Abstract
Adequate responses by our innate immune system toward invading pathogens were of vital importance for surviving infections, especially before the antibiotic era. Recently, a polymorphism in Mal (Ser180Leu, TIRAP rs8177374), an important adaptor protein downstream of the Toll-like receptor (TLR) 2 and 4 pathways, has been described to provide protection against a broad range of infectious pathogens. We assessed the functional effects of this polymorphism in human experimental endotoxemia, and we demonstrate that individuals bearing the TIRAP 180L allele display an increased, innate immune response to TLR4 and TLR2 ligands, but not to TLR9 stimulation. This phenotype has been related to an increased resistance to infection. However, an overshoot in the release of proinflammatory cytokines by TIRAP 180L homozygous individuals suggests a scenario of balanced evolution. We have also investigated the worldwide distribution of the Ser180Leu polymorphism in 14 populations around the globe to correlate the genetic makeup of TIRAP with the local infectious pressures. Based on the immunological, clinical, and genetic data, we propose that this mutation might have been selected in West Eurasia during the early settlement of this region after the out-of-Africa migration of modern Homo sapiens. This combination of functional and genetic data provides unique insights to our understanding of the pathogenesis of sepsis.
Keywords: Innate immunity, TLR4, TLR2, evolution, cytokines
The nonsynonymous single nucleotide polymorphism Ser180Leu (S180L) of the TIR domain-coding adaptor protein (TIRAP) gene, also known as adaptor protein Mal (1), was found to be protective in heterozygous individuals for a broad range of infectious diseases such as malaria, tuberculosis, bacteremia, and pneumococcal disease (1). Mal is involved in the signaling pathways of Toll-like receptors (TLR) 2 and 4, which are important innate immune receptors for the recognition of a broad range of pathogenic Gram-negative and Gram-positive bacteria (2). Recognition by TLRs eventually leads to activation of NF-κB and the transcription of proinflammatory genes that activate the mechanisms of host defense and the clearance of the pathogens. However, little is known about the in vivo functional consequences of the S180L polymorphism or the possible differences in the geographic distribution of the S180L polymorphism in relation to infectious pressure.
In this study we hypothesized that the difference in the frequency of TIRAP 180L between the different geographical regions was due to the effects of the mutation on the function of TLR signaling, and was shaped by the pressure exerted by different infections, either during the out-of-Africa migration of early modern humans during the early Upper Paleolithic period (60–40kya) (3) or the Neolithic demic expansion from the Near East into Europe (≈10,000kya) (4, 5). This pressure may have resulted in an increase of the frequency of TIRAP S180L heterozygous individuals and thereby a strong rise of the frequency of the TIRAP 180L allele in West-Eurasia populations.
Results and Discussion
In Vivo Models of Sepsis That Determine the Phenotypic Effect of the S180L Genotype.
Because of the high prevalence of the TIRAP S180L polymorphism in the European population, we tested the hypothesis that under severe bacterial pressure there is a pathophysiological advantage that could favor selection of the TIRAP 180L allele variant. Therefore, we evaluated the functional consequences of the heterozygous TIRAP S180L genotype during sepsis, a life-threatening infection accompanied by a systemic inflammatory response (6). TLR4 is mainly involved in the recognition of the lipopolysaccharide (LPS) of Gram-negative bacteria (7), inducing intracellular signals through the adaptor protein TIRAP/Mal (8). To avoid limitations of artifacts between in vitro and in vivo results, we determined the inflammatory phenotype in the most relevant in vivo model of sepsis: that of experimental endotoxemia of human volunteers by i.v. challenge with LPS.
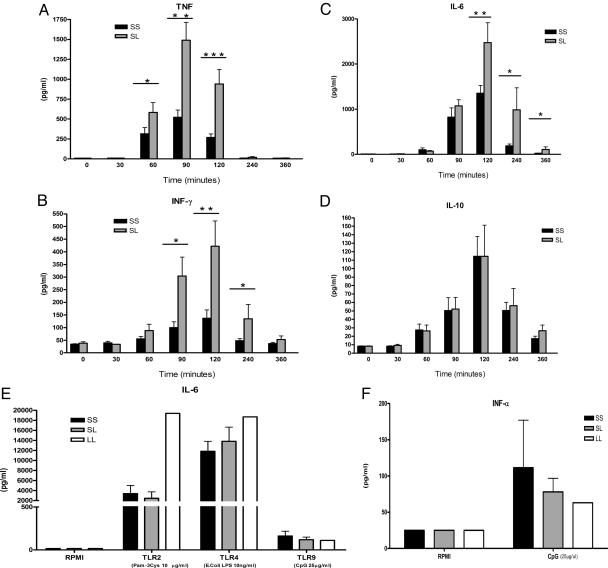
Before and after exposure to LPS, blood was drawn at several time points for proinflammatory (TNF, IFN-γ, IL-6) and anti-inflammatory cytokine (IL-10) measurements. We found that, compared with the normal homozygous TIRAP S180 individuals, the heterozygous TIRAP S180L volunteers had a produced significantly more proinflammatory cytokines after challenge with LPS (Fig. 1 A–D). The higher in vivo production rate of proinflammatory cytokines in individuals bearing the 180L allele was accompanied by increased cytokine production when cells isolated from these individuals were stimulated with the TLR2 ligand Pam3Cys, but not when they were stimulated with the TLR9 ligand 3′5′-cytidylylguanosine (CpG) DNA (Fig. 1E). Although the cells isolated from the individuals bearing the 180L did not release significantly more cytokines in vitro after LPS stimulation, it is interesting to observe that the TIRAP 180L homozygous individuals produced the highest amount of IL-6 after both Pam3Cys and LPS stimulation, but not after CpG DNA. Both TLR2 and TLR4 signal through the TIRAP adaptor molecule and therefore it is expected that the effect of the 180L polymorphism should be observed in both. In contrast, TLR9 does not need the TIRAP adaptor molecule pathway and therefore showed no difference between the various TIRAP genotypes. TIRAP-mediated signals are involved in LPS-induced NF-κB activation, but not for the IFN regulatory factor 3 (IRF3)-induced, type I IFN release. In line with the hypothesis that type I IFN release would not be effected by the 180L TIRAP allele, we found its production after cell stimulation with CpG DNA was not increased in individuals bearing this polymorphism (Fig. 1F). LPS alone was not able to induce production of type I IFN.
Fig. 1.
Cytokine production capacity in individuals screened for the TIRAP S180L polymorphism. (A–D)Serum concentrations of proinflammatory cytokines TNF, IFN-γ, IL-6 (A–C) and the anti-inflammatory cytokine IL-10 (D) after 2 ng/kg LPS injection. Concentrations are given as mean (pg/mL) and SD for TIRAP S180 homozygous (SS, n = 14) and S180L heterozygous (SL, n = 6) individuals. (E) To study the effect of the TIRAP 180L polymorphism on other TLRs, PBMCs were stimulated with TLR2 (10 μg/mL Pam3Cys), TLR4 (10 ng/mL LPS), and TLR9 (25 μg/mL CpG) ligands. (F) IFN-α release by PBMC of patients with or without polymorphisms was also assessed. Concentrations are given as mean (pg/mL) and SEM for TIRAP S180 homozygous (SS, n = 4), S180L heterozygous (SL, n = 5) individuals, and one 180L homozygous individual. *, P = 0.05; **, P = 0.01; ***, P = 0.001 (Mann-Whitney U test).
These data, clearly demonstrating an increased cytokine production in primary cells of individuals bearing the S180L polymorphism, are in contrast to the functional in vitro analyses of Khor et al. (1), based on transfection studies of the mutated TIRAP allele, who reported diminished proinflammatory cytokine induction in cells transfected with the mutated allele. It is possible that endogenously expressed 180L modulates 180S in a positive way, and that the transfection-based experiments do not provide an accurate reflection of the in vivo data.
The increased cytokine production in individuals bearing the TIRAP S180L genotype may have important consequences for the susceptibility and severity of infections. It has been proposed that individuals with relatively low cytokine production would be protected against the effects of systemic inflammation. This idea is based on the assumption that the high cytokine release during sepsis is the cause of systemic inflammatory response syndrome (SIRS), which ultimately leads to septic shock and death. Although this may be the case for individuals already in a septic state, it is unlikely to be true for the initial susceptibility to infections. During the first stage of infections, the proinflammatory response of the host has to be strong and adequate for a good clearance of the pathogenic microorganism (9, 10). Therefore, a moderately increased cytokine release during the primary stage of infection, as would be expected in heterozygous S180L individuals, is likely beneficial for the outcome of the infection, and the early clearance of bacteria prevents those individuals from developing sepsis that could lead to SIRS. However, if the cytokine release is reaching a certain threshold leading to systemic inflammatory effects, which is suggested to take place in homozygous 180L individuals, then this could ultimately prove deleterious to the host and preclude the fixation of the mutation in the population. This hypothesis is supported by a study performed on 166 patients with Gram-negative bacterial sepsis and ventilator-associated pneumonia (VAP). Individuals homozygous for the 180L polymorphism had a significantly more severe infection compared with the other TIRAP genotypes (P = 0.038 concerning clinical pulmonary infection score) and a tendency toward a decreased oxygenation of the tissues (Table 1).
Table 1.
Clinical characteristics of 160 patients with VAP and sepsis enrolled in the study
| Characteristics | SS | SL | LL |
|---|---|---|---|
| n (Male/female) | 121 (91/30) | 43 (31/12) | 2 (2/0) |
| Age in years, mean ± SD | 61.4 ± 18.1 | 55.9 ± 12.2 | 48.5 ± 27.6 |
| CPIS, mean ± SD* | 7.76 ± 1.11 | 7.74 ± 1.31 | 9.50 ± 0.70 |
| SAPS II score, mean ± SD | 38.8 ± 13.8 | 37.4 ± 13.5 | 39.0 ± 0 |
| White blood cells per mL, mean ± SD | 13,312.1 ± 7,013.4 | 12,809.8 ± 4,921.8 | 10,030.1 ± 5,600.3 |
| Septic shock: no (%)† | 65 (53.7) | 25 (58.1) | 1 (50?) |
| Septic shock: yes (%)† | 56 (46.3) | 18 (41.9) | 1 (50) |
| pO2/FiO2, mean ± SD | 226.1 ± 117.1 | 222.3 ± 100.2 | 190.5 ± 13.4 |
Polymorphism between genotypes are in Hardy–Weinberg equilibrium. SS, wild type, i.e. patients without Mal mutations; SL, mutant Mal, i.e. patients carrying the Ser180Leu SNP; LL, homozygous mutants, i.e., patines only carrying the 180Leu SNP; CPIS, clinical pulmonary infection score. pO2/FiO2 ratios are given as an index of the oxygentation.
*P = 0.038 between WT and homozygotes; P = 0.039 between heterozygotes and homozygotes (P values after Bonferroni correction).
†Total numbers of patients in the subgroups comparing patients with and without septic shock. There were no differences between groups.
Distribution of the TIRAP Polymorphism in Different Populations.
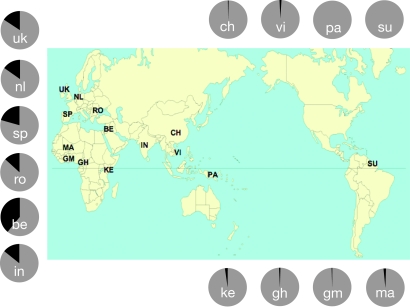
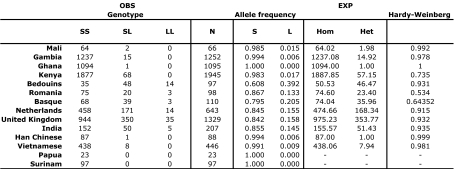
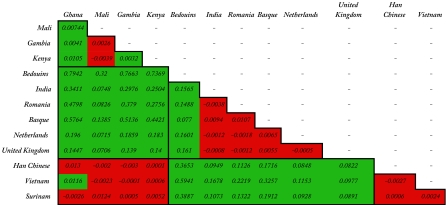
We determined the distribution of the TIRAP polymorphism frequencies in 14 populations originating from Africa, West-Eurasia, East-Eurasia, and the Americas (Fig. 2 and Table 2). Distribution of the TIRAP S180 and 180L allele showed a high frequency of 180L in West-Eurasian compared with African and East-Asian populations (Fig. 1). To gain insight in the allelic distribution of the TIRAP 180 polymorphism, all populations were compared pairwise by using both an exact test for population differentiation (11) and the Fst (12) statistics (Table 3). Combining the population differentiation results with the Fst test reveals that the populations are broadly divided according to these 3 main geographic regions (Africa, East Eurasia, and West Eurasia; see Table 2). The apparent difference in TIRAP allelic frequencies within populations in Africa is most likely caused by the low frequencies of the TIRAP 180L allele among these populations (Table 2). Our data also revealed that the highest frequencies of the TIRAP 180L allele is found in the Middle-Eastern Bedouins from the Negev desert, which results in the highest differentiation and dissimilarity with all other studied populations. Two scenarios may explain this distribution of the mutation. In one of the scenarios, the mutation arose in Africa, where it persisted in low prevalence after spreading due to human migration in the Middle-East and Europe; and in the other scenario, the mutation arose in West-Eurasia and was brought back later in Africa through the gene transfer by the European colonists. However, the high prevalence of the mutation in the Middle-East and Europe suggests that if any selection has influenced TIRAP 180L prevalence, it most likely took place in the West-Eurasian region.
Fig. 2.
World distribution frequency of TIRAP S180 (gray) and 180L (black) alleles among 14 populations. Populations of the African continent are Mali (MA), Gambia (GA), Ghana (GH), and Kenya (KE). Eurasia populations are the Bedouins (BE). European populations are Romania (RO), Basque (SP), the Netherlands (NL), and United Kingdom (UK). Asian and New World populations are India (IN), Han Chinese (CH), Vietnam (VI), Papua New Guinea (PA), and Trio Indians from Surinam (SU). The number of individuals of each population is given in Table 1.
Table 2.
Frequencies of the TIRAP S180L polymorphism (SL) in all screened populations
OBS, observed frequencies; EXP, expected frequency if populations would be in Hardy–Weinberg equilibrium (HW); Hom, homozygotes (SS and LL); Het, heterozygotes (SL). P values for the HW represent values for the HW probability test. P < 0.0035, corrected with the Bonferroni method, was considered significant.
Table 3.
Population differentiation test outcomes
Outcomes are presented in red if there was no significant differences (P > 0.05) between the populations, meaning that the allelic compositions of the populations do not differ. Outcomes are presented in green if the allelic composition differed (P < 0.05 was considered to be significant). Numbers indicate the pairwise Fst values between populations. Squares indicate populations that came from the same continent.
Dating the Selection on the 180L TIRAP Allele.
The S180L TIRAP polymorphism is conspicuously low in the African population and absent in the Asian population. This similarity between the African and East-Asian populations could indicate that the selection on TIRAP 180L did not take place during the first human Paleolithic migration out of Africa, because higher frequencies of the TIRAP 180L allele would then also be expected in Asia. Therefore, we tested the hypothesis that selection on TIRAP 180L occurred during the more recent Neolithic period when farming settlement of Europe took place (1, 4, 5, 13). To test this, we performed an extended haplotype homozygosity (EHH) test on the European panel from HapMap (14). The EHH test, which can discern recent selection until a maximum of ≈30,000 years (15, 16), revealed no statistically significant difference (3′ end borders, significance P = 0.053; 5′ end, P = 0.438), indicating no strong recent selection on the TIRAP 180L allele in the West-Eurasian population sample (individuals with Northern and Western European ancestry). Although this could indicate that the observed differences between the populations of West-Eurasia and other continents was the result of genetic drift, an alternative explanation is that selection could also have taken place during the period between the out-of-Africa migration [60–40 kya (3)] and the first successful colonization of Europe [40–45 kya (3)]. This time scale allows for recombination to have had enough time to make it difficult to discover an evolutionary pressure by using the EHH test (17). The hypothesis of an old ancestry of the mutation in West-Eurasia is also supported by the presence of the mutation in India, most likely due to the Indo-European origin of the population tested. India clusters with other West-Eurasian populations, rather than East-Asian neighbors.
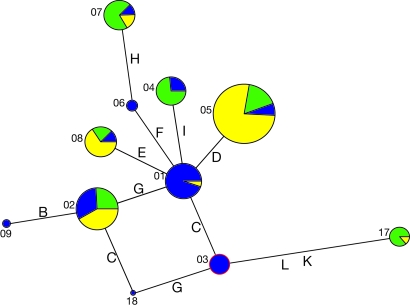
With the use of HapMap (14), we investigated the distribution pattern of the haplotype allele network of the TIRAP gene between African, West-Eurasian, and East-Eurasian populations. Within these 3 HapMap populations, we detected 17 different haplotypes based on 12 polymorphisms (see Fig. 3 and Table S1). Modeling established that Haplotype 3 is the ancient haplotype of TIRAP, also to be found in the chimpanzee. Most haplotypes form a star-like pattern around Haplotype 1, which is a direct descendant of Haplotype 3. The exception is Haplotype 17, which is the only haplotype containing the TIRAP 180L mutation. Haplotype 17 is closely linked to the ancient Haplotype 3, representing a different branching than the Haplotype 1 cluster (Fig. 3). In the HapMap database, no African individuals with Haplotype 17 were found, although our own screening revealed the presence of this haplotype in a small proportion of the African individuals tested (Fig. 2 and Table 2). It is likely that this is a result of recent genetic flow between the modern human populations. This modeling shows that all haplotypes, with the exception of Haplotype 17, are equally spread among the different populations. The lack of the TIRAP 180L mutation within the African HapMap database supports the scenario of the origin of the mutation in a population already located outside of Africa.
Fig. 3.
TIRAP gene allele haplotype network of the HapMap (14). Numbers stand for the different alleles, and letters between each circle are the different polymorphisms included (see Table S1). Area of each circle correlates with the amount of alleles.
Conclusion
These combined genotypic and phenotypic studies have shown that a potent cytokine response, as a marker of an adequate innate immune response, is associated with a lower susceptibility to infections. Our findings form a link between this hypothesis and support of the protective effects of heterozygosity for TIRAP S180L reported by Khor and colleagues (1). Moreover, in an evolutionary context this suggests that before the dispersal of modern humans into Western Eurasia [40–45 kya (3)], those individuals bearing the TIRAP S180L heterozygous phenotype would have had greater resistance to infections in a harsh environment. Furthermore, they would have been more likely to survive should an infection become established. However, the deleterious effects during severe sepsis in homozygous individuals, due to even-higher proinflammatory cytokine production, prevented fixation of this haplotype in the entire population.
In summary, our study confirms the finding that TIRAP S180L heterozygous individuals have a distinctive phenotype compared with the TIRAP S180 homozygous bearers, with the 180L allele associated with a more pronounced inflammatory response and a lower risk of developing septic shock. The frequency of the TIRAP 180L allele is highly elevated in West-Eurasia, with selection due to infectious pressure in this region being a likely explanation. We could not confirm that this selection has taken place recently during the Neolithic period from the Near East into Europe. More likely, the selection of the advantageous TIRAP variants that has shaped the West-Eurasian populations has taken place during the early phase of the dispersal of modern humans into West-Eurasia. Finally, the present study demonstrates that, by determining the evolutionary path of genes involved in the innate immune system, and complementing genotype analysis with the functional phenotype, one can provide new insights in our understanding of the pathogenesis of severe infections such as sepsis.
Materials and Methods
Human Endotoxemia Model.
Twenty healthy volunteers were infused with highly purified LPS at a dose of 2 mg/kg, as previously described (18). Before and after 30, 60, 90, 120, 240, and 360 min, blood was collected and plasma separated by centrifugation. Circulating concentrations of TNF, IL-6, IFN-γ, and IL-10 were measured by specific LUMINEX kit (BioRad). Among the volunteers, 14 were homozygote for the wild-type TIRAP allele, whereas 6 individuals were heterozygous for the TIRAP 180L allele.
Peripheral Blood Mononuclear Cell (PBMC) Stimulation.
PBMCs were isolated by density gradient centrifugation on Ficoll, washed 3 times in cold RPMI medium 1640, counted, and resuspended in complete culture medium. PBMCs (final concentration 2.5 × 106/mL) were stimulated with Pam3Cys (10 μg/mL), LPS (10 ng/mL), or CpG DNA (25 μg/mL) in 96-well, round-bottom plates for 24 h at 37 °C. After stimulation, supernatants were collected and directly used for cytokine measurement.
Sepsis Patients.
One-hundred-sixty-six patients were enrolled in a prospective study conducted over the period of June 2004 to September 2005 at the Department of Critical Care of the Evangelismos General Hospital and in the 2nd Department of Critical Care of the Attikon University Hospital of Athens, Greece. The study was approved by the Ethics Committee of both hospitals. All enrolled patients were intubated for at least 48 h before diagnosis of sepsis and they were all aged above 18 years. Exclusion criteria were the presence of (i) neutropenia (<500 neutrophils per mm3), (ii) HIV infection, and (iii) oral intake of corticoids at a dose ≥1 mg/kg of equivalent prednisone for a period greater than 1 month. Enrolled patients were followed-up for 28 days. Inclusion criteria were the concomitant presence of (i) VAP and (ii) sepsis, severe sepsis, or septic shock. The patients were selected based on infection with a Gram-negative microorganism. The most commonly cultured microorganisms were Acinetobacter baumannii, Pseudomonas aeruginosa, Klebsiella pneumoniae, and Enterobacter spp. Septic shock was defined as sepsis with sepsis-induced hypotension requiring vasopressor therapy despite adequate fluid challenge, along with the presence of hypoperfusion and organ dysfunction.
DNA Samples and TIRAP 180 Screening.
DNA samples from the Han people came from Corriell (Cat No. HD1000CHI). Other included populations are described elsewhere (1, 19). Informed consent and written approval from the relevant ethical review boards has been granted from all sample collections. From most populations, blood was drawn and the DNA was extracted by using the isolation kit Puregene (Gentra Sytems). Genotyping of the Mal/TIRAP S180L polymorphism was performed by using TaqMan, SNP Genotyping Assay (Applied Biosystems). PCRs were set up according to the manufacturer's instructions. Thermal cycling was performed on a fast optical 96-well reaction plate on the 7500 Fast Real-Time PCR system (Applied Biosystems) as follows: Initial denaturation and enzyme activation at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s, and annealing/extension at 60 °C for 1 min. Mal SNP polymorphism was determined by performing an Allelic Discrimination Assay run on the 7500 Fast Real-Time PCR system (Applied Biosystems). SNP polymorphism results were verified by using positive controls (20).
Statistics.
The distributions between the TIRAP alleles in the different populations, the Hardy Weinberg equilibrium, and Fst were preformed by means of GENEPOP (21). The differentiation analysis is based on the unbiased estimation of the P value of a log-likelihood-based (G) exact test, which is based on the same principles as the Fisher exact test.
Cytokine and sepsis statistics were preformed by using the Statistical Package for the Social Sciences package, and differences were tested by the Mann–Whitney U test. P < 0.05 was considered to represent a statistically significant difference.
As a neutrality test, we applied the EHH test (36) to the phased genotypic information available from HapMap Data Rel. 21a/phaseII Jan 07, on NCBI NB35 assembly, dbSNP b125. We used as the core haplotype that haplotype formed by allele G at rs3802813 and allele A at rs3802814 as proxies for rs8177374, which is absent in the phased haplotype display of this HapMap release. As a summary statistic we used the extent for with the EHH for this core haplotype was 1 in the HapMap Caucasian population within a genomic region that extended 100 kb in each of the directions from the core. To test whether the distance observed was longer than expected under neutrality, we ran coalescent simulations that took into account the heterogeneity in recombination rates across these regions (obtained from the HapMap link) by means of msHOT (22). We used demographic parameters reflecting the Out-of-Africa model of human evolution by slightly modifying the model by Schaffner et al. (23). Simulations were run for both 100kb downstream from the core and for 100kb upstream from the core. The number of SNPs was adjusted to that observed in the HapMap sample for that region. We added 3 filters to the neutral simulations: (i) We only considered those simulations that resulted in a number of different haplotypes equal to that observed in the HapMap sample (as determined by the core SNPs rs3802813 and rs3802814), in the simulations the first 2 SNPs were considered the core set; (ii) in addition, one of the simulated core haplotypes had to show a similar frequency to that core haplotype under examination (obtained from the HapMap Caucasian SNP information and defined by core SNPS, (allele G at rs3802813 and allele A at rs3802814); (iii) the ancestral/derived states of the core SNPs in this preselected, simulated haplotype had to also be identical to the ancestral/derived states of the core SNPs at the core haplotype under examination. By comparison with the orthologous genome regions of the Chimp and Rhesus Macaca, rs3802813 “G” and rs3802814 “A” were defined as “ancestral” and “derived”, respectively. Simulations satisfying these conditions were kept, but only the information on the core haplotype was used for the EHH analysis. Finally, the extent from the core SNPs for which EHH was 1 was recorded for the observed HapMap data and this was tested against the distribution obtained from the filtered simulations.
Network analysis was preformed by using the program Network 4.5.0.0 from Fluxus-engineering (http://www.fluxus-engineering.com) using the median joining methodology (24). For information regarding the composing of the allele haplotypes and weighting of the different SNPs, see Table S1.
Supplementary Material
Acknowledgments.
We thank Garret Hellenthal (University of Oxford, UK) for kindly proving a modified version of msHOT, and the Servicios Generales de Investigacion/Ikerkuntzarako Zerbitsu Orokorrak-Servicios Generales-Ikerkuntza (SGI/IZO-SGIker), Universidad de País Vasco/Euskal Herriko Unibertsitatea (supported by the National Program for the Promotion of Human Resources within the National Plan of Scientific Research, Development and Innovation—Fondo Social Europeo, MCyT, and Basque Government) for generous allocation of computational resources. This work was supported by a Vidi grant of the Netherlands Organisation for Scientific Research (to M.G.N.) and Science Foundation Ireland (L.A.J.O.N.), and Basque Government Grant GIC07/42-IT-453-07 (to S.A., N.I., and C.d.l.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.A. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811273106/DCSupplemental.
References
- 1.Khor CC, et al. A Mal functional variant is associated with protection against invasive pneumococcal disease, bacteremia, malaria and tuberculosis. Nat Genet. 2007;39:523–528. doi: 10.1038/ng1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 3.Mellars P. Going East: New genetic and archaeological perspectives on the modern human colonization of Eurasia. Science. 2006;313:796–800. doi: 10.1126/science.1128402. [DOI] [PubMed] [Google Scholar]
- 4.Simoni L, Calafell F, Pettener D, Bertranpetit J, Barbujani G. Geographic patterns of mtDNA diversity in Europe. Am J Hum Genet. 2000;66:262–278. doi: 10.1086/302706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Underhill PA, et al. Y chromosome sequence variation and the history of human populations. Nat Genet. 2000;26:358–361. doi: 10.1038/81685. [DOI] [PubMed] [Google Scholar]
- 6.Cohen J. The immunopathogenesis of sepsis. Nature. 2002;420:885–891. doi: 10.1038/nature01326. [DOI] [PubMed] [Google Scholar]
- 7.Russel JA. Management of Sepsis. N Engl J Med. 2006;355:1699–1713. doi: 10.1056/NEJMra043632. [DOI] [PubMed] [Google Scholar]
- 8.Mansell A, Brint E, Gould JA, O'Neill LA, Hertzog PJ. Mal interacts with tumor necrosis factor receptor-associated factor (TRAF)-6 to mediate NF-kappaB activation by toll-like receptor (TLR)-2 and TLR4. J Biol Chem. 2004;279:37227–37230. doi: 10.1074/jbc.C400289200. [DOI] [PubMed] [Google Scholar]
- 9.Netea MG, van der Meer JW, van Deuren M, Kullberg BJ. Proinflammatory cytokines and sepsis syndrome: Not enough, or too much of a good thing? Trends Immunol. 2003;24:254–258. doi: 10.1016/s1471-4906(03)00079-6. [DOI] [PubMed] [Google Scholar]
- 10.Westendorp RG, et al. Genetic influence on cytokine production and fatal meningococcal disease. Lancet. 1997;349:170–173. doi: 10.1016/s0140-6736(96)06413-6. [DOI] [PubMed] [Google Scholar]
- 11.Raymond M, Rousset F. An exact test for population differentiation. Evolution. 1995;49:1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- 12.Weird BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 13.Cavalli-Sforza LL, Menozzi P, Piazza A. The history and geography of human genes. Princeton: Princeton Univ Press; 1996. [Google Scholar]
- 14.The International HapMap Consortium. The international HapMap project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 15.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 16.Sabeti PC, et al. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 17.Bamshad M, Wooding SP. Signatures of natural selection in the human genome. Nat Rev Genet. 2003;4:99–111. doi: 10.1038/nrg999. [DOI] [PubMed] [Google Scholar]
- 18.van Eijk LT, et al. Gender differences in the innate immune response and vascular reactivity following the administration of endotoxin to human volunteers. Crit Care Med. 2007;35:1464–1469. doi: 10.1097/01.CCM.0000266534.14262.E8. [DOI] [PubMed] [Google Scholar]
- 19.Ferwerda B, et al. TLR4 polymorphisms, infectious diseases, and evolutionary pressure during migration of modern humans. Proc Natl Acad Sci USA. 2007;104:16645–16650. doi: 10.1073/pnas.0704828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheedy FJ, Marinou I, O'Neill LA, Wilson AG. The Mal/TIRAP S180L and TLR4 G299D polymorphisms are not associated with susceptibility to, or severity of, rheumatoid arthritis. Ann Rheum Dis. 2008;67:1328–1331. doi: 10.1136/ard.2007.083337. [DOI] [PubMed] [Google Scholar]
- 21.Raymond M, Rousset F. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J Heredity. 1995;86:248–249. [Google Scholar]
- 22.Hellenthal G, Stephens M. msHOT: Modifying Hudson's ms simulator to incorporate crossover and gene conversion hotspots. Bioinformatics. 2007;23:520–521. doi: 10.1093/bioinformatics/btl622. [DOI] [PubMed] [Google Scholar]
- 23.Schaffner SF, et al. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 2005;15:1576–1583. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.