Abstract
Genome-wide association studies (GWAS) have successfully identified common single nucleotide polymorphisms (SNPs) associated with a wide variety of complex diseases, but do not address gene function or establish causality of disease-associated SNPs. We recently used GWAS to identify SNPs in a genomic region on chromosome 4 that associate with serum urate levels and gout, a consequence of elevated urate levels. Here we show using functional assays that human ATP-binding cassette, subfamily G, 2 (ABCG2), encoded by the ABCG2 gene contained in this region, is a hitherto unknown urate efflux transporter. We further show that native ABCG2 is located in the brush border membrane of kidney proximal tubule cells, where it mediates renal urate secretion. Introduction of the mutation Q141K encoded by the common SNP rs2231142 by site-directed mutagenesis resulted in 53% reduced urate transport rates compared to wild-type ABCG2 (P < 0.001). Data from a population-based study of 14,783 individuals support rs2231142 as the causal variant in the region and show highly significant associations with urate levels [whites: P = 10−30, minor allele frequency (MAF) 0.11; blacks P = 10−4, MAF 0.03] and gout (adjusted odds ratio 1.68 per risk allele, both races). Our data indicate that at least 10% of all gout cases in whites are attributable to this causal variant. With approximately 3 million US individuals suffering from often insufficiently treated gout, ABCG2 represents an attractive drug target. Our study completes the chain of evidence from association to causation and supports the common disease-common variant hypothesis in the etiology of gout.
Keywords: causal variant, genome-wide association study, uric acid
Urate is the end product of purine metabolism in humans. Humans and higher primates have much higher serum urate levels than other species because they lack the enzyme uricase, which converts urate into its breakdown product allantoin (1). Serum urate levels are highly heritable, suggesting a strong genetic component (2). Reduced excretion of urate by the kidney is the main cause for elevated urate levels (1), which can lead to gout, a painful condition affecting approximately 3 million individuals in the U.S. (3). Renal urate transport is complex and still poorly understood (1); although multiple renal urate transporters have been characterized in model systems, their role in human disease is mostly unclear. In a genome-wide association study (GWAS) of serum urate levels, we recently identified multiple SNPs in a genomic region on chromosome 4 containing the ATP-binding cassette subfamily G member 2 (ABCG2) gene as associated with urate levels and prevalence of gout (4). ABCG2 was first identified as a multidrug resistance protein (5) and has been shown to transport a wide range of structurally and functionally diverse substrates such as chemotherapeutics (6). Yet, the physiological substrate and the roles of ABCG2 in vivo have remained elusive.
Results and Discussion
ABCG2 Is a Urate Transporter.
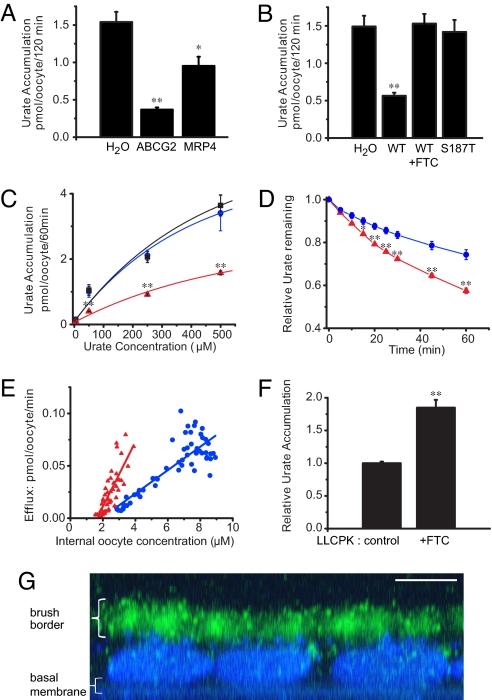
To investigate whether ABCG2 is a hitherto unknown urate transporter, we expressed human ABCG2 in Xenopus oocytes. Accumulation of radiolabeled urate in oocytes and urate efflux rates from oocytes were measured. Urate accumulation was significantly decreased by 75.5% in oocytes expressing ABCG2 compared with water-injected control oocytes (Fig. 1A). In addition, ABCG2-expressing oocytes showed lower urate concentrations than oocytes expressing the known urate efflux transporter MRP4 (7). The reduced urate accumulation in ABCG2-expressing oocytes was absent in the presence of fumitremorgin C (FTC), a specific ABCG2 inhibitor, or after introduction of a mutation in ABCG2 (187T) that is known to disrupt transport of chemotherapeutic agents (5, 6) (Fig. 1B). ABCG2-expressing oocytes showed significantly lower urate accumulation over a wide range of extracellular concentrations compared with control cells or cells expressing the loss-of-function mutation ABCG2 187T (Fig. 1C). The reduced urate accumulation was due to ABCG2-mediated export of urate out of the cells as shown in experiments monitoring the decrease of the intracellular urate concentration over time in oocytes preloaded with radiolabeled urate (Fig. 1D). Furthermore, ABCG2-mediated urate efflux was dependent on the intracellular urate concentration and significantly higher in ABCG2-expressing oocytes than in control oocytes (Fig. 1E). Together, these data show that ABCG2 is a urate efflux transporter.
Fig. 1.
ABCG2 is a urate transporter. (A) C-14 urate accumulation data from Xenopus oocytes injected with either H2O or mRNA coding for MRP4 or ABCG2. (B) Urate accumulation in oocytes injected with H2O, ABCG2 WT mRNA (incubated with or without 5 μM FTC), or the nonfunctional ABCG2 mutant S187T (N = 10 samples each and n = 20 oocytes each for all accumulation experiments). (C) Urate accumulation is dependent on the extracellular urate concentration (H2O [blue circles]; S187T [black squares]; WT [red triangles]; N = 10 and n = 20 each). (D) Urate efflux in oocytes incubated overnight in 500 μM C-14 urate as relative efflux over time (H2O [blue circles]; ABCG2 [red triangles]; N = 5 and n = 50 each). (E) Urate efflux depends on the intracellular urate concentration (H2O [blue circles]: slope = 0.01, N = 55; ABCG2 [red triangles]: slope = 0.03, N = 55; P < 0.001). (F) FTC (5 μM) inhibits urate export in native LLC-PK1 renal proximal tubule cells as shown by increased urate accumulation in FTC-treated cells compared to non-treated control cells (N = 6). (G) LLC-PK1 cells express endogenous ABCG2 at the apical brush border membrane. Merged Z sections of LLC-PK1 cells stained with BCRP1 antibody (green) and nuclear DAPI stain (blue; scale bar, 5 μm). Mean ± SEM; **, P < 0.001; *, P < 0.01.
ABCG2 in Renal Epithelial Cells.
In mammals, the proximal tubule is the major site of renal urate handling (1). To study the urate transport capacity of endogenous ABCG2 in polarized renal epithelial cells, we measured urate accumulation in native LLC-PK1 cells. Inhibition of ABCG2 by FTC resulted in a significant impairment of urate export as shown by significantly higher intracellular urate accumulation compared with control cells (Fig. 1F). To investigate whether ABCG2 mediates renal reabsorption or secretion of urate, we studied its subcellular localization. In polarized LLC-PK1 cells, ABCG2 was localized to the apical brush border membrane (Fig. 1G), which is consistent with the recent demonstration of ABCG2 in the brush border of human proximal tubule cells (8). These data therefore establish that ABCG2 is a secretory urate transporter in the proximal tubule. Consequently, mutations in ABCG2 that increase serum urate concentrations must be loss-of-function mutations.
Impaired Urate Transport of ABCG2 Q141K.
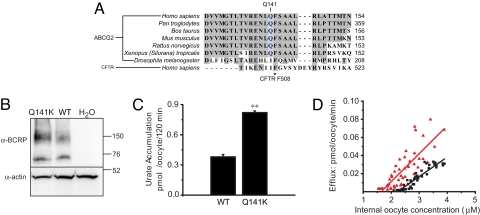
The most significant SNP in our recent GWAS of serum urate levels was the non-synonymous coding SNP rs2231142 in exon 5 of ABCG2 (4). The glutamine (Q) residue at ABCG2 position 141, encoded by the rs2231142 G allele, is highly conserved across species (Fig. 2A). Interestingly, Q141 is located in the nucleotide-binding domain of ABCG2 right next to the corresponding amino acid F508 in the nucleotide-binding domain of CFTR, a residue commonly mutated in patients with cystic fibrosis. To test whether this SNP is not only statistically associated but causally related to elevated urate levels, we introduced the mutation Q141K encoded by the rs2231142 T allele by site-directed mutagenesis. Wild-type and Q141K mutant ABCG2 showed similar expression levels at the cell surface when expressed in Xenopus oocytes (Fig. S1). Notably, Q141K-expressing oocytes showed 54% reduced urate transport rates compared with those expressing wild-type ABCG2 at similar levels (Fig. 2 B and C), and decreased urate efflux across a range of intracellular urate concentrations (Fig. 2D). Our evidence for Q141K as a causal loss-of-function variant is supported by data showing reduced transport of chemotherapeutic agents by this mutation (6).
Fig. 2.
The ABCG2 Q141K mutation results in reduced urate transport. (A) Across-species comparison of the ABCG2 protein sequence. Sequence is also compared with the cystic fibrosis transmembrane conductance regulator (CFTR). Note the close proximity of Q141 in ABCG2 to the mutational hot spot F508 in CFTR. (B) Western blot of oocytes expressing ABCG2 WT or Q141K (monomers and dimers of ABCG2 are detected). Actin was used as a loading control. (C) Accumulation rates in oocytes expressing ABCG2 WT or Q141K normalized to ABCG2 expression level. (D) Efflux rates for ABCG2 WT and Q141K. (WT [red triangles]: slope = 0.01, N = 55; Q141K [black squares]: slope = 0.02, N = 55). Mean ± SEM; **, P < 0.001.
Relevance in the General Population.
To investigate the genomic context and to quantify the effect of this common causal variant in the general population, we studied 10,902 self-reported white and 3,881 black participants of the Atherosclerosis Risk in Communities (ARIC) Study (9) who had serum urate measured and self-reported information on gout. Information on the SNP rs2231442, encoding the Q141K variant, was available as an imputed SNP from genome-wide Affymetrix 6.0 data along with 601 other SNPs in a 600-kb region containing ABCG2, and also genotyped directly using the TaqMan assay.
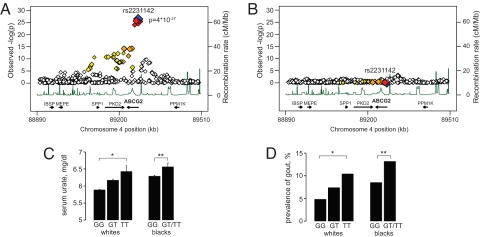
In a subset of 8,092 white participants with available GWAS data, the rs2231142 T allele showed highly significant association with higher urate levels in multivariable adjusted association analyses (P = 4 × 10−27; Fig. 3A). Other strongly associated SNPs were located in introns or downstream of ABCG2 and were in perfect or high linkage disequilibrium with rs2231142. After repeating the analyses conditional on genotype at rs2231142, no significant associations in the region remained (Fig. 3B), which supports the presence of a single common causal variant in the region.
Fig. 3.
Analysis of the ABCG2 locus and rs2231142 in population-based samples. (A) Association of urate levels and 602 SNPs in the ABCG2 region. Color legend: rs2231142 (blue), other SNPs coded by LD with rs2231142 based on HapMap CEU: red (r2 with rs2231142 0.8–1.0), orange (r2 = 0.5–0.8), yellow (r2 = 0.2–0.5), and white (r2<0.2). Gene annotations are based on Build 36.1, and arrows correspond to direction of transcription and gene size. (B) Analyses as in A, conditional on genotype at rs2231142. (C) Mean serum urate levels by genotype at rs2231142 in white and black participants. *, P trend = 2 × 10−17, **, P trend = 0.015. (D) Prevalence of gout by genotype at rs2231142. *, P trend = 4 × 10−6, **, P trend = 0.04. Gout cases/overall sample in whites: GG (327/6792), GT (118/1601), TT (10/96); blacks: GG (187/2198), GT/TT (20/152).
In both white and black study participants, serum urate levels showed a graded and significant increase across genotypes at the directly genotyped rs2231142 variant (Fig. 3C). The association became more significant when adjusting for known correlates of serum urate levels: age, sex, BMI, alcohol consumption, and antihypertensive medication intake (P trend = 3 × 10−30 for whites and P = 7 × 10−4 for blacks). Adjustment for sex explained the largest part of this difference, with the genetic effect in men significantly stronger than in women (4), which is in agreement with higher rates of renal ABCG2 expression in men compared with women (10). Similarly, the prevalence of gout was significantly different across genotypes in both whites and blacks (Fig. 3D). Of note, the prevalence of gout was 40% among black TT carriers, but genotypes GT and TT were pooled because of the low minor allele frequency of 3% in blacks. The multivariable-adjusted relative odds of gout per each copy of the T allele were 1.68 in both races (P = 2 × 10−7 in whites and P = 0.05 in blacks); the consistency of the effect size in groups of different ancestry further supports causality of rs2231142. In this context, it is of interest that the T allele frequency at rs2231142 among Asian populations (HapMap CHB, and JPT) is approximately 30% compared with 11% in white ARIC participants, and gout prevalence in U.S. individuals of Asian ancestry is about 3 times higher compared with individuals of European ancestry (11). Among the white study participants, population-attributable risk calculations yielded that a conservatively estimated 10% of gout cases could be attributed to the Q141K mutation.
Our findings are of substantial clinical interest because of the high prevalence of the Q141K mutation in individuals of European and Asian ancestry and the substantial population attributable risk. Gout has a high prevalence of approximately 3 million affected individuals in the U.S.; the treatment is often insufficient, with only 21% of gout patients who receive the most common urate-lowering treatment, reaching optimal serum urate levels in clinical trials (12, 13). ABCG2 therefore represents a potential drug target.
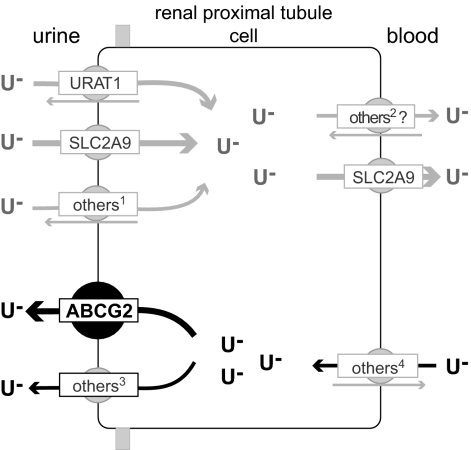
Fig. 4 summarizes and integrates our findings: based on the urate efflux capacity of ABCG2, its apical cellular localization, and the identification of a loss-of-function mutation that causes increased serum urate levels and gout, we postulate that ABCG2 is a secretory urate transporter in the proximal renal tubule. The physiological importance of ABCG2 in humans is illustrated by the sizable differences in serum urate levels and the prevalence of gout caused by genetic variation in ABCG2.
Fig. 4.
Model of urate handling by human renal proximal tubule cells. The physiological relevance of urate transporters (named by their gene symbols) in humans is established by genetic variation causing hyper-/hypouricemia and gout for URAT1 (23) and SLC2A9 (4, 24–28). Transporters expressed in human kidney and shown to transport urate in model systems: 1OAT4; 2OAT1, OAT3; 3MRP4; 4OAT1, OAT3 (1, 29, 30). Arrows indicate the direction of urate transport (gray, reabsorption; black, secretion). Abbreviations: U−, urate.
Conclusion.
In summary, our study overcomes key limitations of GWAS, namely establishing gene function and determining causality of an associated genetic variant (14, 15). We show that ABCG2 is a urate efflux transporter and rs2231142 (Q141K) a loss-of-function mutation that causes hyperuricemia and gout. This completes the chain of evidence from association to causation and supports the common disease-common variant hypothesis in the etiology of gout.
Materials and Methods
Functional Studies.
Xenopus laevis oocytes.
Oocytes were removed from female pigmented Xenopus laevis (Xenopus 1 Inc.) and de-folliculated using collagenase A (Roche) in Ca2+-free OR-2 ringer solution. Stage V-VI oocytes were selected and injected (Nanoinject II, Drummond Scientific) with 50 nL of either mRNA or H2O (as a control). All experiments were performed on the fourth day after injection. Oocytes after injection were cultured in a modified L-15 media (OR-3) and kept at 15–20 °C. mRNA was prepared using the SP6 mMessage mMachine (Ambion Inc.) according to the manufactures protocol. The Q141K mutant was created using site directed mutagenesis with the primers: 5′-GGT GAG AGA AAA CTT AAA GTT CTC AGC AGC TC 3-′ and 5′-GAG CTG CTG AGA ACT TTA AGT TTT CTC TCA CC-3′ and completed using the Quick-Change Lightning kit (Stratagene) according to the manufacturer's protocol.
C-14 uric acid.
Radiolabeled C-14 uric acid (American Radio-labeled Chemicals) was dissolved in a stock solution of 2 mM NaOH with a final urate concentration of 2 mM (either 2 mM hot-radio labeled urate or 1 mM hot and 1 mM cold urate). For oocyte transport experiments, the radio labeled C-14 urate was included at various concentrations in ND96 ringers solution (in mM: 96 NaCl, 2 KCl, 2 MgCl2, 5 Hepes, 1.8 CaCl2, pH 7.5, and 5 mM glucose for overnight incubation experiments).
Oocyte transport experiments.
For accumulation studies, oocytes were incubated either for 1 or 2 h at room temperature in 50 μM C-14 urate or overnight in 500 μM C-14 urate at 15 °C. After incubation the oocytes were washed in ice-cold ND96 solution 3–5 times to remove any superficial, residual C-14 urate from the oocytes. Oocytes were then pooled for scintillation counting. For the 1–2-h accumulation studies, after incubation in the C-14 urate solution, 2 oocytes were placed in each scintillation tube (N) with lysis buffer of 1 N NaOH; a total of 20 oocytes (n) was used for each treatment. For the overnight accumulation studies, after incubation, 10 oocytes were pooled into 1 scintillation vial (N) with the lysis buffer; a total of 50 oocytes (n) was used for each treatment. For the efflux studies, 50 oocytes expressing each of the different proteins of interest were incubated overnight in 500 μM C-14 urate at 15–20 °C, washed 5 times in ice cold ND96 solution, and then, in groups of 10, placed into a well with 500 μL room temp ND96. After 5 min, the bath was collected and replaced with a new bath solution. Bath collections continued for 2–3 h. After the final collection, the oocytes were also collected and placed into scintillation vials (10 per vial) containing 1N NaOH lysis buffer. All samples in scintillation vials were mixed with 5 mL scintillation fluid and their cpm determined using a scintillation counter (LS600 LL, Beckman Coulter Inc.). To confirm that after 24 h the accumulated intracellular urate had not been metabolized by the uricase enzyme, we probed oocytes as well as other tissues for the presence of uricase mRNA using RT-PCR (Fig. S2). Total RNA was extracted from Xenopus laevis oocytes, kidneys, and liver using the Qiagen RNeasy Mini Kit (Qiagen Inc.) according to the manufacturer's protocol. Thirty de-folliculated oocytes were pooled for the RNA preparation. For both reverse transcriptase and PCR, the Qiagen One Step RTPCR kit (Qiagen Inc.) was used according to the manufacturer's protocol. The reaction mixtures were run for 35 cycles. As a control for genomic DNA contamination, we added the One Step enzyme mixture (containing the reverse transcriptase) only after the mixtures reached 94 °C, inactivating the reverse transcriptase. The following primers were used for the detection of uricase RNA: 5′-ACC ATC TAT GCC TTG ACT AAA CTC-3′ and 5′-TAA GGA CCA GCA AAT GTA TCA AG-3′. For the actin 5 control, we used: 5′-CTGAGTTCATGAAGGATCAC-3′ and 5′-AAA TTT ACA GGT GTA CCT GC-3′.
For each batch of counting, a set of control vials of known volume and concentration of C-14 urate was included. Based on the counts measured in any of the experimental samples and assuming that stage V and VI oocytes have an approximate average volume of 1 μL (16), this allowed for the calculation of intracellular C-14 urate concentrations. Statistical comparisons were performed using non-paired Student t-tests and linear regression analyses. Exponential and linear fits were performed using Origin 8 software (OriginLab Corp.).
LLC-PK1 experiments.
LLC-PK1 porcine renal epithelial cells were cultured in Dulbecco's Modified Eagle medium (DMEM) supplemented with 10% FBS. LLC-PK1 cells were seeded at a high density into a 6-well plate and accumulation experiments were performed after the cells polarized. The cells were incubated in DMEM containing 500 μM cold urate and 25 μM hot urate with or without 5 μM FTC (US Biologicals); the incubation lasted 120 min. The cells were then washed four times with ice-cold DMEM and lysed with 500 μL of 1M NaOH. The lysate was collected and placed in a scintillation tubule along with 5 mL scintillation fluid and counted.
For immunofluorescence, LLC-PK1 cells were cultured on Transwell Permeable Supports (Corning Inc.). Only polarized cells were imaged, and polarization occurred 5–7 days after cells reached confluence. The cells were fixed using 4% paraformaldehyde and permeablized using 0.1% Triton X 100. The cells were then blocked with 5% goat serum and the primary antibody was applied (anti-BCRP; Millipore Corp., clone BXP-21 at 1:100). An anti-mouse Cy3 (Sigma) secondary antibody was used (1:200) as well as a DAPI nuclear stain (Vector Labs). The cells were imaged using a confocal laser scanning microscope (Zeiss model LSM510) at 40 ×. Z slices were created from stacked images using the LSM Image Browser (Zeiss), decimated, exported to Image J (National Institutes of Health) and flattened into one 2D image; finally the artifact raster lines resulting from the decimation were reduced using a 16 pixel 90° motion blur filter in Adobe Photoshop (Adobe Inc.).
Western blot and surface biotinylation.
Oocytes were pooled 4 days after injection with either mRNA or H2O (10 per treatment). Ice-cold oocyte lysis buffer was added (in mM: 20 Tris-HCl, 140 NaCl, 2% Triton X 100, and protease inhibitors), and the oocytes were incubated on ice for 30 min. Then the oocytes were homogenized and spun at 7500 rpm at 4 °C. The cell lysate was then stored at −80 °C until used for the western blot. A BCRP antibody was used to probe the blot (anti-BCRP; Millipore Corp., clone BXP-21 at 1:200) and an anti-actin antibody (1:1,000) was used as a loading control. Measurement of mutant and wild-type protein expression was done by densitometry of the homodimer bands on the western blot, performed using the Multi Gauge V3.1 program (Fugi Film). For surface biotinylation experiments, 30 oocytes from each treatment were pooled and washed with ice-cold ND96 at 4 °C. The oocytes were treated with sulfo-NHS-SS-biotin ND96 solution (1 mg/ml) (Pierce Biotechnology) for 1 h at 4 °C, then washed in a 192 mM glycine ND96 biotin quenching solution and lysed as described above. A portion of the lysate was retained as the total protein fraction, the remainder was added to UltraLink Immobilized NeurtrAvidin Protein beads (Pierce Biotechnology) and incubated overnight at 4 °C. The following day the beads were washed, eluted, and run on a SDS-page gel.
Population-Based Study Data.
Study population.
The Atherosclerosis Risk in Communities Study (ARIC) is an ongoing population-based prospective study initiated to study cardiovascular disease and its risk factors. From 1987–1989, 15,792 mostly self-reported white and black study participants were recruited by probability sampling from 4 U.S. communities (visit 1) (9). Participants returned for 3 subsequent visits approximately every 3 years, and participants are contacted yearly by telephone. Written informed consent was obtained from all study participants, and Institutional Review Boards of the participating institutions approved the study protocols.
Genotyping.
Individuals were excluded from all genotyping for non-consent (n = 53) or self-reported race other than black or white (n = 47). Genotyping was conducted using the TaqMan assay to genotype the SNP rs2231142 as described previously (4) and genome-wide association data were obtained from the Affymetrix 6.0 array.
For TaqMan genotyping of the rs2231142 variant, 11,440 white and 4,252 black participants were genotyped using the functionally tested TaqMan Drug Metabolizing Enzyme (DME) assay (Applied Biosystems). PCR product was amplified using 0.9 μM of each of the forward and reverse primers, 0.2 μM of each of the sequence-specific probes, 3 ng DNA, and 1 × TaqMan Universal PCR Master Mix containing AmpliTaq Gold DNA Polymerase in a 5.5-μL reaction volume. Primer and sequence-specific probes are proprietary to Applied Biosystems. After an initial step of 10 min at 95 °C to activate the AmpliTaq Gold, the products were amplified using 50 cycles of 15 s at 92 °C and 90 s at 60 °C. Allele detection and genotype calling were performed using the ABI 7900HT and the Sequence Detection System software (Applied Biosystems). Genotyping was successful in 97.3%. Reproducibility in a set of 313 blind duplicates was >99% with a kappa-coefficient of 0.96 indicating excellent agreement; internal lab reproducibility was 100%. Genotype distributions conformed to Hardy-Weinberg expectations in both white and black study participants (P > 0.1, exact test).
GWAS genotyping was performed using the Affymetrix 6.0 genotyping assay on 8,861 white study participants. Genomic DNA was hybridized in accordance with the manufacturer's standard recommendations, and genotypes were determined using the Birdseed clustering algorithm. Individual samples were filtered to ensure genotyping quality, and 734 out of 8,861 samples were removed in data-cleaning steps for sex mismatch, discordance with previously genotyped markers, first-degree relative of an included individual, and genetic outlier based on allele sharing and principal components analyses (17, 18).
Finally, individuals were excluded from analyses for missing outcome or covariates, and the final study sample for rs2231142 genotyped by TaqMan consisted of 10,902 white and 3,881 black individuals for the urate analyses and 8,489 white and 2,350 blacks for the analyses of gout. In the GWAS analyses, both genotype information on 2,557,232 genotyped and imputed SNPs and phentoype information was available for 8,092 white individuals for the urate analyses and 6,540 for gout. Of the 8,092 individuals with GWAS data, 7,768 had data on rs2231142 by TaqMan available. Concordance of genotypes at rs2231142 obtained by TaqMan or imputed from the GWAS data was 98%.
Genotype imputation.
For imputation, 602,642 high-quality SNPs with call rate greater than or equal to 95%, minor allele frequency greater than or equal to 1%, and conformation to Hardy-Weinberg expectations (P ≥ 10−5) were used. Imputation was conducted using a Hidden Markov Model as implemented in MACH to a set of 2,557,232 high-quality polymorphic HapMap SNPs. For imputation, genotype data from each sample were combined with the HapMap CEU samples. The software MACHv1.0.16 was used to infer unobserved genotypes probabilistically based on identification of shared haplotype stretches between the study sample and the HapMap CEU reference individuals [release 21 (build 35)]. Imputation results are then summarized for each genotype as an “allele dosage,” a fractional value between 0 and 2, corresponding to the expected number of copies of the minor allele at that SNP.
Outcome measures.
Uric acid was measured in serum at the baseline visit using an uricase method (19). Measurements of uric acid in 40 individuals were repeated greater than or equal to 1 week apart; the reliability coefficient was 0.91 and the coefficient of variation was 7.2% (20). Gout was self-reported at study visit 4 in answer to the question “were you ever told you had gout.” At a physiologic pH of 7.4, 98% of uric acid is present in its ionized form, urate. We therefore refer to serum urate levels rather than serum uric acid levels throughout the manuscript.
Statistical analyses.
For the analyses of rs2231142 genotyped by TaqMan, differences in mean urate levels by genotype were evaluated using simple linear regression. Due to the low minor allele frequency of 3%, the genotype groups GT and TT were pooled among black participants. Adjusted differences in mean urate levels were tested using multivariable adjusted linear regression, assuming an additive genetic model (P trend). Prevalence of gout was graphed and crude and multivariable adjusted odds of gout were evaluated using logistic regression in both whites and blacks.
For analyses of GWAS data, mean serum urate levels were modeled using multivariable adjusted residuals in linear regression. Covariates in all regression models were age, sex, study site, BMI, antihypertensive treatment (yes/no), and alcohol consumption (g/week) at the time of uric acid measurement or ascertainment of gout (4). All analyses were conducted separately by self-reported race and used an additive genetic model. The genomic control parameter for the GWAS analyses of serum urate levels across the entire genome was 1.02, indicating no substantial inflation of the test statistics.
Given the a priori hypothesis and prior evidence (4), the statistical significance threshold for the association analyses of rs2231142, and serum urate levels and gout was set at α = 0.05. In the analyses of the GWAS data, 601 additional SNPs at 88.9–89.5 Mb (Build 36) on chromosome 4 were analyzed, and the analyses were repeated conditional on genotype at rs2231142. The significance threshold for these analyses was set at α = 8 × 10−5, which corresponds to a Bonferroni correction for testing 601 independent markers. Population attributable risk (PAR) was calculated using the formula PAR (%) = [Σ(pi)(RRi-1)/[1+Σ(pi)(RRi − 1)]] × 100, where pi corresponds to the proportion of individuals exposed to the risk allele (separate groups for heterozygotes and risk homozygotes) among those without gout, and RRi is estimated by the respective multivariable adjusted odds ratio of gout (21). As gout is a common disease, analyses were repeated using the prevalence ratio rather than the odds ratio of gout to obtain a conservative estimate of PAR which was 10% as compared to 13% using the odds ratio.
Single SNP analyses were conducted using Stata, and GWAS analyses were performed using the ProbABEL package from the ABEL set of programs (http://mga.bionet.nsc.ru/yurii/ABEL/) (22) and the statistical software R.
Supplementary Material
Acknowledgments.
The authors thank the staff and participants of the Atherosclerosis Risk in Communities (ARIC) Study for their important contributions, Douglas D. Ross (University of Maryland, Baltimore, MD) and Thomas Lang (University of Mainz, Mainz, Germany) for the kind gift of plasmids, and Andrew Bahn for expert advice. The ARIC Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute Contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022, R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute Contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and National Institutes of Health Roadmap for Medical Research. The functional studies were supported by R01DK32753. Anna and Michael Köttgen dedicate this work to the late Rainer Greger, a pioneer in the field of renal transport physiology and an inspiring mentor and teacher.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901249106/DCSupplemental.
References
- 1.Anzai N, Kanai Y, Endou H. New insights into renal transport of urate. Curr Opin Rheumatol. 2007;19:151–157. doi: 10.1097/BOR.0b013e328032781a. [DOI] [PubMed] [Google Scholar]
- 2.Yang Q, et al. Genome-wide search for genes affecting serum uric acid levels: The Framingham Heart Study. Metabolism. 2005;54:1435–1441. doi: 10.1016/j.metabol.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Lawrence RC, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part II Arthritis Rheum. 2008;58:26–35. doi: 10.1002/art.23176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dehghan A, et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372:1953–1961. doi: 10.1016/S0140-6736(08)61343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doyle LA, et al. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci USA. 1998;95:15665–15670. doi: 10.1073/pnas.95.26.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polgar O, Robey RW, Bates SE. ABCG2: structure, function and role in drug response. Expert Opin Drug Metab Toxicol. 2008;4:1–15. doi: 10.1517/17425255.4.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Van Aubel RA, Smeets PH, van den Heuvel JJ, Russel FG. Human organic anion transporter MRP4 (ABCC4) is an efflux pump for the purine end metabolite urate with multiple allosteric substrate binding sites. Am J Physiol Renal Physiol. 2005;288:F327–333. doi: 10.1152/ajprenal.00133.2004. [DOI] [PubMed] [Google Scholar]
- 8.Huls M, et al. The breast cancer resistance protein transporter ABCG2 is expressed in the human kidney proximal tubule apical membrane. Kidney Int. 2008;73:220–225. doi: 10.1038/sj.ki.5002645. [DOI] [PubMed] [Google Scholar]
- 9.The Atherosclerosis Risk in Communities (ARIC) Study: Design and objectives. The ARIC investigators. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 10.Krishnamurthy P, Schuetz JD. Role of ABCG2/BCRP in biology and medicine. Annu Rev Pharmacol Toxicol. 2006;46:381–410. doi: 10.1146/annurev.pharmtox.46.120604.141238. [DOI] [PubMed] [Google Scholar]
- 11.Krishnan E, Lienesch D, Kwoh CK. Gout in ambulatory care settings in the United States. J Rheumatol. 2008;35:498–501. [PubMed] [Google Scholar]
- 12.Becker MA, et al. Febuxostat compared with allopurinol in patients with hyperuricemia and gout. N Engl J Med. 2005;353:2450–2461. doi: 10.1056/NEJMoa050373. [DOI] [PubMed] [Google Scholar]
- 13.Schumacher HR, Jr, et al. Effects of febuxostat versus allopurinol and placebo in reducing serum urate in subjects with hyperuricemia and gout: A 28-week, phase III, randomized, double-blind, parallel-group trial. Arthritis Rheum. 2008;59:1540–1548. doi: 10.1002/art.24209. [DOI] [PubMed] [Google Scholar]
- 14.Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–731. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- 15.McCarthy MI, et al. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 16.Zeuthen T, Zeuthen E, Klaerke DA. Mobility of ions, sugar, and water in the cytoplasm of Xenopus oocytes expressing Na(+)-coupled sugar transporters (SGLT1) J Physiol. 2002;542:71–87. doi: 10.1113/jphysiol.2001.014530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 19.Iribarren C, Folsom AR, Eckfeldt JH, McGovern PG, Nieto FJ. Correlates of uric acid and its association with asymptomatic carotid atherosclerosis: the ARIC Study. Atherosclerosis Risk in Communities Ann Epidemiol. 1996;6:331–340. doi: 10.1016/s1047-2797(96)00052-x. [DOI] [PubMed] [Google Scholar]
- 20.Eckfeldt JH, Chambless LE, Shen YL. Short-term, within-person variability in clinical chemistry test results. Experience from the Atherosclerosis Risk in Communities Study. Arch Pathol Lab Med. 1994;118:496–500. [PubMed] [Google Scholar]
- 21.Yates JR, et al. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357:553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- 22.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 23.Enomoto A, et al. Molecular identification of a renal urate anion exchanger that regulates blood urate levels. Nature. 2002;417:447–452. doi: 10.1038/nature742. [DOI] [PubMed] [Google Scholar]
- 24.Li S, et al. The GLUT9 Gene Is Associated with Serum Uric Acid Levels in Sardinia and Chianti Cohorts. PLoS Genet. 2007;3:e194. doi: 10.1371/journal.pgen.0030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallace C, et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am J Hum Genet. 2008;82:139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doring A, et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat Genet. 2008;40:430–436. doi: 10.1038/ng.107. [DOI] [PubMed] [Google Scholar]
- 27.Vitart V, et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat Genet. 2008;40:437–442. doi: 10.1038/ng.106. [DOI] [PubMed] [Google Scholar]
- 28.Matsuo H, et al. Mutations in Glucose Transporter 9 Gene SLC2A9 Cause Renal Hypouricemia. Am J Hum Genet. 2008;83:744–751. doi: 10.1016/j.ajhg.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enomoto A, Endou H. Roles of organic anion transporters (OATs) and a urate transporter (URAT1) in the pathophysiology of human disease. Clin Exp Nephrol. 2005;9:195–205. doi: 10.1007/s10157-005-0368-5. [DOI] [PubMed] [Google Scholar]
- 30.Rizwan AN, Burckhardt G. Organic anion transporters of the SLC22 family: biopharmaceutical, physiological, and pathological roles. Pharm Res. 2007;24:450–470. doi: 10.1007/s11095-006-9181-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.