when lipid efflux from the liver is not favored (29). In yeast, the Doa10 ubiquitin ligase also targets the cytoplasmic MATα2 repressor for degradation in the absence of its MATa1-binding partner (14, 30). However, Pca1 is a plasma membrane protein that is targeted to the ERAD pathway by a degron in a substrate-dependent manner. There is accumulating evidence for expressional regulation of other plasma membrane transporters and ion channels in the secretory pathway, although the implication of ERAD in this process has not been established. For instance, it was shown that ubiquitination and proteasome-dependent degradation affects the expression of aquaporin (31), acetylcholine receptor (32), ATP sensitive K+ channels (33), and opioid receptors (34). Intriguingly, under normal conditions only 40% of newly synthesized human delta opioid receptors ever reach the cell surface; however membrane-permeable opioid ligands facilitate maturation and ER export (35). It is of interest to determine whether this opioid dependent degradation of its receptor occurs by a similar mechanism that we have characterized for Pca1.
It is unknown whether Doa10 directly recognizes the Pca1 degron or whether other accessory factor(s) recruit Pca1 for ubiquitination. The requirement of the Pca1 N terminus in the co-precipitation of Doa10 suggests a physical interaction between the Pca1 degron and Doa10, although it is not certain yet whether this interaction is mediated by other factor(s). In the case of Ste6*, cytosolic Hsp70 chaperones aid in this process (23). However, our data did not indicate a significant role for cytosolic Hsp70 chaperones in Pca1 ERAD (Fig. S5A), although expression levels of CL1 degron-fused GFP (36) were higher when Hsp70 chaperones were inactivated (Fig. S5B).
The recognition determinants of the Pca1 degron and that of other ERAD substrates remain unknown. The current hypothesis is that misfolded proteins display normally buried hydrophobic residues to the cytosolic surface, which serves as a targeting signal for degradation. The Deg1 degron of MATα2 repressor is predicted to form an amphipathic helix of which hydrophobic residues are crucial for its instability (30). However, there is no obvious similarity in the primary sequence of Deg1 and the Pca1 degron. Structural characterization of the Pca1 metal-sensing degron in apo- and metal-bound forms would define the features that attract the ERAD machinery. This study would also provide useful information for the identification of other proteins that may possess a similar type of degradation signal. While the Pca1 degron does not have significant sequence identity with known proteins, we could identify potential metal-binding cysteine and histidine-rich extensions in predicted Pca1 family members of plants (25). We are currently determining whether they are targeted to the ERAD pathway in a regulated manner.
The identification of the molecular factors involved in Pca1 turnover reveals a regulatory mechanism by which yeast cells can selectively control the expression of a plasma membrane protein at an early step in its synthesis. Small molecules, including substrates or metabolites, or signaling pathways may actively regulate protein secretion at the ER in a target specific manner as demonstrated for Pca1.
Materials and Methods
(For additional materials and methods see SI Materials and Methods.)
Selection of Yeast Mutants Constitutively Expressing Pca1.
An expression construct of functional Pca1 fused with green fluorescent protein (GFP) at the N terminus was transformed into a yeast S. cerevisiae pool of 4,848 strains containing individual deletions of all nonessential genes (Open Biosystems). Approximately 400,000 transformed colonies were collected by re-suspending in sterilized water and then diluted to A600 = 1. Highly fluorescent cells that are defective in GFP-Pca1 turnover were sorted by flow cytometry and then plated on SC media selecting the GFP-Pca1 expression plasmid. Strong GFP signals in cells of growing colonies were confirmed by confocal microscopy, and gene deletions were identified using a primer set for PCR amplification and sequencing of a unique 20-base “tag” sequence (10).
Cycloheximide Chase Analysis.
Protein synthesis was inhibited by the addition of 100 μg/mL cycloheximide to logarithmically growing cultures. Cells (A600 = 15) were collected into equal volumes of ice-cold kill buffer (PBS containing 15 mM NaN3) at the indicated time points. At time 0, cells were collected before addition of cycloheximide. Cells were collected by centrifugation and stored at −80 °C until preparation of cell extracts for SDS/PAGE and immunoblotting (6). Pca1 was detected by chemiluminescence using anti-HA antibodies and horseradish peroxidase (HRP)-conjugated anti-rabbit IgG antibodies. Phosphoglycerate kinase (PGK) was probed as a loading control using anti-PGK antibodies and HRP-conjugated anti-mouse IgG antibodies. Signals were quantified using Total Lab TL100 software and then normalized to PGK.
64Cu Blotting and Autoradiography.
Triple HA epitope-tagged Pca1 (250-350) or Pca1 (250-350)ΔCys where all cysteine residues were mutated to alanine were immunoprecipitated from Δdoa10 cell lysates using anti-HA antibody-conjugated beads (Pierce). Samples were subjected to SDS/PAGE and electroblotted onto a nitrocellulose membrane. Membranes were equilibrated in metal binding buffer (24), probed with 10 μCi 64Cu (≈1 μM CuCl2) (Isotrace Technique) for 1 h, and then washed extensively before autoradiography and protein staining (MemCode) (Pierce). Competition with 64Cu binding was determined in the presence of 5-fold excess of non-radioactive competitor ions.
Limited Trypsin Proteolysis of Pca1 (250-350).
Cytosolic fractions were obtained from Δdoa10 cells expressing triple HA epitope-tagged Pca1 (250-350) or Pca1 (250-350)ΔCys cultured with or without cadmium (50 μM CdCl2, 1 h). Lysates were incubated with trypsin (Sigma) for 10 min on ice before addition of 0.2 μg/mL soybean trypsin inhibitor (Fluka BioChemika) for an additional 15 min on ice. Proteolysis patterns were visualized by anti-HA immunoblotting.
Supplementary Material
Acknowledgments.
We thank Dr. Susan Michaelis (Johns Hopkins University) for sharing the Ste6* construct and the Δubc6 and Δubc6 Δubc7 yeast stains, Drs. Randy Schekman (University of California Berkeley), Tom Rapoport (Harvard University) and Jeffrey Brodsky (University of Pittsburgh) for providing the sec23-1, cdc48-3 and Doa10-myc strains. This work was supported by National Institutes of Health Grants P20-RR-17675 (Nebraska Redox Biology Center), ES16337 and DK79209 (to J. L.), and funds from the Hatch Act in the University of Nebraska Agricultural Research Division.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812114106/DCSupplemental.
References
- 1.De Domenico I, McVey Ward D, Kaplan J. Regulation of iron acquisition and storage: Consequences for iron-linked disorders. Nat Rev Mol Cell Biol. 2008;9:72–81. doi: 10.1038/nrm2295. [DOI] [PubMed] [Google Scholar]
- 2.Madsen E, Gitlin JD. Copper and iron disorders of the brain. Annu Rev Neurosci. 2007;30:317–337. doi: 10.1146/annurev.neuro.30.051606.094232. [DOI] [PubMed] [Google Scholar]
- 3.Kuhlbrandt W. Biology, structure, and mechanism of P-type ATPases. Nat Rev Mol Cell Biol. 2004;5:282–295. doi: 10.1038/nrm1354. [DOI] [PubMed] [Google Scholar]
- 4.Adle DJ, Sinani D, Kim H, Lee J. A cadmium-transporting P1B-type ATPase in yeast Saccharomyces cerevisiae. J Biol Chem. 2007;282:947–955. doi: 10.1074/jbc.M609535200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jarup L, Berglund M, Elinder C-G, Nordberg G, Vahter M. Health effects of cadmium exposure-a review of the literature and risk estimate. Scand J Work Environ Health. 1998;24:1–52. [PubMed] [Google Scholar]
- 6.Adle DJ, Lee J. Expressional control of a cadmium-transporting P1B-type ATPase by a metal sensing degradation signal. J Biol Chem. 2008;283:31460–31468. doi: 10.1074/jbc.M806054200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 2007;18:716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: The long road to destruction. Nat Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 9.Sayeed A, Ng DT. Search and destroy: ER quality control and ER-associated protein degradation. Crit Rev Biochem Mol Biol. 2005;40:75–91. doi: 10.1080/10409230590918685. [DOI] [PubMed] [Google Scholar]
- 10.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 11.Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- 12.Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- 13.Sommer T, Jentsch S. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature. 1993;365:176–179. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- 14.Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- 16.Novick P, Ferro S, Schekman R. Order of events in the yeast secretory pathway. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- 17.Huyer G, et al. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J Biol Chem. 2004;279:38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- 18.Casagrande R, et al. Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol Cell. 2000;5:729–735. doi: 10.1016/s1097-2765(00)80251-8. [DOI] [PubMed] [Google Scholar]
- 19.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 20.Pagant S, Kung L, Dorrington M, Lee MC, Miller EA. Inhibiting endoplasmic reticulum (ER)-associated degradation of misfolded Yor1p does not permit ER export despite the presence of a diacidic sorting signal. Mol Biol Cell. 2007;18:3398–3413. doi: 10.1091/mbc.E07-01-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Epping EA, Moye-Rowley WS. Identification of interdependent signals required for anterograde traffic of the ATP-binding cassette transporter protein Yor1p. J Biol Chem. 2002;277:34860–34869. doi: 10.1074/jbc.M202987200. [DOI] [PubMed] [Google Scholar]
- 22.Ravid T, Kreft SG, Hochstrasser M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 2006;25:533–543. doi: 10.1038/sj.emboj.7600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DiDonato M, Narindrasorasak S, Forbes JR, Cox DW, Sarkar B. Expression, purification, and metal binding properties of the N-terminal domain from the wilson disease putative copper-transporting ATPase (ATP7B) J Biol Chem. 1997;272:33279–33282. doi: 10.1074/jbc.272.52.33279. [DOI] [PubMed] [Google Scholar]
- 25.Williams LE, Mills RF. P(1B)-ATPases-an ancient family of transition metal pumps with diverse functions in plants. Trends Plants Sci. 2005;10:491–502. doi: 10.1016/j.tplants.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 26.Schubert U, et al. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 27.Yewdell JW, Reits E, Neefjes J. Making sense of mass destruction: Quantitating MHC class I antigen presentation. Nat Rev Immunol. 2003;3:952–961. doi: 10.1038/nri1250. [DOI] [PubMed] [Google Scholar]
- 28.Hampton RY. Proteolysis and sterol regulation. Annu Rev Cell Dev Biol. 2002;18:345–378. doi: 10.1146/annurev.cellbio.18.032002.131219. [DOI] [PubMed] [Google Scholar]
- 29.Fisher EA, Ginsberg HN. Complexity in the secretory pathway: The assembly and secretion of apolipoprotein B-containing lipoproteins. J Biol Chem. 2002;277:17377–17380. doi: 10.1074/jbc.R100068200. [DOI] [PubMed] [Google Scholar]
- 30.Johnson PR, Swanson R, Rakhilina L, Hochstrasser M. Degradation signal masking by heterodimerization of MATalpha2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell. 1998;94:217–227. doi: 10.1016/s0092-8674(00)81421-x. [DOI] [PubMed] [Google Scholar]
- 31.Leitch V, Agre P, King LS. Altered ubiquitination and stability of aquaporin-1 in hypertonic stress. Proc Natl Acad Sci USA. 2001;98:2894–2898. doi: 10.1073/pnas.041616498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christianson JC, Green WN. Regulation of nicotinic receptor expression by the ubiquitin-proteasome system. EMBO J. 2004;23:4156–4165. doi: 10.1038/sj.emboj.7600436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan FF, Lin CW, Cartier EA, Shyng SL. Role of ubiquitin-proteasome degradation pathway in biogenesis efficiency of {beta}-cell ATP-sensitive potassium channels. Am J Physiol Cell Physiol. 2005;289:1351–1359. doi: 10.1152/ajpcell.00240.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petaja-Repo UE, et al. Newly synthesized human delta opioid receptors retained in the endoplasmic reticulum are retrotranslocated to the cytosol, deglycosylated, ubiquitinated, and degraded by the proteasome. J Biol Chem. 2001;276:4416–4423. doi: 10.1074/jbc.M007151200. [DOI] [PubMed] [Google Scholar]
- 35.Petaja-Repo UE, et al. Ligands act as pharmacological chaperones and increase the efficiency of delta opioid receptor maturation. EMBO J. 2002;21:1628–1637. doi: 10.1093/emboj/21.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metzger MB, Maurer MJ, Dancy BM, Michaelis S. Degradation of a cytosolic protein requires endoplasmic reticulum-associated degradation machinery. J Biol Chem. 2008;283:32302–32316. doi: 10.1074/jbc.M806424200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.