Abstract
Background
World-wide phylogeographic distribution of human complete mitochondrial DNA sequences suggested a West Asian origin for the autochthonous North African lineage U6. We report here a more detailed analysis of this lineage, unraveling successive expansions that affected not only Africa but neighboring regions such as the Near East, the Iberian Peninsula and the Canary Islands.
Results
Divergence times, geographic origin and expansions of the U6 mitochondrial DNA clade, have been deduced from the analysis of 14 complete U6 sequences, and 56 different haplotypes, characterized by hypervariable segment sequences and RFLPs.
Conclusions
The most probable origin of the proto-U6 lineage was the Near East. Around 30,000 years ago it spread to North Africa where it represents a signature of regional continuity. Subgroup U6a reflects the first African expansion from the Maghrib returning to the east in Paleolithic times. Derivative clade U6a1 signals a posterior movement from East Africa back to the Maghrib and the Near East. This migration coincides with the probable Afroasiatic linguistic expansion. U6b and U6c clades, restricted to West Africa, had more localized expansions. U6b probably reached the Iberian Peninsula during the Capsian diffusion in North Africa. Two autochthonous derivatives of these clades (U6b1 and U6c1) indicate the arrival of North African settlers to the Canarian Archipelago in prehistoric times, most probably due to the Saharan desiccation. The absence of these Canarian lineages nowadays in Africa suggests important demographic movements in the western area of this Continent.
Background
Attested presence of Caucasian people in Northern Africa goes up to Paleolithic times. From the archaeological record it has been proposed that, as early as 45,000 years ago (ya), anatomically modern humans, most probably expanded the Aterian stone industry from the Maghrib into most of the Sahara [1]. More evolved skeletal remains indicate that 20,000 years later the Iberomaurusian makers, replaced the Aterian culture in the coastal Maghrib. Several hypothesis have been forwarded concerning the Iberomaurusian origin. They can be resumed in those which propose an arrival, from the East, either from the Near East or Eastern Africa, and those which point to west Mediterranean Europe, either from the Iberian Peninsula, across the Gibraltar Strait, or from Italy, via Sicily, as their most probable homeland [2]. Between 10,000 and 6,000 ya the Neolithic Capsian industry flourished farther inland. The historic penetration in the area of classical Mediterranean cultures, ending with the Islamic domination, supposed a strong cultural influx. However, it seems that the demic impact was not strong enough to modify the prehistoric genetic pool.
Linguistic research suggests that the Afroasiatic phylum of languages could have originated and extended with these Caucasians, either from the Near East or Eastern Africa and that posterior developments of the Capsian Neolithic in the Maghrib might be related to the origin and dispersal of proto-Berber speaking people into the area [3]. Nowadays, the Berber speakers, scattered throughout Northwest Africa from the Atlantic to the Lybic desert and from the Mediterranean shores to the south of the Sahel, are considered the genuine descendants of those prehistoric colonizers. Some important issues are pending of resolution to clarify the past and present of the North African Caucasians: To which extent the Neolithic waves substituted the Paleolithic recipients? Which is the most probable origin of these prehistoric occupants? Did they come from Europe, East Africa, Southwest Asia or are they a result of an "in situ" evolution? Is there a correspondence between the Afroasiatic diversification and the spread of Caucasians?
Recently, molecular genetic research on North African populations has contributed new data to test the major issues proposed on archaeological, anthropological and linguistic grounds. The studies based on uniparental genetic markers have been particularly informative. Both, mitochondrial DNA (mtDNA) sequences [4,5], and Y-chromosome binary markers [6,7] detected specific North African haplotypes that confirm an ancient human colonization for this area and a sharp discontinuity between Northwest Africa and the Iberian Peninsula. From a mtDNA point of view, the most informative of these genetic markers is the North African clade U6. On the basis of complete mtDNA sequences, it has been proposed that U6 lineages, mainly found in North Africa, are the signatures of a return to Africa around 39,000–52,000 ya [8]. This stresses the importance of its detailed study in order to trace one of the earliest Caucasian arrivals to Africa. Although in moderate frequencies, the geographic range of this clade extends from the Near East to the Canary Islands, along the Atlantic shores of Northwest Africa and from the Sahel belt, including Ethiopia, to the southern Mediterranean rim. Out of this area, U6 has only been spotted in the Iberian Peninsula [9-12], Sicily [13], in the north European Ashkenazic Jews [14], and in Ibero-America. The presence in the latter is, most probably, the result of the Spanish and Portuguese colonization [15,16].
In order to construct an unambiguous phylogeny for this clade and infer precise ages for the whole group and for its derivatives, we have fully sequenced eleven mitochondrial lineages representing the main branches of U6. Subsequently, we analyzed the geographic distribution range and relative diversity of these subclades, to deduce their most probable expansion origins based on sequence information of the first hypervariable segment (HVSI) of the mitochondrial control region and on new RFLPs, discovered to be diagnostic for them.
Results
A new sublineage for U6
Haplogroup U splits from R by mutations 11467, 12308 and 12372. Three branches sprout from this root: U5 (3197, 9477, 13617 and 16270), U6 (3348 and 16172) and the rest of the U clade defined by mutation 1811 [8,17,18]. For this reason, a representative of U5 was chosen as an outgroup.
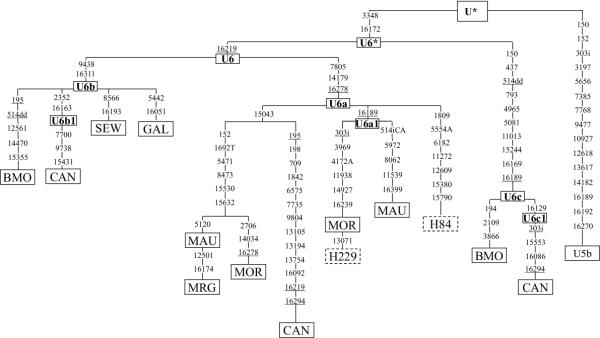
The phylogenetic tree based on complete mtDNA U6 sequences, confirms that this clade is defined by mutations 3348 and 16172 (Fig. 1). The former can be detected by RFLP analysis using MboI [15]. The existence of three subgroups is also evident. U6a was defined by the presence of HVSI mutations 16172, 16219 and 16278 [4] and now by 7805 and 14179 in the coding region, that can be tested by RFLPs -7802 MaeI and +14179 AccI, respectively. Subgroup U6b was characterized by HVSI mutations 16172, 16219 and 16311 [4], to which mutation 9438 (detectable by RFLP -9438 HaeIII) can now be added. The new clade U6c is defined by HVSI mutations 16169, 16172 and 16189 and at least by mutations 4965 and 5081, that can be tested by RFLPs +4963 Aci I and -5079 Tsp509 I, respectively. In addition, a subgroup, U6a1, has been detected within U6a characterized by the addition of HVSI mutation 16189 [4]. In the same way, HVSI mutation 16163 classifies subgroup U6b1, autochthonous of the Canary Islands [19]. Within the coding region, this subgroup can be further defined by RFLP + 2349 MboI.
Figure 1.
Phylogenetic tree based on complete U6 mtDNA genome sequences. A U5b individual has been added in order to root the tree. Nomenclature of individuals as in Table 2. Numbers along links refer to nucleotide positions; suffixes are transversions; i insertions and dd deletion of two nucleotides. Underlining indicates recurrent mutations in U6 subhaplogroup. U presents the following mutations with respect to rCRS: 73, 263, 311i, 750, 1438, 2706, 4769, 7028, 8860, 11467, 11719, 12308, 12372, 14766 and 15326.
From Fig. 1, an important question rises about the constant mutation rate in the coding region. The mean number of substitutions accumulated in U6b lineages (Table 1) is significantly smaller than those in U6a (P = 0.013) and is near significance in U6c (P = 0.058). These differences are mainly due to the number of mutations accumulated in the coding region. Following others [20], we used the likelihood-ratio test [21] to asses whether the mutations accumulated on the different branches were compatible or not with a uniform rate. The difference between the values obtained for the uniform clock model (L0 = -23060.25) and for the variable rate model (L1 = -23032.22), was statistically significant at the 5% level. So, the simpler clock-like tree was rejected. On the other hand, the substitution ratio between coding vs. HVSI region is double in U6a than in U6b or U6c (Table 1). Furthermore, taking into account the ratio of synonymous vs. non-synonymous substitutions in the coding region, again the U6a value doubles that of U6b or U6c, reaching a significant level (P = 0.0237, in a two-tailed Fisher exact test). Both selection and stochastic processes have to be invoked to satisfactorily explain these data. A bias in lineage sampling is the most probable cause of the different substitution ratios between D-loop and coding regions: the U6b and U6c lineages were chosen for their different geographic origin and, comparatively, large divergence in HVSI, whilst for U6a we chose central representatives of the different subclusters excepting that of the Canary Islands. In relation to the differences in synonymous vs. non-synonymous ratios, they could be attributed to the action of purifying selection, having a stronger effect on the older U6a lineages. From this, we deduced that both U6b and U6c spread more recently. Finally, the apparent differences in substitution rates between U6b and U6a or U6c could better be the result of genetic drift, so that the founder lineage that originated the U6b subgroup was less evolved than those that originated U6a and U6c. However, we have to point out that in a similar case, in which significant differences were found in the number of mutations accumulated on two clades of haplogroup L2, selection was suggested as the most probable cause [20].
Table 1.
Mean number of substitutions, from the base of U6, for the three subgroups of U6 calculated for the twelve complete sequences.
| REGIONS | ||||||
| N | Total | Coding | HVSI | Coding/HVSI | Syn./nonsyn. | |
| U6b | 4 | 6.3 | 3.3 | 3.0 | 1.1 | 0.8 |
| U6a | 6 | 12.8 | 8.5 | 4.3 | 2.0 | 2.3 |
| U6c | 2 | 12.5 | 6.5 | 6.0 | 1.1 | 1.0 |
Geographic distribution of U6 lineages
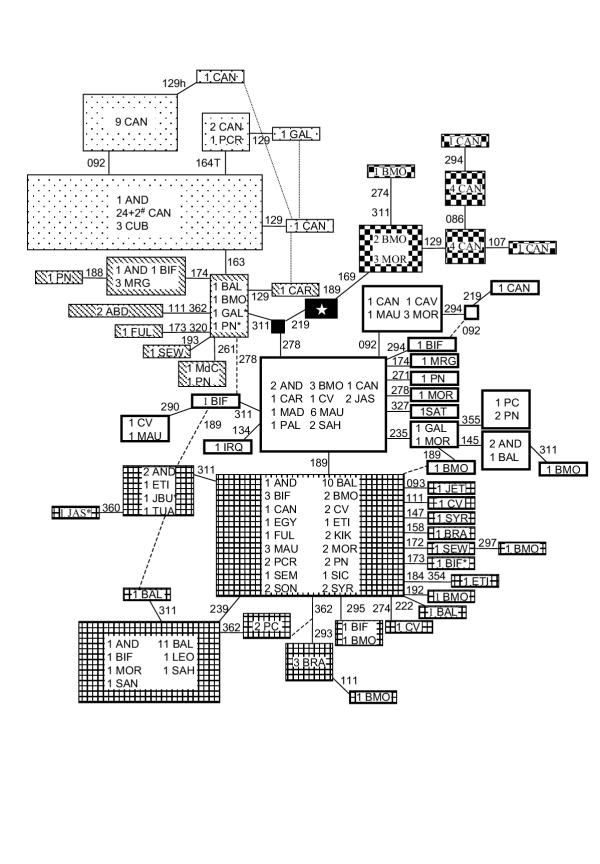
Fig. 2 shows the reduced median network obtained from the 56 U6 haplotypes found for the HVSI region between positions 16086–16370. The basal motif for haplogroup U6 has varied as new data have been added. Algerian sequences [9] suggested that the ancestral sequence harbored mutations 16172 16189. Additional data [4] considered 16172 16219 as the most probable ancestral motif. However, the complete sequence of the individual with this motif relocates it in U6a, presenting a back mutation in HVSI position 16278. Our data points to 16172 as the only substitution present in the basal motif. Unfortunately, the high recurrence of this mutation makes it insufficient to diagnose this haplogroup. The highest frequencies for haplogroup U6 as a whole are found in Northwest Africa (Table 2), with a maximum of 29% in the Algerian Berbers [9]. Subgroup U6a and its derivative U6a1 present the widest geographic distribution, from the Canary Islands in the West, to Syria and Ethiopia in the East, and from the Iberian Peninsula in the North, to Kenya in the South. In contrast, U6b shows a more limited and patched distribution, restricted to western populations. In the Iberian Peninsula, U6b is more frequent in the North whilst U6a is prevalent in the South. In Africa, it has been sporadically found in Morocco and Algeria in the North, and Senegal and Nigeria in the South, pointing to a wider distribution in the past, or to gene flow from a geographic focus which has still not been sampled. Curiously, two Arab Bedouins [22] with the same haplotype (16111 16172 16219 16311 16362), are the only Eastern representatives classified as U6b. It would be very interesting to test the 9438 HaeIII restriction enzyme to confirm this classification. Furthermore, subgroup U6b1 characterized by mutation 16163, is restricted to the Canarian Archipelago and the Iberian Peninsula. The geographic distribution of the new subgroup U6c, characterized by the basic motif 16169 16172 16189, is even more localized. It has only been found in the Canary Islands and Morocco. It could also be present in Algeria, if the two individuals with haplotype 16172 16189 16234 16311 [9], classified as U* by RFLP analysis [5], belong to this subgroup. Like for U6b, an autochthonous U6c subcluster (characterized by mutation 16129) was also detected in the Canarian Archipelago.
Figure 2.
Reduced median network relating HVSI sequences of subhaplogroup U6. The central motif (star) differs from rCRS [55,56] at position 16172. Population codes as in Table 2. Numbers along links refer to nucleotide positions minus 16000; suffix indicates a transversion. Individuals marked with * and # additionally present mutations 051 and 048, respectively. Dotted boxes correspond to subgroup U6b1; transversals to U6b; white to U6a; grids to U6a1, and checkered to U6c. Size of boxes is proportional to the number of individuals included. Dotted links represent less probable paths.
Table 2.
List of populations used in this study. Population codes are given, as well as sample size, number of U6 haplotypes detected, percentage of U6 in the sample, and the relative frequency (%) of the three U6 sub-groups.
| Group | Population | Name | sample | HapU6 | U6(%) | U6a(%) | U6b(%) | U6c(%) | References |
| EAST (E) | |||||||||
| Arab Bedouin | ABD | 29 | 1 | 2 (7) | - | 2 (100) | - | 22 | |
| Egyptians | EGY | 67 | 1 | 1 (1) | 1 (100) | - | - | 46 | |
| Ethiopian | ETI | 74 | 3 | 3 (4) | 3 (100) | - | - | 14 | |
| Iraqi | IRQ | 116 | 1 | 1 (1) | 1 (100) | - | - | 47 | |
| Ashkenazic Jew | JAS | 78 | 2 | 3 (4) | 3 (100) | - | - | 14 | |
| Bukharan Jew | JBU | 33 | 1 | 1 (3) | 1 (100) | - | - | 14 | |
| Ethiopian Jew | JET | 48 | 1 | 1 (2) | 1 (100) | - | - | 14 | |
| Kikuyu | KIK | 24 | 1 | 2 (8) | 2 (100) | - | - | 48 | |
| Palestinian | PAL | 109 | 1 | 1 (1) | 1 (100) | - | - | 47 | |
| Syrians | SYR | 69 | 2 | 3 (4) | 3 (100) | - | - | 47 | |
| Total E | 647 | 9 | 18 (3) | 16 (89) | 2 (11) | - | |||
| WEST (W) | |||||||||
| Europe | |||||||||
| Med. Center | MdC | 302 | 1 | 1 (0) | - | 1 (100) | - | 47 | |
| Sicilian | SIC | 106 | 1 | 1 (1) | 1 (100) | - | - | 13 | |
| Iberian Peninsula (IP) | |||||||||
| Andalusian | AND | 397 | 7 | 10 (3) | 8 (80) | 2 (20) | - | 9, 11, a | |
| Cat and Aragón | CAR | 118 | 2 | 2 (2) | 1 (50) | 1 (50) | - | 49 | |
| Castilla | CAV | 38 | 1 | 1 (3) | 1 (100) | - | - | 11 | |
| León | LEO | 61 | 1 | 1 (2) | 1 (100) | - | - | 11 | |
| Galician | GAL | 135 | 3 | 3 (2) | 1 (33) | 2 (67) | - | 12, 50 | |
| Maragatos | MRG | 49 | 2 | 4 (8) | 1 (25) | 3 (75) | - | 11 | |
| Portugal Center | PC | 162 | 2 | 3 (2) | 3 (100) | - | - | 10, 12 | |
| Portuguese | PCR | 54 | 2 | 3 (6) | 2 (67) | 1 (33) | - | a | |
| North Portugal | PN | 184 | 6 | 8 (4) | 5 (63) | 3 (38) | - | 10, 12 | |
| Santander | SAN | 88 | 1 | 1 (1) | 1 (100) | - | - | 38 | |
| Total IP | 1286 | 18 | 36 (3) | 24 (67) | 12 (33) | - | |||
| Islands | |||||||||
| Canarian | CAN | 300 | 11 | 53(18) | 4 (8) | 39 (74) | 10 (19) | 19 | |
| Cabo Verde | CV | 315 | 5 | 6 (2) | 6 (100) | - | - | 51, a | |
| Madeira | MAD | 96 | 1 | 1 (1) | 1 (100) | - | - | a | |
| Sao Tomé | SAT | 50 | 1 | 1 (2) | 1 (100) | - | - | 52 | |
| Total Islands | 761 | 15 | 61 (8) | 12 (20) | 39 (64) | 10 (16) | |||
| West Africa (WA) | |||||||||
| Algerian Berbers | BAL | 86 | 5 | 25 (29) | 24 (96) | 1 (4) | - | 9, a | |
| Ifrane Berbers | BIF | 46 | 7 | 9 (20) | 8 (89) | 1 (11) | - | 14 | |
| Moroccan Ber. | BMO | 213 | 11 | 15 (7) | 11 (73) | 1 (7) | 3 (20) | 4, 14, 53, a | |
| Fulbe | FUL | 60 | 2 | 2 (3) | 1 (50) | 1 (50) | - | 48 | |
| Mauritanian | MAU | 64 | 4 | 11 (17) | 11 (100) | - | - | 4, a | |
| Moroccan | MOR | 56 | 6 | 11 (20) | 8 (73) | - | 3 (27) | 4, a | |
| Saharan | SAH | 37 | 2 | 3 (8) | 3 (100) | - | - | 4, a | |
| Mandenka | SEM | 119 | 1 | 1 (1) | 1 (100) | - | - | 54 | |
| Wolof | SEW | 48 | 2 | 2 (4) | 1 (50) | 1 (50) | - | 4 | |
| Songhai | SON | 10 | 1 | 2 (20) | 2 (100) | - | - | 48 | |
| Tuareg | TUA | 23 | 1 | 1 (4) | 1 (100) | - | - | 48 | |
| Total WA | 762 | 27 | 82 (11) | 71 (87) | 5 (6) | 6 (7) | |||
| AMERICA | |||||||||
| Brazilian | BRA | 247 | 2 | 4 (2) | 4 (100) | - | - | 16 | |
| Cuban patients | CUB | 4 | 1 | 3 (75) | - | 3 (100) | - | 15 | |
| TOTAL | 56 | 206 | 128(62) | 62 (30) | 16 (8) | ||||
athis study
Relationships between areas
Linearized FST values distinguished three significantly differentiated geographical areas: Continental Africa, the Iberian Peninsula and the Canary Islands (Table 3). Nucleotide diversities within areas (Table 3) ranged from 3.253 in the Iberian Peninsula to 2.059 in East Africa. At first sight, it is striking that diversities are larger in the Canary Islands and Iberia than in Africa. We think that demographic processes are responsible of this situation. In Africa, the geographic and social isolation of the different Berber groups [23], could have promoted a loss of diversity by genetic drift. On the contrary, the presence in the Canary Islands and Iberia of representatives of all, or nearly all, U6 subclades, some of them not detected nowadays in the Continent, strongly point to the existence of several migratory waves from Africa, possibly at different times, which have increased their variability. This explanation is reinforced when the number of segregating sites (S) are taken into account. This value is larger in West Africa (5.10 ± 1.5) than in the Canaries (2.60 ± 1.0) and the Iberian Peninsula (3.90 ± 1.4), but East Africa presents a lower value (3.2 ± 1.4). The fact that U6b and U6c have a restricted western distribution undoubtedly contributes to this Continental difference. However, the younger U6a1 branch contradicts this general trend. For this subclade, East and West Africa are statistically differentiated (P = 0.016), and the former presents a higher nucleotide diversity (1.55 ± 1.11) than the latter (0.98 ± 0.75). Geographic distributions and diversity values of U6 are congruent with a western origin and radiation for all subclades excepting U6a1 that, most probably, had an eastern origin.
Table 3.
Linearized FST values between areas (below diagonal) and π diversities within areas (on diagonal).
| Canarian | Iberian | East Africa | West Africa | |
| Canarian | 2.867 | |||
| Iberian | 0.386*** | 3.253 | ||
| East Africa | 0.714*** | 0.060* | 2.059 | |
| West Africa | 0.738*** | 0.077*** | 0.048 | 2.284 |
Radiation ages
Radiation ages for U6 and its subclades have been estimated on the basis of complete coding and HVSI sequences (Table 4). In general, ages obtained from HVSI are larger than those deduced from the coding region. Both approaches present inconveniences for the time estimates. It has been demonstrated that the coding region has evolved at a roughly constant rate [24]. However, as relatively few clades are fully sequenced, stochastic and/or intentional sampling may bias the representation of the chosen lineages. On the other hand, HVSI estimations are based on a large number of individuals minimizing sampling errors. However, we deal with a short sequence that has not evolved at a constant rate across all human lineages [24]. Furthermore, from the phylogeny of complete U6 sequences (Fig. 1), it has been deduced, once more, that empirical time estimations are not independent of the demographic history of the population sampled. Therefore, we have taken coalescence ages only as rough time frames into which lineage expansions could have occurred. Adopting a conservative position we have used ages based on the coding region whenever possible.
Table 4.
Estimated ages (years) for different subgroups of U6 haplogroup, based on coding and HVSI regions.
| Coding Region | HVSI | ||||
| Subgroup | N | Age[24] | Age[45] | N | Age |
| U6b1 | - | 45 | 5,830 ± 4,551 | ||
| U6b | 4 | 8,568 ± 2,856 | 11,560 ± 3,853 | 62 | 24,411 ± 15,200 |
| U6a | 8 | 24,277 ± 4,831 | 32,754 ± 6,518 | 128 | 27,590 ± 13,576 |
| U6a1 | 3 | 15,232 ± 5,677 | 20,552 ± 7,659 | 82 | 13,289 ± 5,470 |
| U6c | 2 | 5,712 ± 3,298 | 7,707 ± 4,450 | 16 | 17,658 ± 12,862 |
| U6 | 14 | 25,297 ± 4,543 | 34,130 ± 6,130 | 206 | 66,222 ± 25,269 |
Discussion
African U6 origin and expansions
Discarding the Canary Islands, because the most ancient human settlement seems to be no earlier than 2,500 ya [25], and the Iberian Peninsula, because there are no consistent traces of U6 lineages in Europe, Northwest Africa is left as the most probable place from where the African U6 subclades radiated. Another point is to decide whether the proto-U6 ancestor was also of African origin. Although it cannot be completely excluded, this hypothesis seems highly improbable even invoking strong bottlenecks in African populations. It is clear that the whole haplogroup U is an offshoot of macrohaplogroup N. This lineage, together with macrohaplogroup M, were the only ones that, belonging to the star radiation of L3 in Africa, left this continent to colonize Eurasia. Five mutations separate N from the root of the African L3 [8], and there are only late evolved N lineages in Africa, whereas representatives of the full N radiation are present in Eurasia. Thus, this continent would be the logical homeland of the proto-U6 that came back to Africa and spread in its northwest area around 30,000 ya (Table 4). Its most probable route had to be through East Africa. So, the loss of variability in this area is puzzling, although posterior demic expansions affecting East Africa might be the cause. This date roughly corresponds to the Paleolithic occupation of the Maghrib by the Iberomaurusian culture and to the age of the evolved Homo sapiens sapiens skeletons found in this area. Only one of the three U6 subclades, U6a, experienced a great geographic radiation spreading west to the Atlantic shores and east, crossing Africa, to the Near East. A posterior offshoot of this clade, U6a1, has a similar distribution. The upper bound for these expansions are around 28,000 and 17,000 ya, respectively (Table 4). Genetic diversities are congruent with a west to east expansion for U6a and a more probable east to west expansion for U6a1. Furthermore, the absence of U6b and U6c lineages in the East suggests that the population from which the U6a colonizers originated also lacked these lineages or presented them in very low frequencies. The fact that 5 of the 8 U6a haplotypes detected in the Near East are unique of this area (Fig. 2), points to prehistoric demic movements as the most probable cause of the U6a Africa to Asia migration, although historic events cannot be completely ruled out. In frame with the estimated age of U6a are archaeological data supporting early migrations from Africa into the Near East [26]. The expansion of Caucasians in Africa has been correlated with the spread and diversification of Afroasiatic languages. There are different hypothesis to explain the Afroasiatic origin. For some, it would be the result of a Neolithic demic diffusion from the Near East to Africa [27,28]. For others, the Afroasiatic originated in Africa and had a posterior demic spread to West Asia [29,30]. A third possibility is that Afroasiatic languages spread mostly through cultural contacts either from Africa or from Asia [31]. Only demic diffusions could be correlated with U6 expansions detected here. Since an upper bound of 15,000 ya has been estimated for the proto-Afroasiatic origin, it seems that the coalescence age for U6a predates by far the origin of the Afroasiatic phylum. However, the recent spread of U6a1 is more in frame with the emergence of a proto-Afroasiatic language. This U6a1 expansion would favor an East African origin for the Afroasiatic and a posterior expansion to West Africa and West Asia. However, a Near Eastern origin, most probably predating the Neolithic expansion, cannot be ruled out.
Iberian U6 origin and expansions
In Europe, U6 lineages have been consistently sampled only in the Iberian Peninsula. It has been mentioned that U6 nucleotide diversity is higher in Iberia than in Africa [12]. This has been confirmed here (Table 3). However, S is greater in West Africa. Considering the isolation of the different Berber groups we think that, in this case, the latter is a better diversity measure. The absence of U6 representatives in the rest of Europe, is also an argument against the hypothesis that these lineages could have migrated to North Africa from Europe. Naturally, this does not exclude that other mitochondrial lineages could have followed this route. Most probably, the presence of these African lineages in Iberia is the result of northward expansions from Africa. The time of this expansion has been predominantly attributed to either the Arab/Berber occupation that lasted seven centuries [10] or to prehistoric immigrations of North Africans to Iberia [12]. Both processes could have contributed to model the U6 landscape in Iberia. First, haplotype matches show that 10 of the 19 U6 lineages detected in Iberia are not present in Africa (Fig. 2), which points against only one recent immigration. Second, the geographic distribution of the U6 lineages in Iberia is puzzling. Whereas the U6b lineages, nowadays very scarce in Africa, are mainly detected in the Northwest, the U6 lineages found in highest frequencies in Africa are predominant in the south, where the Islamic rule lasted longer. At the light of these results we propose that U6b in Iberia is the signal of a prehistoric North African immigration that could have also brought some U6a lineages. Its actual northern range could be the result of a forced retreat due to the arrival of new southern incomers to Iberia. However, the U6a distribution is better explained as the result of more recent gene flow from North Africa. The age of U6b (approx. 10,000 ya) might be considered as an upper bound for the prehistoric wave. Curiously, around this time the Iberomaurusians began to be displaced by the incoming Capsian culture in the Maghrib. On archaeological grounds, it has been proposed that Iberomaurusians slowly retreated towards the Atlantic coast from where they sailed to the Canary Islands and southwards to the Malinese Sahara [2]. Coincidentally, these are the same places where the U6b lineages have been spotted (Fig. 2).
Canary Islands U6 origin and expansions
At a genetic level, the Berber origin of the Guanches, the aboriginal population of the Canary Islands, and their survival after the Spanish occupation, has been inferred from the high frequency of U6 lineages in its modern population (Table 2), similar to that of North Africa [19,32]. This fact has been recently confirmed in a mtDNA sequence study on aboriginal remains [33]. It was found that in the Guanche maternal gene pool, U6b1 and U6a were present at frequencies of 8.22% and 1.37%, respectively. U6c was probably also present in the aboriginal pool as a haplotype (16129 16169 16172 16189), now known to belong to subhaplogroup U6c, was proposed as a probable Canarian founder type [19]. As in Northwest Iberia, U6b was the dominant U6 subclade carried by the North African settlers of the islands. All three subclades are present in the modern Canarian population at frequencies of 1.3%, 13.0% and 3.3% for U6a, U6b and U6c, respectively, which is indicative of a broad aboriginal component in the present maternal pool. Perhaps, the comparatively higher frequency of U6a lineages might be attributed to an additional Berber input as result of the slave trade after the Spanish conquest [34,35]. What remains enigmatic of the indubitable North African prehistoric colonization of the Archipelago is that it was carried out by people whose U6 lineages mainly belonged to the U6b subclade that has only been spotted in very low frequencies in the modern African populations of Morocco, Algeria, Senegal and Nigeria (Table 2). Moreover, the U6b and U6c insular haplotypes belong to the autochthonous U6b1 and U6c1 branches differing by substitutions 16163 and 16129, respectively, from all their African counterparts. As the most probable arrival of the first prehistoric Canarian settlers was around 2,500 ya, it is highly improbable that these mutations occurred on the islands. Therefore, we expected to find these Canarian lineages in some place of Africa. However, after extensive sampling they have still not been detected. It is possible that they are present somewhere in low frequencies but, in any case, this phylogeographic distribution suggests that Northwest Africa suffered important demic displacements in the past.
Besides U6, other genetic markers such as 110(-) haplotype of the CD4/Alu system [36], and the M81 Y-chromosome binary marker [6,7], point to an ancient and autochthonous human presence in Northwest Africa. An eastward decline in M81 frequencies has been detected, regrettably the lack of extensive intra-M81 microsatellite diversity studies in Africa precludes phylogeographic comparisons as those done with mtDNA. There are other coincidences between mtDNA data and other systems. For instance, using classical genetic markers, it was found that the Iberian Peninsula showed smaller genetic distances with East Africa than with West Africa [37]. The same pattern was observed for Y-chromosome studies [7], both in line with our results (Table 3). More studies with other genetic markers are necessary to corroborate, complement or even contradict the proposed U6 landscape.
In summary, the phylogeography, nucleotide diversity, and coalescence ages of U6 lineages show that this clade came back to Africa in Paleolithic times. Its most probable origin was the Near East and not Europe, and since then, its presence in North Africa has been permanent. The focus of the first African expansion, detected by the spread of U6a, was Northwest Africa reaching the Near East also in the Paleolithic. The posterior U6a1 radiation most probably occurred in Northeast Africa again extending to the Near East. This movement is correlated in time with the attributed origin and expansion of Afroasiatic languages. This U6a1 wave also arrived to the Maghrib, the Northwest African margin, where the more localized U6b and U6c lineages were spreading. This movement is in time frame with the Capsian culture. Based on archaeological and anthropological grounds, it has been speculated that these incomers slowly pushed away the aboriginal residents [2]. It could be in that time when U6b reached the south of the Iberian Peninsula from where it was displaced to the north where it persists today. The U6b and U6c diaspora also reached the Atlantic fringe from where they sailed to the Canary Islands. Two autochthonous U6 lineages (U6b1 and U6c1), present today in the islands, attest the survival of those aboriginal North Africans until nowadays. The fact that these Canarian lineages have not been detected in Africa and that, in contrast to the ubiquitous U6a and U6a1, the U6b and U6c lineages are scarcely spotted in present African populations, may be clues of past important demographic movements in this western area.
Methods
Complete mtDNA lineages
We have fully sequenced eleven mitochondrial lineages belonging to different subclades of the North African subhaplogroup U6. DNA extraction, amplification and manual sequencing methods have already been described [8].
HVSI and RFLPs
A sample of 332 Andalusians, 1 Algerian Berber, 89 Moroccan Berbers, 23 individuals from Cape Verde, 96 from Madeira, 34 Mauritanians, 24 Moroccans and 12 Saharans were sequenced for the HVSI region from 16000 to 16390, and RFLP characterized as detailed [11,12]. In order to distinguish putative U6 members, all these subjects and the U individuals from a sample of 1059 previously published [4,11,12,19,38], were amplified with primers L3073/H3670 [8], and tested for the presence of the 3348 MboI site [15], that characterizes all U6 members.
Phylogenetic analyses
Phylogenetic relationships among complete mtDNA sequences were established using the reduced median network algorithm [39]. In addition to our eleven sequences, four lineages were added: U6 and U5b [8] (Accession numbers: AF382008 and AF381980, respectively) and for the coding region, H84 and H229 [18].
U6 phylogeographic analyses
In addition to our 611 samples, 41 populations where U6 haplotypes have been detected were included in our phylogeographic analysis (Table 2). Relationships among the different U6 haplotypes were inferred using the reduced median network algorithm [39]. To resolve reticulations, the highly recurrent mutations 16129, 16189, 16311 and 16362 were less weighted.
Differences in accumulated mutations among U6 branches
The non-parametric test, resampling probability estimates for the difference between the means of two independent samples (http://faculty.vassar.edu/lowry/VassarStats.html), was used to calculate the significance level of accumulated mutations between the different U6 subclades. The likelihood-ratio test, as implemented in TREE-PUZZLE [21], was used to check between a uniform clock or variable site model in the U6 tree.
U6 diversity and differentiation within and between areas
Arlequin package [40] was used to evaluate the U6 diversity within areas using nucleotide diversity (π) and segregating sites (S). Affinities between areas were obtained by means of linearized FST [41].
Time estimates
For HVSI, the age of clusters or expansions was calculated as the mean divergence ρ from inferred ancestral sequence types [42] and converted into time by assuming that one transition within np 16090–16365 corresponds to 20,180 years [43]. The standard deviation of the ρ estimator was calculated as previously described [44].
For the complete sequences only substitutions in the coding region (15,447 nucleotides), excluding indels, were taken into account. The mean number of substitutions per site to the most recent common ancestor of each clade (ρ) was estimated, and converted into time using two substitution rates: 1.7 × 10-8 [24] and 1.26 × 10-8 [45].
Supplementary material
The eleven complete mitochondrial DNA sequences are registered under GenBank accession numbers: AY275527 to AY275537.
Acknowledgments
Acknowledgements
This research was supported by grants of Ministry of Science and Technology (BMC2001-3511) and Gobierno de Canarias (COF2002/015) to V.M. Cabrera.
Contributor Information
Nicole Maca-Meyer, Email: nmacame@ull.es.
Ana M González, Email: amglez@ull.es.
José Pestano, Email: jpestano@dbbf.ulpgc.es.
Carlos Flores, Email: cflores@ull.es.
José M Larruga, Email: jlarruga@ull.es.
Vicente M Cabrera, Email: vcabrera@ull.es.
References
- Alimen H. Evolution du climat et des civilisations depuis 40000 ans du nord au sud du Sahara occidental. (Premières conceptions confrontées aux données récentes) Bull L'Assoc Franç L'Êtude Quarternaire. 1987;4:215–227. [Google Scholar]
- Ferembach D. On the origin of the Iberomaurusians (Upper Palaeolithic: North Africa). A new hypothesis. J Hum Evol. 1985;14:393–397. [Google Scholar]
- Newman JL. The peopling of Africa: a geographic interpretation. Yale University Press, New Haven; 1995. [Google Scholar]
- Rando JC, Pinto F, González AM, Hernández M, Larruga JM, Cabrera VM, Bandelt HJ. Mitochondrial DNA analysis of Northwest African populations reveals genetic exchanges with European, Near-Eastern, and sub-Saharan populations. Ann Hum Genet. 1998;62:531–550. doi: 10.1046/j.1469-1809.1998.6260531.x. [DOI] [PubMed] [Google Scholar]
- Macaulay V, Richards M, Hickey E, Vega E, Cruciani F, Guida V, Scozzari R, Bonné-Tamir B, Sykes B, Torroni A. The emerging tree of west Eurasian mtDNAs: A synthesis of control-region sequences and RFLPs. Am J Hum Genet. 1999;64:232–249. doi: 10.1086/302204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch E, Calafell F, Comas D, Oefner PJ, Underhill PA, Bertranpetit J. High-resolution analysis of human Y-Chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian Peninsula. Am J Hum Genet. 2001;68:1019–1029. doi: 10.1086/319521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores C, Maca-Meyer N, Pérez JA, Hernández M, Cabrera VM. Y-Chromosome differentiation in northwest Africa. Hum Biol. 2001;73:513–524. doi: 10.1353/hub.2001.0046. [DOI] [PubMed] [Google Scholar]
- Maca-Meyer N, González AM, Larruga JM, Flores C, Cabrera VM. Major genomic mitochondrial lineages delineate early human expansions. BMC Genetics. 2001;2:13–20. doi: 10.1186/1471-2156-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côrte-Real HBSM, Macaulay VA, Richards MB, Hariti G, Issad MS, Cambon-Thomsen A, Papiha S, Bertranpetit J, Sykes B. Genetic diversity in the Iberian Peninsula determined from mitochondrial sequence analysis. Ann Hum Genet. 1996;60:331–350. doi: 10.1111/j.1469-1809.1996.tb01196.x. [DOI] [PubMed] [Google Scholar]
- Pereira L, Prata MJ, Amorim A. Diversity of mtDNA lineages in Portugal: Not a genetic edge of European variation. Ann Hum Genet. 2000;64:491–506. doi: 10.1046/j.1469-1809.2000.6460491.x. [DOI] [PubMed] [Google Scholar]
- Larruga JM, Díez F, Pinto FM, Flores C, González AM. Mitochondrial DNA characterisation of European isolates: The maragatos from Spain. Eur J Hum Genet. 2001;9:708–716. doi: 10.1038/sj.ejhg.5200693. [DOI] [PubMed] [Google Scholar]
- González AM, Brehm A, Pérez JA, Maca-Meyer N, Flores C, Cabrera VM. Mitochondrial DNA affinities at the Atlantic fringe of Europe. Am J Phys Anthropol. 2003;120:391–404. doi: 10.1002/ajpa.10168. [DOI] [PubMed] [Google Scholar]
- Cali F, Le Roux MG, D'Anna R, Flugy A, De Leo G, Chiavetta V, Ayala GF, Romano V. MtDNA control region and RFLP data for Sicily and France. Int J Leg Med. 2001;114:229–231. doi: 10.1007/s004140000169. [DOI] [PubMed] [Google Scholar]
- Thomas MG, Weale ME, Jones AL, Richards M, Smith A, Redhead N, Torroni A, Scozzari R, Gratrix F, Tarekegn A, Wilson JF, Capelli C, Bradman N, Goldstein D. Founding Mothers of Jewish Communities: Geographically separated Jewish groups were independently founded by very few female ancestors. Am J Hum Genet. 2002;70:1411–1420. doi: 10.1086/340609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Cruciani F, Rengo C, Sellitto D, López-Bigas N, Rabionet R, Govea N, López de Munain A, Sarduy M, Romero L, Villamar M, del Castillo I, Moreno F, Estivill X, Scozzari R. The A1555G mutation in the 12S rRNA gene of human mtDNA: recurrent origins and founder events in families affected by sensorineural deafness. Am J Hum Genet. 1999;65:1349–1358. doi: 10.1086/302642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves-Silva J, da Silva Santos M, Guimarães PEM, Ferreira ACS, Bandelt HJ, Pena SDJ, Ferreira Prado V. The ancestry of Brazilian lineages. Am J Hum Genet. 2000;67:444–461. doi: 10.1086/303004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnilä S, Lehtonen S, Majamaa K. Phylogenetic network for European mtDNA. Am J Hum Genet. 2001;68:1475–1484. doi: 10.1086/320591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrnstadt C, Elson JE, Fahy E, Preston G, Turnbull DM, Anderson C, Ghosh SS, Olefsky JM, Beal MF, Davis RE, Howell N. Reduced-Median-Network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian and European haplogroups. Am J Hum Genet. 2002;70:1151–1171. doi: 10.1086/339933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando JC, Cabrera VM, Larruga JM, Hernández M, González AM, Pinto F, Bandelt HJ. Phylogeographic patterns of mtDNA reflecting the colonization of the Canary Islands. Ann Hum Genet. 1999;63:413–428. doi: 10.1046/j.1469-1809.1999.6350413.x. [DOI] [PubMed] [Google Scholar]
- Torroni A, Rengo C, Guida V, Cruciani F, Sellitto D, Coppa A, Calderon FL, Simionati B, Valle G, Richards M, Macaulay V, Scozzari R. Do the four clades of the mtDNA haplogroup L2 evolve at different rates? Am J Hum Genet. 2001;69:1348–1356. doi: 10.1086/324511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haessler A. TREE-PUZZLE v51. 1999–2002. [DOI] [PubMed]
- Di Rienzo A, Wilson AC. Branching pattern in the evolutionary tree for human mitochondrial DNA. Proc Natl Acad Sci USA. 1991;88:1597–1601. doi: 10.1073/pnas.88.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett M, Fentress E. The Berbers. Oxford: Blackwell; 1996. [Google Scholar]
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- Onrubia-Pintado J. Les cultures préhistoriques des Îles Canaries: État de la question. L'Anthropologie. 1987;91:653–78. [Google Scholar]
- Bar-Yosef O. Pleistocene connections between Africa and southwest Asia. African Archaeological Review. 1987;5:29–38. [Google Scholar]
- Cavalli-Sforza LL. The Basque population and ancient migrations in Europe. Munibe. 1988;6:129–137. [Google Scholar]
- Renfrew C. Before Babel: Speculations on the origins of linguistic diversity. Cambridge Archaeol J. 1991;1:3–23. [Google Scholar]
- Greenberg J. The languages of Africa. Mouton, The Hague; 1966. [Google Scholar]
- Ehret C. An African Classical Age:Eastern and Southern Africa in World History,1000 BC to AD 400. Oxford: J. Currey; 1998. [Google Scholar]
- Barbujani G, Pilastro A, De Dominico S, Renfrew C. Genetic variation in North Africa and Eurasia: neolithic demic diffusion vs. Paleolithic colonisation. Am J Phys Anthropol. 1994;95:137–154. doi: 10.1002/ajpa.1330950203. [DOI] [PubMed] [Google Scholar]
- Pinto F, González AM, Hernández M, Larruga JM, Cabrera VM. Genetic relationship between the Canary Islanders and their African and Spanish ancestors inferred from mitochondrial DNA sequences. Ann Hum Genet. 1996;60:321–330. doi: 10.1111/j.1469-1809.1996.tb01195.x. [DOI] [PubMed] [Google Scholar]
- Maca-Meyer N, Arnay M, Rando JC, Flores C, González AM, Cabrera VM, Larruga JM. Ancient mtDNA analysis and the origin of the Guanches. Eur J Hum Genet. 2003. [DOI] [PubMed]
- Marrero M. La esclavitud en Tenerife a raíz de la Conquista. Instituto de Estudios Canarios, La Laguna, Tenerife; 1966. [Google Scholar]
- Suárez JJ, Rodríguez F, Quintero CL. In: Historia Popular de Canarias. Centro de la Cultura Popular Canaria, editor. Conquista y Colonización; 1988. Volume 2. [Google Scholar]
- Flores C, Maca-Meyer N, González AM, Cabrera VM. Northwest African distribution of the CD4/Alu microsatellite haplotypes. Ann Hum Genet. 2000;64:321–327. doi: 10.1046/j.1469-1809.2000.6440321.x. [DOI] [PubMed] [Google Scholar]
- Bosch E, Calafell F, Pérez-Lezaun A, Comas D, Mateu E, Bertranpetit J. Population history of North Africa: Evidence from classical genetic markers. Hum Biol. 1997;69:295–311. [PubMed] [Google Scholar]
- Maca-Meyer N, Sánchez-Velasco P, Flores C, Larruga JM, González AM, Oterino A, Leyva-Cobián F. Y chromosome and mitochondrial DNA characterization of Pasiegos, a human isolate from Cantabria (Spain) Ann Hum Genet. 2003;67:329–339. doi: 10.1046/j.1469-1809.2003.00045.x. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Schneider S, Roessli D, Excoffier L. Arlequin ver 2000: A software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland; 2000. [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morral N, Bertranpetit J, Estivill X, Nunes V, Casals T, Giménez J, Reis A, Varon-Mateeva R, Macek M, Kalaydjieva L, Angelicheva D, Dancheva R, Romeo G, Russo MP, Garnerone S, Restagno G, Ferrari M, Magnani C, Claustres M, Desgeoges M, Schwartz M, Dallapiccola B, Novelli G, Ferec C, de Arce M, Nemeti M, Kere J, Anvret M, Dahl N, Kadasi L. The origin of the major cystic fibrosis mutation (ΔF508) in European populations. Nature Genetics. 1994;7:169–175. doi: 10.1038/ng0694-169. [DOI] [PubMed] [Google Scholar]
- Forster P, Harding R, Torroni A, Bandelt HJ. Origin and evolution of Native American mtDNA variation: a reappraisal. Am J Hum Genet. 1996;59:935–945. [PMC free article] [PubMed] [Google Scholar]
- Saillard J, Forster P, Lynnerup N, Bandelt H-J, Norby S. mtDNA variation among Greenland eskimos: the edge of the Beringian expansion. Am J Hum Genet. 2000;67:718–726. doi: 10.1086/303038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, Sukernik RI, Olckers A, Wallace DC. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings M, Salem AH, Bauer K, Geisert H, Malek AK, Chaix L, Simon C, Welsby D, Di Rienzo A, Utermann G, Sajantila A, Pääbo S, Stoneking M. mtDNA analysis of Nile River Valley populations: A genetic corridor or a barrier to migration? Am J Hum Genet. 1999;64:1166–1176. doi: 10.1086/302314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, Sellitto D, Cruciani F, Kivisild T, Villems R, Thomas M, Rychkov S, Rychkov O, Rychkov Y, Golge M, Dimitrov D, Hill E, Bradley D, Romano V, Cali F, Vona G, Demaine A, Papiha S, Triantaphyllidis C, Stefanescu G. Tracing European founder lineages in the Near Eastern mtDNA pool. Am J Hum Genet. 2000;67:1251–1276. [PMC free article] [PubMed] [Google Scholar]
- Watson E, Forster P, Richards M, Bandelt HJ. Mitochondrial footprints of human expansions in Africa. Am J Hum Genet. 1997;61:691–704. doi: 10.1086/515503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespillo M, Luque JA, Paredes M, Fernández R, Ramírez E, Valverde JL. Mitochondrial DNA sequences for 118 individuals from northeastern Spain. Int J Legal Med. 2000;114:130–132. doi: 10.1007/s004140000158. [DOI] [PubMed] [Google Scholar]
- Salas A, Comas D, Lareu M, Bertranpetit J, Carracedo A. MtDNA analysis of the Galician population: a genetic edge of European variation. Eur J Hum Genet. 1998;6:365–375. doi: 10.1038/sj.ejhg.5200202. [DOI] [PubMed] [Google Scholar]
- Brehm A, Pereira L, Bandelt HJ, Prata MJ, Amorin A. Mitochondrial portrait of the Cabo Verde archipelago: the Senegambian outpost of Atlantic slave trade. Ann Hum Genet. 2002;66:49–60. doi: 10.1017/S0003480001001002. [DOI] [PubMed] [Google Scholar]
- Mateu E, Comas D, Calafell F, Pérez-Lezaun A, Abade A, Bertranpetit J. A tale of two islands: population history and mitochondrial DNA sequence variation of Bioko and São Tomé, Gulf of Guinea. Ann Hum Genet. 1997;61:507–518. doi: 10.1017/S0003480097006544. [DOI] [PubMed] [Google Scholar]
- Brakez Z, Bosch E, Izaabel H, Akhayat O, Comas D, Bertranpetit J, Calafell F. Human mitochondrial DNA sequence variation in the Moroccan population of the Souss area. Ann Hum Biol. 2001;28:295–307. doi: 10.1080/030144601300119106. [DOI] [PubMed] [Google Scholar]
- Graven L, Passarino G, Semino O, Boursit P, Santachiara-Benerecetti AS, Langaney A, Excoffier L. Evolutionary correlation between control region sequence and restriction polymorphisms in the mitochondrial genome of a large Senegalese Mandenka sample. Mol Biol Evol. 1995;12:334–345. doi: 10.1093/oxfordjournals.molbev.a040206. [DOI] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, Bruijn MHLDE, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young IG. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]