Abstract
The current paradigm for tuning synthetic biological systems is through re-engineering system components. Biological systems designed with the inherent ability to be tuned by external stimuli will be more versatile. We engineered Escherichia coli cells to behave as an externally tunable band-pass filter for enzyme activity and small molecules. The band's location can be positioned within a range of 4 orders of magnitude simply by the addition of compounds to the growth medium. Inclusion in the genetic network of an enzyme-substrate pair that functions as an attenuator is a generalizable strategy that enables this tunability. The genetic circuit enabled bacteria growth to be patterned in response to chemical gradients in nonintuitive ways and facilitated the isolation of engineered allosteric enzymes. The application of this strategy to other biological systems will increase their utility for biotechnological applications and their usefulness as a tool for gaining insight into nature's underlying design principles.
Keywords: biological circuit, biological engineering, pattern formation, protein switch, synthetic biology
External control over cellular processes is invaluable for research and technology. For example, inducible promoters allow one to control the amount of gene transcription from the outside the cell, obviating the need to engineer different cell strains with different promoters for each level of transcription desired. Moreover, inducible promoters enable control of transcription levels as a function of time and position. They consist of a simple genetic circuit that has a set input-output relation between inducer concentration and transcription level. This input-output relation cannot be conveniently altered from outside the cell. Thus, although the inducer can be said to “tune” the level of transcription, the circuit itself is not externally tunable. To tune these circuits, researchers must modify system components (e.g., by changing a repressor's affinity for its inducer). This approach to tuning currently predominates in the design of synthetic biological systems. However, such an approach is tantamount to building a unique biological system for each desired behavior and precludes the possibility of dynamic tuning or spatially patterned tuning. For example, Basu et al. (1) created a multicellular system that functioned as a band-detect filter for a small molecule. They demonstrated how the position of the band could be shifted to different concentration ranges by changing a repressor's affinity for a signaling molecule and by altering gene copy number. Tuning was achieved through building a different biological system for each desired band position. However, electronic band-detect filters can be built such that any desired range can be set dynamically through adjusting knobs from outside the device. The ability to create analogous externally tuneable biological systems would greatly expand their versatility. We describe how proper inclusion of an enzyme-substrate pair in the network results in an externally tunable genetic circuit and that such a circuit enables cellular behavior that cannot be readily achieved by traditional tuning.
Results and Discussion
Design of the Band-Pass Filter.
We have constructed a single biological system (Fig. 1A) that functions as a band-pass filter for cellular β-lactamase activity in which the position and width of the band can be tuned externally. External tunability necessitated genetic circuitry with a different architecture than band-detect systems that lack this feature (1–3). Our circuitry, like that of previous band-detect systems, can be conceptualized as an incoherent type 3 feed-forward loop (ref. 4; Fig. 1B). Our design is a combination of positive and negative genetic selections for β-lactamase activity and analogous to an electronic band-pass filter constructed from low-pass and high-pass filters in series (Fig. 1C). β-Lactamases catalyze the hydrolysis of penicillin antibiotics such as ampicillin (Amp), which inhibit cell wall synthesis. Our positive selection is growth in the presence of Amp. Only cells with β-lactamase activity above a certain level (the high-pass cutoff) are able to grow in the presence of Amp. Our negative selection is growth in the presence of tetracycline (Tet). The tetracycline resistance gene (tetC) and the green fluorescent protein gene (gfp) are under the control of the ampicillin resistance gene (ampC) promoter, which is repressed by AmpR. Both ampC and ampR are from Citrobacter freundii and naturally control the induction of its β-lactamase (5). The cell wall intermediate aM-pentapeptide (L1,6-anhydro-N-acetylmuramyl-L-alanyl-D-glutamyl-meso-diaminopimelic acid-D-alanyl-D-alanine) accumulates in the presence of low levels of β-lactams and induces the ampC promoter via interactions with AmpR (6, 7). Cells with too much β-lactamase activity degrade the β-lactams and prevent the accumulation of aM-pentapeptide, leaving the cells sensitive to Tet. Only cells with less than a certain level of β-lactamase activity (the low-pass cutoff) will proliferate in the presence of Tet. Provided the low-pass cutoff is set above the high-pass cutoff (Fig. 1C), only cells with intermediate levels of β-lactamase activity should proliferate in the presence of Amp and Tet.
Fig. 1.
Design of the bacterial band-pass filter. (A) Schematic depiction of the essential components of the band-pass filter. Plasmids pTS1 and pDIM-C8 are compatible. In the absence of sufficient cellular β-lactamase (BLA) activity for hydrolysis of Amp, cell wall synthesis is compromised and cells cannot proliferate. In addition, cell wall breakdown results in the accumulation of aM-pentapeptide (aM-Pp), which induces the ampC promoter via interactions with AmpR (6, 7), resulting in the production of TetC (which confers Tet resistance) and GFP. However, the level of Amp necessary to induce ampC is lower than the level that prevents the growth of E. coli cells (7). BLA gene expression is regulated through IPTG induction of the tac promoter, which is repressed by LacI. Complete plasmid maps are provided in Fig. S1. (B) Conceptualization of the genetic circuit as an incoherent type 3 feed-forward loop (4). In order for growth to occur in the presence of Tet, the Amp concentration must not be above the MICAmp but TetC must be present as a result of induction by Amp (Kind, Amp). The values of MICAmp and Kind, Amp are approximate and for when BLA is absent. BLA serves to attenuate the Amp signal entering the feed-forward loop. (C) Schematic depiction of how the combination of positive and negative genetic selection results in band-pass behavior. Band-pass behavior results when the high-pass cutoff (f1) is set at a lower enzyme activity than the low-pass cutoff (f2).
Band-Pass Filter Is Externally Tunable.
To test our system, we constructed strains with 3 different β-lactamase genes that conferred different levels of Amp resistance (Table 1). These genes were placed under the control of the isopropyl β-D-1-thiogalactopyranoside (IPTG)–inducible tac promoter on pDIM-C8. The strains' level of β-lactamase activity as a function of IPTG was quantified by determining the minimum inhibitory concentration of Amp (MICAmp) (Fig. 2A and Table 1). We verified Amp induction of the ampC promoter by measuring cell fluorescence (i.e., GFP expression) as a function of Amp (Fig. 2B).
Table 1.
MICAmp of bacterial strains*
| Strain | Protein expressed from tac promoter | MICAmp (μg/mL) in presence of |
|
|---|---|---|---|
| 0 μM of IPTG | 300 μM of IPTG | ||
| RH01 | P99 β-lactamase | 2 | 64 |
| RH05 | MBP (negative control) | 1 | 1 |
| RH06 | TEM1 β-lactamase | 32 | 2,048 |
| RH09 | TEM1 β-lactamase with W208G mutation | 1 | 16 |
*These are SNO301 strains that contain plasmids pTS1 and pDIM-C8. They differ only in the protein expressed from the tac promoter on pDIM-C8.
Fig. 2.
External tuning of the position and bandwidth of the band-pass filter. (A) Relation between IPTG and the MICAmp for strains RH06 and RH09 in liquid LB at 37 °C in a 96-deep-well format. (B) Induction of the ampC promoter by Amp. Total culture fluorescence (i.e., induction of GFP) was measured as a function of Amp for RH05 cells in liquid LB at 37 °C in a 96-deep-well format. Fluorescence (arbitrary units) was normalized by the culture's OD600. The values are the mean of the ratio (n = 3), and the error bars reflect the SD. The value for cells in the absence of Amp was 120 ± 20. The value is plotted as 0 for Amp ≥1 μg/mL because the cells did not grow. (C) The growth of cells in liquid LB containing 20 μg/mL Tet was detected by measuring the OD600 of the liquid culture and is presented in the form of a heat map as a function of Amp concentration (y axis) and cellular β-lactamase activity (x axis). The level of cellular β-lactamase is controlled by the concentration of IPTG (see A) and expressed as MICAmp. Strain RH09 was used to observe the behavior of cells with very low levels of β-lactamase activity. (D) Selected data from the heat map illustrate how cellular β-lactamase activity (controlled by IPTG and quantified as MICAmp) can be used to tune the position of the band-pass filter for Amp. (E) Selected data from the heat map illustrate how Amp can be used to tune the position of the band-pass filter for β-lactamase activity. Values in C–E are the mean (n = 3), and error bars reflect the SD. (F) The bandwidth can be externally tuned by the concentration of Tet. The growth of RH06 cells in liquid media containing IPTG (300 μM), different amounts of Tet (different lines), and Amp (x axis) was detected by measuring the OD600 of the liquid culture. The concentrations of Tet were 0, 5, 6, 7, 8, 9, 10, 15, and 20 μg/mL. The OD600 is normalized to the maximum value for each Tet concentration tested. The detailed conditions for C–E can be found in Table S3.
To test for band-pass behavior, we measured growth as a function of Amp, MICAmp, and Tet concentration. The system functioned as designed (Fig. 2 C–F). In the presence of Amp and Tet, cells would grow only in a narrow concentration range of Amp determined by their IPTG-dependent level of β-lactamase activity (Fig. 2C). Above the high end of this concentration range, the cells lacked enough β-lactamase activity to hydrolyze Amp sufficiently. Below the low end of this concentration range, the cells hydrolyzed Amp too efficiently and the ampC promoter was not sufficiently induced, leaving the cells susceptible to Tet. This concentration range shifted to higher levels of Amp as cellular β-lactamase activity was increased. Likewise, an increase in the level of Amp required an increase in the level of β-lactamase activity for the strain to grow. Thus, the position of our band-pass filter for Amp can be externally tuned by adding IPTG to the medium (Fig. 2D), and our band-pass filter for β-lactamase activity can be externally tuned by adding Amp to the medium (Fig. 2E). The observed behavior was attributable to differences in cellular β-lactamase activity and not to some unanticipated consequence of varying the concentration of IPTG, because strains engineered to exhibit different levels of β-lactamase activity at the same IPTG concentration had equivalent behavior predictable based on their MICAmp (Fig. S2). The bandwidth could be broadened about 4-fold (in terms of Amp concentration) by decreasing the concentration of Tet from 20 to 8 μg/mL (Fig. 2F). Decreasing Tet concentration further resulted in behavior intermediate between a band-pass filter and the low-pass filter behavior observed in the absence of Tet.
Our single system achieves a continuum of behaviors that would have previously required the nontrivial construction of a plethora of different biological systems. In addition to convenience and versatility, external tunability offers temporal and spatial control over circuit characteristics, something that is not possible through traditional tuning. Such control is useful for gaining an understanding of basic biological phenomena and for biotechnological applications. We demonstrate this through the patterning of bacterial growth using chemical gradients and the engineering of allosteric enzymes.
Spatial-Dependent Tuning: Patterning of Bacterial Growth.
Specific cellular responses to chemical gradients are important for natural development processes and for engineering desired growth and differentiation patterns for applications such as biomaterials and tissue engineering. Band-detect systems that lack external tuning can be used to pattern cells using gradients of the detected molecule (1). However, cell patterning cannot be altered independent of the gradient (i.e., for a given cell type with a given gradient, only a single pattern is possible), and the patterns are limited to those that follow the gradient's contour lines. An externally tunable system overcomes both of these limitations.
We initially verified that cells spread evenly on agar containing Tet and a gradient of Amp radiating from the center of the plate produced the expected rings of growth and fluorescence. In the absence of Tet, cells grew everywhere except at the high Amp concentrations near the disc; however, fluorescence was observed only in the expected ring pattern (Fig. 3A). As the Tet concentration increased, growth became restricted to the same ring pattern. The ring of growth is thinnest at the highest Tet concentration, presumably because of the bandwidth's dependence on Tet concentration (Fig. 2F). The radius of the ring could be controlled through the level of Amp (Fig. 3B). These patterns are expected because cells will grow in the presence of Tet only if the Amp concentration falls within a narrow range. This range depends on the level of cellular β-lactamase activity. Thus, a mixture of 3 cell types possessing different levels of β-lactamase activity formed 3 concentric rings of growth around an Amp-soaked disc in which each ring corresponded to cells with a different level of β-lactamase activity (Fig. 3D). This single-step separation of a mixture of cells into a rank order based on their cellular enzyme activity was verified by analyzing the cells in each ring by DNA sequencing (Fig. S3). Because β-lactamase activity can be controlled using IPTG (Fig. 2A), IPTG can be used to create different-sized rings using identical cells and the same concentration gradient of Amp (Fig. 3C). Such external control over the position of “cell fate” within the gradient is useful in achieving the desired positional dependence without system reengineering. In addition, such control would be invaluable in situations in which the gradient cannot be easily controlled, such as controlling differentiation patterns in response to cell-produced signaling molecules.
Fig. 3.
Bacterial pattern formation using the band-pass filter circuit. Liquid cultures were spread evenly on 86-mm-diameter round or 91-mm square LB-agar plates containing Tet and/or IPTG. Ninety minutes after plating, filter paper discs were placed in the center or at the indicated locations and were spotted with Amp, IPTG, or Tet. Plates were incubated at 37 °C. Visible light and fluorescent images are shown. The detailed conditions for all plates can be found in Table S4 and Fig. S5. (A) Cells on IPTG plates with different concentrations of Tet and an Amp disc in the center. (B) Cells on Tet/IPTG plates with Amp discs of different concentrations in the center. (C) Cells on Tet plates with different concentrations of IPTG and an Amp disc in the center. (D) A 1:1:1 mixture of RH01, RH05, and RH06 cells (strains with different levels of β-lactamase activity) on Tet/IPTG plates with an Amp disc in the center. (E) Overlapping radial concentration gradients of 2 compounds. The black line comprising all points equidistant from the 2 point sources marks the location where the ratio of the 2 compounds' concentrations is a constant. (F) Cells on Tet plates with discs of Amp, IPTG, or Tet at the indicated positions.
Whereas it is intuitive how circular patterns of growth might arise from radially symmetrical concentration profiles, it is not intuitive how such profiles can impose patterns that lack radial symmetry (e.g., linear patterns). However, external tuning enables position-dependent tailoring of the genetic circuit and provides a mechanism for establishing patterns that do not follow the gradient's contour lines. Our cells require a particular ratio of Amp to β-lactamase activity for growth (Fig. 2C). Because IPTG controls the level of β-lactamase activity, our cells require a particular ratio of Amp to IPTG for growth (in the regimen in which the relation between IPTG concentration and β-lactamase activity is approximately linear). When 2 compounds (e.g., Amp, IPTG) diffuse from point sources near each other, the ratio of the 2 compounds' concentrations will be a constant along the line comprising all points equidistant from the point sources (Fig. 3E), provided that the compound's diffusivities are the same. Thus, by placing the right amount of Amp and IPTG on discs at different locations, we specify that our cells grow only on the line equidistant from the 2 discs—the location at which their ratio is the same and at a value that allows growth at some critical time. Thus, our cells interpret overlapping, opposing radial concentration profiles of Amp and IPTG, and produce extended linear patterns (Fig. 3F). This patterning is enabled by IPTG's spatial-dependent tuning of the position of the band-pass filter for Amp. The key for obtaining these patterns is to determine the amount of Amp and IPTG to place at the points located a set distant apart. The details of the dynamics did not need to be modeled explicitly to determine these amounts. Determination of the necessary amount of Amp and IPTG to add to the plate was facilitated by a model for ring size as a function of Amp that used Fick's second law of diffusion (ref. 8; Fig. S4) and was fine-tuned by trial and error.
A Synthetic Model System for Development.
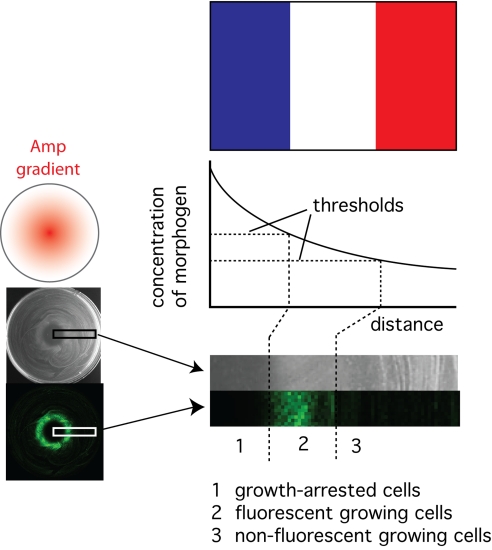
Both Amp and IPTG can be thought of as “morphogens” that are affecting the “fate” of our bacteria through changes in gene expression and cell physiology. Morphogens are substances in eukaryotes that induce different cell fates at different concentrations. Morphogens are especially important in development because emission of a morphogen from a source can lead to the formation of different cell types in a defined spatial relation to the source and to each other (9). The concept is succinctly stated in Wolpert's French flag model of positional information (ref. 10; Fig. 4). In this highly influential framework for testing and understanding pattern formation processes, cells have 2 or more threshold concentrations for a morphogen that elicit different cell behavior. A pattern of cell differentiation (represented by the 3 stripes on the French flag) forms when the cells are presented with a morphogen gradient.
Fig. 4.
A synthetic model system for morphogen-induced pattern formation in development. RH01 cells were plated with an Amp disc in the center (conditions for plating as in Fig. 3A). An Amp gradient elicits a growth pattern that fits the French flag model (10) and consists of 3 different cell types with a defined spatial relation with each other and the Amp source.
Using a simple band-pass filter paradigm, we have succeeded in engineering Escherichia coli into a simple model of developmental patterning that fits the French flag model (Fig. 4). When a lawn of our cells is challenged to grow in the presence of an Amp gradient, cells have differential fates depending on their distance from the Amp source. This fate is directly attributable to Amp. Near the Amp source, Amp inhibits cell proliferation. Further away, a threshold is crossed beyond which cells can proliferate and Amp causes the induction of GFP. Further away still, a second threshold is crossed in which the Amp concentration is no longer sufficient to induce GFP expression but the cells can still proliferate.
The study of natural morphogens during development is challenging because of the complexity of in vivo systems and the extremely low concentrations of most morphogens (9). Our synthetic system offers a convenient well-defined model system for fundamental studies on how morphogen gradients can affect cells and lead to pattern formation. When the French flag model was proposed (10), morphogens were purely theoretical, but the model provided experimentalists with a conceptual framework with specific molecular mechanisms from which to search for morphogens. Similarly, a simple experimental system like ours can quickly test our understanding and lead to hypotheses on morphogen-induced pattern formation. For example, our patterning of cell fate using overlapping gradients of different morphogens (Fig. 3F) predicts that such a mechanism could be used in natural development to form linear structures between 2 morphogen sources.
Dynamic Tuning: Genetic Selection of Protein Switches.
As a second example of an application that benefits from external tunability, we identified allosteric protein switches from combinatorial libraries by exploiting the ability of our band-pass selection system to be tuned to different settings as a function of time. Protein switches hold promise as selective protein therapeutics and biosensors (11, 12). We have previously created maltose-activated β-lactamase switches by the nonhomologous recombination of the genes encoding maltose-binding protein (MBP) and BLA and subjecting the resulting libraries to positive genetic selection for β-lactamase activity in the presence of maltose, followed by a time-consuming screen for maltose-dependent β-lactamase activity (13, 14). For example, from Library 7, in which random circular permutations of bla were inserted at a specific site in the gene encoding MBP, we identified 3 switches (MBP317–347, MBP317–639, and MBP317–694) by screening 1,056 library members that survived the positive selection (13). These switches have low β-lactamase activity in the absence of maltose and high β-lactamase activity in the presence of maltose. Ligand-activated β-lactamases have potential use as cancer-specific prodrug-activating enzymes and as genetically encoded biosensors for the in vitro, live-cell, and in vivo quantification of biological molecules.
Here, we re-examined Library 7 using our band-pass system, performing a 2-tiered selection designed to identify switches with high β-lactamase activity in the presence of maltose and low β-lactamase activity in the absence of maltose. This 2-tiered selection makes use of the ability of our band-pass filter for β-lactamase activity to be tuned to different positions at different points in time. In the first tier, cells were challenged to grow at a high level of Amp in the presence of maltose (i.e., the circuit was set to high-pass mode for cells with high β-lactamase activity). In the second tier, library members surviving the first tier were challenged to grow in the presence of Tet and low levels of Amp but in the absence of maltose (i.e., the circuit was set to band-pass mode for cells with low β-lactamase activity).
After the second tier of the selection, the lysates of 4 of the 34 surviving library members exhibited the desired maltose-dependent β-lactamase activity. DNA sequencing revealed that 2 were MBP317–347 and 2 were MBP317–694. Thus, the second tier of the band-pass selection enriched for allosteric switches 35-fold. Interestingly, many of the remaining 30 library members exhibited the desired switching phenotype despite the lack of maltose-dependent β-lactamase activity in their lysates. Four of 5 library members examined had >8-fold higher MICAmp in the presence of maltose compared with its absence. Expression studies on 2 of these library members indicated that periplasmic production of the fusion protein increased substantially when the cells were grown in the presence of maltose, perhaps as a result of ligand-induced stabilization (15) of the fusion protein. Such proteins represent an important class of protein switches different from our allosteric switches—ones for which our band-pass genetic selection (but not our original screen) has the ability to identify.
General Design Principles for Externally Tunable Genetic Circuits.
Our system's characteristic property (the location of the band-pass region) is tunable from outside the cell using Amp, IPTG, and Tet. The inclusion of an enzyme-substrate pair (BLA and Amp), which functions as an attenuator in the network, enabled this tunability. With few exceptions (16), synthetic gene circuit analogues of electronic circuits have not incorporated enzymes as essential components. However, their use as a mechanism to produce or consume key network molecules, as we have done with our system, is a generalizable strategy for creating externally tunable systems that allow temporal and spatial control over genetic circuit behavior. For example, a simple inducible promoter could be made externally tunable by engineering control over the expression of an enzyme that degrades (or produces) the inducer.
Recently, the period of a synthetic gene oscillator was observed to vary with inducer concentrations (17). The inducer controlled production of a repressor protein. Based on modeling, the investigators proposed that the period depended on the time required for the repressor protein to decay (i.e., the larger the burst of repressor protein, which depended on the inducer concentration, the longer it took for the repressor protein to decay). This system obeys our design principle, with a cellular protease serving as the enzyme and the repressor protein serving as the substrate.
Genetic circuits designed with the inherent ability to be tuned from outside offer a continuum of circuit settings in a system and allow time- and position-dependent tuning. Aside from making more versatile cells, the ability to tune externally facilitates system optimization as well as experiments aimed at understanding network behavior. Externally tunable systems with band-pass characteristics such as ours should enable more precise and complex growth and gene expression patterning for applications such as tissue engineering and biomaterials as well as improve our understanding of mechanisms by which differentiation occurs during development.
Methods
Strains.
Strains and their properties are provided in Table S1. E. coli SNO301 (ampD1, ampA1, ampC8, pyrB, recA, and rpsL) was used to observe band-pass behavior in liquid culture and on plates. The ampD1 mutation allows β-lactam hyperinducible transcription of genes under the control of the ampC promoter (7). The rpsL mutation confers streptomycin (Sm) resistance to this host, the same resistance marker used to maintain pTS1. However, we observed that pTS1 was stably maintained in SNO301 cells in the presence of 100 μg/mL Sm. Multiple aliquots of frozen cell stocks for plating and growth experiments were prepared from overnight LB cultures at a final OD600 of 0.5 and contained 15% (wt/vol) glycerol. These stocks were stored at −80 °C. E. coli strain RH22 was constructed and used for evaluation of the β-lactamase switch library because it had much better transformation efficiency than SNO301. RH22 is derived from ElectroMAX DH5α-E E. coli cells (Invitrogen) and carries pTS1. The ampD1 mutation was introduced into DH5α-E by homologous recombination facilitated by phage λ Red recombinase (18) as described in SI Text.
Plasmids.
Plasmid descriptions and maps are provided in Table S2 and Fig. S1. The ampR gene and the ampC promoter were PCR-amplified from plasmid pNU305 (5) such that EcoRI and NdeI restriction enzyme sites were added at the 5′ and 3′ ends, respectively. GFPmut2 was PCR-amplified from pDB2-GFP (19) such that NdeI and HindIII restriction enzyme sites were added at the 5′ and 3′ ends, respectively. TetC was PCR-amplified from pBR322 such that HindIII and XhoI restriction enzyme sites were added at the 5′ and 3′ ends, respectively. These 3 components were inserted between the EcoR1 and XhoI sites of pCDFDuet-1 (Novagen) such that the gfp and tetC genes were expressed polycistronically under the control of the ampC promoter. This reporter plasmid (pTS1) has Sm resistance and carries the CloDF13 replication origin. The β-lactamase expression plasmid pDIM-C8 has chloramphenicol (Cm) resistance and a p15a replication origin, which is compatible with the CloDF13 replicon. All genes cloned into pDIM-C8 are under the control of the IPTG-inducible tac promoter whose leaky expression was suppressed by LacI from pTS1.
Liquid Culture Growth Experiments.
Glycerol stocks were thawed on ice, and a working stock was prepared by adding 15 μL of the frozen stock to 1.2 mL of LB. In each well of a 96-deep-well plate, 5 μL of working stock (corresponding to about 6,000 cfu) was added to 500 μL of LB medium containing Sm (100 μg/mL) and Cm (50 μg/mL) as well as various concentrations of Amp, IPTG, and Tet. The plate was incubated at 37 °C in a shaker incubator. After 17 h of incubation, 200 μL was transferred to black-walled, clear-bottomed, 96-well plates and the OD600 was determined using a Molecular Devices SpectraMax plus. The MICs for Amp (Fig. 2A and Table 1) were determined in the absence of Tet using this growth method. The MIC was defined as the lowest concentration of Amp at which the OD600 was <3% of the OD600 in the absence of Amp. The characterization of the ampC promoter (Fig. 2B) was performed in the absence of Tet and IPTG. Total cell fluorescence was measured at 538 nm using a Molecular Devices Gemini XPS plate reader with excitation at 485 nm.
LB-Agar Growth Experiments.
Ten-fold dilutions of frozen stocks (150 μL for 86-mm round plates and 206 μL for 91-mm square plates) were spread evenly over the entire LB-agar [1.5% (wt/vol)] plates containing Sm (100 μg/mL), Cm (50 μg/mL), and various concentrations of IPTG and Tet. For the plate in Fig. 3D, 50 μL of each of the 3 strains was mixed before plating. Agar volume was 20 mL for 86-mm round plates and 27.5 mL for 91-mm square plates. Ninety minutes after plating, dry 1-cm-diameter paper discs (Whatman Grade 1) were placed at the desired position using a schematic map laid underneath the plate to ensure reproducible placement. The discs were spotted with Amp, IPTG, or Tet as indicated. The detailed conditions for each pattern are provided in Table S4 and Fig. S5. The plates were incubated at 37 °C for 21 h for rings with radii <2.5 cm. Plates producing growth rings with radii >2.5 cm were incubated at 37 °C for 24 h and further incubated at room temperature (23 °C) for 16 h to produce clearly visible growth. After disc removal, growth images were acquired by a CCD camera under white light reflection conditions using either DC290 (Kodak) or ChemiDoc XRS (BioRad). Fluorescence images were acquired with a Typhoon 9410 imager (GE Lifescience) using 350-V 488-nm laser excitation; 526-nm short pass filter detection with 200-μm resolution; and agar top focusing (+3 mm). Grayscale images were converted to green, and the contrast was adjusted by using Image Quant TL software (GE Lifescience).
Selection of Maltose-Activated β-Lactamases from a Combinatorial Library.
Plasmid DNA comprising Library 7 (13) was transformed into RH22 cells. These cells were subjected to a 2-tiered selection process designed to identify switches. In the first tier, cells were spread on LB-agar plates containing Amp (200 μg/mL), Cm (50 μg/mL), Sm (50 μg/mL), IPTG (300 μM), and maltose (10 mM), which were incubated overnight at 37 °C. Colonies that formed were recovered en masse in LB. In the second tier, cells surviving the first tier were spread on LB-agar plates containing Amp (8 μg/mL), Cm (50 μg/mL), Sm (50 μg/mL), Tet (60 μg/mL), and IPTG (300 μM), which were incubated at 37 °C overnight. Colonies that formed were screened for maltose-dependent nitrocefin hydrolysis activity as described (14).
Supplementary Material
Acknowledgments.
We thank Jef D. Boeke, Jeffrey J. Gray, and Monica Berrondo for discussions. We thank Matti Karp, University of Turku (Finland), for strain SNO301 and Susanne Lindquist, Umeå University (Sweden), for plasmid pNU305. This work was supported by funding from the National Institutes of Health and a gift from the Asahi Kasei Corporation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901246106/DCSupplemental.
References
- 1.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 2.Weber W, Bacchus W, Daoud-El Baba M, Fussenegger M. Vitamin H-regulated transgene expression in mammalian cells. Nucleic Acids Res. 2007;35:e116. doi: 10.1093/nar/gkm466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Entus R, Aufderheide B, Sauro HM. Design and implementation of three incoherent feed-forward motif based biological concentration sensors. Syst Synth Biol. 2007;1:119–128. doi: 10.1007/s11693-007-9008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindberg F, Westman L, Normark S. Regulatory components in Citrobacter freundii ampC beta-lactamase induction. Proc Natl Acad Sci USA. 1985;82:4620–4624. doi: 10.1073/pnas.82.14.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dietz H, Pfeifle D, Wiedemann B. The signal molecule for beta-lactamase induction in Enterobacter cloacae is the anhydromuramyl-pentapeptide. Antimicrob Agents Chemother. 1997;41:2113–2120. doi: 10.1128/aac.41.10.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valtonen SJ, Kurittu JS, Karp MT. A luminescent Escherichia coli biosensor for the high throughput detection of beta-lactams. J Biomol Screen. 2002;7:127–134. doi: 10.1177/108705710200700205. [DOI] [PubMed] [Google Scholar]
- 8.Humphrey JH, Lightbown JW. A general theory for plate assay of antibiotics with some practical applications. J Gen Microbiol. 1952;7:129–143. doi: 10.1099/00221287-7-1-2-129. [DOI] [PubMed] [Google Scholar]
- 9.Gurdon JB, Bourillot PY. Morphogen gradient interpretation. Nature. 2001;413:797–803. doi: 10.1038/35101500. [DOI] [PubMed] [Google Scholar]
- 10.Wolpert L. Positional information and the spatial pattern of cellular differentiation. J Theor Biol. 1969;25:1–47. doi: 10.1016/s0022-5193(69)80016-0. [DOI] [PubMed] [Google Scholar]
- 11.Dueber JE, Yeh BJ, Bhattacharyya RP, Lim WA. Rewiring cell signaling: The logic and plasticity of eukaryotic protein circuitry. Curr Opin Struct Biol. 2004;14:690–699. doi: 10.1016/j.sbi.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Ostermeier M. Engineering allosteric protein switches by domain insertion. Protein Eng, Des Sel. 2005;18:359–364. doi: 10.1093/protein/gzi048. [DOI] [PubMed] [Google Scholar]
- 13.Guntas G, Mansell T, Kim JR, Ostermeier M. Directed evolution of protein switches and their application to the creation of ligand-binding proteins. Proc Natl Acad Sci USA. 2005;102:11224–11229. doi: 10.1073/pnas.0502673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guntas G, Mitchell SF, Ostermeier M. A molecular switch created by in vitro recombination of nonhomologous genes. Chem Biol. 2004;11:1483–1487. doi: 10.1016/j.chembiol.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Kohn JE, Plaxco KW. Engineering a signal transduction mechanism for protein-based biosensors. Proc Natl Acad Sci USA. 2005;102:10841–10845. doi: 10.1073/pnas.0503055102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fung E, et al. A synthetic gene-metabolic oscillator. Nature. 2005;435:118–122. doi: 10.1038/nature03508. [DOI] [PubMed] [Google Scholar]
- 17.Stricker J, et al. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durai S, Bosley AD, Bridgeman A, Chandrasegaran S, Ostermeier M. A bacterial one-hybrid selection system for interrogating zinc finger-DNA interactions. Comb Chem High Throughput Screening. 2006;9:301–311. doi: 10.2174/138620706776843147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.