Abstract
Leishmania species of the subgenus Viannia and especially Leishmania braziliensis are responsible for a large proportion of New World leishmaniasis cases. The reproductive mode of Leishmania species has often been assumed to be predominantly clonal, but remains unsettled. We have investigated the genetic polymorphism at 12 microsatellite loci on 124 human strains of Leishmania braziliensis from 2 countries, Peru and Bolivia. There is substantial genetic diversity, with an average of 12.4 ± 4.4 alleles per locus. There is linkage disequilibrium at a genome-wide scale, as well as a substantial heterozygote deficit (more than 50% the expected value from Hardy−Weinberg equilibrium), which indicates high levels of inbreeding. These observations are inconsistent with a strictly clonal model of reproduction, which implies excess heterozygosity. Moreover, there is large genetic heterogeneity between populations within countries (Wahlund effect), which evinces a strong population structure at a microgeographic scale. Our findings are compatible with the existence of population foci at a microgeographic scale, where clonality alternates with sexuality of an endogamic nature, with possible occasional recombination events between individuals of different genotypes. These findings provide key clues on the ecology and transmission patterns of Leishmania parasites.
Keywords: clonality, microsatellites, population genetics, endogamyl, heterozygote defiency
Leishmaniases are worldwide vector-borne diseases of humans and domestic animals, caused by protozoan parasites of the genus Leishmania. These parasitoses are a serious public health problem, with about 350 million persons at risk and 2,357,000 new cases per year (1). Leishmaniases occur on all continents except Antarctica. There are more than 20 described species causing human infections (review in ref. 2). Clinical symptoms range from asymptomatic, cutaneous, and mucocutaneous to visceral forms, depending on the Leishmania species. Visceral leishmaniasis is mainly caused by species from the Leishmania donovani complex; cutaneous and mucocutaneous forms are associated with species from the Viannia and Leishmania subgenera (3–5). L. braziliensis causes cutaneous and mucocutaneous leishmaniases in South America, where these are a severe public health problem.
Despite numerous studies and recent advances in the molecular genetics of these organisms, the reproductive mode of these parasites remains unsettled. Tibayrenc and Ayala (6) proposed that all (or most) Leishmania species are clonal. Other authors have challenged this hypothesis, based on pulse field gel electrophoresis (PFGE) data, and argued that some Leishmania species are potentially automictic, with frequent genetic exchanges (7). Several studies suggest that recombination may occur in Leishmania, and that other complexities may exist (see review in ref. 2). For example, based on evidence from PFGE analyses, Bañuls et al. (8) have proposed the occurrence of pseudorecombination in Leishmania populations. Moreover, several genetic studies indicate genetic recombination between Leishmania individuals, despite lack of evidence for a sexual stage (9–16). In any case, the molecular data suggest that, after a hybridization event, hybrids propagate clonally in natural populations (9, 12).
The prevailing hypothesis is that Leishmania displays a clonal mode of reproduction with occasional pseudorecombination and intragenic recombination, which mimic sexual reproduction processes, and that infrequent genetic exchanges take place in wild populations. Nevertheless, much remains to be elucidated as this interpretation is challenged by certain data, such as the absence of large excess in heterozygosity, as expected in clonal diploids (17, 18), and the lack of a clear structure in individualized lineages at the intraspecific level (2). Indeed, in a clonal model, an excess of heterozygotes and significant linkage disequilibrium are expected. Thus, the known results have failed to resolve the issue of clonality vs. sexuality in these protozoan parasites. Improved knowledge of the population structure and reproductive strategy of Leishmania parasites would provide a better understanding of their transmission patterns, as well as useful information for diagnostic purposes, epidemiological surveys, and drug and vaccine development.
Microsatellite loci are highly polymorphic, codominant, abundant throughout the genome, and relatively easy to assay (19, 20). In Leishmania, microsatellite studies are relatively recent; a small number of polymorphic microsatellites have been described for Leishmania species of the Viannia subgenus and especially for L. braziliensis (21). We analyze the population structure of L. braziliensis in several natural populations from South America (Peru and Bolivia), based on 12 microsatellite loci previously described (22). Peru and Bolivia are 2 of 7 world countries that report 90% of cutaneous leishmaniasis cases. Our population genetics analysis may be the first study of this kind for this Leishmania species. It reveals an unexpectedly high level of inbreeding within local samples, a large part of which is explained by local heterogeneity (Wahlund effect), probably due to a microgeographic population substructure, but also to the occurrence of mixed-mating events that include a significant contribution of endogamy (i.e., recombination between 2 genetically identical cells).
Results
We analyzed 124 human strains of L. braziliensis from 4 samples: 2 from the Pilcopata department in Peru, isolated in either 1993 or 1994, and 2 from Chapare Natural Park in Bolivia, isolated in either 1994 or 1998 (Tables 1 and 2). Both sites are located in the Amazonian forest and extend over large areas of great faunal and floral diversity.
Table 1.
Genetic diversity at 12 microsatellite loci in 124 strains of Leishmania braziliensis strains from 4 populations
| Locus | GenBank accession no. | Allele size, bp | N | Hs | FIS |
|---|---|---|---|---|---|
| AC01 | AF139110 | 198–212 | 8 | 0.707 | 0.576 |
| AC16 | AF139112 | 147–161 | 11 | 0.754 | 0.341 |
| AC52 | AF139111 | 098–126 | 22 | 0.914 | 0.501 |
| ARP | AF045249 | 121–157 | 18 | 0.874 | 0.441 |
| ITSbraz | AJ300483 | 100–108 | 6 | 0.603 | 0.923 |
| Ibh3 | AF044682 | 116–136 | 9 | 0.584 | 0.599 |
| LRC | BX544585 | 118–134 | 15 | 0.826 | 0.424 |
| CAK | BX544561 | 152–170 | 13 | 0.743 | 0.676 |
| EMI | BX541508 | 183–189 | 14 | 0.809 | 0.645 |
| LBA | BX539885 | 168–180 | 14 | 0.803 | 0.225 |
| GO9 | BX539509 | 148–168 | 10 | 0.673 | 0.466 |
| E11 | BX542509 | 096–108 | 9 | 0.715 | 0.618 |
| Mean ± SE | 12.4 ± 4.4 | 0.750 ± 0.100 | 0.537 ± 0.040 |
N, number of alleles; Hs, Nei's unbiased genetic diversity within subsamples (23); FIS, deviation from panmixia.
Table 2.
Data set with each sample code, the country, and the year of collection and all genotypes obtained at each locus by PCR
| Sample code | Country | Year | Loci |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AC01 | AC16 | AC52 | ARP | ITSbraz | Ibh3 | LRC | CAK | EMI | LBA | G09 | E11 | |||
| LC1568 | Peru | 1993 | 202–202 | 149–161 | 104–104 | 139–139 | 102–102 | 130–130 | 124–124 | 162–162 | 165–165 | 180–180 | 150–152 | 102–102 |
| LC2231 | Peru | 1994 | 202–202 | 149–155 | 084–098 | 139–145 | 106–106 | 130–130 | 132–132 | 162–162 | 189–189 | 178–178 | 150–154 | 096–096 |
| LC2280 | Peru | 1994 | 198–210 | 151–161 | 084–084 | 137–139 | 104–106 | 130–130 | 132–132 | 158–158 | 191–191 | 176–182 | 150–162 | 098–102 |
| LC2282 | Peru | 1994 | 206–212 | 149–149 | 100–100 | 151–153 | 102–102 | 128–128 | 120–126 | 162–162 | 165–165 | 176–180 | 156–156 | 096–096 |
| LC2291 | Peru | 1994 | 202–202 | 151–151 | 120–120 | 133–149 | 102–106 | 130–130 | 128–128 | 162–162 | 175–185 | 174–182 | 148–148 | 100–100 |
| LC2292 | Peru | 1994 | 204–210 | 149–149 | 084–120 | 139–139 | 104–104 | 130–130 | 132–132 | 160–160 | 189–189 | 174–180 | 148–152 | 096–096 |
| LC2293 | Peru | 1994 | 202–202 | 149–151 | 098–098 | 133–149 | 102–106 | 130–130 | 128–128 | 164–168 | 175–185 | 174–174 | 150–152 | 100–102 |
| LC2308 | Peru | 1994 | 204–210 | 151–169 | 100–100 | 149–149 | 100–102 | 116–130 | 120–128 | 160–160 | 185–185 | 174–180 | 154–154 | 098–100 |
| LC2310 | Peru | 1994 | 204–210 | 159–159 | 084–094 | 137–139 | 102–102 | 130–130 | 132–132 | 160–160 | 189–189 | 176–182 | 148–156 | 100–100 |
| LC2315 | Peru | 1994 | 202–210 | 151–161 | 108–108 | 141–141 | 102–102 | 130–130 | 130–134 | 162–162 | 189–189 | 176–182 | 154–154 | 100–104 |
| LC2316 | Peru | 1994 | 200–204 | 147–161 | 088–088 | 135–135 | 100–100 | 118–130 | 130–138 | 160–160 | 185–185 | 176–180 | 148–154 | 098–098 |
| LC2318 | Peru | 1994 | 202–202 | 149–167 | 092–108 | 139–139 | 102–102 | 130–130 | 130–130 | 162–162 | 177–195 | 174–182 | 152–152 | 100–100 |
| LC2319 | Peru | 1994 | 200–200 | 149–157 | 104–110 | 141–147 | 104–108 | 130–130 | 124–124 | 164–164 | 193–193 | 174–184 | 152–152 | 100–100 |
| LC2320 | Peru | 1994 | 202–210 | 149–155 | 118–118 | 155–155 | 104–104 | 128–128 | 132–132 | 162–162 | 189–189 | 174–182 | 150–154 | 102–102 |
| LC2321 | Peru | 1994 | 208–208 | 149–149 | 086–106 | 149–155 | 100–100 | 128–128 | 122–130 | 162–162 | 189–189 | 176–176 | 148–156 | 098–106 |
| LC2352 | Peru | 1994 | 202–212 | 149–161 | 110–110 | 143–143 | 102–104 | 130–130 | 126–130 | 162–162 | 191–191 | 178–180 | 150–154 | 098–098 |
| LC2353 | Peru | 1994 | 202–212 | 149–161 | 084–094 | 129–139 | 102–102 | 128–128 | 132–132 | 158–162 | 191–191 | 176–180 | 150–152 | 098–098 |
| LC2355 | Peru | 1994 | 210–210 | 149–159 | 098–122 | 129–137 | 102–102 | 128–128 | 132–132 | 158–160 | 189–189 | 176–180 | 150–150 | 100–100 |
| LC2367 | Peru | 1994 | 202–202 | 149–161 | 084–098 | 131–139 | 102–106 | 130–130 | 132–132 | 158–164 | 191–191 | 174–180 | 148–148 | 098–098 |
| LC2368 | Peru | 1994 | 202–202 | 149–161 | 084–098 | 135–141 | 102–104 | 130–130 | 132–132 | 158–162 | 191–191 | 174–180 | 148–148 | 100–100 |
| LC2369 | Peru | 1994 | 200–210 | 149–161 | 096–096 | 135–141 | 102–106 | 130–130 | 126–132 | 158–164 | 189–189 | 174–182 | 150–150 | 098–098 |
| LC2371 | Peru | 1994 | 200–200 | 151–161 | 084–098 | 135–141 | 100–106 | 130–130 | 132–132 | 158–164 | 189–189 | 174–180 | 148–148 | 098–098 |
| LC2373 | Peru | 1994 | 202–202 | 151–161 | 084–098 | 133–139 | 102–104 | 130–130 | 132–132 | 162–162 | 191–191 | 174–180 | 150–150 | 100–100 |
| LC2284 | Peru | 1994 | 210–210 | 151–161 | 086–116 | 139–139 | 102–102 | 130–130 | 130–130 | 158–160 | 189–189 | 174–180 | 150–152 | 100–102 |
| LC2322 | Peru | 1994 | 202–202 | 149–161 | 094–110 | 149–149 | 104–104 | 130–130 | 130–130 | 160–160 | 191–193 | 174–182 | 154–154 | 100–102 |
| CH12B | Bolivia | 1994 | 204–206 | 149–159 | 126–126 | 155–157 | 102–102 | 116–130 | 124–130 | 168–168 | 183–183 | 174–180 | 154–154 | 100–100 |
| CH15 | Bolivia | 1994 | 206–206 | 147–157 | 124–124 | 155–157 | 102–102 | 116–130 | 122–134 | 164–164 | 189–189 | 168–174 | 154–154 | 100–106 |
| CH17 | Bolivia | 1994 | 202–202 | 147–161 | 096–096 | 131–131 | 102–102 | 130–130 | 132–132 | 164–164 | 185–191 | 168–168 | 154–154 | 098–102 |
| CH25B | Bolivia | 1994 | 202–204 | 149–149 | 100–100 | 121–121 | 102–102 | 130–130 | 124–134 | 158–164 | 179–187 | 168–172 | 152–168 | 100–102 |
| CH29B | Bolivia | 1994 | 204–206 | 149–159 | 126–126 | 125–157 | 102–102 | 128–128 | 130–134 | 158–166 | 183–185 | 174–180 | 154–154 | 100–100 |
| CUM106 | Bolivia | 1994 | 204–206 | 147–159 | 100–100 | 129–131 | 102–102 | 128–128 | 124–128 | 164–164 | 185–189 | 174–174 | 152–154 | 100–100 |
| CUM107 | Bolivia | 1994 | 202–202 | 149–149 | 092–092 | 121–121 | 106–106 | 120–120 | 118–120 | 156–156 | 189–189 | 170–170 | 144–144 | 090–090 |
| CUM31 | Bolivia | 1994 | 206–206 | 149–159 | 126–126 | 135–137 | 102–102 | 116–128 | 122134 | 162–172 | 187189 | 168–174 | 154–154 | 102–108 |
| CUM38 | Bolivia | 1994 | 202–204 | 149–149 | 098–104 | 129–135 | 102–102 | 130–130 | 114134 | 160–160 | 185–185 | 168–168 | 154–154 | 102–102 |
| CUM41 | Bolivia | 1994 | 202–202 | 149–149 | 114–120 | 131–131 | 102–102 | 118–128 | 126–130 | 160–160 | 185187 | 168–172 | 154–154 | 100–102 |
| CUM50 | Bolivia | 1994 | 206–206 | 149–159 | 100–110 | 129–147 | 102–102 | 116–128 | 122–124 | 164–164 | 185–187 | 174–174 | 154–154 | 100–102 |
| CUM52 | Bolivia | 1994 | 206–206 | 149–149 | 100–110 | 129–147 | 102–102 | 116–128 | 122–124 | 164–164 | 185–187 | 174–174 | 154–154 | 100–102 |
| CUM53 | Bolivia | 1994 | 206–206 | 149–149 | 112–112 | 129–147 | 104–104 | 116–116 | 122–132 | 164–170 | 185–189 | 174–174 | 154–154 | 098–104 |
| CUM55 | Bolivia | 1994 | 206–206 | 149–149 | 126–126 | 137–137 | 102–102 | 128–130 | 114–130 | 158–170 | 187–189 | 162–168 | 154–154 | 098–102 |
| CUM59 | Bolivia | 1994 | 206–206 | 149–159 | 124–124 | 129–129 | 102–102 | 136–136 | 122–134 | 162–174 | 187–189 | 168–174 | 154–154 | 102–108 |
| CUM65 | Bolivia | 1994 | 202–202 | 149–159 | 114–114 | 129–137 | 102–102 | 116–128 | 122–130 | 164–164 | 179–185 | 174–174 | 154–154 | 102–102 |
| CUM67 | Bolivia | 1994 | 208–208 | 149–149 | 088–108 | 149–155 | 102–106 | 128–128 | 122–130 | 160–166 | 187–193 | 176–178 | 150–156 | 100–108 |
| CUM68 | Bolivia | 1994 | 202–206 | 151–151 | 128–128 | 129–129 | 102–108 | 128–128 | 122–130 | 164–168 | 187–189 | 168–174 | 152–154 | 102–102 |
| CUM82 | Bolivia | 1994 | 202–206 | 149–149 | 098–112 | 131–131 | 100–102 | 136–136 | 124–132 | 164–164 | 179–189 | 180–180 | 154–154 | 098–100 |
| CUM84 | Bolivia | 1994 | 206–206 | 149–149 | 096–096 | 131–131 | 100–100 | 116–130 | 130–130 | 158–166 | 181–189 | 174–174 | 152–152 | 100–100 |
| CUM97 | Bolivia | 1994 | 206–206 | 149–159 | 100–110 | 129–147 | 102–102 | 116–128 | 122–124 | 164–164 | 185–187 | 174–174 | 154–154 | 100–102 |
| CUM96 | Bolivia | 1994 | 204–204 | 149–161 | 084–098 | 129–137 | 102–102 | 128–128 | 130–130 | 162–172 | 191–193 | 168–182 | 154–154 | 100–102 |
| CUM99 | Bolivia | 1994 | 204–204 | 149–159 | 088–088 | 121–121 | 102–102 | 122–122 | 132–132 | 176–176 | 185–191 | 162–162 | 150–156 | 098–098 |
| CUM32 | Bolivia | 1994 | 204–204 | 147–161 | 124–124 | 129–147 | 102–102 | 118–130 | 120–134 | 164–172 | 189–189 | 168–174 | 152–152 | 100–106 |
| CUM46 | Bolivia | 1994 | 202–202 | 149–149 | 118–118 | 127–127 | 102–102 | 136–136 | 120–128 | 152–152 | 185–187 | 168–182 | 154–154 | 100–100 |
| CUM30 | Bolivia | 1994 | 200–200 | 149–149 | 090–090 | 129–131 | 104–104 | 122–122 | 114–130 | 164–164 | 179–179 | 170–170 | 150–150 | 096–096 |
| CUM24 | Bolivia | 1994 | 204–204 | 149–149 | 110–110 | 129–129 | 102–102 | 132–132 | 124–134 | 158–164 | 189–189 | 174–174 | 166–166 | 102–102 |
| CUM42 | Bolivia | 1994 | 204–204 | 149–159 | 098–110 | 129–147 | 100–100 | 116–128 | 124–124 | 164–164 | 183–183 | 174–174 | 152–154 | 100–100 |
| CUM26 | Bolivia | 1994 | 202–204 | 151–151 | 118–118 | 129–137 | 102–102 | 116–116 | 120–132 | 164–164 | 183–183 | 174–180 | 152–152 | 102–102 |
| CUM39 | Bolivia | 1994 | 204–204 | 151–151 | 098–110 | 135–137 | 100–100 | 116–128 | 120–122 | 164–164 | 189–189 | 172–172 | 152–152 | 098–102 |
| CH22B | Bolivia | 1994 | 204–204 | 149–159 | 118–118 | 129–129 | 102–102 | 128–128 | 120–128 | 162–162 | 185–187 | 168–182 | 154–154 | 102–102 |
| CUM49 | Bolivia | 1994 | 206–206 | 151–151 | 122–122 | 129–129 | 102–102 | 130–130 | 122–124 | 164–164 | 185–187 | 174–174 | 154–154 | 100–102 |
| CUM51 | Bolivia | 1994 | 202–204 | 151–161 | 098–118 | 135–137 | 102–102 | 116–130 | 132–134 | 162–162 | 187–191 | 168–174 | 152–154 | 100–102 |
| CUM251 | Bolivia | 1998 | 200–200 | 149–149 | 092–092 | 143–143 | 102–102 | 122–122 | 128–128 | 166–166 | 189–189 | 172–172 | 152–152 | 096–096 |
| CUM252 | Bolivia | 1998 | 200–200 | 153–153 | 116–116 | 141–141 | 106–106 | 122–122 | 128–128 | 166–166 | 183–183 | 170–172 | 152–152 | 096–096 |
| CUM263 | Bolivia | 1998 | 204–204 | 161–161 | 100–116 | 131–147 | 102–102 | 116–130 | 122–122 | 162–162 | 185–185 | 176–182 | 154–154 | 100–100 |
| CUM266 | Bolivia | 1998 | 200–200 | 149–149 | 116–116 | 145–145 | 106–106 | 122–122 | 116–130 | 164–164 | 183–189 | 168–172 | 152–152 | 098–098 |
| CUM271 | Bolivia | 1998 | 200–200 | 153–153 | 098–118 | 143–143 | 106–106 | 122–122 | 116–128 | 164–164 | 181–181 | 166–170 | 152–152 | 096–096 |
| CUM274 | Bolivia | 1998 | 200–200 | 153–157 | 098–108 | 139–139 | 106–106 | 122–122 | 116–118 | 164–164 | 183–183 | 170–172 | 152–152 | 096–096 |
| CUM275 | Bolivia | 1998 | 200–200 | 151–151 | 092–092 | 143–143 | 106–106 | 122–122 | 120–120 | 164–164 | 181–181 | 170–172 | 152–152 | 096–096 |
| CUM276 | Bolivia | 1998 | 200–200 | 151–151 | 092–092 | 143–143 | 106–106 | 122–122 | 116–130 | 164–164 | 183–183 | 172–172 | 152–152 | 096–096 |
| CUM277 | Bolivia | 1998 | 204–206 | 151–151 | 100–110 | 131–147 | 102–102 | 130–130 | 116–126 | 162–162 | 185–185 | 168–174 | 154–154 | 100–100 |
| CUM279 | Bolivia | 1998 | 200–200 | 151–161 | 094–094 | 145–145 | 100–102 | 122–122 | 116–128 | 164–164 | 183–183 | 170–172 | 152–152 | 096–096 |
| CUM280 | Bolivia | 1998 | 200–200 | 151–151 | 092–092 | 145–145 | 106–106 | 122–122 | 128–128 | 164–164 | 183–183 | 174–174 | 154–154 | 096–096 |
| CUM281 | Bolivia | 1998 | 200–200 | 149–149 | 092–116 | 145–145 | 106–106 | 124–124 | 132–132 | 164–164 | 181–181 | 187–189 | 150–150 | 096–096 |
| CUM282 | Bolivia | 1998 | 200–200 | 149–159 | 090–090 | 139–139 | 106–106 | 122–122 | 130–130 | 164–164 | 181–181 | 180–180 | 152–152 | 100–100 |
| CUM285 | Bolivia | 1998 | 200–200 | 151–151 | 092–092 | 139–149 | 106–106 | 122–122 | 130–130 | 164–164 | 183–183 | 170–170 | 152–152 | 096–096 |
| CUM286 | Bolivia | 1998 | 200–200 | 151–151 | 092–092 | 141–149 | 106–106 | 122–122 | 130–130 | 164–164 | 183–183 | 170–170 | 152–152 | 100–100 |
| CUM288 | Bolivia | 1998 | 200–200 | 149–149 | 092–092 | 139–139 | 106–106 | 122–122 | 120–120 | 164–164 | 183–183 | 172–172 | 152–152 | 096–096 |
| CUM289 | Bolivia | 1998 | 200–200 | 161–161 | 092–092 | 141–149 | 102–102 | 122–122 | 130–130 | 166–166 | 183–189 | 170–170 | 152–152 | 098–098 |
| CUM290 | Bolivia | 1998 | 200–200 | 149–159 | 092–092 | 145–145 | 106–106 | 122–122 | 118–128 | 166–166 | 183–183 | 172–172 | 152–152 | 096–096 |
| CUM291 | Bolivia | 1998 | 200–200 | 149–159 | 092–092 | 145–145 | 106–106 | 116–130 | 132–132 | 164–164 | 187–187 | 170–170 | 152–152 | 096–096 |
| CUM294 | Bolivia | 1998 | 202–202 | 149–149 | 088–088 | 147–147 | 102–102 | 122–122 | 130–130 | 174–174 | 189–189 | 164–164 | 150–150 | 094–094 |
| CUM295 | Bolivia | 1998 | 202–202 | 151–151 | 088–088 | 131–131 | 102–102 | 122–122 | 132–132 | 164–164 | 191–191 | 164–164 | 152–152 | 096–096 |
| CUM310 | Bolivia | 1998 | 202–202 | 151–151 | 112–112 | 129–147 | 102–102 | 116–130 | 124–132 | 162–162 | 185–189 | 174–180 | 154–156 | 102–102 |
| CUM311 | Bolivia | 1998 | 200–200 | 149–149 | 100–108 | 131–131 | 102–102 | 116–128 | 132–132 | 162–162 | 189–189 | 174–174 | 152–152 | 098–100 |
| CUM323 | Bolivia | 1998 | 206–208 | 159–159 | 100–110 | 131–137 | 102–102 | 130–130 | 124–130 | 164–164 | 187–187 | 174–182 | 154–154 | 098–102 |
| CUM324 | Bolivia | 1998 | 206–206 | 161–161 | 098–098 | 131–137 | 100–100 | 128–128 | 132–132 | 166–168 | 185–185 | 174–182 | 154–154 | 096–096 |
| CUM327 | Bolivia | 1998 | 204–204 | 151–161 | 104–108 | 137–137 | 102–102 | 128–128 | 132–132 | 162–162 | 181–181 | 178–180 | 150–152 | 102–102 |
| CUM329 | Bolivia | 1998 | 204–204 | 149–149 | 116–116 | 145–145 | 102–102 | 116–130 | 124–128 | 162–162 | 185–185 | 174–180 | 154–154 | 100–102 |
| CUM331 | Bolivia | 1998 | 200–200 | 149–149 | 098–112 | 145–145 | 106–106 | 122–122 | 116–130 | 164–164 | 181–189 | 170–170 | 152–152 | 096–096 |
| CUM346 | Bolivia | 1998 | 204–204 | 151–161 | 104–108 | 131–137 | 102–102 | 130–130 | 132–132 | 162–162 | 185–185 | 174–180 | 152–152 | 098–098 |
| CUM381 | Bolivia | 1998 | 200–200 | 149–153 | 100–110 | 145–145 | 106–106 | 122–122 | 116–118 | 166–166 | 183–183 | 168–168 | 152–152 | 096–096 |
| CUM384 | Bolivia | 1998 | 198–204 | 149–159 | 116–116 | 129–135 | 102–102 | 116–130 | 122–122 | 162–162 | 185–185 | 174–180 | 154–154 | 100–102 |
| CUM388 | Bolivia | 1998 | 200–200 | 149–149 | 098–110 | 131–137 | 100–102 | 116–116 | 122–132 | 162–162 | 183–183 | 172–180 | 150–150 | 098–098 |
| CUM389 | Bolivia | 1998 | 204–212 | 151–151 | 118–118 | 131–137 | 102–102 | 116–116 | 122–132 | 162–162 | 183–183 | 172–180 | 152–152 | 098–098 |
| CUM393 | Bolivia | 1998 | 200–200 | 153–153 | 098–098 | 149–149 | 106–106 | 128–128 | 130–130 | 164–164 | 185–185 | 168–172 | 148–156 | 100–100 |
| CUM396 | Bolivia | 1998 | 200–200 | 151–153 | 116–116 | 143–143 | 102–102 | 122–122 | 116–118 | 164–164 | 183–183 | 170–170 | 152–152 | 096–096 |
Genetic Diversity and Heterozygote Deficiency.
We obtained clear electrophoregrams for all genotypes at all 12 loci investigated, with only 1 or 2 alleles per strain at each locus, which excludes events of aneuploidy (for which we would have expected individuals with 3 or 4 alleles). There is considerable genetic diversity, with an average number of alleles per locus of 12.4 ± 4.4, ranging from 6 (ITSbraz) to 22 (AC52), and a mean genetic diversity Hs = 0.750 ± 0.100 (Table 1).
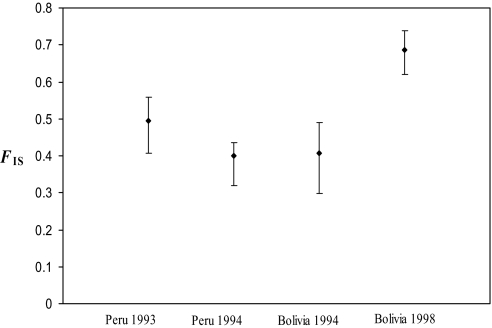
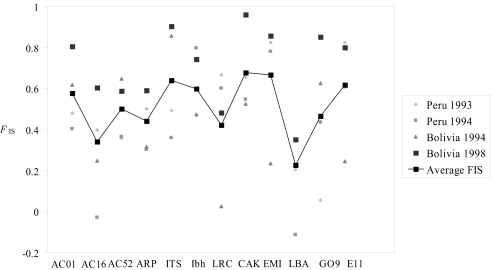
There is large deficiency of heterozygotes compared with Hardy–Weinberg expectations in each population, for both the multilocus data and each locus separately. The population FIS ranges from FIS = 0.396 in Bolivia 1994, to FIS = 0.687 in Bolivia 1998 (Fig. 1). For individual loci, the average values range from FIS = 0.225 for LBA to FIS = 0.676 for CAK (Fig. 2). The overall mean value is FIS = 0.504 (95% CI = 0.427–0.577). All FIS values are significantly different from zero (P ≤ 0.001).
Fig. 1.
FIS and 95% confidence intervals obtained by bootstrap over loci, for the 124 human Leishmania braziliensis strains collected in 2 different countries in 2 different years. FIS measures the local deficiency of heterozygous genotypes due to nonrandom mating. There is a large heterozygote deficiency in each population, shown because FIS is significantly greater than zero.
Fig. 2.
FIS for each of 12 microsatellite loci in the 4 populations (and their mean) of Leishmania braziliensis collected in Peru and Bolivia. There is a large heterozygote deficiency at each locus.
The selfing rate (s) required to account for the heterozygote deficiency observed over all samples and loci is s = 0.67.
Wahlund Effect.
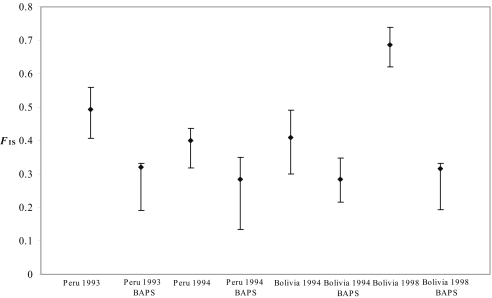
The heterozygote deficiency could possibly result from the Wahlund effect—that is, population subdivision within each subsample. This can be investigated with the Bayesian analysis of genetic population structure (BAPS) software. The 2 populations from Bolivia collected in 1994 and 1998 are composed of 15 (with probability PBAPS = 0.46) and 13 clusters (PBAPS = 0.69), respectively. The other 2 populations from Peru collected in 1993 and 1994 are composed of 18 (PBAPS = 0.60) and 11 clusters (PBAPS = 0.63), respectively. In each partition identified by BAPS in the 4 subsamples, the heterozygote deficit was calculated again. There was a decrease in FIS with respect to the initial data set. However, FIS_C = 0.307 (CI = 0.227–0.584) remains significant (P ≤ 0.001; Fig. 3). Moreover, analyses with the Wilcoxon test showed a significant decrease (Z = 1.657, P = 0.0488). Thus, ≈40% of total FIS can be explained by a Wahlund effect. The selfing rate required to account for this remaining FIS is high, sc = 0.47.
Fig. 3.
FIS for Leishmania braziliensis strains in each population and within their subdivisions as identified by BAPS. The decrease of FIS in the subdivision suggests a Wahlund effect. However, the residual FIS values are still high, which suggests the persistence of nonrandom mating (due, for example, to selfing).
Population Differentiation.
The genetic differentiation between Peru and Bolivia for both 1994 collections was small but significant (FST = 0.092, P < 0.001). There was also a small temporal differentiation between 1993 and 1994 in Peru (FST = 0.004, P < 0.001) and an apparently larger one between 1994 and 1998 in Bolivia (FST = 0.114, P < 0.001).
The differentiation between BAPS clusters, using the HIERFSTAT software, was very high (FCluster/Country = 0.31), as expected. The remaining variation between countries was smaller but significant (FCountry/Total = 0.07, P < 0.002).
Linkage Disequilibrium.
Linkage disequilibrium for all populations is significant for 46 of the 66 pairs of loci (70%), which is much higher than the 5% (about 3 loci pairs) expected by chance. After sequential Bonferroni correction, 13 pairs remain in significant linkage disequilibrium, so that each of the 12 loci is involved in at least one significant linkage. This cannot be attributed to close physical linkage between loci, as the 12 loci are distributed on different chromosomes (22). These findings indicate strong linkage at a genome-wide scale.
Discussion
Numerous studies published since 1990 suggest that Leishmania species may have a predominantly clonal mode of reproduction associated with rare sexual recombination events. The majority of these studies are, however, based on databases that may not be suitable to reach that conclusion. Clonality is mainly inferred from analyses of strong linkage disequilibria observed across loci (24). Yet, computer simulations show that linkage disequilibrium is not a reliable measure of the proportion of clonal versus sexual reproduction in a population (25) because it is too sensitive to population demographic parameters (see also refs. 26–28). Moreover, the genetic markers used (such as multilocus enzyme electrophoresis [MLEE], random amplified polymorphic DNA [RAPD], restriction fragment length polymorphism [RFLP], and pulse field gel electrophoresis [PFGE]) have inherent limitations for inferences on the population genetic structure. These molecular markers have little resolution power (MLEE or RFLP), are dominant (RAPD), or multifactorial (reflecting global genomic organization in PFGE). Thus, even if these approaches can give valuable information on the evolutionary history of Leishmania species, they do not allow definitive conclusions about the population structure and the mode of reproduction of such organisms.
Our findings reveal a strong deficiency of heterozygotes (as well as linkage disequilibrium), theoretically incompatible with a strictly clonal reproduction model. Theoretical studies have shown that diploid clones are expected to accumulate heterozygosity at every locus over time (29–33). Clonal diploids should therefore exhibit negative FIS values (17). There are several nonexclusive hypotheses that could account for heterozygote deficiency. They include the presence of null alleles, natural selection, genic conversion, the Wahlund effect, and inbreeding.
Null alleles are often encountered in population genetics studies. They may be frequent in allozymes (34, 35) and in such DNA markers as microsatellites (36–38). In our data, there is relatively little FIS variation across loci, and those loci displaying the strongest FIS variance are not necessarily those with the highest FIS (see Fig. 2), which is what would be expected if null alleles were present. Moreover, no blank has ever been observed in the genotypes (no missing data; i.e., all individuals were amplified at all loci), which, given the high FIS, makes the null allele explanation unlikely.
Selection can strongly affect allele and genotypic frequencies. Underdominance, which decreases the fitness of heterozygous individuals, would result in deficiency of heterozygous genotypes relative to Hardy–Weinberg expectations. Underdominance, however, is not expected to be frequently encountered in nature because it is highly unstable (the rarest allele tends to disappear). We have observed similar FIS patterns across all 12 (dinucleotide, noncoding) microsatellite loci. Widespread, almost genome-wide, underdominance would be required to fit our data, which does not seem reasonable.
Parasites of the Trypanosomatidae family, such as Leishmania species, are characterized by genetic plasticity, so that they can use different pathways to generate genetic diversity (e.g., gene conversion; refs. 39 and 40), a process of unidirectional transfer of genetic material between members of a multigenic family (41). Gene conversion generates a transition from the heterozygous stage to the homozygous stage, and thus can result in substantial heterozygote deficiency (40). Given that we obtained similar findings across the 12 independent microsatellite loci, gene conversion could account for our findings only if it occurred among all of the loci studied, across the entire genome, which seems unlikely. If gene conversion significantly affected microsatellite loci heterozygosity, a negative relationship would be expected between differences in allele size in heterozygous individuals and the number of such heterozygous individuals in the data set. This is because heterozygotes recently aroused through mutation have less chance of being immediately converted again compared with older heterozygotes, and with microsatellite loci in clonal organisms, old heterozygotes are expected to carry the most distant alleles in terms of size (30, 31). If gene conversion occurs frequently, microsatellite loci should restore heterozygosity through mutation and thus between alleles that are close in length. We obtained a significant negative relative relationship between the size difference in bases between alleles (Δ) and the number of heterozygous individuals, NHz, only for locus E11 (see Fig. S1 and SI Text). This locus is located 60 bp before the trifunctional enzyme alpha subunit mitochondrial precursor-like gene, and this observation might reflect frequent conversion in this genomic region. If we exclude E11 from our data, the overall high FIS values persist.
Clustering within each sample, as performed by BAPS, results in a substantial (40%) decrease in FIS values. This is consistent with the existence of a strong Wahlund effect within each subsample. Considering the large areas investigated (100–200 km2), it is not unreasonable to expect geographic subdivision within our Leishmania samples. In addition to geographic barriers that could influence parasite distribution, the biology of vectors and reservoirs may strongly interfere with the homogeneous spread of genotypes across both regions sampled. We note, for example, that the overall flight distance traveled by a sandfly over its entire lifetime is estimated to be ≈1 km (42); the scale at which our samples were collected is far above this limit. The Wahlund effect we have detected indicates that the Bolivian and Peruvian samples are probably each composed of several strongly differentiated subpopulations. This substructure may result in very large global effective population sizes, as shown by the weak temporal differentiation we observed, and thus may contribute to maintaining the genetic diversity at the scale of each geographic population. This could provide Leishmania populations with an advantage in adaptability to environmental differences and changes. The modest but significant differentiation observed between countries (most of the variance is contained within each country) reflects that there is little migration between countries, as well as between subpopulations within each country, where most of the variance occurs (see SI).
Heterozygote deficiency remains high (above 0.307) for every sample and for every locus, even within the clusters defined by BAPS. These findings, together with the high linkage disequilibrium observed, support the idea that these parasites, known to reproduce by clonal fission, also often sexually cross with individuals from the same strain (endogamy), unless our sampling did not allow us to detect a more nested population structure than is apparent. To evaluate this possibility, we studied the distribution of the heterozygous loci in each individual and found a random distribution (adjusted to a Poisson distribution, Kolmogorov–Smirnoff test). A small simulation study undertaken with Easypop v. 2.0.1 (43) suggests that our data are compatible with partially clonal populations. More especially, it seems likely to correspond to very small subpopulations, well structured at a scale much smaller than what can be detected even with clustering procedures (see Figs. S2 and S3 and SI Text).
On the basis of the previous considerations, we propose that Leishmania parasites use alternative modes of reproduction: clonality in both the vertebrate host and the insect vector and occasional sexual fusion in the vector, as has been shown to occur for other kinetoplastid parasites, such as Trypanosoma brucei s.l. (44). However, in Leishmania, this fusion may frequently involve genetically related parasites or even genetically identical members of the same strain, given the very low incidence of Leishmania parasites generally observed in the sandfly vectors (45–47).
We are not aware of any previous evidence of such strong inbreeding in Leishmania. This changes the assumption that its mode of reproduction is overwhelmingly clonal. This finding is an important step toward an understanding of leishmaniasis epidemiology. Reproductive mode influences the distribution of alleles within individuals and impacts the rate of selection of recessive or dominant alleles.
An important observation is the high overall genetic diversity observed in each sample. Published studies suggest that there is a link between the genetic polymorphism of circulating strains of Leishmania and environmental diversity (48). The large diversity observed in the present samples may be related to the extremely diversified ecosystem (various host and vector species; ref. 49) of the Amazonian forest.
To conclude this detailed population genetics study of L. braziliensis, which we believe may be the first of its kind, it seems that these parasites alternate clonal and sexual, although endogamic, reproduction, with infrequent recombination events between different individuals. In addition, our findings show the existence of strong genetic heterogeneity within each country (Wahlund effect), suggesting a substantial population structure at a microgeographic scale. In future studies, it will be important to work at finer geographic scales to detect and delimit this substructuring. The approach used here needs to be applied to other species of Leishmania to ascertain the generality of our findings. In vitro experiments could explore whether sexual recombination readily occurs within the sandfly vectors.
Materials and Methods
Parasite Culture and DNA Extraction.
One hundred twenty-four human isolates of Leishmania (Viannia) braziliensis were cultured. Promastigote cultures were maintained at 26 °C by weekly subpassages in RPMI1640 medium, buffered with 25 mM Hepes, 2 mM NaHCO3, and supplemented with 20% heat-inactivated FCS, 2 mM glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. Cultures were harvested by centrifugation and stored at −80 °C until DNA extraction. Fifty-six strains from Peru and 68 from Bolivia were isolated in the Laboratory of Biochemistry, the Alexander Von Humbolt Institute of Tropical Medicine of the University of Lima (Peru), and at the University of Cochabamba Medical Center (Bolivia). We characterized the 124 strains as L. braziliensis using the MLEE technique as described in ref. 50. DNA was extracted from parasite mass cultures (promastigotes), using the classical phenol/chloroform extraction technique (51).
Genotyping.
The 12 microsatellite loci investigated are listed in Table 1 (see ref. 22). A 30-μL reaction mix was made of 1.2 μL of each primer (10 μM), with the forward primer being labeled with fluorochrome, 100 ng template DNA, 0.9 μL dNTP mix (5 mM), 3 μL buffer 10× and 0.3 μL Taq polymerase (5 UI/μL; Roche Diagnostics). Amplifications were carried out in a thermal cycler: 30 cycles of 94 °C for 30 s, annealing temperature of each locus for 1 min at 72 °C, final extension at 72 °C for 7 min. The reaction products were visualized on a 1.5% agarose gel stained with ethidium bromide. Fluorescence-labeled PCR products were sized on Applied Biosystems Prism 310, with a Genescan 500 LIZ internal size standard. All 124 isolates were genotyped at all 12 loci.
Statistical Analyses.
Data were analyzed with the software FSTAT (version 2.9.3.2; ref. 52), which computes estimates and tests the significance of various population genetics parameters. Genetic polymorphism was measured by the number of alleles per locus (N) and by Nei's unbiased genetic diversity within subsamples Hs (23). We estimated Wright's F statistics (53) with Weir and Cockerham's method (54): FIS measures the relative inbreeding of individuals due to the local nonrandom union of gametes in each subpopulation; FIS_C measures the relative inbreeding of individuals clustered; and FST measures the relative inbreeding in subpopulations attributable to the subdivision of the total population into subpopulations of limited size. FST thus also measures genetic differentiation between subpopulations. FIS ranges between −1 and 1. A negative value corresponds to an excess of heterozygotes, a positive value to heterozygote deficiency; 0 is expected under panmixia. FST varies between 0, when genetic identity between individuals is independent from the subpopulation (no differentiation) and 1, when all individuals of the same subpopulation are homozygous for the same allele but differ from individuals of different subpopulations. The significance of the departure from 0 was tested by 10,000 randomizations of alleles within subpopulations (for FIS) and of individuals between subpopulations (for FST). For FIS, the statistic used was Weir and Cockerham's estimator f; for FST, the statistic used was the log-likelihood ratio G (55) summed over all loci. Confidence intervals were estimated by bootstrapping over loci or jackknifing over populations with FSTAT. From the FIS parameter, a potential selfing rate s was inferred using the formula s = (2 * FIS)/(1 + FIS) (e.g., ref. 29).
Linkage disequilibrium between pairs of loci (nonrandom association of alleles at different loci) was assessed with a randomization test (genotypes at 2 loci are associated at random a number of times). The statistic used was the log likelihood ratio G summed over all subpopulations. Because this procedure was repeated on all pairs of loci, we applied the sequential Bonferroni correction (56) to the P values (P value × number of tests).
The #3.2 software identifies a hidden structure within populations through a Bayesian analysis. It clusters individuals into genetically distinguishable groups based on allele frequencies. This software was used to detect possible Wahlund effects and has been successfully applied to other parasites (16, 57). The BAPS software used stochastic optimization to infer the posterior mode of the genetic structure. To obtain the best distribution of the 4 populations under study, we ran the program many times to obtain the number of clusters. We also checked that nonstructured populations would not give the same results as ours. This was done by running BAPS on populations simulated with EASYPOP (version 2.0.1). Each of the 4 samples was submitted to a clustering exploration by BAPS with 160 runs with a maximum number of clusters set to 20. FIS was recalculated in each best distribution identified by BAPS and compared FIS_C with the initial FIS using a unilateral Wilcoxon signed-rank test for paired data, the pairing units being the 12 loci. If FIS_C is lower than FIS, it is probable that the initial subsamples were composed of several genetically distinct entities (e.g., geographical microstructure or subpopulations).
To estimate the contribution of macrogeography (between Bolivia and Peru) corrected for the effect of the subpopulation structure (between BAPS clusters), we used HIERFSTAT (version 0.03–2) software (58). This test uses the same statistics as those used for FST analyses, but the permutation procedure takes into account the hierarchy of the population structure. Differentiation between clusters within countries, FCluster/Country, is tested by randomization of individuals between clusters of the same country. FCountry/Total, the fixation index due to the distribution of clusters into different countries, is tested by randomizing clusters (including all individuals) between countries.
Supplementary Material
Acknowledgments.
The authors acknowledge F. Kjellberg, F. Renaud, M. Choisy, and F. Prugnolle for helpful discussions and for their assistance in the analysis and interpretation of the results. We also thank 2 anonymous referees who considerably helped improve the manuscript. We are grateful to the Institut de Recherche pour le Développement and the Centre National de la Recherche Scientifique for financial support. The strains were isolated as part of a European Community STD3 project (n8TS3*-CT92–0129). This work was also supported in a framework of a French National Project ANR SEST.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904420106/DCSupplemental.
References
- 1.World Health Organization. Leishmaniases. 2002 Available at http://www.who.int/zoonoses/diseases/leishmaniasis/en/
- 2.Bañuls AL, Hide M, Prugnolle F. Leishmania and the leishmaniases: A parasite genetic update and advances in taxonomy, epidemiology and pathogenicity in humans. Adv Parasitol. 2007;64:1–109. doi: 10.1016/S0065-308X(06)64001-3. [DOI] [PubMed] [Google Scholar]
- 3.Desjeux P. Leishmaniasis: Current situation and new perspectives. Comp Immunol Microbiol Infect Dis. 2004;27:305–318. doi: 10.1016/j.cimid.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Lainson RS, Shaw JJ. In: The Leishmaniasis in Biology and Medicine. Peters W, Killick-Kendrick R, editors. New York: Academic; 1987. pp. 1–120. [Google Scholar]
- 5.WHO. Leishmania, geography. 1998 Available at http://www.who.int/leishmaniasis/leishmaniasis_maps/en/index.html.
- 6.Tibayrenc M, Ayala FJ. The clonal theory of parasitic protozoa: 12 years on. Trends Parasitol. 2002;18:405–410. doi: 10.1016/s1471-4922(02)02357-7. [DOI] [PubMed] [Google Scholar]
- 7.Bastien P, Blaineau C, Pages M. Leishmania: Sex, lies and karyotype. Parasitol Today. 1992;8:174–177. doi: 10.1016/0169-4758(92)90016-u. [DOI] [PubMed] [Google Scholar]
- 8.Bañuls AL, et al. Is Leishmania (Viannia) peruviana a distinct species? A MLEE/RAPD evolutionary genetics answer. J Eukaryot Microbiol. 2000;47:197–207. doi: 10.1111/j.1550-7408.2000.tb00039.x. [DOI] [PubMed] [Google Scholar]
- 9.Bañuls AL, et al. Evidence for hybridization by multilocus enzyme electrophoresis and random amplified polymorphic DNA between Leishmania braziliensis and Leishmania panamensis/guyanensis in Ecuador. J Eukaryot Microbiol. 1997;44:408–411. doi: 10.1111/j.1550-7408.1997.tb05716.x. [DOI] [PubMed] [Google Scholar]
- 10.Belli AA, Miles MA, Kelly JM. A putative Leishmania panamensis/Leishmania braziliensis hybrid is a causative agent of human cutaneous leishmaniasis in Nicaragua. Parasitology. 1994;109(Pt 4):435–442. doi: 10.1017/s0031182000080689. [DOI] [PubMed] [Google Scholar]
- 11.Da-Cruz AM, Machado ES, Menezes JA, Rutowitsch MS, Coutinho SG. Cellular and humoral immune responses of a patient with American cutaneous leishmaniasis and AIDS. Trans R Soc Trop Med Hyg. 1992;86:511–512. doi: 10.1016/0035-9203(92)90089-u. [DOI] [PubMed] [Google Scholar]
- 12.Dujardin JC, et al. Karyotype plasticity in neotropical Leishmania: An index for measuring genomic distance among L. (V.) peruviana and L. (V.) braziliensis populations. Parasitology. 1995;110(Pt 1):21–30. doi: 10.1017/s0031182000081002. [DOI] [PubMed] [Google Scholar]
- 13.Evans DA, et al. Hybrid formation within the genus Leishmania? Parassitologia. 1987;29:165–173. [PubMed] [Google Scholar]
- 14.Hide M, Bañuls AL. Species-specific PCR assay for L. infantum/L. donovani discrimination. Acta Trop. 2006;100:241–245. doi: 10.1016/j.actatropica.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 15.Kelly JM, Law JM, Chapman CJ, Van Eys GJ, Evans DA. Evidence of genetic recombination in Leishmania. Mol Biochem Parasitol. 1991;46:253–263. doi: 10.1016/0166-6851(91)90049-c. [DOI] [PubMed] [Google Scholar]
- 16.Ravel C, et al. First report of genetic hybrids between two very divergent Leishmania species: Leishmania infantum and Leishmania major. Int J Parasitol. 2006;36:1383–1388. doi: 10.1016/j.ijpara.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 17.Balloux F, Lehmann L, de Meeûs T. The population genetics of clonal and partially clonal diploids. Genetics. 2003;164:1635–1644. doi: 10.1093/genetics/164.4.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Meeûs T, Lehmann L, Balloux F. Molecular epidemiology of clonal diploids: A quick overview and a short DIY (do it yourself) notice. Infect Genet Evol. 2006;6:163–170. doi: 10.1016/j.meegid.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Chambers GK, MacAvoy ES. Microsatellites: Consensus and controversy. Comp Biochem Physiol B Biochem Mol Biol. 2000;126:455–476. doi: 10.1016/s0305-0491(00)00233-9. [DOI] [PubMed] [Google Scholar]
- 20.Lehmann T, et al. Genetic differentiation of Anopheles gambiae populations from East and west Africa: Comparison of microsatellite and allozyme loci. Heredity. 1996;77(Pt 2):192–200. doi: 10.1038/hdy.1996.124. [DOI] [PubMed] [Google Scholar]
- 21.Russell R, et al. Intra and inter-specific microsatellite variation in the Leishmania subgenus Viannia. Mol Biochem Parasitol. 1999;103:71–77. doi: 10.1016/s0166-6851(99)00117-6. [DOI] [PubMed] [Google Scholar]
- 22.Rougeron V, et al. A set of 12 microsatellite loci for genetic studies of. Leishmania braziliensis. Mol Ecol Notes. 2008;8:351–353. doi: 10.1111/j.1471-8286.2007.01953.x. [DOI] [PubMed] [Google Scholar]
- 23.Nei M, Chesser RK. Estimation of fixation indices and gene diversities. Ann Hum Genet. 1983;47:253–259. doi: 10.1111/j.1469-1809.1983.tb00993.x. [DOI] [PubMed] [Google Scholar]
- 24.Tibayrenc M, Kjellberg F, Ayala FJ. A clonal theory of parasitic protozoa: The population structures of Entamoeba, Giardia, Leishmania, Naegleria, Plasmodium, Trichomonas, and Trypanosoma and their medical and taxonomical consequences. Proc Natl Acad Sci USA. 1990;87:2414–2418. doi: 10.1073/pnas.87.7.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Meeûs T, Balloux F. Clonal reproduction and linkage disequilibrium in diploids: A simulation study. Infect Genet Evol. 2004;4:345–351. doi: 10.1016/j.meegid.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Bartley D, Bagley M, Gall G, Bentley B. Use of linkage disequilibrium data to estimate effective size of hatchery and natural fish populations. Conserv Biol. 1992;6:365–375. [Google Scholar]
- 27.Vitalis R, Couvet D. Estimation of effective population size and migration rate from one- and two-locus identity measures. Genetics. 2001;157:911–925. doi: 10.1093/genetics/157.2.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waples RS, Do S. LDNE: A program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour. 2008;8:753–756. doi: 10.1111/j.1755-0998.2007.02061.x. [DOI] [PubMed] [Google Scholar]
- 29.De Meeûs T, et al. Population genetics and molecular epidemiology or how to “debusquer la bete/”. Infect Genet Evol. 2007;7:308–332. doi: 10.1016/j.meegid.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 30.Mark Welch DB, Meselson M. Evidence for the evolution of bdelloid rotifers without sexual reproduction or genetic exchange. Science. 2000;288:1211–1215. doi: 10.1126/science.288.5469.1211. [DOI] [PubMed] [Google Scholar]
- 31.Mark Welch DB, Meselson MS. Rates of nucleotide substitution in sexual and anciently asexual rotifers. Proc Natl Acad Sci USA. 2001;98:6720–6724. doi: 10.1073/pnas.111144598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pamilo P. Heterozygosity in apomictic organisms. Hereditas. 1987;107:95–101. doi: 10.1111/j.1601-5223.1987.tb00272.x. [DOI] [PubMed] [Google Scholar]
- 33.Suomalainen E, Saura A, Lokki J. Evolution of parthenogenetic insects. Evol Biol. 1976;9:209–257. [Google Scholar]
- 34.Gaffney D, Campbell RA. A PCR based method to determine the Kalow allele of the cholinesterase gene: The E1k allele frequency and its significance in the normal population. J Med Genet. 1994;31:248–250. doi: 10.1136/jmg.31.3.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nébavi F, et al. Clonal population structure and genetic diversity of Candida albicans in AIDS patients from Abidjan (Cote d'lvoire) Proc Natl Acad Sci USA. 2006;103:3663–3668. doi: 10.1073/pnas.0511328103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brookfield J. Population genetics. Curr Biol. 1996;6:354–356. doi: 10.1016/s0960-9822(02)00493-1. [DOI] [PubMed] [Google Scholar]
- 37.Paetkau D, Strobeck C. The molecular basis and evolutionary history of a microsatellite null allele in bears. Mol Ecol. 1995;4:519–520. doi: 10.1111/j.1365-294x.1995.tb00248.x. [DOI] [PubMed] [Google Scholar]
- 38.Pemberton JM, Slate J, Bancroft DR, Barrett JA. Nonamplifying alleles at microsatellite loci: A caution for parentage and population studies. Mol Ecol. 1995;4:249–452. doi: 10.1111/j.1365-294x.1995.tb00214.x. [DOI] [PubMed] [Google Scholar]
- 39.Mauricio IL, Gaunt MW, Stothard JR, Miles MA. Glycoprotein 63 (gp63) genes show gene conversion and reveal the evolution of Old World Leishmania. Int J Parasitol. 2007;37:565–576. doi: 10.1016/j.ijpara.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 40.Regis-da-Silva CG, et al. Characterization of the Trypanosoma cruzi Rad51 gene and its role in recombination events associated with the parasite resistance to ionizing radiation. Mol Biochem Parasitol. 2006;149:191–200. doi: 10.1016/j.molbiopara.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 41.Jackson JA, Fink GR. Gene conversion between duplicated genetic elements in yeast. Nature. 1981;292:306–311. doi: 10.1038/292306a0. [DOI] [PubMed] [Google Scholar]
- 42.Morrison AC, Ferro C, Morales A, Tesh RB, Wilson ML. Dispersal of the sand fly Lutzomyia longipalpis (Diptera: Psychodidae) at an endemic focus of visceral leishmaniasis in Colombia. J Med Entomol. 1993;30:427–435. doi: 10.1093/jmedent/30.2.427. [DOI] [PubMed] [Google Scholar]
- 43.Balloux F. EASYPOP (version 1.7): A computer program for population genetics simulations. J Hered. 2001;92:301–302. doi: 10.1093/jhered/92.3.301. [DOI] [PubMed] [Google Scholar]
- 44.Tait A, MacLeod A, Tweedie A, Masiga D, Turner CMR. Genetic exchange in Trypanosoma brucei: Evidence for mating prior to metacyclic stage development. Mol Biochem Parasitol. 2007;151:133–136. doi: 10.1016/j.molbiopara.2006.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin-Sanchez J, Gallego M, Baron S, Castillejo S, Morillas-Marquez F. Pool screen PCR for estimating the prevalence of Leishmania infantum infection in sandflies (Diptera: Nematocera, Phlebotomidae) Trans R Soc Trop Med Hyg. 2006;100:527–532. doi: 10.1016/j.trstmh.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 46.Rogers ME, Bates PA. Leishmania manipulation of sand fly feeding behavior results in enhanced transmission. PLoS Pathog. 2007;3:e91. doi: 10.1371/journal.ppat.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akopyants NS, et al. Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science. 2009;324:265–268. doi: 10.1126/science.1169464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Botilde Y, et al. Comparison of molecular markers for strain typing of Leishmania infantum. Infect Genet Evol. 2006;6:440–446. doi: 10.1016/j.meegid.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 49.Rotureau B, et al. Diversity and ecology of sand flies (Diptera: Psychodidae: Phlebotominae) in coastal French Guiana. Am J Trop Med Hyg. 2006;75:62–69. [PubMed] [Google Scholar]
- 50.Ben Abderrazak S, et al. Isoenzyme electrophoresis for parasite characterization. Methods Mol Biol. 1993;21:361–382. doi: 10.1385/0-89603-239-6:361. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook J, Fitsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 52.Goudet J. FSTAT: A program to estimate and test gene diversities and fixation indices. Version 2.9.3.2. 2002 Available at http://www.unil.ch/izea/softwares/fstat.html.
- 53.Wright S. The interpretation of population structure by F-statistics with special regard to system of mating. Evolution. 1965;19:395–420. [Google Scholar]
- 54.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 55.Goudet J, Raymond M, De Meeûs T, Rousset F. Testing differentiation in diploid populations. Genetics. 1996;144:1933–1940. doi: 10.1093/genetics/144.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- 57.Chevillon C, et al. Direct and indirect inferences on parasite mating and gene transmission patterns. Pangamy in the cattle tick Rhipicephalus (Boophilus) microplus. Infect Genet Evol. 2007;7:298–304. doi: 10.1016/j.meegid.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 58.Goudet J. HIERFSTAT, a package for R to compute and test hierarchical F-statistics. Mol Ecol Notes. 2005;5:184–186. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.