Abstract
MicroRNAs (miRNAs) are a class of evolutionarily conserved small noncoding RNAs that are increasingly being recognized as important regulators of gene expression. The ribonuclease III enzyme Dicer is essential for the processing of miRNAs. CD1d-restricted invariant natural killer T (iNKT) cells are potent regulators of diverse immune responses. The role of Dicer-generated miRNAs in the development and function of immune regulatory iNKT cells is unknown. Here, we generated a mouse strain with a tissue-specific disruption of Dicer, and showed that lack of miRNAs after the deletion of Dicer by Tie2-Cre (expressed in hematopoietic cells and endothelial cells) interrupted the development and maturation of iNKT cells in the thymus and significantly decreased the number of iNKT cells in different immune organs. Thymic and peripheral iNKT cell compartments were changed in miRNA-deficient mice, with a significantly increased frequency of CD4+CD8+ iNKT cells in the thymus and a significantly decreased frequency of CD4+ iNKT cells in the spleen. MiRNA-deficient iNKT cells display profound defects in α-GalCer-induced activation and cytokine production. Bone marrow (BM) from miRNA-deficient mice poorly reconstituted iNKT cells compared to BM from WT mice. Also, using a thymic iNKT cell transfer model, we found that iNKT cell homeostasis was impaired in miRNA-deficient recipient mice. Our data indicate that miRNAs expressed in hematopoietic cells and endothelial cells are potent regulators of iNKT cell development, function, and homeostasis.
Keywords: T cell, thymus, galactosylceramide, bone marrow
Although only a small number of genetic transcripts (2–3%) code for proteins in higher organisms, a large fraction of the genome is transcribed. MicroRNAs (miRNAs) are a class of 21–25 nt single-stranded non-coding small RNAs that are transcribed from DNA but are not translated into protein. miRNAs are increasingly being recognized as important regulators of gene expression through the inhibition of effective mRNA translation via imperfect base pairing with the 3′-untranslated region (3′ UTR) of target mRNAs in animals. Primary miRNAs are transcribed from DNA segments and are then cleaved by Drosha (a nuclear enzyme) to form a premiRNA that is actively transported out of the nucleus. In the cytoplasm, the ribonuclease III enzyme Dicer cuts the hairpin loop to form the mature miRNA, which is incorporated into the RNA-induced silencing complex (RISC) to impede mRNA translation into protein (1–3). Dicer is required for the processing of mature and functional miRNAs. Therefore, the deletion of Dicer provides a genetic test for the relevance of miRNAs in mammalian development. Emerging evidence suggests that miRNA-mediated gene regulation represents a fundamental layer of genetic programming at the posttranscriptional level and has diverse functional roles in animals, including development, differentiation, and homeostasis (4–10).
Defining the role of Dicer-generated miRNAs in mammalian development is complicated by the embryonic lethality of constitutive Dicer knockouts in mice (10). The overall importance of miRNAs during hematopoiesis has been investigated by specific disruption of steps in miRNA biogenesis (4, 6, 7, 11–13). Using Cre-loxP tissue-specific Dicer deletion, Cobb et al. (11) and Muljo et al. (7) each reported that deletion of Dicer in early T cell development results in reduced thymocyte number and increased thymocyte susceptibility to cell death. Dicer-deficient helper T cells preferentially expressed IFN-γ, the hallmark effector cytokine of the Th1 lineage. Using a similar mouse model, Cobb et al. (4) further reported that deletion of Dicer in the thymus interrupted CD4+CD25+Foxp3+ regulatory T (Treg) cell development as well as reduced in vitro CD4+CD25+Foxp3+ Treg cell induction by TGF-β. In addition, Dicer-deficient mice spontaneously developed colitis and other inflammatory diseases, indicating a role for Dicer in regulation of the immune system. More recently, 2 groups reported that depletion of Dicer within the CD4+CD25+Foxp3+ Treg cell lineage resulted in fatal systemic autoimmune diseases, and that Dicer-deficient CD4+CD25+Foxp3+ Treg cells lose suppression activity in vivo (6, 13). Thus, the role of Dicer in immune regulation has clearly demonstrated that miRNAs are important regulators of the development and regulatory function of CD4+CD25+Foxp3+ Treg cells.
Natural killer T (NKT) cells comprise another major subset of regulatory T cells in mice and humans. NKT cells possess the properties of both T cells and NK cells as they co-express a rearranged T cell receptor (TCR) and several NK cell receptors, including NK1.1. Most NKT cells are restricted to the non-classical MHC-I like molecule CD1d and preferentially use an invariant TCR consisting predominantly of the Vα14-Jα18/Vβ8 pair in mice (14–16). These invariant NKT (iNKT) cells are almost uniformly reactive to the synthetic glycolipid ligand α-galactosylceramide (α-GalCer) presented by CD1d, and they can be identified using α-GalCer-loaded CD1d tetramers. iNKT cells are potent regulators of diverse immune responses, including the onset of cancer, infection, and autoimmune diseases (14–19). The role of miRNAs in iNKT cell development is currently unknown. Here, we tested the role of miRNAs in iNKT cell development by generating a mouse strain with Tie2 Cre mediated tissue-specific disruption of Dicer (Tie2 is expressed in hematopoietic progenitors and endothelial cells). We found that deficiency of miRNAs during hematopoiesis resulted in significantly fewer iNKT cells in the thymus, spleen, and liver, and interrupted the development and maturation of iNKT cells in the thymus. miRNA-deficient peripheral iNKT cells display profound defects in α-GalCer-induced activation and cytokine production. Bone marrow (BM) from miRNA-deficient mice poorly reconstituted iNKT cells compared to BM from wild-type (WT) mice. Further more, using a thymic iNKT cell transfer model, we found that WT iNKT cell homeostasis was impaired in miRNA-deficient recipient mice. Our data indicate that miRNAs expressed in hematopoietic cells and endothelial cells are potent regulators of iNKT cell development, function, and homeostasis.
Results and Discussion
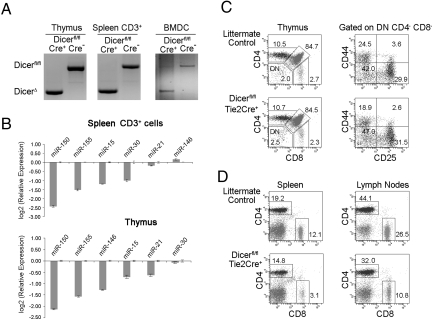
Ablation of Dicer Expression in Hematopoietic Stem Cells.
To investigate the role of miRNAs in immunological function, we generated a hematopoietic specific Dicer mutation mouse by mating loxP-flanked Dicer gene mutation Dicerfl/fl mice (9) with Tie2-Cre transgenic mice (20). Tie2 kinase is specifically expressed by hematopoietic progenitors and endothelial cells. The Tie2 gene promoter driving Cre expression was used to delete the genes in bone marrow and endothelial cells (20). We obtained mice homozygous for Dicerfl/fl with Tie2-Cre expression, which are conditional Dicer knockout (KO) mice. These mice are designated as Dicerfl/fltie2cre+ (Dicer KO) mice. Littermate Dicerfl/fltie2cre− mice were used as wild-type (WT) controls. There was substantial deletion of Dicer in hematopoietic cells, including thymocytes, CD3+ splenocytes, and bone marrow-derived dendritic cells in these mice (Fig. 1A). Taqman real-time PCR further indicated that miRNA gene expression was significantly reduced in the thymus and spleen CD3+ T cells, including miR-150, mir-155, and miR-15 (Fig. 1B). Dicerfl/fltie2cre+ mice were viable and fertile. We then examined the development of B cells, macrophages, and dendritic cells in the bone marrow. Dicerfl/fltie2cre+ mice showed no significant defects in the number and frequencies of macrophages, different stages of B cells, and bone marrow-derived dendritic cells compared to WT littermate controls. Analyses of T cell development in the thymus showed that the total number of thymocytes in Dicerfl/fltie2cre+ mice was comparable to that in Dicerfl/flTie2cre− WT littermate controls. Flow cytometry analyses demonstrated that the percentages of the thymic subsets, CD4 and CD8 single-positive (SP) T cells, CD4+CD8+ double-positive (DP) and CD4−CD8− double-negative (DN) T cells, were also normal compared to littermate controls (Fig. 1C Left). Early T cell precursors, CD4−CD8− DN T cells, progress through the CD44+CD25− (DN1) and the CD44+CD25+ (DN2) stages to the CD44−CD25+ (DN3) stage. To further dissect the early T cell development profile in Dicerfl/flTie2cre+ mice, we gated DN thymocytes and analyzed their CD44 and CD25 expression profiles. As shown in Fig. 1C Right, the Dicerfl/flTie2cre+ mouse showed a DN thymocyte developmental profile comparable to the WT littermate control. Previous studies indicated that expression of Dicer in the thymus is required for the optimal maturation and homeostasis of peripheral T cells, particularly CD8+ T cells (4, 7, 11). Therefore, we next analyzed peripheral lymphocytes. As shown in Fig. 1D, lymphocytes in spleen and lymph nodes showed a marked reduction of CD8+ T cells (4- and 2-fold reduction, respectively) with a smaller reduction in CD4+ cells in Dicerfl/flTie2cre+ mice. Thus, our data lend further support to previous studies (4, 7, 11), as deletion of Dicer in hematopoietic progenitors did not significantly affect thymoctye development but did influence peripheral CD8 T cell homeostasis. In addition, we found that Dicerfl/flTie2cre+ mice had significantly reduced CD4+CD25+Foxp3+ Treg cells in the thymus, spleen and lymph nodes compared to WT mice. This observation supports recent findings that miRNAs are required for CD4+CD25+Foxp3+ Treg cell development (4, 6, 13). Taken together, the immune phenotypes in Dicerfl/flTie2cre+ mice are very similar to Dicerfl/flCD4cre+ or Dicerfl/flLckcre+ mice (4, 7, 11).
Fig. 1.
Ablation of dicer expression in hematopoietic stem cells. (A) PCR typing of genomic DNA isolated from thymocytes, CD3+ T cells, and bone marrow-derived dendritic cells (BMDC) of Dicerfl/flTie2cre+ and Dicerfl/flTie2cre− mice. The deletion Dicer allele produced a 471-bp PCR product whereas the WT allele resulted in a 1,300-bp product. (B) Micro RNA gene expressions in thymus and spleen CD3+ cells by Taqman real-time PCR. Results were the average measured in duplicate and normalized to a control gene (snoRNU 202). Error bars are SD. The expression of miRNAs in thymus and splenic T cells was significantly reduced in Dicerfl/flTie2cre+ mice compared to Dicerfl/flTie2cre− mice. (C) Flow cytometric analysis of thymocytes from Dicerfl/flTie2cre+ and Dicerfl/flTie2cre− littermates. Contour plots depict CD4 (y axis) versus CD8 (x axis) staining profiles in thymus cells (Left), CD44 (y axis) versus CD25 (x axis) staining profile in gated CD4−CD8− DN thymus cells (Right). (D) Flow cytometric analyses of spleen (Right) and lymph nodes (Left) from Dicerfl/flTie2cre+ and Dicerfl/flTie2cre− littermates. Percentages of cells in each quadrant are indicated. Data are representative of 3 independent experiments (2–3 mice per group).
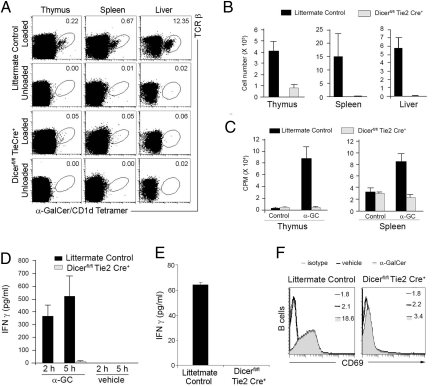
Number of iNKT Cells Is Reduced in Dicerfl/flTie2cre+ Mice.
To assess the role of miRNAs in iNKT cell development, we first examined the frequency and number of iNKT cells in the different hematopoietic organs of Dicerfl/flTie2cre+ mice and WT littermate controls by flow cytometry analysis. The percentages of iNKT cells stained by anti-TCRβ and α-GalCer-loaded CD1d tetramer in the liver, spleen, and thymus from Dicerfl/flTie2cre+ mice were severely decreased compared with littermate controls, with an almost complete absence in the liver (Fig. 2A). We further confirmed that the observed defect is due neither to Cre expression in hematopoeitic progenitor cells nor to Dicer-loxp insertion (Fig. S1). Consistent with the decreased proportions of iNKT cells in Dicerfl/flTie2cre+ mice, the absolute numbers of iNKT cells in the liver, spleen, and thymus were drastically reduced in Dicerfl/flTie2cre+ mice compared to WT littermate controls (Fig. 2B). Thus, Dicerfl/flTie2cre+ mice lack CD1d-restricted iNKT cells.
Fig. 2.
The number of iNKT cells is reduced in Dicerfl/flTie2re+ mice. iNKT cells were analyzed by flow cytometory in the thymus, spleen, and liver of Dicerfl/flTie2cre+ and WT littermates (Dicerfl/flTie2cre−) (2 to 3 mice per group, 4–8 weeks old). (A) Cells isolated from the indicated tissues of Dicerfl/flTie2cre+ and littermate control mice were co-stained with α-GalCer-loaded CD1d tetramers and a TCRβ-specific mAb. The numbers of iNKT cells are shown as a percentage of gated B220− lymphocytes from spleen and liver, and of total lymphocytes from thymus. (B) Absolute numbers of iNKT cells. Using the total cell count obtained from each organ and the proportion of iNKT cells (as shown in A), the absolute numbers of iNKT lymphocytes were determined. Data (mean ± SD) are representative of 4 separate experiments in which 5 to 8 mice of each strain were analyzed. (C) Defective proliferation of thymic and splenic iNKT cells. Thymocytes and splenocytes from Dicerfl/flTie2cre+and littermate control mice were cultured with 100 ng/mL α-GalCer for 48 h. Proliferation of cells was assessed by [3H]thymidine incorporation. Spontaneous proliferation in the absence of α-GalCer was similar with both splenocytes and thymocytes from Dicerfl/flTie2cre+and littermate control mice. Data are mean ± SD, representative of 3 independent experiments. (D) Impaired α-GalCer-dependent iNKT cell responses in vivo in Dicerfl/flTie2cre+ mice. Dicerfl/flTie2cre+ and littermate control mice were injected with 2 μg of α-GalCer or vehicle. Serum was collected at 2 and 5 h for detection of IFN-γ by ELISA. (E) Defective cytokine production of thymocytes from Dicerfl/flTie2cre+ mice. Thymocytes from Dicerfl/flTie2cre+ and littermate control mice were cultured with 100 ng/mL α-GalCer. The culture supernatant was collected at 24 h for detection of IFN-γ by ELISA. (F) The expression of CD69 by splenic B cells was analyzed 5 h after α-GalCer injection. CD69 mean fluorescence intensity (MFI) is indicated.
iNKT cells are known to proliferate and produce IFN-γ and IL-4 upon engagement of their invariant TCR with CD1d-presented α-GalCer (17, 21). Stimulation of splenocytes and thymocytes from WT littermate control mice with α-GalCer resulted in robust cell proliferation. In contrast, no significant cell proliferation was observed with Dicerfl/flTie2cre+ thymocytes and splenoytes (Fig. 2C). Consistent with the proliferation results, robust production of IFN-γ was induced after α-GalCer stimulation in vivo and in vitro from WT littermate control mice, but not from Dicerfl/flTie2cre+ mice (Fig. 2 D and E). Up-regulation of CD69 has been demonstrated in B cells, a consequence of the iNKT-induced cytokine storm after α-GalCer stimulation (22). However, we did not detect significant up-regulation of CD69 in B cells from Dicerfl/flTie2cre+ mice compared to WT littermate control mice (Fig. 2F). Taken together, these results further indicate that the compartment of iNKT cells was functionally impaired in the absence of miRNAs in hematopoietic stem cells.
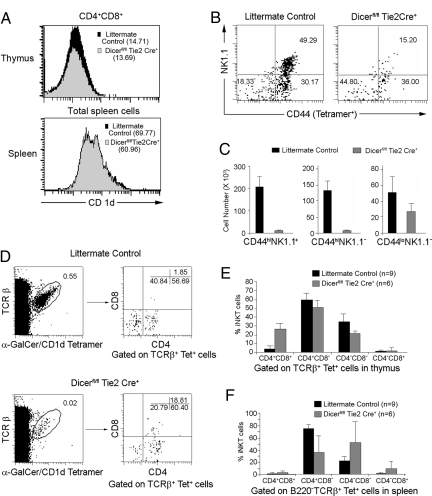
The iNKT Cell Development and Maturation Are Impaired in Dicerfl/flTie2cre+ Mice.
During iNKT cell development in the thymus, CD1d-restricted iNKT cell precursors primarily acquire the V14α-Jα18/Vβ8 TCR that allows their subsequent selection by the CD1d-presented self-glycolipid expressed by CD4+CD8+ thymocytes (23). To ascertain that the defect of iNKT cells was not due to defective CD1d expression, thymocytes and splenocytes from Dicerfl/flTie2cre+ mice and littermate controls were compared for CD1d expression and were found to be almost equivalent (Fig. 3A). Thus, we conclude that loss of CD1d expression does not account for the defect of iNKT cells observed in Dicerfl/flTie2cre+ mice. After positive selection, iNKT precursors progress through CD44lo and CD44hi stages and lastly acquire NK1.1 expression during their final maturation (23–25). Therefore, the CD44/NK-1.1 profiles of tetramer+ thymocytes were analyzed to determine if iNKT cell development and maturation were defective in Dicerfl/flTie2cre+ mice. The frequencies of mature CD44hiNK-1.1+ iNKT cells were significantly reduced by >3-fold in Dicerfl/flTie2cre+ mice, while the percentage of immature CD44loNK-1.1− iNKT cells was increased by 2-fold (Fig. 3B). Differences between Dicerfl/flTie2cre+ mice and littermate controls were even larger when the absolute numbers of the different subpopulations were compared (Fig. 3C). A previous study indicated that the most immature iNKT cells found in the thymus express heat stable antigen (HAS)high, while mature iNKT cells are NK1.1+Tet+HSAlow (26). As shown in Fig. S2, the majority of thymic iNKT cells in Dicerfl/flTie2cre+ mice were indeed immature Tet+HSAhigh, and few mature NK1.1+Tet+HSAlow iNKT cells were detected in Dicerfl/flTie2cre+ mice. Collectively, these results identify a severe impairment in iNKT cell development and maturation at the CD44low to CD44hi and NK1.1− to NK1.1+ checkpoints in Dicerfl/flTie2cre+ mice.
Fig. 3.
Abnormal development of iNKT cells in Dicerfl/flTie2cre+ mice. (A) CD1d expression on CD4CD8 DP thymocytes and total splenocytes from Dicerfl/flTie2cre+ and littermates were compared. CD1d MFI is shown in parentheses. Data are representative of 3 independent experiments. (B) CD44 and NK1.1 expression in thymic iNKT cells. Events shown are gated on TCRβ and CD1d tetramer double-positive cells. (C) Absolute number of iNKT cell subpopulations (mean ± SD) based on their CD44 and NK1.1 expression patterns. (D–F) The iNKT cell subset distribution in thymus and spleen are abnormal in Dicerfl/flTie2cre+ mice. TCRβ and CD1d tetramer double-positive cells were analyzed in the thymus (D and E) and spleen (F) of Dicerfl/flTie2cre+ and littermates and the relative frequencies of subsets defined by CD4 and CD8 expression were assessed (mean ± SD).
Thymic and peripheral iNKT cell compartments are composed of well-defined subsets, and the majority are CD4+ and CD4−CD8− DN iNKT cells. The existence of very rare α-Galcer CD1d tetramer-positive DP thymocytes is controversial (26, 27). We next examined these subsets on gated α-GalCer CD1d tetramer-positive iNKT cells in the thymus and spleen. Surprisingly, in the thymus (Fig. 3 D and E), a dramatic increase in frequency of CD4+CD8+ iNKT cells (26.64 ± 6.67%, n = 6) was seen in Dicerfl/flTie2cre+ mice compared to littermate controls (4.18 ± 3.2%, n = 9), P = 0.00016, while a significantly decreased frequency of CD4−CD8− iNKT cells (21.16 ± 3.5% vs. 34.95 ± 8.49%, P = 0.0011) was observed. An increase of CD4+CD8+ iNKT cells in the thymus is further suggestive of an early block in iNKT cell development (26). As shown in Fig. S2, most immature Tet+HSAhigh iNKT cells were CD4+CD8+ and the CD4−CD8− iNKT cells were significantly reduced in Tet+HSAlow iNKT cells in Dicerfl/flTie2cre+ mice, suggesting a defect in iNKT cell development from CD4+CD8+ to CD4+ iNKT cells and from CD4+ to CD4−CD8− iNKT cells. A higher frequency of CD4+CD8+ iNKT cells in Dicerfl/flTie2cre+ mice was also seen in the spleen (Fig. 3F, P = 0.0001). Strikingly, there was more than a 2-fold reduction of splenic CD4+ iNKT cells in Dicerfl/flTie2cre+ mice compared to control mice (P = 1.9E−10). Together, these results suggest that the development of iNKT cell subsets in the thymus and their maintenance in the periphery are affected by the absence of miRNAs in the hematopoietic progenitors.
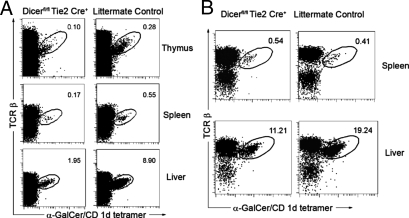
Micro RNAs Expressed in Bone Marrow and Endothelial Cells Regulate iNKT Cell Development and Homeostasis.
Accumulated studies suggest that iNKT cell development and function are regulated by both the bone marrow and microenvironments. Although the findings from the Dicerfl/flTie2cre+ mice are suggestive of a cell-intrinsic (hematopoietic cell) defect in iNKT development, these do not exclude the possible role of subtle yet important defects in the microenvironment of the Dicerfl/flTie2cre+ mouse, since the Tie2 gene promoter drives Cre expression in both bone marrow and endothelial cells. Recent studies indicate that the maintenance and regulation of endogenous miRNA levels via Dicer mediated processing is also critical for endothelial cell gene expression and function (28, 29). The reduced number of iNKT cells in the Dicerfl/flTie2cre+ mouse may reflect either a defect in iNKT cell precursors, positive selection and maturation of iNKT cells, and/or a block in iNKT cell homeostasis in the peripheral tissues, which is regulated by both the bone marrow and microenvironments. Initially, we used a bone marrow transfer model to further investigate if a Tie2-mediated Dicer deficiency influences iNKT cell development via a stem cell-intrinsic mechanism. Lethally irradiated normal C57/B6 mice were reconstituted with donor Dicerfl/flTie2cre+ mouse or Dicerfl/flTie2cre− littermate bone marrow. As shown in Fig. 4A, the bone marrow from Dicerfl/flTie2cre+ mice poorly reconstituted iNKT cells in the thymus, spleen, and liver compared to bone marrow from littermate controls, suggesting a cell-intrinsic defect in Dicerfl/flTie2cre+ mice. Interestingly, slightly recovered iNKT cells were detected in mice that received Dicerfl/flTie2cre+ BM compared to unmanipulated Dicerfl/flTie2cre+ mice, particularly in the liver, indicating the miRNA expression in endothelial cells may also regulate iNKT cell trafficking and/or homeostasis. Recent studies indicate that lymphocyte trafficking to lymphoid organs or non-lymphoid tissues is governed in large part by the interactions of selectively expressed adhesion receptors and chemokine receptors on the surface of diverse lymphocytes with ligands on the endothelium, including CXCR6/CXCL16 interaction for iNKT trafficking (30). To further test an extrinsic defect on iNKT cell trafficking in Dicerfl/flTie2cre+ mice, a thymic iNKT cell transfer experiment was performed in which CD8-depleted thymus cells (iNKT cells enriched up to 10-fold) from B6.CD45.1 congenic mice were transferred to irradiated Dicerfl/flTie2cre+ mice and WT littermate (CD45.2 receipts). The CD45.1+ lymphocytes from the spleen and liver were analyzed for their frequency of iNKT cells at day 5 after transfer. As shown in Fig. 4B, the iNKT cells were moderately reduced in the liver but not in the spleen from Dicerfl/flTie2cre+ recipient mice compared to their WT littermates. Interestingly, CD4+ iNKT cells were reduced in spleen from Dicerfl/flTie2cre+ recipient mice (Fig. S3), which may provide a possible explanation why CD4+ iNKT cell frequency was largely decreased in the spleen of Dicerfl/flTie2cre+ mice (Fig. 3F), while a relatively normal population of CD4+ iNKT cells is still present in the thymus (Fig. 3E). These data suggest that a lack of miRNAs in endothelial cells may affect iNKT cell homeostasis. Therefore, the defective iNKT cells observed in Dicerfl/flTie2cre+ mice may result from both intrinsic (hematopoietic cells) and extrinsic (endothelia cells) defects.
Fig. 4.
The defect of iNKT cells in Dicer deletion mice are both cell intrinsic and extrinsic. (A) B6 lethally irradiated hosts (4 to 5 mice per group) were reconstituted with Dicerfl/flTie2cre+ or littermate control bone marrow cells. Eight weeks later, host thymus, spleen and liver were examined by staining with α-GalCer/CD1d tetramers and anti-TCRβ antibody. (B) CD8-depleted thymocytes (up to 4% of recovered thymocytes were Tetramer+iNKT cells) from CD45.1-congenic mice were transferred to irradiated 6 week-old Dicerfl/flTie2cre+ or littermate control mice (2 to 3 mice per group). The CD45.1+ lymphocytes from the spleen and liver were analyzed for iNKT cells at day 5 after transferring. The percentages of iNKT cells are shown. Data are representative of 2–3 experiments.
Impaired iNKT Cell Function in Dicerfl/flTie2cre+ Mice.
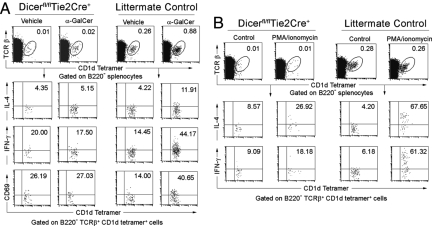
One unique feature of iNKT cells is the prompt production of large amounts of cytokines in response to TCR signaling. The lack of proliferation and cytokine response to α-GalCer in Dicerfl/flTie2cre+ mice could be explained by the markedly reduced numbers of iNKT cells. In addition, Dicerfl/flTie2cre+ iNKT cells may have an intrinsic defect precluding them from activation and producing cytokines. To test the latter possibility, we again injected mice i.v. with α-GalCer and examined the production of cytokines by iNKT cells with intracellular cytokine staining. As shown in Fig. 5A, the number of splenic iNKT cells that express intracellular IL-4 and IFN-γ were increased 2- to 3-fold in WT littermate controls after in vivo α-GalCer stimulation (40 min), while splenic iNKT cells from Dicerfl/flTie2cre+ mice had almost no response to α-GalCer stimulation. The decreased cytokine production response to α-GalCer stimulation in Dicer-deficient mice could result from either a defect in cytokine synthesis or TCR signaling, thereby affecting the activation of iNKT cells. We therefore further investigated whether Dicer-deficient iNKT cells are activated normally upon i.v. α-GalCer stimulation. As shown in Fig. 5A Bottom, α-GalCer stimulation resulted in up-regulation of CD69 expression, a downstream marker of TCR signaling, in iNKT cells from WT mice. In contrast, α-GalCer stimulation did not significantly up-regulate CD69 expression in Dicer-deficient iNKT cells. Those observations strongly argue that miRNA deficiency leads to a defect in TCR signaling. To further localize the block in TCR-mediated signal transduction targeted by miRNAs, we stimulated Dicerfl/flTie2cre+ iNKT cells in vitro with phorbol myristate acetate (PMA) and ionomycin, which bypass proximal TCR-mediated signaling events and activates cells most likely at a stage proximal to protein kinase C and calcium flux (31). Three hours after stimulation with PMA and ionomycin, >60% of splenic iNKT cells from WT control mice stained positive for IL-4 and IFN-γ, respectively, while only 26.9% and 18.2% of Dicerfl/flTie2cre+ iNKT cells stained positive for IL-4 and IFN-γ, respectively (Fig. 5B). Taken together, miRNAs seem to be critical for iNKT cell activation and cytokine production.
Fig. 5.
Impaired iNKT cell function upon in vivo α-GalCer treatment and in vitro PMA/ionomycin stimulation. (A) IL-4 and IFN-γ production by splenic iNKT cells was analyzed after α-GalCer or vehicle injection by intracellular cytokine staining. Events shown are gated on B220-negative, TCRβ-positive, CD1d tetramer-positive events. α-GalCer or vehicle-stimulated iNKT cells were also analyzed for the expression of the activation marker CD69. (B) Whole splenocytes of Dicerfl/flTie2cre+ and littermate control mice were treated with or without PMA and ionomycin for 3 h in vitro. The production of IL-4 and INF-γ by splenic iNKT cells were analyzed with intracellular cytokine staining. Data are representative of 3 experiments.
In summary, depletion of miRNAs by eliminating Dicer at the bone marrow stem cell stage significantly reduces the frequencies of iNKT cells, but seems to be dispensable for early thymocyte development and CD4/CD8 lineage commitment. Dicer-generated miRNAs in hematopoietic cells are required for iNKT cell development, maturation, and function. Defects in the number and function of iNKT cells may also contribute to autoimmunity in Dicerfl/flTie2cre+ mice. In addition, miRNAs expression in endothelial cells regulates iNKT cell homeostasis in the liver. The identification of specific miRNAs differentially required for the proper development, function, and homeostasis of iNKT cells and the downstream genes targeted by miRNAs may further unravel the immunological and molecular mechanisms underlying iNKT cell development, and may also facilitate the development of new intervention strategies for cancer and autoimmune diseases related to iNKT cell function.
Materials and Methods
Mice.
Mice carrying a conditional floxed allele of Dicer (Dicerflox) (9) were backcrossed onto the C57BL/6 background for 5 generations and then mated to C57BL/6 mice carrying the Tie2 Cre allele (obtained from The Jackson Laboratory) (20) to generate Dicerflox/flox Tie-2cre/+ conditional knockout mice, designated as Dicerfl/flTie2cre+. All WT mice (Dicerfl/flTie2cre−), unless indicated otherwise, are littermate controls of Dicerfl/flTie2cre+ mice. Experiments were conducted at 4–8 weeks of age, unless otherwise indicated. Mice were housed in a specific pathogen-free barrier unit. Handling of mice and experimental procedures were in accordance with requirements of the Institutional Animal Care and Use Committee.
Genotyping.
Offspring were genotyped using the following PCR primer pairs: for Cre, 5′-TGATGAGGTTCGCAAGAACC-3′ and 5′-CCATGAGTGAACGAACCTGG-3′ (product size: 420 bp); and for Dicer, 5′-CCTGACAGTGACGGTCCAAAG-3′ (DicerF1) and 5′-CATGACTCTTCAACTCAAACT-3′ (product sizes: 420 bp from the Dicerflox allele and 351 bp from the wild-type Dicer allele). The deletion allele was genotyped using primers DicerF1 and DicerDel (5′-CCTGAGCAAGGCAAGTCATTC-3′). The deletion allele produced a 471-bp PCR product whereas the WT allele resulted in a 1,300-bp product.
Flow Cytometry.
Single-cell suspensions were washed twice with staining buffer (PBS, 2% FCS) and incubated with Fc Block (clone 2.4G2). Cells were stained with α-GalCer/CD1d tetramers as described (17). The following conjugated monoclonal antibodies (mAbs) were used: NK1.1 (PK136), TCRβ (H57–597), CD44 (IM7), CD1d (1B1), FoxP3 (FJK-16s), CD69 (H1.2F3), CD8 (53–6.7), CD4 (RM4–5), CD25 (PC61), IL-4 (11B11), and IFN-γ (XMG1.2). All mAbs were from BD Biosciences or eBioscience. Data were analyzed using CELLQuest Pro software (BD Biosciences).
Cell Proliferation Assay.
Spleen and thymus cell suspensions were incubated in complete medium supplemented with or without 100 ng/mL of α-GalCer (Alexis). After 30 h in culture, [3H]thymidine was added and the culture continued for an additional 18 h. Cells were then harvested and counted in a microbetaplate counter (Wallac).
In Vivo αGalCer-Induced Activation Assays.
Two micrograms of α-GalCer or vehicle in 100 μL of PBS were injected into the tail vein. For serum cytokine determination, mice were euthanized and bled at 2 and 5 h. The blood was allowed to clot, and the serum was separated from the clot by centrifugation. IL-4 and IFN-γ were detected by sandwich ELISA using capture and biotinylated antibodies (Abs) from R & D Systems. For secondary stimulation of B cells, splenocytes were collected 5 h after α-GalCer injection and stained with anti-CD69 and anti-B220. For intracellular cytokine staining, splenocytes were collected at 40 min after injection and cultured in T cell medium (RPMI 1640 with 10% FCS, Hepes, penicillin and streptomycin, pyruvate, nonessential amino acids, L-glutamine, and 2-ME). Monensin was added to a final concentration of 3 μM, and the cells were incubated for an additional hour. Cells were extracellularly stained with anti-TCRβ and CD1d-tetramer. After washing and fixing with 2% paraformaldehyde, cells were permeabilized with 0.1% saponin and stained with anti-IFN-γ and anti-IL4 Abs and analyzed by flow cytometry.
Bone Marrow and iNKT Cell Transfer Experiments.
To generate bone marrow chimeras, 6–8 week-old female recipient mice (C57/B6) were lethally irradiated initially with 900 rads. Donor bone marrow was harvested from 4 to 6 week-old Dicerfl/flTie2cre+ or littermate control mice by flushing with a syringe containing sterile tissue culture medium. After erythrocyte lysis, mature T cells were depleted by biotin-conjugated anti-mouse CD3 (BD Biosciences) mAbs and anti-biotin magnetic beads (Miltenyi Biotec) using an AutoMACS sorter (Miltenyi Biotec.). Depletion of >90% mature T cells was confirmed by flow cytometry. Donor bone marrow cells after T cell depletion (5 × 106 cells per mouse, in a volume of 100 μL) were then injected into irradiated recipients by the intraocular vein in the retro-orbital plexus. The chimeras were analyzed 8 weeks after reconstitution for iNKT cell development. For iNKT cell transfer, CD8 depleted thymocytes (3–4% of recovered thymocytes were Tetramer+ iNKT cells) from CD45.1-congenic mice were transferred to irradiated 6 week-old Dicerfl/flTie2cre+ or littermate control mice. The CD45.1+ lymphocytes from the thymus, spleen, and liver were analyzed for iNKT cells at day 5 after transfer.
Real-Time RT-PCR for miRNA Expression.
Total thymic cells and splenic CD3+ T cell RNA from 4 to 6 week-old Dicerfl/flTie2cre+ or littermate control mice, respectively, were harvested using the Ambion mirVana miRNA isolation kit (Ambion) according to the manufacturer's instructions. The expression levels of miR-150, miR-155, miR-15, miR-30, miR-21, and miR-146 were examined using the Applied Biosystems TaqMan MicroRNA Assay kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. The kit uses gene-specific stem-loop reverse transcription primers and TaqMan probes to detect mature miRNA transcripts. PCR amplification was carried on the Applied Biosystems 7900 Real-Time PCR system. The assay was run in duplicate for each case to allow for assessment of technical variability. snoRNU202 was used as endogenous control. Relative quantitation using the ΔΔct method in Dicerfl/flTie2cre+ versus littermate control mice was carried out and fold changes were calculated for each gene.
Statistics.
Data were analyzed by the 2-tailed Student's t test. The differences were considered significant with a P < 0.05.
Supplementary Material
Acknowledgments.
We thank Dr. T. L. Delovitch for critical reading of the manuscript, Dr. M. McManus (University of California, San Francisco, CA) for Dicer-loxP mice, and the National Institutes of Health tetramer facility for CD1d-tetramer. Q.-S.M. was supported by Juvenile Diabetes Research Foundation International Grants 1-2005-039, 5-2006-918, and 5-2006-403, and American Diabetes Association Grant 7-05-JF-30.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811119106/DCSupplemental.
References
- 1.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Gupta M, Brewer G. MicroRNAs: New players in an old game. Proc Natl Acad Sci USA. 2006;103:3951–3952. doi: 10.1073/pnas.0601268103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen CZ, Lodish HF. MicroRNAs as regulators of mammalian hematopoiesis. Semin Immunol. 2005;17:155–165. doi: 10.1016/j.smim.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Cobb BS, et al. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuellar TL, et al. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc Natl Acad Sci USA. 2008;105:5614–5619. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liston A, et al. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med. 2008;205:1993–2004. doi: 10.1084/jem.20081062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muljo SA, et al. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otsuka M, et al. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 9.Harfe BD, et al. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein E, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 11.Cobb BS, et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–1373. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asirvatham AJ, et al. MicroRNA targets in immune genes and the Dicer/Argonaute and ARE machinery components. Mol Immunol. 2008;45:1995–2006. doi: 10.1016/j.molimm.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X, et al. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med. 2008;205:1983–1991. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godfrey DI, Berzins SP. Control points in NKT-cell development. Nat Rev Immunol. 2007;7:505–518. doi: 10.1038/nri2116. [DOI] [PubMed] [Google Scholar]
- 15.Kronenberg M, Engel I. On the road: Progress in finding the unique pathway of invariant NKT cell differentiation. Curr Opin Immunol. 2007;19:186–193. doi: 10.1016/j.coi.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 16.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 17.Sharif S, et al. Activation of natural killer T cells by alpha-galactosylceramide treatment prevents the onset and recurrence of autoimmune Type 1 diabetes. Nat Med. 2001;7:1057–1062. doi: 10.1038/nm0901-1057. [DOI] [PubMed] [Google Scholar]
- 18.Miyake S, Yamamura T. NKT cells and autoimmune diseases: Unraveling the complexity. Curr Top Microbiol Immunol. 2007;314:251–267. doi: 10.1007/978-3-540-69511-0_10. [DOI] [PubMed] [Google Scholar]
- 19.Van KL. NKT cells: T lymphocytes with innate effector functions. Curr Opin Immunol. 2007;19:354–364. doi: 10.1016/j.coi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Koni PA, et al. Conditional vascular cell adhesion molecule 1 deletion in mice: Impaired lymphocyte migration to bone marrow. J Exp Med. 2001;193:741–754. doi: 10.1084/jem.193.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharif S, et al. Regulation of autoimmune disease by natural killer T cells. J Mol Med. 2002;80:290–300. doi: 10.1007/s00109-002-0332-8. [DOI] [PubMed] [Google Scholar]
- 22.Kitamura H, et al. alpha-galactosylceramide induces early B-cell activation through IL-4 production by NKT cells. Cell Immunol. 2000;199:37–42. doi: 10.1006/cimm.1999.1602. [DOI] [PubMed] [Google Scholar]
- 23.Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol. 2001;2:971–978. doi: 10.1038/ni710. [DOI] [PubMed] [Google Scholar]
- 24.Matsuda JL, Gapin L. Developmental program of mouse Valpha14i NKT cells. Curr Opin Immunol. 2005;17:122–130. doi: 10.1016/j.coi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 25.Townsend MJ, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 26.Benlagha K, et al. Characterization of the early stages of thymic NKT cell development. J Exp Med. 2005;202:485–492. doi: 10.1084/jem.20050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pellicci DG, et al. A natural killer T (NKT) cell developmental pathway iInvolving a thymus-dependent NK1.1(-)CD4(+) CD1d-dependent precursor stage. J Exp Med. 2002;195:835–844. doi: 10.1084/jem.20011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79:581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- 29.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 30.Geissmann F, et al. Intravascular immune surveillance by CXCR6+ NKT cells patrolling liver sinusoids. PLoS Biol. 2005;3:e113. doi: 10.1371/journal.pbio.0030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim PJ, et al. GATA-3 regulates the development and function of invariant NKT cells. J Immunol. 2006;177:6650–6659. doi: 10.4049/jimmunol.177.10.6650. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.