Abstract
A large body of evidence strongly suggests that the p53 tumor suppressor pathway is central in reducing cancer frequency in vertebrates. The protein product of the haploinsufficient mouse double minute 2 (MDM2) oncogene binds to and inhibits the p53 protein. Recent studies of human genetic variants in p53 and MDM2 have shown that single nucleotide polymorphisms (SNPs) can affect p53 signaling, confer cancer risk, and suggest that the pathway is under evolutionary selective pressure (1–4). In this report, we analyze the haplotype structure of MDM4, a structural homolog of MDM2, in several different human populations. Unusual patterns of linkage disequilibrium (LD) in the haplotype distribution of MDM4 indicate the presence of candidate SNPs that may also modify the efficacy of the p53 pathway. Association studies in 5 different patient populations reveal that these SNPs in MDM4 confer an increased risk for, or early onset of, human breast and ovarian cancers in Ashkenazi Jewish and European cohorts, respectively. This report not only implicates MDM4 as a key regulator of tumorigenesis in the human breast and ovary, but also exploits for the first time evolutionary driven linkage disequilibrium as a means to select SNPs of p53 pathway genes that might be clinically relevant.
Keywords: evolutionary selection pressure, linkage disequilibrium, p53 pathway, tumorigenesis, single nucleotide polymorphisms
The p53 signaling pathway mediates a cellular stress response that is activated on stresses such as DNA damage and oncogene activation. This signaling pathway initiates cellular responses such as DNA repair, cell cycle arrest, cell death (apoptosis), and senescence (5) and has been demonstrated to be crucial both in reducing cancer frequency in vertebrates and in mediating the response of commonly used cancer therapies (6–8). The protein product of the mouse double minute 2 (MDM2) oncogene binds to, and inhibits, the p53 protein (9). It has been shown in multiple mouse models that even a modest change in the expression level can affect p53-dependent tumor suppression in mice (10, 11). It was reasoned that such observations allow for the possibility that changing just a few (or even just one) base pair(s) in regulatory regions of the gene could alter the levels of MDM2 activity enough to affect the p53 pathway and, therefore, cancer in humans (1). Extensive study of a single nucleotide polymorphism (SNP) in the promoter of MDM2 (MDM2 SNP309, rs2279744, T/G) has lent support to this hypothesis (1–3).
The MDM2 SNP309 locus results in either a thymine (T) or a guanine (G) in the intronic promoter/enhancer region of the MDM2 oncogene (1). The G-allele of MDM2 SNP309 was shown to increase the affinity of the transcription factor Sp1, which resulted in the increased transcription and expression of the MDM2 protein (1). In concordance to these initial observations, the G-allele of MDM2 SNP309 has been shown to associate with significantly higher levels of MDM2 expression in tumor-derived cell lines (1, 12, 13), in renal cell carcinoma (14), in normal esophageal tissue (15), and in B-cell chronic lymphocytic leukemia (16). In tumor-derived cell lines, the elevated levels of MDM2 associated with the G-allele have been shown in multiple studies to result in the attenuation of the p53 apoptotic response after exposure to multiple types of chemotherapeutic agents, compared with cells containing the T-allele (1, 12, 17). It was proposed that the high levels of MDM2 resulting from the G-allele attenuate the p53 stress response, resulting in a higher mutation rate, poorer DNA repair processes, reduced apoptosis, and senescence, leading to faster and more frequent tumor formation (1). Support for the model has come from the association of MDM2 SNP309 with differences in the onset of, or the risk for, cancer, as well as survival (2, 3, 18). For example, earlier ages of onset associated with individuals possessing the G-allele have been demonstrated in soft-tissue sarcomas (1, 19, 20), lymphoma (21), leukemia (22), melanoma (23), head, neck, and oral squamous cell carcinomas (24, 25), and cancers of the colon (26), breast (21, 27, 28), bladder (29), ovary (30), brain (31), and liver (32).
Recently, the MDM2 haplotype structure was determined in humans from several different demographic groups (33). It was noted that the frequency of the haplotype harboring the functional G-allele of MDM2 SNP309 deviated significantly from the standard assumptions of models of selective neutrality by using multiple selection tests, including an entropy-based selection test that compares both the frequency and long-range correlations of an allele with a simulated neutral model, where the allele is selectively neutral. These observations suggested that the human p53 pathway could be acted on by an evolutionary selection pressure. Indeed, it had been previously and subsequently shown that a well-regulated p53 pathway is crucial for proper murine embryonic development, embryonic implantation, pregnancy rates, and litter sizes (33, 34), and also human embryonic implantation (35). Thus, the p53 pathway not only plays a role in genomic error correction in the lifetime of humans, but also determines the fate of viable genetic transmission on the timescale of generations. From this observation, we reasoned that other SNPs in the p53 pathway with signatures of natural selection might also alter cancer risk in humans.
To this end, we chose to focus on the haplotype structure of MDM4, a structural homolog of MDM2 and a key inhibitor of p53 (36–38), in the hope of detecting functional genetic variants that alter p53 pathway activity and, ultimately, are associated with risk of human tumorigenesis. Like MDM2, its structural homolog, MDM4, can also bind directly to p53 and inhibit its ability to function as a transcriptional activator, as well as regulate its stability, most likely through its interactions with MDM2 (37, 38). Its central role in regulating p53 activity and human cancer has been highlighted by many observations. For example, MDM4 is an essential gene in murine development, and knockout embryos die in utero. This lethal phenotype can be rescued by knocking out the p53 gene (36). In human cancer, MDM4 was found to be amplified or over-expressed in a large subset of human tumors (38–40). Terzian et al. recently demonstrated that similar to Mdm2, Mdm4 haploinsufficiency leads to increased p53 activity exhibited as increased sensitivity to DNA damage, decreased transformation potential, and tumor development (41). These data suggest that changing just a few (or even just one) base pair(s) in MDM4 could alter the levels of MDM4 enough to affect the p53 pathway and, therefore, cancer in humans. In this report, data are presented that both support this hypothesis as well as implicate MDM4 as a key regulator of tumorigenesis in human breast and ovary.
Results
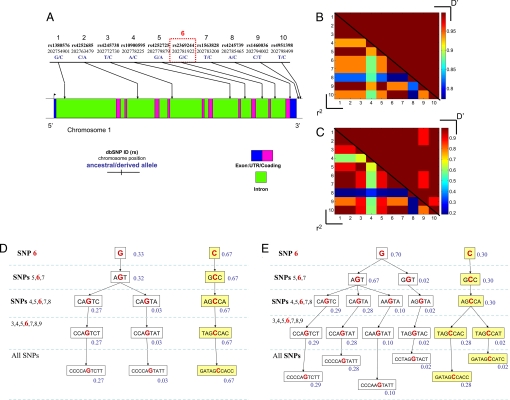
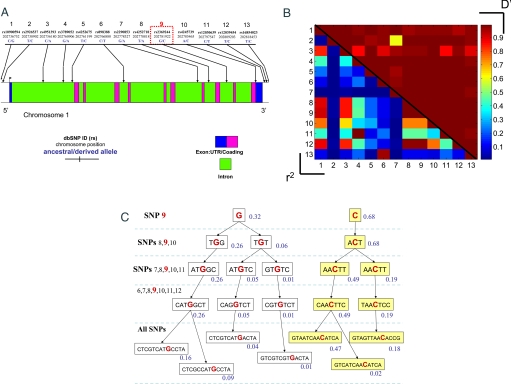
Genotype data for SNPs across MDM4 were collected from 3 sources: (i) a collection of 84 lymphoblastoid cell lines from the Coriell Institute for Medical Research Diversity Cell line panel (46 from African Americans and 38 from Caucasians), of which 10 SNPs were chosen to characterize the haplotype diversity across the largest isoform of the transcript (Fig. 1A); (ii) the Caucasian (CEU) and African (Yoruba, YRI) populations of the International HapMap project, of which 46 SNPs were selected across the gene including those lying 10 kb 5′ of the first exon and 10 kb 3′ of the terminal exon; and (iii) a cohort of 299 Caucasian females of Ashkenazi Jewish (AJ) ancestry (see Materials and Methods), of which 13 SNPs mapping within and immediately surrounding the MDM4 gene were selected (Fig. 2A).
Fig. 1.
Haplotype structures of the MDM4 oncogene in a Caucasian and in an African American population. (A) A schematic diagram of the MDM4 gene and the SNPs genotyped in the present study. Pairwise linkage disequilibriums are shown for the Caucasian (B), and the African American (C) populations. The upper right triangles report the D′ measures, and the lower left triangles report the r2 measures. The inferred haplotype frequencies in the Caucasian (D) and African American (E) populations centered around a particular SNP (SNP-6, rs2369244; denoted with a red box in A), where G(C) is the ancestral (derived) allele.
Fig. 2.
Haplotype structure of the MDM4 oncogene in a Caucasian population of AJ ethnicity. (A) A schematic diagram of the MDM4 gene and the SNPs genotyped in the present study. (B) Pairwise linkage disequilibriums for the Caucasian population of AJ ethnicity. The upper right triangles report the D′ measures, and the lower left triangles report the r2 measures. (C) The inferred haplotype frequencies in the population centered on a particular SNP (SNP-9, rs2369244; denoted with a red box in A), where G(C) is the ancestral (derived) allele.
After the genotypes were computationally phased into haplotypes (42), we began determining the linkage patterns of SNPs as quantified by the pair-wise correlation statistics D′ and r2 (see Fig. 1 B and C for Coriell Caucasians and African Americans and Fig. 2B for AJ Caucasians). We noted that many SNPs are tightly correlated along the extent of the gene, more so in the Caucasian population than in the African and African American datasets (see Fig. 1B vs. 1C for the linkage patterns in the Coriell datasets and Figs. 2B and Fig. S1 in AJ), most likely because of the reduced number of recombination events in the genealogical history of the Caucasian population. The common haplotypes in the Caucasian population, from the Coriell dataset and the Hapmap dataset, exhibit similar frequencies to those in the AJ population (Figs. 1D and 2C). Analysis of the marginal haplotypes of the 10 SNPs in the Coriell dataset revealed that there is a dominant haplotype in 67% of the Caucasian population, and the same haplotype (Fig. 1D), when extended to the 46 SNPs of the public Hapmap dataset, constitutes 50% of the CEU population. The African American dataset from Coriell and the YRI dataset from Hapmap also include this haplotype, but at lower frequencies (Fig. 1E). The marginal haplotype frequencies from the Coriell dataset inferred from this calculation are shown in Fig. 1 D and E, centered around a particular SNP (SNP-6, rs2369244), where G(C) is the ancestral (derived) allele. The high frequency and large extent of the derived haplotype and the low diversity in all of the populations suggest the possibility that the derived haplotype could have arisen because of natural selection.
To test the possibility of natural selection, we applied a battery of classical tests of neutrality (Fu and Li's F, Fu and Li's D, Tajima's D) that implicitly invoke different assumptions about the expected frequency distribution of alleles across a set of neutral SNPs. However, these tests suffer from 3 main deficits: (i) they ignore multiallelic correlations and thereby reduce their power to detect a recent reduction in haplotype diversity, i.e., selective sweep; (ii) historical episodes of reduced population sizes (population bottlenecks) can give rise to a distribution of allele frequencies that appear to be nonneutral; and (iii) these classical tests do not explicitly pinpoint which allele or haplotype is under selection pressure. In an attempt to overcome these deficits, we also performed a previously described selection test for each SNP, based on the entropy of the marginal haplotype distributions (33). Interestingly, the entropy-haplotype selection tests and the majority of the classical selection tests suggest a significant departure from neutrality of the MDM4 oncogene in the Caucasian, African American, and Ashkenazi-Jewish populations (Table 1). In particular, the entropy-haplotype test rejects the null hypothesis of neutrality for several tightly linked SNPs using the conventional significance threshold of 0.05, adjusted with the conservative Bonferroni correction for multiple hypotheses testing. In Figs. 1D, 1E, and 2C, we present the marginal haplotypes associated with one of these selected SNPs (i.e., SNP-6, rs2369244). The evidence for selection in the Yoruba population from the Hapmap seems less compelling than the others, and the Yoruba is the only population that is not significant under the entropy-haplotype test.
Table 1.
Results of various selection tests
| Selection test | Coriell, Caucasian | Coriell, African American | Hapmap,CEU | Hapmap,YRI | MSKCC,AJ |
|---|---|---|---|---|---|
| Fu and Li's F | 0.05 < P < 0.1 | 0.05 < P < 0.1 | 0.05 < P < 0.1 | P < 0.02 | 0.05 < P < 0.1 |
| Fu and Li's D | P < 0.02 | P < 0.02 | P < 0.02 | P < 0.05 | P < 0.02 |
| Tajima's D | P < 0.01 | P < 0.01 | P < 0.01 | P > 0.1 | P < 0.01 |
| Entropy haplotype | P < 0.02 | P < 0.05 | P < 0.05 | 0.05 < P < 0.1 | P < 0.01 |
The above-described analysis suggests that the genetic variants in the MDM4 oncogene have undergone a selective sweep. As this scenario is similar to the haplotype containing the functional SNP309 in the MDM2 oncogene (33), it was hypothesized that the MDM4 haplotype distribution might also harbor one or more functional SNPs and therefore also demonstrate allelic differences in human cancer populations. To test this hypothesis, a set of SNPs was used, each of which can differentiate the nonneutral major MDM4-haplotype (the major alleles) from the other neutral MDM4 haplotypes, and it was determined whether their genotypes were associated with either altered cancer risk in case/control studies for familial and sporadic breast cancer or age-dependent incidence in case studies for both familial and sporadic ovarian cancers.
Functional p53 pathway polymorphisms and somatic genetic alterations of p53 pathway genes have been shown to affect breast cancer incidence, progression, and/or survival (2, 43, 44). Therefore, we sought allelic differences in cancer risk for the MDM4 haplotype, first by using data generated in a 2-stage genome-wide association study (GWAS) performed on an Affymetrix 500K (Affy 500K) platform that has identified breast cancer susceptibility loci in the AJ population (45). We used the data that originated from the first stage of the GWAS that was carried out in 250 AJ Caucasian women affected with breast cancer, with a family history of 3 or more breast cancers in a single lineage, and who tested negative for the 3 AJ breast cancer (BRCA) founder mutations, and 300 healthy AJ controls. We then sought confirmation of noted allelic differences in an additional dataset that originated from an unpublished GWAS, constituted of a cohort of 240 sporadic breast cancer patients unselected for family history or BRCA1/2 status and also genotyped by using an Affy 500K array. Among the SNPs that differentiated the nonneutral, major MDM4-haplotype from the other MDM4 haplotypes, 3 MDM4 SNPs were also present on the array and genotyped with good quality [in Fig. 2A: SNP-1 (rs10900594), SNP-9 (rs2369244), SNP-12 (rs12039454)]. Similarly to the above-described analysis, all 3 SNPs were in high LD (D′ > 0.9) in the AJ controls and patients (Fig. S1). Interestingly, statistically significant allelic associations for all 3 loci in both groups (familial and sporadic breast cancer cohorts) of patients were observed when compared with the 300 AJ controls (Fig. S1). The distributions of the genotypes for all 3 SNPs varied significantly between the cases and controls as indicated in Table 2. For all 3 SNPs, these differences resulted in an enrichment of the minor allele homozygotes in the cases and the subsequent decrease of the major allele homozygotes (Table 2 and Table S1).
Table 2.
Significant enrichments of the minor allele homozygotes suggest increased familial and sporadic breast cancer risk
| Study | SNP | Genotype | Number |
Percent |
Odds ratio | 95% CI* | P value† | ||
|---|---|---|---|---|---|---|---|---|---|
| Controls | Cases | Controls | Cases | ||||||
| Familial breast cancer | rs10900594 | GG | 134 | 94 | 45 | 39 | 1 | ||
| CG | 140 | 108 | 47 | 45 | |||||
| CC | 23 | 36 | 8 | 15 | 2.2 | 1.24–4.01 | 0.0065 | ||
| rs2369244 | CC | 130 | 95 | 44 | 39 | 1 | |||
| CG | 144 | 114 | 48 | 47 | |||||
| GG | 24 | 36 | 8 | 15 | 2.1 | 1.15–3.67 | 0.01408 | ||
| rs12039454 | CC | 133 | 95 | 44 | 38 | 1 | |||
| CT | 143 | 114 | 48 | 46 | |||||
| TT | 23 | 39 | 8 | 16 | 2.4 | 1.3–4.2 | 0.00294 | ||
| Sporadic breast cancer | rs10900594 | GG | 126 | 83 | 47 | 35 | 1 | ||
| CG | 119 | 119 | 45 | 50 | |||||
| CC | 21 | 35 | 8 | 15 | 2.5 | 1.38–4.65 | 0.00231 | ||
| rs2369244 | CC | 121 | 80 | 45 | 34 | 1 | |||
| CG | 123 | 117 | 46 | 50 | |||||
| GG | 22 | 37 | 8 | 16 | 2.5 | 1.40–4.63 | 0.00187 | ||
| rs12039454 | CC | 124 | 82 | 46 | 34 | 1 | |||
| CT | 122 | 119 | 46 | 50 | |||||
| TT | 21 | 37 | 8 | 16 | 2.7 | 1.46–4.88 | 0.00118 | ||
*Using the approximation of Woolf.
†Two-sided Fisher's Exact Test.
We then sought to confirm these observations by genotyping SNP-9 (rs2369244) in another, independent cohort of 654 AJ breast cancer cases and 1,085 AJ controls. Similar to the findings in the GWAS cohorts, the minor allele homozygotes were significantly enriched in the cases compared with the controls (P = 0.01, Table S1). Furthermore, a meta-analysis of all 3 cohorts, controlling for age, suggests that a SNP, which can differentiate the nonneutral, major MDM4 haplotype from the other neutral MDM4 haplotypes (SNP-9, rs2369244) demonstrates significant associations with altered AJ breast cancer risk (Table 3). Specifically, the minor allele homozygotes (G/G), representing the neutral MDM4 haplotypes, associate with an increased breast cancer risk with an OR of 1.52 (95% CI 1.18–1.95, P = 0.00085) and C/G with an OR of 1.2 (95% CI 1.01–1.42, P = 0.037). It is important to note that similar associations were sought in non-AJ Caucasian breast cancer studies (46) (Table S2) and not observed, suggesting potential ethnic differences in these results.
Table 3.
AJ meta-analysis
| rs2369244 | Cases | Controls | OR (95% CI) | P value, chi square |
|---|---|---|---|---|
| CC | 404 (35) | 557 (41) | 1 | |
| CG | 549 (48) | 631 (47) | 1.2 (1.01–1.42) | 0.037 |
| GG | 180 (16) | 163 (12) | 1.52 (1.18–1.95) | 0.00085 |
| Total | 1,133 | 1,351 |
Together, the results of the AJ familial and both AJ sporadic breast cancer studies significantly support the hypothesis that MDM4 haplotypes might also harbor functional SNP(s), as in all 3 studies the minor allele homozygotes of SNPs that can differentiate the neutral from the nonneutral haplotypes were enriched in those with cancer compared with controls. However, to further test the hypothesis, allelic differences for the MDM4 haplotype were explored in a different tumor type and by using a different experimental design. As functional p53 pathway polymorphisms and somatic genetic alterations of p53 pathway genes have also been shown to affect ovarian cancer (30, 43, 44), potential allelic differences in the age of diagnosis were tested in case studies of both familial and sporadic ovarian cancer in Caucasian populations of non-AJ ethnicity.
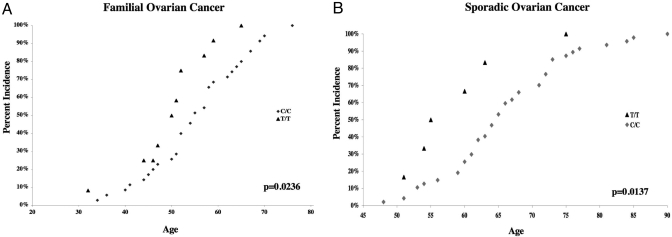
The first ovarian cancer case study consists of 244 individuals that are members of families with extremely high rates of breast and ovarian cancer. In this cohort, 95 women have been diagnosed with ovarian cancer and were genotyped for a MDM4 SNP that can differentiate the nonneutral, major MDM4-haplotype from the other MDM4 haplotypes in the Caucasian population (SNP-7, rs1563828; see Fig. 1 A, B, and D). Interestingly, there are significant allelic differences in the age of diagnosis between the genotypes. In this population, the median age of diagnosis was very young at 52 years of age. However, the women homozygous for the major allele were diagnosed on average at 56 years of age, with those heterozygous or homozygous for the minor allele diagnosed on average either 4 or 5 years earlier, respectively (P = 0.0236, Mann–Whitney Test) (Fig. 3A).
Fig. 3.
A tagSNP that differentiates the nonneutral MDM4-haplotype from the neutral haplotypes demonstrates significant allelic differences in the age of diagnosis of both familial and sporadic ovarian cancer. The cumulative incidence of ovarian cancer for both the women T/T in genotype (black triangles) and C/C in genotype (gray diamonds) for rs1563828 is plotted as a function of age for familial (A) and sporadic (B) disease. The p-values on both panels were calculated by comparing the medians of both genotypes by using a Mann–Whitney Test.
To confirm and extend these observations, this same SNP was analyzed in a second, independent case study of 110 individuals diagnosed with invasive ovarian carcinomas. As expected, this sporadic population was diagnosed on average much later than the familial cohort, with a median age of diagnosis of 64 years (P < 0.0001, unpaired t test). Interestingly, the minor allele homozygotes were also diagnosed 9 years earlier than the major allele homozygotes (P = 0.0137, Mann–Whitney) (Fig. 3B). Together, these data provide evidence that genetic variants in MDM4 can affect tumor suppression in familial and sporadic ovarian cancers, whereby the minor allele carriers associate with the development of tumors earlier in life.
Discussion
In summary, we have first shown that the human MDM4 gene harbors a haplotype that deviates from standard assumptions of selective neutrality, using a variety of selection tests, in 3 different populations of Caucasians of different ethnic backgrounds and in 1 population of African Americans. Secondly, we have demonstrated that the minor allele homozygotes, and therefore the neutral haplotypes, are associated with both a greater risk of developing breast tumors in 3 independent AJ case cohorts and developing ovarian tumors up to 9 years earlier in life, in both familial and sporadic ovarian cancer in non-AJ Caucasian cohorts. These data support the initial hypothesis that haploinsufficient MDM4, like haploinsufficient MDM2, harbors a genetic variant that affects human cancer. With what is known of MDM4 function to date, it is tempting to speculate that a SNP(s) associated with this haplotype would result in allelic differences in MDM4 activity levels. In turn, the differences in MDM4 activity levels would result in differences in the p53 stress response, resulting in a higher mutation rate, poorer DNA repair processes, reduced cell cycle arrest, apoptosis, and senescence leading to faster and more frequent tumor formation (Fig. S2).
As mentioned in the introduction, it was previously shown that the nonneutral haplotype of the MDM2 oncogene harbors the G-allele of MDM2 SNP309 and associates with an attenuation of the p53 pathway, altered MDM2 expression, and enhanced early onset of, or increased risk for, tumorigenesis (33). In this report, we demonstrate that in the case of the MDM4 oncogene, it is the neutral haplotype that associates with increased breast cancer risk and an earlier age of onset of ovarian cancer, which could suggest a weaker p53 pathway. In both cases, the deviations that form the standard assumptions of selective neutrality are more readily explained by models of positive selection. This observation could suggest that both a weaker and stronger p53 pathway could be under positive selection pressure. Interestingly, both too much p53 activity, as well as too little, has been demonstrated to be correlated with reduced fecundity in mice and could help explain these observations. For example, in a recent report, significant decreases in embryonic implantation, pregnancy rate, and litter size were observed in matings with p53-null female mice, but not with p53-null male mice, suggesting that the absence of p53 in the mother is deleterious toward successful mating (34). In contrast, multiple studies have shown that the absence of the key negative regulators of p53, MDM2 and MDM4, in embryos results in early embryonic lethality, which is rescued by knocking out the p53 gene (8, 36). These data suggest that too much p53 activity in the embryo is deleterious for successful embryonic development. Hence, more p53 activity in mothers could be advantageous for successful propagation and therefore selected, whereas less p53 activity in embryos could be advantageous and therefore selected.
Of course, the observations reported here remain to be validated in other patient cohorts, and it will be important to determine the precise regulatory changes associated with the MDM4 SNP(s) and their effect on MDM4 activity and p53 signaling. However, these data strongly suggest that MDM4 plays a significant regulatory role in the development of breast and ovarian cancers and that the Mdm4 gene harbors genetic variants that can affect human cancer and may be under evolutionary selection pressure. Lastly, these data also suggest that evolutionary driven linkage disequilibrium can be used as a means to select SNPs of p53 pathway genes that might be clinically relevant.
Methods
Haplotype and Selection Analysis.
A description of the populations and genotyping can be found in SI Methods.
Selection test.
The entropy selection test was previously described in detail (33), and a brief description can be found in SI Methods.
AJ Breast Cancer Cases and Controls.
Populations.
The case and control populations studied in the GWAS portion of this study were described previously in detail (45), and a brief description can be found in SI Methods.
Familial and Sporadic Ovarian Cancer.
Familial ovarian cancer population.
Breast and ovarian cancer families were identified through the Padua Hereditary Breast/Ovarian Cancer Center of the Istituto OncologicoVeneto (Padova, Italy). Family selection was based on published operational criteria (47). Ninety-five of the 131 affected probands were diagnosed with ovarian cancer. Among them, 33 were carriers of BRCA1 (26 patients) or BRCA2 (7 patients) germline pathogenic mutations. Blood samples were obtained from each proband or family member after informed consent as to the aims of the research project. The study was performed in accordance with the principles embodied in the Declaration of Helsinki and was approved by the Oncology Centre Ethical and Technical Scientific Committees.
Sporadic ovarian cancer population.
This patient population was previously described in great detail (30), and a brief description can be found in SI Methods.
Supplementary Material
Acknowledgments.
This work was supported by the Ludwig Institute for Cancer Research, the Simons Foundation, the Leon Levy Foundation, the Breast Cancer Research Foundation, the Robert and Kate Niehaus Clinical Genetics Research Initiative, the Deutsche Krebshilfe, and the Carmel Research Fund.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901298106/DCSupplemental.
References
- 1.Bond GL, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 2.Bond GL, Levine AJ. A single nucleotide polymorphism in the p53 pathway interacts with gender, environmental stresses and tumor genetics to influence cancer in humans. Oncogene. 2007;26:1317–1323. doi: 10.1038/sj.onc.1210199. [DOI] [PubMed] [Google Scholar]
- 3.Hu Z, et al. MDM2 promoter polymorphism SNP309 contributes to tumor susceptibility: Evidence from 21 case-control studies. Cancer Epidem Biomar. 2007;16:2717–2723. doi: 10.1158/1055-9965.EPI-07-0634. [DOI] [PubMed] [Google Scholar]
- 4.Hu W, Feng Z, Atwal GS, Levine AJ. p53: A new player in reproduction. Cell Cycle. 2008;7:848–852. doi: 10.4161/cc.7.7.5658. [DOI] [PubMed] [Google Scholar]
- 5.Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol. 2008;9:702–712. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- 6.Haupt S, Haupt Y. Importance of p53 for cancer onset and therapy. Anticancer Drugs. 2006;17:725–732. doi: 10.1097/01.cad.0000217422.52208.fa. [DOI] [PubMed] [Google Scholar]
- 7.Johnstone RW, Ruefli AA, Lowe SW. Apoptosis: A link between cancer genetics and chemotherapy. Cell. 2002;108:153–164. doi: 10.1016/s0092-8674(02)00625-6. [DOI] [PubMed] [Google Scholar]
- 8.Lozano G, Zambetti GP. What have animal models taught us about the p53 pathway? J Pathol. 2005;205:206–220. doi: 10.1002/path.1704. [DOI] [PubMed] [Google Scholar]
- 9.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 10.Lane DP. Exploiting the p53 pathway for the diagnosis and therapy of human cancer. Cold Spring Harbor Symp Quant Biol. 2005;70:489–497. doi: 10.1101/sqb.2005.70.049. [DOI] [PubMed] [Google Scholar]
- 11.Poyurovsky MV, Prives C. Unleashing the power of p53: Lessons from mice and men. Genes Dev. 2006;20:125–131. doi: 10.1101/gad.1397506. [DOI] [PubMed] [Google Scholar]
- 12.Arva NC, et al. A chromatin-associated and transcriptionally inactive p53-Mdm2 complex occurs in mdm2 SNP309 homozygous cells. J Biol Chem. 2005;280:26776–26787. doi: 10.1074/jbc.M505203200. [DOI] [PubMed] [Google Scholar]
- 13.Hu W, et al. A single nucleotide polymorphism in the MDM2 gene disrupts the oscillation of p53 and MDM2 levels in cells. Cancer Res. 2007;67:2757–2765. doi: 10.1158/0008-5472.CAN-06-2656. [DOI] [PubMed] [Google Scholar]
- 14.Hirata H, et al. MDM2 SNP309 polymorphism as risk factor for susceptibility and poor prognosis in renal cell carcinoma. Clin Cancer Res. 2007;13:4123–4129. doi: 10.1158/1078-0432.CCR-07-0609. [DOI] [PubMed] [Google Scholar]
- 15.Hong Y, et al. The role of P53 and MDM2 polymorphisms in the risk of esophageal squamous cell carcinoma. Cancer Res. 2005;65:9582–9587. doi: 10.1158/0008-5472.CAN-05-1460. [DOI] [PubMed] [Google Scholar]
- 16.Gryshchenko I, et al. MDM2 SNP309 is associated with poor outcome in B-cell chronic lymphocytic leukemia. J Clin Oncol. 2008;26:2252–2257. doi: 10.1200/JCO.2007.11.5212. [DOI] [PubMed] [Google Scholar]
- 17.Nayak MS, Yang JM, Hait WN. Effect of a single nucleotide polymorphism in the murine double minute 2 promoter (SNP309) on the sensitivity to topoisomerase II-targeting drugs. Cancer Res. 2007;67:5831–5839. doi: 10.1158/0008-5472.CAN-06-4533. [DOI] [PubMed] [Google Scholar]
- 18.Murphy ME. Polymorphic variants in the p53 pathway. Cell Death Differ. 2006;13:916–920. doi: 10.1038/sj.cdd.4401907. [DOI] [PubMed] [Google Scholar]
- 19.Bougeard G, et al. Impact of the MDM2 SNP309 and p53 Arg72Pro polymorphism on age of tumour onset in Li-Fraumeni syndrome. J Med Genet. 2006;43:531–533. doi: 10.1136/jmg.2005.037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruijs MW, et al. The single-nucleotide polymorphism 309 in the MDM2 gene contributes to the Li-Fraumeni syndrome and related phenotypes. Eur J Hum Genet. 2007;15:110–114. doi: 10.1038/sj.ejhg.5201715. [DOI] [PubMed] [Google Scholar]
- 21.Bond GL, et al. MDM2 SNP309 Accelerates tumor formation in a gender-specific and hormone-dependent manner. Cancer Res. 2006;66:5104–5110. doi: 10.1158/0008-5472.CAN-06-0180. [DOI] [PubMed] [Google Scholar]
- 22.Swinney RM, Hsu SC, Hirschman BA, Chen TT, Tomlinson GE. MDM2 promoter variation and age of diagnosis of acute lymphoblastic leukemia. Leukemia. 2005;19:1996–1998. doi: 10.1038/sj.leu.2403941. [DOI] [PubMed] [Google Scholar]
- 23.Firoz EF, et al. Association of MDM2 SNP309, age of onset, and gender in cutaneous melanoma. Clin Cancer Res. 2009;15:2573. doi: 10.1158/1078-0432.CCR-08-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang SF, et al. Combined effects of MDM2 SNP 309 and p53 mutation on oral squamous cell carcinomas associated with areca quid chewing. Oral Oncol. 2008;45:16–22. doi: 10.1016/j.oraloncology.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Nakashima M, et al. Impact of MDM2 single nucleotide polymorphism on tumor onset in head and neck squamous cell carcinoma. Acta Otolaryngol. 2008;128:808–813. doi: 10.1080/00016480701724904. [DOI] [PubMed] [Google Scholar]
- 26.Menin C, et al. Association between MDM2-SNP309 and age at colorectal cancer diagnosis according to p53 mutation status. J Natl Cancer Inst. 2006;98:285–288. doi: 10.1093/jnci/djj054. [DOI] [PubMed] [Google Scholar]
- 27.Wasielewski M, et al. MDM2 SNP309 accelerates familial breast carcinogenesis independently of estrogen signaling. Breast Cancer Res Treat. 2007;104:153–157. doi: 10.1007/s10549-006-9407-5. [DOI] [PubMed] [Google Scholar]
- 28.Yarden RI, et al. MDM2 SNP309 accelerates breast and ovarian carcinogenesis in BRCA1 and BRCA2 carriers of Jewish-Ashkenazi descent. Breast Cancer Res Treat. 2008;111:497–504. doi: 10.1007/s10549-007-9797-z. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez-Carbayo M, et al. A polymorphism in HDM2 (SNP309) associates with early onset in superficial tumors, TP53 mutations, and poor outcome in invasive bladder cancer. Clin Cancer Res. 2007;13:3215–3220. doi: 10.1158/1078-0432.CCR-07-0013. [DOI] [PubMed] [Google Scholar]
- 30.Bartel F, et al. Both germ line and somatic genetics of the p53 pathway affect ovarian cancer incidence and survival. Clin Cancer Res. 2008;14:89–96. doi: 10.1158/1078-0432.CCR-07-1192. [DOI] [PubMed] [Google Scholar]
- 31.Khatri RG, Navaratne K, Weil RJ. The role of a single nucleotide polymorphism of MDM2 in glioblastoma multiforme. J Neurosurg. 2008;109:842–848. doi: 10.3171/JNS/2008/109/11/0842. [DOI] [PubMed] [Google Scholar]
- 32.Yoon YJ, et al. MDM2 and p53 polymorphisms are associated with the development of hepatocellular carcinoma in patients with chronic hepatitis B virus infection. Carcinogenesis. 2008;29:1192–1196. doi: 10.1093/carcin/bgn090. [DOI] [PubMed] [Google Scholar]
- 33.Atwal GS, et al. Haplotype structure and selection of the MDM2 oncogene in humans. Proc Natl Acad Sci USA. 2007;104:4524–4529. doi: 10.1073/pnas.0610998104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu W, Feng Z, Teresky AK, Levine AJ. p53 regulates maternal reproduction through LIF. Nature. 2007;450:721–724. doi: 10.1038/nature05993. [DOI] [PubMed] [Google Scholar]
- 35.Kay C, Jeyendran RS, Coulam CB. p53 tumour suppressor gene polymorphism is associated with recurrent implantation failure. Reprod Biomed Online. 2006;13:492–496. doi: 10.1016/s1472-6483(10)60635-9. [DOI] [PubMed] [Google Scholar]
- 36.Marine JC, et al. Keeping p53 in check: Essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ. 2006;13:927–934. doi: 10.1038/sj.cdd.4401912. [DOI] [PubMed] [Google Scholar]
- 37.Toledo F, Wahl GM. MDM2 and MDM4: p53 regulators as targets in anticancer therapy. Int J Biochem Cell Biol. 2007;39:1476–1482. doi: 10.1016/j.biocel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wade M, Wahl GM. Targeting Mdm2 and Mdmx in cancer therapy: Better living through medicinal chemistry? Mol Cancer Res. 2009;7:1–11. doi: 10.1158/1541-7786.MCR-08-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laurie NA, et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006;444:61–66. doi: 10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- 40.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 41.Terzian T, et al. Haploinsufficiency of Mdm2 and Mdm4 in tumorigenesis and development. Mol Cell Biol. 2007;27:5479–5485. doi: 10.1128/MCB.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pietsch EC, Humbey O, Murphy ME. Polymorphisms in the p53 pathway. Oncogene. 2006;25:1602–1611. doi: 10.1038/sj.onc.1209367. [DOI] [PubMed] [Google Scholar]
- 44.Zambetti GP. The p53 mutation “gradient effect” and its clinical implications. J Cell Physiol. 2007;213:370–373. doi: 10.1002/jcp.21217. [DOI] [PubMed] [Google Scholar]
- 45.Gold B, et al. Genome-wide association study provides evidence for a breast cancer risk locus at 6q22.33. Proc Natl Acad Sci USA. 2008;105:4340–4345. doi: 10.1073/pnas.0800441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Federico M, et al. Identification of families with hereditary breast and ovarian cancer for clinical and mammographic surveillance: The Modena Study Group proposal. Breast Cancer Res Treat. 1999;55:213–221. doi: 10.1023/a:1006192230332. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.