Abstract
Imprinting of the PWS/AS 2.4 Mb domain in the human is controlled by a paternally active imprinting center (PWS-IC). PWS-IC on the maternal allele is methylated and inactivated by an 880-bp sequence (AS-IC) located 30 kb upstream. In this communication, we report the identification of 7 cis acting elements within AS-IC. The elements: DMR, DNS, 2 OCTA sequences, SOX, E1, and E2 bind specific proteins that form at least 2 protein complexes. Using variants of an imprinted transgene, mutated at the elements each at a time, we show that (i) all 7 elements are involved in the methylation and inactivation of the maternal PWS-IC; (ii) the OCTA and SOX elements that bind a protein complex, and the E1 and E2 elements, function in establishing the primary imprint that constitutes an active and unmethylated AS-IC in the oocyte; (iii) DNS and DMR bind a multiprotein complex that may facilitate interaction between AS-IC and PWS-IC, mediating the inactivation in cis of PWS-IC; and (iv) all 7 elements participate in maintaining an unmethylated PWS-IC in the oocyte, which is essential for its maternal methylation later in development. Altogether, the above observations imply that the cis acting elements on AS-IC display diverse functions in establishing the imprints at both AS-IC and PWS-IC in the oocyte. A postulated epigenetic mark imprints the PWS-IC in the oocyte and maintains its inactive status during development before it is translated into maternal methylation.
Keywords: cis elements, DNA methylation, imprinting, protein factors
A number of imprinted genes are clustered within a 2.4-Mb domain on human chromosome 15q11-q13. Genetic aberrations in this domain result in 2 clinically distinct neurobehavioral disorders: Prader-Willi (PWS) and Angelman (AS) syndromes. PWS is a result of molecular defects that bring about silencing of the paternally expressed genes, while AS is caused by molecular defects that result in the loss of expression of the UBE3A gene on the maternal allele. Imprinting of the PWS/AS domain is regulated by a bipartite control center (IC) composed of a 4.3-kb sequence (PWS-IC) and a 880-bp sequence (AS-IC), which are separated by 30 kb (1–3).
PWS-IC, which includes the SNRPN gene promoter, functions on the paternal allele as a bidirectional activator that controls the expression of the paternally expressed genes and indirectly controls the maternally expressed UBE3A gene. On the maternal allele, PWS-IC is inactivated and methylated by AS-IC (4). Deletion or loss of function of AS-IC on the maternal allele leads to an abnormally unmethylated and active PWS-IC. That in turn causes an aberrant activation of the paternally expressed genes on the maternal allele. The recognition of the central role played by AS-IC in the imprinting process prompted us to characterize cis elements within AS-IC and the trans-acting factors that are involved in establishing the primary imprint in the gametes and in the repression of PWS-IC on the maternal allele.
Results
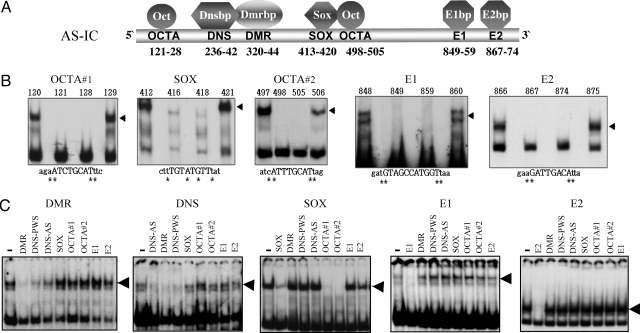
First, to identify elements on AS-IC, we performed band-shift experiments with nuclear extracts of mouse ES cells, using short oligonucleotides that together cover the entire AS-IC sequence. Seven short sequences were found to specifically bind proteins (Fig. 1 A and B), among them DMR (differentially methylated region) and DNS (de novo signal) were reported previously. The DMR sequence that includes 4 CpGs is differentially methylated in the gametes, and binds a protein that is present in parthenogenetic mouse ES cells and absent in androgenetic ones. The DMR binding protein (Dmrbp) does not bind in vitro the methylated DMR sequence (4). the DNS sequence is identical to the cis element of Snrpn that was previously observed in the mouse (5). Interestingly, DMR and DNS, which show a similar protein-binding pattern, compete with each other in binding their respective proteins, suggesting the formation of a protein complex (Fig. 1C). Among the other elements that we found were 2 Octamer (OCTA) sites and a SOX consensus site. Oct-4 and Sox2, which are known to bind these sequences and are expressed in oocytes (6–8), are transcription factors known to be crucial in normal embryonic development, maintaining the pluripotent state of the very early embryo (9) (10). These 2 factors are therefore suitable candidates to serve as imprinting regulators. Binding competition experiments show that the 2 OCTA and SOX elements bind a multiprotein complex, as the OCTA sequences mutually compete with each other and with SOX. This observation is consistent with the notion that OCTA and SOX elements act in concert in gene regulation (11, 12). Two elements, E1 and E2, bind 2 independent nuclear proteins that do not compete with any of the other elements in the binding experiments. These 2 proteins were termed E1bp (E1 binding protein) and E2bp, respectively (Fig. 1 B and C). It should be noted that no specific binding activity could be observed in the 370 bp that lie downstream to the 880 bp AS-IC. Interestingly, this additional 370-bp sequence was initially included in the AS-IC (13) but additional studies minimized the AS-IC size to 880 bp (2). Our results of protein-binding analysis are therefore in accordance with the shortest AS-IC as it was inferred from deletion analyses of Angelman families.
Fig. 1.
Cis elements and protein binding factors in AS-IC. (A) Names and positions of the cis elements and binding proteins in AS-IC (position 1 corresponds to nucleotide 22716614 of chr. 15, according to hg18 at http://genome.ucsc.edu/cgi-bin/hgGateway). (B) Representative band-shift assays used to characterize the binding specificity of each element. All assays were performed with nuclear extracts of mouse ES cells. Numbers above the lanes indicate the position of the mutation in the AS-IC sequence. Mutations were single base replacements G↔ T and C↔ A. The mutated bases are marked by an asterisk. Capital letters indicate bases that are part of the binding sequence. Lowercase letters represent the sequence flanking the element. The shifted bands are marked by arrowheads. Other mutated oligomers, as well as no-extract controls, were also used. (C) Competition band-shift experiments were used to determine binding cooperation between different cis elements. Labeled oligomers for each experiment are listed above each picture. The left lane of each gel represents a no-competition control. Lanes to the right represent the effect of various unlabeled competitors (vertical lettering) that were included in 100× molar excess. Loss of bands (arrowheads) indicates competition. DNS-AS and DNS-PWS represent the DNS elements on AS-IC and PWS-IC, respectively. A reciprocal experiment for OCTA competition was also performed, with results similar to that of SOX.
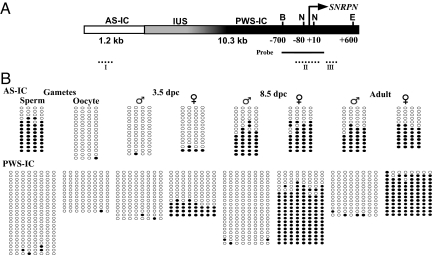
To study in detail the process of imprinting of AS-IC and PWS-IC throughout development, we made use of a transgene designed in our laboratory (AS25I transgene), that includes the human AS-IC and PWS-IC sequences (Fig. 2A). This transgenic sequence was recognized by the mouse imprinting machinery and recapitulated faithfully the human imprint (4). In offspring of this transgenic mouse, the PWS-IC was unmethylated when transmitted paternally and methylated when transmitted maternally (Fig. 2B). This robust transgenic system allowed us to examine methylation in the gametes and during embryogenesis, which is impractical in humans. Thus, we could show that AS-IC acquires an imprinted state in the gametes as judged by the methylated status of its DMR in sperm in contrast to its undermethylation in oocytes. In contrast, the PWS-IC was unmethylated in both sperm and oocytes (Fig. 2B). Our results of methylation of PWS-IC in the oocyte are in accord with the results obtained by El Maarri et al. (14) and the results that were reported previously by us (4). The results of these 2 studies and those reported here are in contrast to those obtained by Geuns et al. (15), who showed SNRPN to be methylated already in the oocyte. The unmethylated state of PWS-IC is maintained post fertilization at the first embryonic cleavages, and its maternal methylation first appears at the blastula stage. The fact that PWS-IC maternal methylation is fully displayed only later in embryo development, when AS-IC had already lost its activity (Fig. 2B), suggests that AS-IC establishes the imprint of PWS-IC in the oocyte by means of an epigenetic mark other than DNA methylation.
Fig. 2.
Methylation analysis of the human AS-25I (WT) transgene. (A) Schematic structure of the transgene: AS-IC (white, positions 22716614–22717869), PWS-IC (black, 22746472–22751856), and IUS (intervening upstream sequence, gray, 22741535–22746471). PWS-IC is based on the PWS-SRO sequence obtained from Prader-Willi patients (3). SNRPN transcription start site (arrow), restriction sites used for Southern blotting [BglII (B), EcoR I (E,) and NotI (N), positions relative to SNRPN start site], probe (solid line), Roman numerals denote regions analyzed by bisulfite sequencing (dotted lines). (B) DNA bisulfite methylation analysis throughout development of AS-IC DMR (region I - positions 322–340 of AS-IC) and PWS-IC DMR [region III - positions (+218)–(+298) relative to SNRPN transcription start site, bottom]. DNA treatment and PCR were carried out as described in Materials and Methods. Empty circles, unmethylated CpGs. Filled circles, methylated CpGs. ♂- paternal transmission, ♀- maternal transmission. Each horizontal line represents a unique clone. At least 5 separate samples of pooled oocytes and 3.5 dpc embryos were treated for each stage and lineage. For adult brain and 8.5 dpc embryos, results represent 3 separate experiments. Similar results for PWS-IC were obtained using primers that encompass region II in positions (−137)–(+101), which includes the NotI sites used in Southern blot.
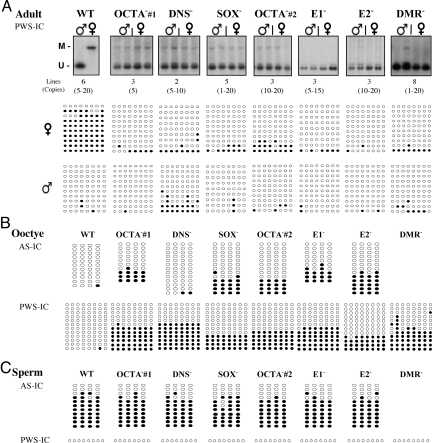
Since cis elements were found to control imprinting at the mouse PWS-IC orthologue (5), we analyzed the role played by each of the protein-binding elements of AS-IC in the process of imprint establishment in a human sequence. We therefore separately mutated each of the AS-IC elements described above in the AS25I transgene, eliminating their protein binding capacity without altering the sequence integrity of AS-IC. For each mutated transgene, we have created several founder lines (Fig. 3A), and examined the methylation status of the DMRs at AS-IC and PWS-IC upon both maternal and paternal transmissions. These experiments revealed that all mutations abrogated the imprinting of PWS-IC in adult tissues. In the mutated transgenes, the maternal PWS-IC was completely unmethylated, as opposed to the WT transgene, yet, upon paternal transmission, methylation of PWS-IC did not vary from the WT (Fig. 3A). Therefore, we conclude that each element plays a role in the exclusive maternal effect of AS-IC on the methylation of PWS-IC. The results were identical with the different transgenic lines of each mutation, ruling out position effects.
Fig. 3.
Methylation analysis of AS-IC and PWS-IC upon mutations in AS-IC. (A) Top: Southern blot analysis of PWS-IC DMR in adult brain transgenic tissues using the methylation sensitive enzyme NotI. M, methylated band. U, unmethylated band. Up to 10 individual samples representing 2 generations were examined from each transgenic line upon both parental transmissions. Representative data are shown. ♂- paternal ♀- maternal. The number of founder lines, and copy number per each mutated transgene are listed at the bottom of the gel. Bottom: Bisulfite sequencing of PWS-IC DMR in adult brain of the various transgenic lines (as described in Fig. 2B). Each horizontal line represents a unique clone. Empty circles, unmethylated CpGs, filled circles, methylated CpGs. All bisulfite data are of a representative line from each mutation. Lines not shown gave similar results. (B) Bisulfite sequencing of AS-IC and PWS-IC DMRs in oocytes of the various mutant transgenic lines (as described in Fig. 2B). At least 5 separate pools of approximately 30 oocytes each were used. Each line represents a unique clone. Empty circles, unmethylated CpGs, filled circles, methylated CpGs. The DMR mutations were not analyzed for AS-IC, as they lack all 4 CpGs. (C) Bisulfite sequencing of AS-IC and PWS-IC DMRs in sperm of the various mutant transgenic lines. For AS-IC, each horizontal line represents a unique clone. For PWS-IC, the single horizontal line represents 12 unique clones that were all completely unmethylated. At least 3 sperm samples per mutated line were used. Southern blot analysis for PWS-IC yielded similar results. Similar results for PWS-IC were also obtained with primers that encompass region II in positions (−137)–(+101).
We next asked whether the cis elements in AS-IC also determine its own methylation pattern and if the differential methylation of the AS-IC DMR in the gametes is crucial for the imprinting process. Analysis of the methylation status of AS-IC in the mutant transgenes revealed that with the exception of the mutation in DNS, all other mutations (2 OCTA, SOX, E1, and E2) resulted in a significant AS-IC DMR methylation in the oocyte (Fig. 3B). Although this methylation occurred in only approximately 30% of the molecules, it is significant since it comprises approximately half of the paternal AS-IC methylation in sperm and adult, where 60% of the molecules are methylated (Fig. 2B). These results imply that a protein complex of Oct and Sox (Fig. 1C) and E1bp and E2bp play a role in determining the methylation status of AS-IC DMR and thereby its activity. Upon fertilization, the abnormal methylation caused by the mutations was effectively lost, and then regained post-implantation upon both maternal and paternal transmissions, as is the case in the wild-type (WT) transgene (Fig. 4B). In the DNS-mutated transgene, AS-IC DMR remained unmethylated in the oocyte, much like the WT transgene (Fig. 3B). This exception indicates that DNS affects the methylation of PWS-IC and not of AS-IC, and supports our model in which the DNS binding protein (Dnsbp) facilitates an interaction between AS-IC and PWS-IC (Fig. 5). This interaction also involves Dmrbp that is in a complex with Dnsbp (Fig. 1C), and perhaps a second Dnsbp that binds to a DNS element in the PWS-IC sequence (Fig. 5) (4). The involvement of a DNS element resembles the mouse orthologue, in which 2 DNS elements were found to be essential to carry out maternal methylation of PWS-IC (5).
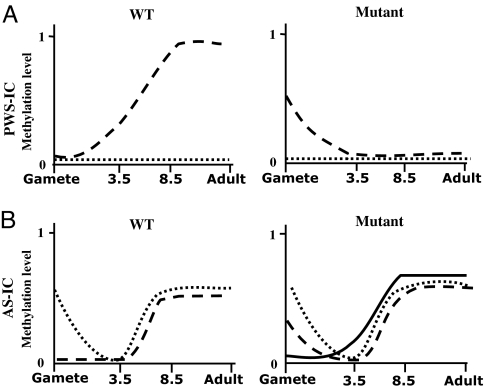
Fig. 4.
Dynamic changes in the methylation levels of the PWS/AS ICs throughout 4 stages of development. Both 3.5 and 8.5 represent the respective dpc (days post coitum) (A) Methylation levels of PWS-IC for the maternal (dashed lines) and paternal (dotted lines) alleles in the AS25I WT transgene (left) and its mutant derivatives (right). Analysis was performed by bisulfite sequencing for all stages, and by Southern blot for PWS-IC in sperm, 8.5 dpc and adult brain, as described in Figs. 2 and 3. (B) Methylation levels of AS-IC for the maternal (dashed lines) and paternal (dotted lines) alleles in the AS25I WT transgene (left) and its mutant derivatives (right). The maternal DNS mutation is drawn separately from the other mutations (solid line).
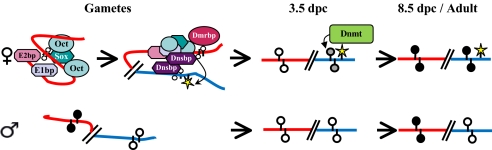
Fig. 5.
A proposed model describing the role of trans-acting factors in the establishment of imprinting in the PWS/AS domain. Top, maternal chromosome: Multiple protein factors bind the AS-IC (red) in the oocyte and protect its DMR from undergoing methylation (empty lollipops). An interaction between AS-IC and PWS-IC (blue) is achieved through separate DNS elements located on each IC. The interaction protects PWS-IC from methylation as well. In parallel, PWS-IC is marked (curved arrow) with an epigenetic modification that is maintained throughout the preimplantation period (yellow star). By implantation, DNA methyltransferase (Dnmt in green box) is recruited to gradually methylate PWS-IC (curved arrow, filled gray and black lollipops). AS-IC becomes methylated by the global wave of post implantation methylation. Bottom, paternal chromosome. Trans-acting factors are either not present, or fail to bind the methylated AS-IC in sperm (filled lollipops). This can be postulated based on the observed absence of Dmrbp in androgenetic ES cells, that express paternally derived genes, and based on its inability to bind a methylated DMR sequence as was demonstrated by in vitro experiments (4). Thus, AS-IC is inactive and leaves PWS-IC, which is a CpG island, unmethylated and active throughout.
An additional effect of the mutations in AS-IC on the methylation of PWS-IC was deduced from the results obtained with the mutated transgenes. In contrast to the unmethylated state of PWS-IC in the WT transgenic oocytes, PWS-IC was substantially methylated in the oocytes harboring the mutated transgenes (Fig. 3B). Paradoxically, this early maternal methylation of PWS-IC in the mutant lines is associated with a maternal unmethylated state of PWS-IC in the adult (Fig. 3A). The aberrant methylation of PWS-IC in the mutated oocytes was erased following fertilization, as is the case for AS-IC. But in contrast to AS-IC, PWS-IC was not remethylated post-implantation (compare Fig. 4 A with B). This is very likely due to the CpG island nature of PWS-IC, and the lack of an epigenetic mark that is normally established on PWS-IC during oogenesis to cause its methylation later in development (Fig. 5). This result implies that AS-IC plays a role in maintaining the unmethylated state of PWS-IC in the oocyte, which is critical for its methylation later in development. It should be emphasized that the effects of the mutations on the methylation of AS-IC and PWS-IC in the gametes are exclusively maternal. In sperm, no difference was observed between the WT and mutant methylation levels in either of the ICs (Fig. 3C).
Interestingly, even though a substantial proportion of AS-IC and PWS-IC molecules are not methylated in the oocyte (Fig. 3B), imprinting of PWS-IC is still not achieved upon mutagenesis (Fig. 3A). That is especially true for the oocytes with a DNS mutation, in which no AS-IC methylation was observed. It is worth mentioning that at the AS-IC the differential methylation and activity are transient, as we find the paternal AS-IC methylation to be erased in the blastula, only to be regained on both parental alleles post implantation (Figs. 2B and 4B). Taken together, these results may indicate that undermethylation of AS-IC and also PWS-IC is not sufficient to support a successful imprinting process, and therefore that differential DNA methylation at AS-IC may not be the determinant hallmark of its activity. Other differential epigenetic modifications have been found at AS-IC in the adult (16), but its gametic activity appears to be dependent on a rigorous design of the binding of trans-acting protein factors.
Still, as our conclusions are drawn from the transgenic experiments, it cannot be ruled out that endogenous methylation of AS-IC in the human plays a role in its activity, since an inactive AS-IC in human adult cells is fully methylated on both alleles (17), as is the case in human sperm. We believe that the observed partial methylation of AS-IC in oocytes of the mutated transgenes, which is extended to PWS-IC, could be a result of a malformed protein complex that binds the mutant AS-IC and allows for its early methylation. This methylation may reflect a defective imprinting process, rather than an active mode of maternal imprint prevention. When AS-IC is intact and carries all its cis elements, both AS-IC and PWS-IC are fully protected from methylation during oogenesis. This protection is apparently required in the maternal lineage only, because in sperm, PWS-IC is not affected by the mutations of the cis elements and remains unmethylated even when AS-IC is mutated (Fig. 3C).
Discussion
We provide here an insight into the mechanism of imprint establishment, as it is reciprocally manifested on 2 subdomains of a bi-partite imprinting center, AS-IC and PWS-IC, and shows its intricacy in the human relative to the mouse. In the mouse, that lacks an AS-IC orthologue, PWS-IC imprinting is completed already in the oocyte (18).
Based on the results of the transgenic experiments described here with the human sequence (summarized in Fig. 4), we propose a model for imprint establishment in the human that takes into consideration the various functions exerted by the trans-acting factors (Fig. 5). On the maternal allele, multiple protein factors, including an Oct-Sox complex, bind AS-IC and protect its DMR from undergoing methylation in the oocyte. A complex of Dmrbp and Dnsbp binds AS-IC and interacts with PWS-IC through its own DNS sequence. This interaction loops out 30 kb of intervening sequence, thus protecting PWS-IC from undergoing early methylation, making it suitable for modification by a yet unknown epigenetic mark that serves later to recruit a DNA-methyltransferase (Dnmt). To postulate such a mark is also essential to explain our previous transgenic experiments in which AS-IC was removed by a Cre-lox system at a very early embryonic stage, without affecting the eventual PWS-IC methylation (4). This is also consistent with a recent report that suggests a mechanism of DNA-methylation-independent memory, where the Zfp-57 protein restored maternal methylation of the mouse Snrpn promoter after it emerged unmethylated from the oocyte in Zfp-57 knocked out mice (19). In addition, Arnaud et al. showed that maternal methylation of the mouse Snrpn can be obtained in the absence of Dnmt3L (20). A possible epigenetic memory mechanism for maintaining the imprint in the absence of DNA methylation is histone H3-K9 methylation. This histone modification is enriched on the adult maternal PWS-IC (16) and was found to be associated with DNA methylation (21). It has also been shown that maintenance of CpG methylation at the PWS-IC in mouse ES cells requires the histone H3-K9 methyltransferase G9a (22).
We have shown that in order for PWS-IC to become methylated in the blastocyst, it has to undergo demethylation in the oocyte. This is known for the mouse sequence, where demethylation in primordial germ cells precedes methylation in the oocyte (18). While the present communication was prepared for publication, it came to our notice that transcription that originates upstream to and goes through the maternally imprinted DMRs of the mouse Gnas locus (on chromosome 2) in growing oocytes is essential for and precedes their eventual methylation (23). In our experiment, with the AS-IC located at the very 5′ end of the transgene, such a mechanism is very unlikely since AS-IC was not found to promote transcription (24). However, a transcription based mechanism could be in accord with the presence of OCTA and SOX elements in AS-IC. These elements are known to enhance and not merely promote transcription in the early embryo (25), and therefore could enhance transcription in a wider, endogenous context. With regard to the mouse, transcripts of the IC that make use of an alternative promoter that lies upstream to Snrpn have been reported in the oocyte (26). The use of alternative promoters and upstream exons of SNRPN is also known in the human (27), and 1 such exon (u5) was found within AS-IC (24). Chotalia et al. propose that transcription serves to ‘open’ the chromatin for DNA methylation (23). This might explain our results, where we found that when imprinting is disrupted by mutations in cis elements, PWS-IC is methylated early nonetheless (Fig. 3B). It is therefore conceivable that this aberrant methylation in the mutated transgene reflects a ‘closed’ chromatin conformation that prevents transcription through PWS-IC at an earlier stage, and consequently its stable methylation at a later stage. We remain open to the possibility that transcription and protein binding to AS-IC in the oocyte are functionally entwined to regulate imprint establishment at the PWS-IC sequence.
By showing the aberrant methylation of PWS-IC in the mutant transgenes in the oocyte, our results shed light on a previous report by Johnstone and colleagues (28). These authors observed that replacement of the mouse PWS-IC with the human endogenous sequence resulted in the human PWS-IC being methylated in the mouse oocyte and losing this methylation in somatic cells. It was suggested that this is due to the divergence of the mechanisms of imprint maintenance between the mouse and the human. We would rather associate this early methylation of PWS-IC and the eventual loss thereof in the Johnstone et al. experiment with the lack of AS-IC in the knocked-in construct, as in our study mutations at AS-IC gave similar results.
If proper protein-binding to the elements of AS-IC is essential for its activity, we can predict that mutations in these proteins could be involved in Angelman syndrome cases that show an imprinting defect (ID) with no genetic alterations in the AS-IC sequence. The report that mutation in the Zfp57 gene in mice results in specific demethylation of the mouse maternal Snrpn (19), strengthens our claim that trans-acting proteins play a critical role in imprint establishment. A possible role for 1 of the cis-elements characterized here (SOX) can be gleaned from a population study that found a 4-bp deletion in Angelman syndrome patients (29). This deletion lies in the core of the SOX element at AS-IC, and gives credence to our methodology of recognizing cis acting elements in vitro. Further work is required to fully characterize the protein factors and their mode of action in the imprinting process.
Materials and Methods
Band-Shift Analysis.
Nuclear extracts were prepared from mouse ES cells as described (30). Labeled oligonucleotides were prepared by end filling using Klenow DNA polymerase I (Roche Industries). The radioactive probe (100 cpm/20–50 pg) was incubated at 30 °C for 30 min, with nuclear extract (≈10 μg protein) in a buffer containing 12 mM Hepes, 60 mM KCl, 0.6 mM Na2-EDTA, 0.6 mM DTT, 5 mM MgCl2, and 1 mg poly[d(I-C)] (or d(G-C) for SOX) in a volume of 20 μL. The reaction mixtures were separated on a 4% polyacrylamide gel, which was dried and exposed to film. Several mutated oligomers were used to characterize each element. Mutations that resulted in loss of a shifted band indicated sequences necessary for protein binding, and vice versa.
Generation of Transgenic Mice.
The mutations in the cis elements were created by PCR mutagenesis of the AS25I construct described previously (4). The primers at the element sites were designed so that a SpeI restriction site replaced the respective element, eliminating the protein binding capacity at that site. Primers used to generate the mutations are available upon request. The integrity of the mutated constructs was evaluated by sequencing and Southern blot. The constructs were injected into the male pronucleus of a fertilized egg derived from (C57BL/6 × BALB/c) F1 mice, and implanted in foster females. Founders were identified by PCR and Southern blot analysis of pup tail DNA. Copy numbers of each line were estimated by Southern blotting, comparing bands of the transgene with an endogenous sample.
Tissue Sample Preparations and DNA Extraction.
Sperm was collected from mouse epididymis and vas deference. Postimplantation mouse embryos were collected from the uteri of pregnant females 8.5 days following mating. The embryo proper and extra-embryonic tissues were separated before DNA extraction. Adult tissues (brain and tail) were also produced. All tissues were lysed overnight at 55 °C in a buffer containing 0.5 mg/mL proteinase K. DNA was extracted by a standard phenol-chloroform procedure.
Isolation of Oocytes and Preimplantation Mouse Embryos.
Three- to 4-week-old (C57BL/6 × BALB/c) transgenic females were superovulated by injection of 10 units pregnant mare's serum gonadotropin (PMSG, sigma) followed 48 h later by 10 units human CG (hCG, sigma). Mature oocytes were collected following 17 h into M2 medium (Sigma) and cleaned of contaminating granulosa cells by repetitive washing.
For the production of preimplantation mouse embryos, superovulated transgenic females were mated overnight with (C57BL/6 × BALB/c) F1 males (maternal transmission), or transgenic males were mated with (C57BL/6 × BALB/c) F1 females (paternal transmission). Zygotes were collected from female oviducts as described above for oocyte isolation and incubated at 37 °C in M16 medium (Sigma) covered with paraffin oil and 5% CO2 for 1–4 days, corresponding to the desired stage of development.
Southern Blot Analysis.
DNA samples (≈10 μg) were digested with EcoRI, BglII and NotI (NEB). Restriction fragments were resolved by gel electrophoresis, alkaline blotted onto a Zeta-Probe GT nylon membrane (Bio-Rad) in 0.4 M NaOH and probed overnight before washing and film exposure. The labeled probe was produced by PCR using 5′-CTCCCCAGGCTGTCTCTTGAGAGAA-3′ as upper and 5′-GGCTCTCCTACTTAGATCTTAGG-3′ as lower primers, and labeled with [α-32P]dCTP using RediPrime labeling system (GE Healthcare).
Bisulfite Methylation Analysis.
DNA samples from mouse brain, tail, sperm, and embryos (8.5 dpc) were prepared as described above were subjected to bisulfite treatment according to a published protocol (31). In treating oocytes and blastocysts we were careful in controlling for contamination from granulosa cells. The medium of each extraction and washing solution served as blanks. Following treatment, the AS-IC and PWS-IC DMRs were amplified by seminested PCR. For AS-IC the primers were 5′-CTAAAACCCAATTTCAATATATAAATAAATA-3′ (forward), 5′-AAATGATGTTTGAGTTTGGTTTTATTGT-3′ (reverse) and 5′-TGGAGGTAATTTGGTTTAGTTATTTTT-3′ (nested reverse). For PWS-IC (region III), primers were 5′-AAGGGAATTTGGGTTTTAAAGTTT-3′ (forward), 5′-TCCCCCTAAAATCCCTATACACTAA-3′ (reverse), and 5′-ATCCCCACACCACAATTAAAAAC-3′ (nested reverse). Primers for PWS-IC region II are available upon request. Reaction conditions for the first round of PCR were 5 cycles of 95 °C 1 min, 52 °C 3 min, 72 °C 3 min followed by 35 cycles of 95 °C 45 s, 52 °C 1 min, 72 °C 1 min. Conditions for the second round were 35 cycles of 95 °C 45 s, 52 °C 1 min, 72 °C 1 min followed by 7 min at 72 °C. Purified PCR products (Roche high pure PCR kit) were cloned into pGEM vectors (Promega) before sequencing. Individual template molecules that showed an identical methylation pattern were distinguished from one another by observation of nonconverted cytosines.
Acknowledgments.
We thank Dr. Ben-Zion Tsuberi for his technical help with embryo culture and Professors Yehudit Bergman and Howard Cedar for their invaluable comments. This study was supported by Israel Science Foundation Grant 104/06 and March of Dimes Foundation Grant 6-FY05–7.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Horsthemke B, Wagstaff J. Mechanisms of imprinting of the Prader-Willi/Angelman region. Am J Med Genet A. 2008;146A:2041–2052. doi: 10.1002/ajmg.a.32364. [DOI] [PubMed] [Google Scholar]
- 2.Buiting K, Lich C, Cottrell S, Barnicoat A, Horsthemke B. A 5-kb imprinting center deletion in a family with Angelman syndrome reduces the shortest region of deletion overlap to 880 bp. Hum Genet. 1999;105:665–666. doi: 10.1007/s004399900196. [DOI] [PubMed] [Google Scholar]
- 3.Ohta T, et al. Imprinting mutation mechanisms in Prader-Willi Syndrome. Am J Hum Genet. 1999;64:397–413. doi: 10.1086/302233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kantor B, Kaufman Y, Makedonski K, Razin A, Shemer R. Establishing the epigenetic status of the Prader-Willi/Angelman imprinting center in the gametes and embryo. Hum Mol Genet. 2004b;13:2767–2779. doi: 10.1093/hmg/ddh290. [DOI] [PubMed] [Google Scholar]
- 5.Kantor B, Makedonski K, Green-Finberg Y, Shemer R, Razin A. Control elements within the PWS/AS imprinting box and their function in the imprinting process. Hum Mol Genet. 2004a;13:751–762. doi: 10.1093/hmg/ddh085. [DOI] [PubMed] [Google Scholar]
- 6.Ambrosetti DC, Scholer HR, Dailey L, Basilico C. Modulation of the activity of multiple transcriptional activation domains by the DNA binding domains mediates the synergistic action of Sox2 and Oct-3 on the fibroblast growth factor-4 enhancer. J Biol Chem. 2000;275:23387–23397. doi: 10.1074/jbc.M000932200. [DOI] [PubMed] [Google Scholar]
- 7.Avilion AA, et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuccotti M, et al. Maternal Oct-4 is a potential key regulator of the developmental competence of mouse oocytes. BMC Dev Biol. 2008;8:97. doi: 10.1186/1471-213X-8-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nichols J, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 10.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 11.Ambrosetti DC, Basilico C, Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol Cell Biol. 1997;17:6321–6329. doi: 10.1128/mcb.17.11.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams DC, Jr, Cai M, Clore GM. Molecular basis for synergistic transcriptional activation by Oct1 and Sox2 revealed from the solution structure of the 42-kDa Oct1. Sox2.Hoxb1-DNA ternary transcription factor complex. J Biol Chem. 2004;279:1449–1457. doi: 10.1074/jbc.M309790200. [DOI] [PubMed] [Google Scholar]
- 13.Ohta T, et al. Molecular mechanism of Angelman syndrome in two large families involves an imprinting mutation. Am J Hum Genet. 1999a;64:385–396. doi: 10.1086/302232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Maarri O, et al. Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat Genet. 2001;27:341–344. doi: 10.1038/85927. [DOI] [PubMed] [Google Scholar]
- 15.Geuns E, De Rycke M, Van Steirteghem A, Liebaers I. Methylation imprints of the imprint control region of the SNRPN-gene in human gametes and preimplantation embryos. Hum Mol Genet. 2003;12:2873–2879. doi: 10.1093/hmg/ddg315. [DOI] [PubMed] [Google Scholar]
- 16.Perk J, et al. The imprinting mechanism of the Prader-Willi/Angelman regional control center. EMBO J. 2002;21:5807–5814. doi: 10.1093/emboj/cdf570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schumacher A, Buiting K, Zeschnigk M, Doerfler W, Horsthemke B. Methylation analysis of the PWS/AS region does not support an enhancer-competition model of genomic imprinting on human chromosome 15. Nat Genet. 1998;19:324–325. doi: 10.1038/1211. [DOI] [PubMed] [Google Scholar]
- 18.Shemer R, Birger Y, Riggs AD, Razin A. Structure of the imprinted mouse Snrpn gene and establishment of its parental-specific methylation pattern. Proc Natl Acad Sci USA. 1997;94:10267–10272. doi: 10.1073/pnas.94.19.10267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X, et al. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 2008;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnaud P, et al. Stochastic imprinting in the progeny of Dnmt3L-/- females. Hum Mol Genet. 2006;15:589–598. doi: 10.1093/hmg/ddi475. [DOI] [PubMed] [Google Scholar]
- 21.Epsztejn-Litman S, et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15:1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xin Z, et al. Role of histone methyltransferase G9a in CpG methylation of the Prader-Willi syndrome imprinting center. J Biol Chem. 2003;278:14996–15000. doi: 10.1074/jbc.M211753200. [DOI] [PubMed] [Google Scholar]
- 23.Chotalia M, et al. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev. 2009;23:105–117. doi: 10.1101/gad.495809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farber C, Dittrich B, Buiting K, Horsthemke B. The chromosome 15 imprinting centre (IC) region has undergone multiple duplication events and contains an upstream exon of SNRPN that is deleted in all Angelman syndrome patients with an IC microdeletion. Hum Mol Genet. 1999;8:337–343. doi: 10.1093/hmg/8.2.337. [DOI] [PubMed] [Google Scholar]
- 25.Botquin V, et al. New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes Dev. 1998;12:2073–2090. doi: 10.1101/gad.12.13.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mapendano CK, et al. Expression of the Snurf-Snrpn IC transcript in the oocyte and its putative role in the imprinting establishment of the mouse 7C imprinting domain. J Hum Genet. 2006;51:236–243. doi: 10.1007/s10038-005-0351-8. [DOI] [PubMed] [Google Scholar]
- 27.Dittrich B, et al. Imprint switching on human chromosome 15 may involve alternative transcripts of the SNPRN gene. Nat Genet. 1996;14:165–170. doi: 10.1038/ng1096-163. [DOI] [PubMed] [Google Scholar]
- 28.Johnstone KA, et al. A human imprinting centre demonstrates conserved acquisition but diverged maintenance of imprinting in a mouse model for Angelman syndrome imprinting defects. Hum Mol Genet. 2006;15:393–404. doi: 10.1093/hmg/ddi456. [DOI] [PubMed] [Google Scholar]
- 29.Zogel C, et al. Identification of cis- and trans-acting factors possibly modifying the risk of epimutations on chromosome 15. Eur J Hum Genet. 2006;14:752–758. doi: 10.1038/sj.ejhg.5201602. [DOI] [PubMed] [Google Scholar]
- 30.Lee KAW, Binderif A, Green MR. A small scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene Anal Tech. 1988;5:22–31. doi: 10.1016/0735-0651(88)90023-4. [DOI] [PubMed] [Google Scholar]
- 31.Engemann S, El-Maarri O, Hajkova P, Oswald J, Walter J. Bisulfite-based methylation analysis of imprinted genes. Methods Mol Biol. 2001;181:217–228. doi: 10.1385/1-59259-211-2:217. [DOI] [PubMed] [Google Scholar]